Abstract
The structural differences among different G-quadruplexes provide an opportunity for site-specific targeting of a particular G-quadruplex structure. However, majority of G-quadruplex ligands described thus far show little selectivity among different G-quadruplexes. In this work, we delineate the design and synthesis of a crescent-shaped thiazole peptide that preferentially stabilizes c-MYC quadruplex over other promoter G-quadruplexes and inhibits c-MYC oncogene expression. Biophysical analysis such as Förster resonance energy transfer (FRET) melting and fluorescence spectroscopy show that the thiazole peptide TH3 can selectively interact with the c-MYC G-quadruplex over other investigated G-quadruplexes and duplex DNA. NMR spectroscopy reveals that peptide TH3 binds to the terminal G-quartets and capping regions present in the 5′- and 3′-ends of c-MYC G-quadruplex with a 2:1 stoichiometry; whereas structurally related distamycin A is reported to interact with quadruplex structures via groove binding and end stacking modes with 4:1 stoichiometry. Importantly, qRT-PCR, western blot and dual luciferase reporter assay show that TH3 downregulates c-MYC expression by stabilizing the c-MYC G-quadruplex in cancer cells. Moreover, TH3 localizes within the nucleus of cancer cells and exhibits antiproliferative activities by inducing S phase cell cycle arrest and apoptosis.
INTRODUCTION
G-quadruplexes are four stranded DNA secondary structures that are hypothesized to be involved in key biological processes such as telomere maintenance and oncogene expression (1–3). G-quadruplex forming sequences are abundant in the promoter region of various proto-oncogenes like c-MYC (4), c-KIT (5), BCL-2 (6) etc. The 27-mer G-quadruplex forming sequence located in the nuclease hypersensitive element (NHE) III1 of the c-MYC promoter region is well studied (7). It has been reported that this sequence exists in equilibrium between transcriptionally active forms (double helical and single stranded) and a silenced form (G-quadruplex) and regulates up to 90% of c-MYC transcription (8). This 27-mer sequence contains five consecutive runs of guanines, with three runs composed of four guanines each and two runs composed of three guanines each (c-MYCPu27). However, only four runs of guanines are required to form G-quadruplex structure. It was reported that the predominant and biologically relevant G-quadruplex conformer of the c-MYC silencer element is a parallel-stranded G-quadruplex with 1:2:1 side loops (formed using the second, third, fourth, and fifth G-runs) (9–10). Small molecules that stabilize the specific c-MYC G-quadruplex structure can regulate expression of c-MYC oncogene at the transcriptional level (11–18). However, all G-quadruplexes contain G-quartets as a common structural feature, making it challenging to develop ligands selective for a particular G-quadruplex.
Most G-quadruplex ligands bind to the terminal G-quartet of G-quadruplex structures via end stacking mode. The groove (19–23) and intermediate regions of G-quadruplexes are different from each other, which may provide an opportunity to target a particular G-quadruplex structure. So far, very few ligands are reported to show selectivity among different G-quadruplex structures. In this work, we report the design and synthesis of novel thiazole peptides and the study of their interaction with four promoter G-quadruplexes (c-MYC, c-KIT1, c-KIT2, BCL-2) and a control duplex DNA. The interaction of these thiazole peptides with G-quadruplexes has been investigated using Förster resonance energy transfer (FRET) melting analysis, fluorescence and NMR spectroscopic studies. Our results show that one molecule of thiazole peptide TH3 interacts with each of the terminal G-quartets and capping structures of 5′- and 3′-ends of c-MYC G-quadruplex. Subsequently, the downstream effect of TH3 has been investigated in human cancer cells using XTT assay, quantitative real time PCR (qRT-PCR), Western blotting, dual luciferase and flow cytometric assay.
MATERIALS AND METHODS
General materials
The general chemicals and labeled DNA sequences were purchased from Sigma-Aldrich. The detailed description of materials used in synthesis has been described in the supporting information section. The labeled DNA sequences of highest purity were purchased for best results. The typical cell culture reagents and the antibodies were purchased from Thermo Fisher Scientific and Merck Millipore, unless stated otherwise. The Annexin V-FITC kit for apoptosis assay was purchased from Life technologies. The c-MYC promoter (Del4) was a gift from Bert Vogelstein (Addgene plasmid # 16604). The BCL-2 luciferase plasmid LB322 (BCL-2 from ATG to –3934) was a gift from Linda Boxer (Addgene plasmid # 15381). The renilla luciferase plasmid pRL-TK was a gift from Dr Susanta Roychoudhury, Indian Institute of Chemical Biology, Kolkata. The dual luciferase reporter assay kit was purchased from Promega.
Synthesis of thiazole peptides
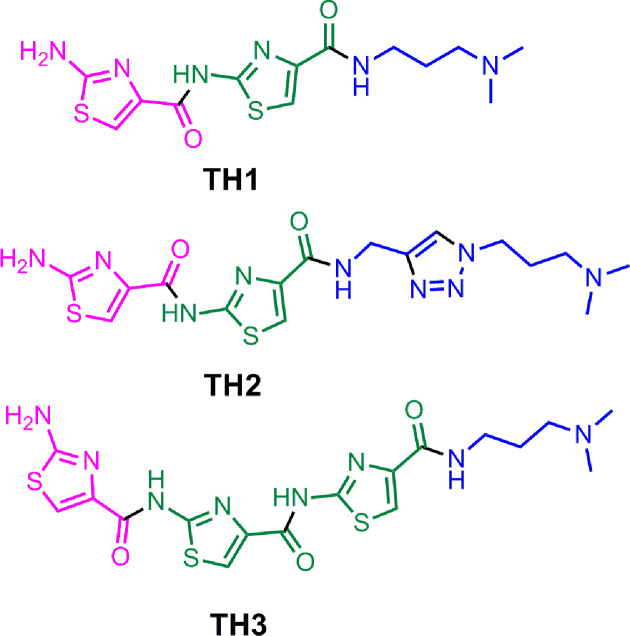
Thiazole peptides (Figure 1) were synthesized using sequential amide bond formation and outlined in Schemes S1-S7 (Supplementary Information). The detailed synthesis and characterization are described in the supplementary information.
Figure 1.
Structure of thiazole peptides (TH 1–3).
FRET melting analysis
FRET melting assay using ligands TH1–TH3 were carried out in a 96-well format on a real-time PCR apparatus (Roche LightCycler® 480 II) (24). Dual labeled DNA sequences with a donor fluorophore 6-carboxyfluorescein (5′-FAM) and an acceptor fluorophore 6-carboxytetramethylrhodamine (3′-TAMRA) were used for the study.
c-MYC14/23: 5′ FAM-d(TGAG3TG3TAG3TG3TA2)-TAMRA 3′
c-MYCPu27: 5′ FAM-d(TG4AG3TG4AG3TG4A2G2 TG4A)-TAMRA 3′
c-KIT1: 5′ FAM-d(G3AG3CGCTG3AG2AG3)-TAMRA 3′
c-KIT2: 5′ FAM-d(G3CG3CGCTAG3AG4)-TAMRA 3′
BCL-2: 5′ FAM-d(G3CGCG3AG2A2T2G3CG3)-TAMRA 3′
hairpin DNA: 5′ FAM-d(TATAGCTATA8TATAGCTATA)-TAMRA 3′
ds26: 5′-d(CA2TCG2ATCGA2T2CGATC2GAT2G)-3′
200 μM stock solutions of peptides TH1–TH3 were prepared in 60 mM potassium cacodylate buffer, or 100 mM KCl, 10 mM Tris•HCl buffer, pH 7.4. Dual labeled oligos were diluted from stock to a concentration of 400 nM in the same buffer. The diluted samples were annealed by heating to 95°C for 5 min followed by gradual cooling at 25°C and incubated overnight at 4°C. Sample solutions were prepared in a 96-well plate (100 μl final volume) by mixing pre-annealed DNA (at 200 nM final concentration) with peptides (1.0 μM final concentration) in respective buffer (60 mM potassium cacodylate, or 100 mM KCl, 10 mM Tris•HCl buffer, pH 7.4). After 1 h incubation, measurements were made in triplicate with excitation at 483 nm and detection at 533 nm. Final analysis of the data was carried out using OriginPro 8.0 (OriginLab Corp.). The detailed procedure for FRET analysis has been included in the supplementary information.
Fluorimetric titration
Fluorescence spectra were recorded on a Horiba JobinYvon Fluorolog instrument at 25°C in a thermostated cell holder using quartz cuvette of 1 cm path-length. The DNA sequences used in the study are described in the supplementary information. Briefly, DNA sequences were pre-annealed in Tris–KCl buffer (100 mM KCl, 10 mM Tris•HCl, pH 7.4). Peptides (TH1, TH2 and TH3) were diluted in filtered and degassed Tris-KCl buffer to a final concentration of 2 μM. Peptide solutions were titrated with the pre-annealed DNA sequences and the emission was recorded from the range of 315–600 nm (λex = 300 nm). The recorded spectral data was used to determine the dissociation constant of the ligands for quadruplexes using the Hill-1 formula:
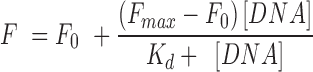 |
F is the fluorescence intensity, Fmax is the maximum fluorescence intensity, F0 is the fluorescence intensity in the absence of DNA and Kd is the dissociation constant.
NMR titration
The c-MYC DNA (c-MYC14/23 and c-MYCPu27) was purchased from Eurofins MWG Operon in HPSF purity grade and further purified with HPLC. During the titration the DNA was provided as a 100 μM solution in 25 mM Tris•HCl buffer (pH 7.4) with 100 mM KCl in 10% d6-DMSO/90% H2O. Small amounts of the ligand stock solution in 100% d6-DMSO were added directly into the NMR tube (12.8% d6-DMSO at the end of the titration). 2,2-dimethyl-2-silapentane-5-sulphonate (DSS) was used as internal reference. Watergate W5 pulse sequence with gradients (25) or jump-return-Echo (26) was used for water suppression. 2D NMR experiments were recorded on the DNA alone (1 mM) and in the complex with 3 equivalents of ligand in 25 mM potassium phosphate pH 7.0 containing 70 mM KCl in 10% d6-DMSO/90% H2O to reduce the conformational diversity according to Yang et al. (10) and to avoid loss of sensitivity due to Tris signal. 1H,13C HSQC (27,28) spectra and 2D 1H,1H NOESY spectra with excitation sculpting as water suppression scheme (29) were recorded.
Cell culture
Human cervical cancer cells (HeLa) and human alveolar basal epithelial cancer cells (A549) were obtained as monolayer culture from the National Centre for Cell Science (NCCS), Pune and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with d-glucose, l-glutamine, penicillin–streptomycin (Invitrogen) and 10% fetal bovine serum (Gibco). HeLa cells were seeded in 6-well plates at a concentration of 1 × 105 cells per well and incubated at 37°C in 5% CO2 incubator to obtain >70% confluency before treatment. Then the cells were treated with ligands and incubated under the same conditions for another 24 h and were harvested for further analysis.
Cell cytotoxicity assay
Cancer Cells (HeLa and A549) and Normal Kidney Epithelial (NKE) were seeded in a 96-well plates (1 × 103 cells/well) and exposed to various concentrations of peptides TH2 and TH3 (Control, 0.5, 1.0, 2.0, 4.0, 5.0, 10.0, 15.0, 20.0, 40.0, 60.0, 80.0 and 100.0 μM). XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) (Thermo Fisher Scientific) stock solution was prepared by mixing 4 ml (1 mg/ml) with 10 μl of the 10 mM phenazine methosulfate (PMS) (Thermo Fisher Scientific) solution. After 24 h treatment with peptide ligands, 25 μl of XTT/PMS stock solution was added to each well and further incubated for 2 h at 37°C. The absorbance was directly recorded at 450 nm by an automated microplate reader (Thermo Fisher Scientific). All experiments were performed in parallel and in triplicate, and the IC50 values were derived from the linear regression parameters using OriginPro 8.0 (OriginLab Corp.).
RNA extraction and qRT-PCR
After HeLa and A549 cells were treated with peptides TH2 and TH3 (2.0 and 5.0 μM) for 24 h, the total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The RNA was quantitated using a Cary Win 300 UV-Vis spectrophotometer and the total RNA was used as a template for reverse transcription using a Verso cDNA synthesis kit (Thermo Fisher Scientific) according to the protocol supplied. The mixtures were incubated at 42°C for 30 min for reverse transcription and then at 95°C for 2 min followed by incubation at 4°C. Afterward, PCR reactions were performed in a Light Cycler® 480 II (Roche) apparatus. A total volume of 20 μl qRT-PCR reaction mixture was prepared containing 10 μl of SYBR green Jump Start Taq Ready Mix (Sigma) reagent, 1 μl each of the forward and reverse primers (500 nM), 2 μl of cDNA (200 nM) and nuclease-free water to make up the volume. Cycle conditions were as follows: pre-incubation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 60 s, then hold at 37°C for 2 min. The primer sequences used for qRT-PCR are:
GAPDH (forward): 5′-GACG2C2GCATCT2CT2GT-3′
GAPDH (reverse): 5′-CACAC2GAC2T2CAC2AT4-3′
18S rRNA (forward): 5′-GAT2C2GTG3TG2TG2TGC-3′
18S rRNA (reverse): 5′-A2GA2GT2G5ACGC2GA-3′
c-MYC (forward): 5′-CTGCGACGAG2AG2AG2ACT-3′
c-MYC (reverse): 5′- G2CAGCAGCTCGA2T3CT2-3′
BCL-2 (forward): 5′-GAG2AT2GTG2C2T2CT3G-3′
BCL-2 (reverse): 5′-GC2G2T2CAG2TACTCAGTC-3′
We used the comparative cycle threshold method (CT method) for relative quantification of gene expression. Further details have been included in supplementary information.
Western blot analysis
HeLa cells were treated with peptide ligands TH2 and TH3 (2.0 and 5.0 μM) for 24 h and subsequently, cells were lysed with cold cell lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA in 1% Triton X-100 and 5% glycerol). Cell lysates were collected and the total protein content was estimated by the Lowry method (30). An aliquot of 70 μg of protein extract was loaded in each lane and separated in a 10% SDS-PAGE gel and electroblotted in 48 mM Tris, 39 mM Glycine, 20% methanol and 0.02% SDS on a nitrocellulose transfer membrane. Membrane was then blocked with 4% BSA in 1× TBS and 0.1% TWEEN®20, washed and probed using antibodies directed against c-MYC, BCL-2 and GAPDH (endogenous loading control) overnight at room temperature. Blots were then washed and incubated with (i) 1:2000 dilution of ALKP conjugated secondary antibody (for c-MYC) (Life technologies), (ii) 1:2000 dilution of HRP secondary antibody (for GAPDH) (Life technologies) and (iii) 1:2000 dilution of HRP secondary antibody (for BCL-2) for 2 h at room temperature. Binding signals were visualized with NBD/BCIP substrate. Relative band intensities were determined by using ImageJ software.
In situ mutagenesis, transfection and dual luciferase assay
The Del4 luciferase reporter plasmid (Addgene plasmid # 16604) was mutagenized by using the Quick Change Site directed Mutagenesis kit (Agilent Technologies, CA, USA) with the following primers:
Forward: 5′-TGAG4CG2AGCTG2C2GCACG3AGA-3′
Reverse: 5′-TCTC3GTGCG2C2AGCTC2GC4TCA-3′
Exponentially growing HeLa cells in DMEM media supplemented with 10% FBS were seeded in a six-well plate. The plasmid used for transfection was Del4 which harbors the 22-mer c-MYC G4 forming sequence in the P1 promoter upstream of the luciferase reporter (Addgene). Del4 mutant plasmid containing mutated G4-forming sequence was used as control. The G4 forming sequences upstream of luciferase reporter gene in the plasmids used in the current study are as following:
Del4: 5′-G4AG3TG4AG3TG4-3′
Mut-Del4: 5′-G4AG3TGAG2AG3TG4-3′ (substitution is underlined)
500 ng plasmid was transfected in HeLa cells growing at >70% confluency using Lipofectamine 2000 as per manufacture's protocol. After 6 h of transfection, cells were washed with PBS and fresh media was added. Cells were treated with peptides TH2 and TH3 (2.0 and 5.0 μM) for 24 h. Then cells were lysed with Cell Culture Lysis Reagent (CCLR) buffer with continuous pipetting at 48°C for 30 min. The homogenate was centrifuged for 5 min at 10,000 g. The supernatant is used for protein estimation by Lowry method (30). Luciferase assay was performed in Orion Microplate Luminometer (Berthold Detection System) for three biological replicates and luciferase activity was normalized by total protein concentration.
Cell cycle analysis
Cell cycle analysis was carried out using propidium iodide (PI) staining by flow cytometry. HeLa cells (1 × 106) per 60 mm petridish (∼80% confluence) were treated with TH3 (2.0 and 5.0 μM) for 24 h in fresh growth medium. Cells were harvested by trypsinization, resuspended in PBS and fixed with 2 ml of ice-cold 70% ethanol for overnight at 4°C. The pellets were collected by centrifugation and resuspended in PBS solution, containing 10 μg/ml PI (Sigma) and 10 μg/ml RNase A (Sigma). After incubation for 30 min in the dark at 37°C, cells were analyzed for DNA content using a FACS flow cytometer (BD Biosciences). Cell distribution among cell cycle phases were evaluated using Cell-Quest Pro software (BD).
Flow cytometric determination of apoptosis
Annexin V–FITC and propidium iodide (PI) were used to determine the percentage of cells undergo apoptosis and necrosis. Briefly, 1 × 106 HeLa cells per 60 mm Petridish (∼80% confluence) were treated with peptide TH3 (2.0 and 5.0 μM) for 24 h in fresh growth medium. Cells were harvested with trypsinization and centrifuged at 700 rpm for 5 min at 4°C. Cell pellet was suspended in 500 μl 1× binding buffer and then treated with 5 μl Annexin V–FITC and 2 μl PI. After incubation for 5 min on ice, each sample was analyzed immediately using fluorescence-activated cell sorter (FACS) analysis (BD Biosciences,Mountain View, CA, USA). Approximately 10 000 HeLa cells were detected for each sample. Cytogram analysis was done using the Cell-Quest Pro software.
Fluorescence microscopy
Cellular localization of thiazole peptide TH3 was monitored by cell imaging. HeLa cells were seeded on glass cover slips placed in 12-well cell culture plates for 24 h followed by incubation with TH3 (5.0 μM) for 2 h in CO2 (5%) incubator at 37°C. After incubation, cells were washed with PBS buffer three times and the cover slips were mounted on glass slides using NucRed (1:1 solution in PBS) and localization of TH3 was viewed under (Olympus IX 81 Confocal Laser Scanning Microscope). At least five fields per slide and three independent sets were examined.
RESULT AND DISCUSSION
Design of thiazole peptides
The five-membered thiazole heterocycle, derived by enzyme-mediated post-translational modification (31–33) of natural amino acid residues is present in numerous cyclic peptides (34–39) that exhibit pharmaceutically useful biological activities. The thiazole moiety is also present in antitumor drugs like bleomycin and tiazofurin (40). Telomestatin, one of the potent G-quadruplex binding ligands contains seven oxazole rings and one thiazoline ring system (41). Inspired from Telomestatin, numerous oxazole macrocycles have been developed for targeting DNA G-quadruplexes (39,42,43). The observation that the triazole containing distamycin and netropsin interacts with DNA minor groove by hydrophobic interaction has led to the development of other selective DNA groove targeting ligands (35). However, distamycin has also been reported to bind to G-quadruplex by specific stacking interaction with terminal G-quartets (44). Interestingly, a 4:1 (ligand:DNA) stoichiometry was reported in both models.
We have anticipated that thiazole peptides, capable of adopting crescent shape, may selectively interact with a particular G-quadruplex. The detailed synthesis of the thiazole peptides (TH 1-3) has been described in the supporting information (Figure 1 and Supplementary Figure S1, Scheme S1–S7). Peptide TH1 contains two thiazole rings whereas peptide TH2 contains two thiazole rings and a triazole ring. In peptide TH3, three thiazole rings are connected through amide bonding. To understand the difference in three dimensional structure of TH2 and TH3, energy minimized structures of TH2 and TH3 were obtained with Gaussian 03 using DFT (Density functional theory) analysis B3LYP/6-31+G(d) level (Supplementary Figures S2 and S3). Unlike TH2, the HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital) orbitals of TH3 are non-overlapping, distributed over a larger area. Therefore, effective conjugation of π-electrons is expected in peptide TH3. On the other hand, the presence of an additional –CH2 linker between thiazole and triazole moieties in peptide TH2 inhibits the π-electron delocalization and provides flexibility to form distorted structure. In comparison, TH3 exists in planar crescent shape as the peptide linkages are less flexible due to their partial double bond character and able to mediate effective π-conjugation. The HOMO-LUMO energy difference values, calculated for peptides TH2 and TH3 were 6.8 and 6.5 eV, respectively. In this study, thiazole peptides (TH 1–3) having different length and three dimensional structures have been investigated as G-quadruplex targeting ligands for chemical regulation of oncogene expression and anticancer therapeutics.
Quadruplex specificity of thiazole peptides
The ability of thiazole peptides to distinguish between quadruplexes was examined by FRET melting assay and fluorimetric titration experiments. We have used four different intramolecular quadruplexes like c-MYC, c-KIT1, c-KIT2 and BCL-2, a control hairpin DNA and a self complementary duplex DNA (ds26). For the c-MYC, we have used a wild-type 27-mer c-MYCPu27 sequence as well as a 22-mer c-MYC14/23 conformer that contains two G-to-T mutations at 14 and 23 positions of the c-MYCPu27 sequence (10).
FRET melting assay
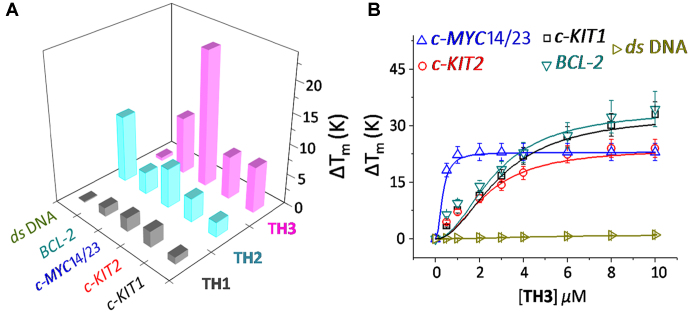
FRET melting analysis was employed to evaluate the ability of thiazole peptides (TH1-TH3) to stabilize the dual labeled (FAM and TAMRA at 5′- and 3′-ends, respectively) G-quadruplexes (c-MYC, c-KIT1, c-KIT2 and BCL-2) and a control hairpin DNA (Figure 2A) (24). The dipeptide TH1 (1.0 μM, 5.0 equiv.) displayed negligible stabilization potential values for G-quadruplexes and hairpin DNA. Although the dipeptide TH2 (1.0 μM, 5.0 equiv.) containing a triazole moiety showed low ΔTm values (2–6°C) for quadruplexes, it could increase the melting temperature of hairpin DNA (ΔTm = 11°C) (Figure 2, Table 1, Supplementary Table S1). Tripeptide TH3 exhibited a high stabilization potential for the c-MYC14/23 G-quadruplex (ΔTm = 22°C) compared to dipeptides TH1 and TH2 at 1.0 μM ligand concentration (5.0 equiv. with respect to DNA). It is further interesting to note that TH3 (1.0 μM, 5.0 equiv.) could preferentially stabilize the c-MYC14/23 G-quadruplex (ΔTm ∼ 22°C) over the c-KIT1, c-KIT2, and BCL-2 quadruplexes (ΔTm ∼ 7 – 9°C). In addition, TH3 did not alter the Tm value of hairpin DNA.
Figure 2.
(A) Stabilization potential (ΔTm) of the ligands TH1, TH2 and TH3 for G-quadruplex and hairpin DNA by FRET melting assay; (B) Thermal shift profiles of TH3 upon interacting with quadruplexes and hairpin DNA in 60 mM K+-cacodylate buffer, pH 7.4.
Table 1. Sequences used in this study and binding data obtained from FRET melting (ΔTm) and fluorescence studies (Kd).
| TH1 | TH2 | TH3 | ||||
|---|---|---|---|---|---|---|
| DNAa | ΔTmb | K d c | ΔTmb | K d c | ΔTmb | K d c |
| c-MYC14/23: 5′-d(TGAG3TG3TAG3TG3TA2)-3′ | 2.5 | 4.81 | 6.5 | 2.82 | 22.0 | 0.25 |
| c-KIT1: 5′-d(G3AG3CGCTG3AG2AG3)-3′ | 1.4 | 5.15 | 3.7 | 3.27 | 9.5 | 1.17 |
| c-KIT2: 5′-d(G3CG3CGCTAG3AG4)-3′ | 1.1 | 20.52 | 2.5 | 3.53 | 7.6 | 1.34 |
| BCL-2: 5′-d(GGGCGCGGGAGGAATTGGGCGGG)-3′ | 2.0 | 9.81 | 3.9 | 4.01 | 7.1 | 1.22 |
| ds DNA: 5′-d(TATAGCTATA8TATAGCTATA)-3′ | 1.0 | n.d | 11.0 | 0.90 | 2.0 | n.d. |
aThe Tm values of the quadruplexes in 60 mM potassium cacodylate buffer, pH 7.4 in the absence of ligands are: c-MYC14/23 (70±1), BCL-2 (70±1), c-KIT1 (57±1), c-KIT2 (69±1), ds DNA (60±1) °C; maximum measurable Tm = 93 °C. Dual FAM-TAMRA labeled sequences were used in the FRET melting experiments.
bΔTm at 1 μM ligand concentration [°C] (ΔTm = ± 1°C).
c K d = ± 10% (expressed in μM).
Next, the FRET melting analysis of promoter quadruplexes and hairpin DNA was carried out with incremental addition of TH3 (0–10 μM) (Figure 2B). A maximum ΔTm value of 22 ± 1°C (i.e. a Tm of 92°C) was observed for the c-MYC14/23 in the presence of 1.0 μM TH3 (Figure 2B), whereas 5–7 fold higher concentrations of TH3 was required to attain similar ΔTm values (ΔTm = 22°C) for c-KIT1, c-KIT2 and BCL-2 suggesting its high affinity towards the c-MYC14/23 quadruplex. TH3 (1.0 μM) could stabilize c-MYCPu27 with a ΔTm value of 21 ± 1°C (i.e. a Tm of 91°C) (Supplementary Figure S4). In addition, peptide TH3 does not significantly alter the melting temperature of hairpin DNA even at 10.0 μM ligand concentration (50.0 equiv.).
A FRET competitive experiment was performed to evaluate the selectivity of TH3 for the c-MYC G-quadruplex over double stranded DNA (Supplementary Figure S5). The melting of 200 nM dual labeled c-MYC14/23 was carried out with TH3 (1 μM) in the presence of a competitor ds26 DNA. The results indicated that the ΔTm value of TH3 for the c-MYC14/23 G-quadruplex was not significantly decreased by adding excess of ds26 DNA, at ds26/c-MYC14/23 ratios up to 50/1 (Supplementary Figure S5).
Fluorimetric titrations
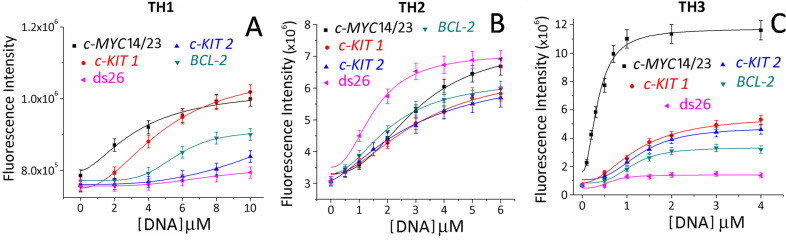
The interaction of thiazole peptides with G-quadruplexes (c-MYC, BCL-2, c-KIT1 and c-KIT2) and ds26 (Figure 3) was investigated using fluorescence spectroscopy. Peptides TH1, TH2 and TH3 exhibited a single broad band with an emission maximum at 430 nm, when excited at 300 nm (λabs = 300 nm) in buffer solution (10 mM Tris•HCl, 100 mM KCl, pH 7.4) (Figure 3, Supplementary Figures S6–S10). The binding of TH3 to the c-MYC14/23 quadruplex resulted in ∼6-fold intensification of the fluorescence at saturation. In contrast, small changes in the fluorescence intensity of TH1 and TH2 were observed upon the addition of DNA quadruplexes. These changes in fluorescence were used to determine affinities for the binding of the peptides to quadruplexes. The dissociation constant (Kd) of ligand TH3 is 0.25 μM for the c-MYC14/23 quadruplex while TH1 and TH2 exhibited Kd values of 4.81 μM and 2.82 μM for the c-MYC14/23 G-quadruplex, respectively. TH3 also binds to c-MYCPu27 (Kd = 0.29 μM) with comparable affinity as c-MYC14/23 (Supplementary Figure S11). These results indicate that TH3 displays strong binding for the c-MYC G-quadruplex with ∼19- and ∼11-fold higher binding affinity compared to TH1 and TH2, respectively. Similar binding titrations with other promoter quadruplexes indicated ∼4–5-fold selectivity of TH3 for the c-MYC G-quadruplex (Kd ∼ 0.25 μM) over c-KIT1, c-KIT2 and BCL-2 quadruplexes (Kd ∼ 1.17–1.34 μM) (Figure 3, Table 1). The Job's plot analysis revealed that the TH3 binds to the c-MYC14/23 G-quadruplex with a 2:1 stoichiometry (Supplementary Figure S12).
Figure 3.
Fluorescence titration of (A) TH1 (2 μM) with c-MYC14/23, c-KIT1, c-KIT2, BCL-2 and ds26 (0–10 equiv.), (B) TH2 (2 μM) with c-MYC14/23, c-KIT1, c-KIT2, BCL-2 and ds26 (0–6 equiv.) and (C) TH3 (2 μM) with c-MYC14/23, c-KIT1, c-KIT2, BCL-2 and ds26 (0–4 equiv.).
Moreover, ligands TH1 and TH3 exhibited negligible changes in fluorescence intensities upon addition of 4 equiv. ds26 duplex DNA. However, an increase in fluorescence intensity of TH2 was observed in the presence of ds26 (Kd = 0.9 μM) indicating its high binding affinity for the ds26. These results indicate that thiazole tripeptide TH3 binds more strongly to quadruplexes than dipeptides TH1 and TH2. Furthermore, TH3 binds to the c-MYC with high specificity over other quadruplexes and ds26.
Binding mode of TH3 with c-MYC G-quadruplex
NMR titrations
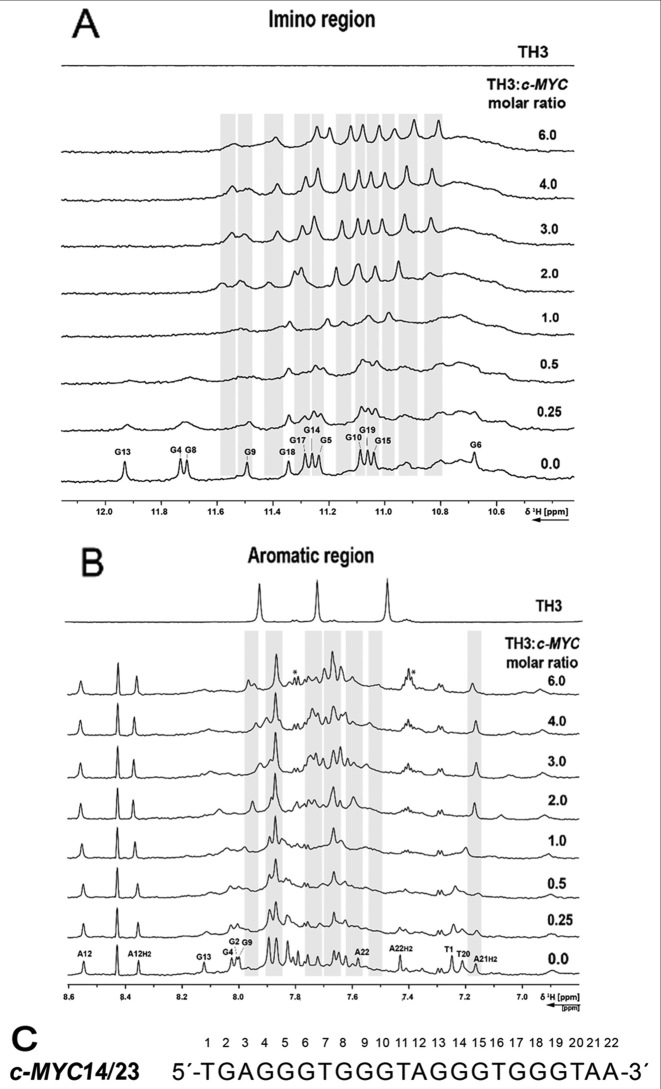
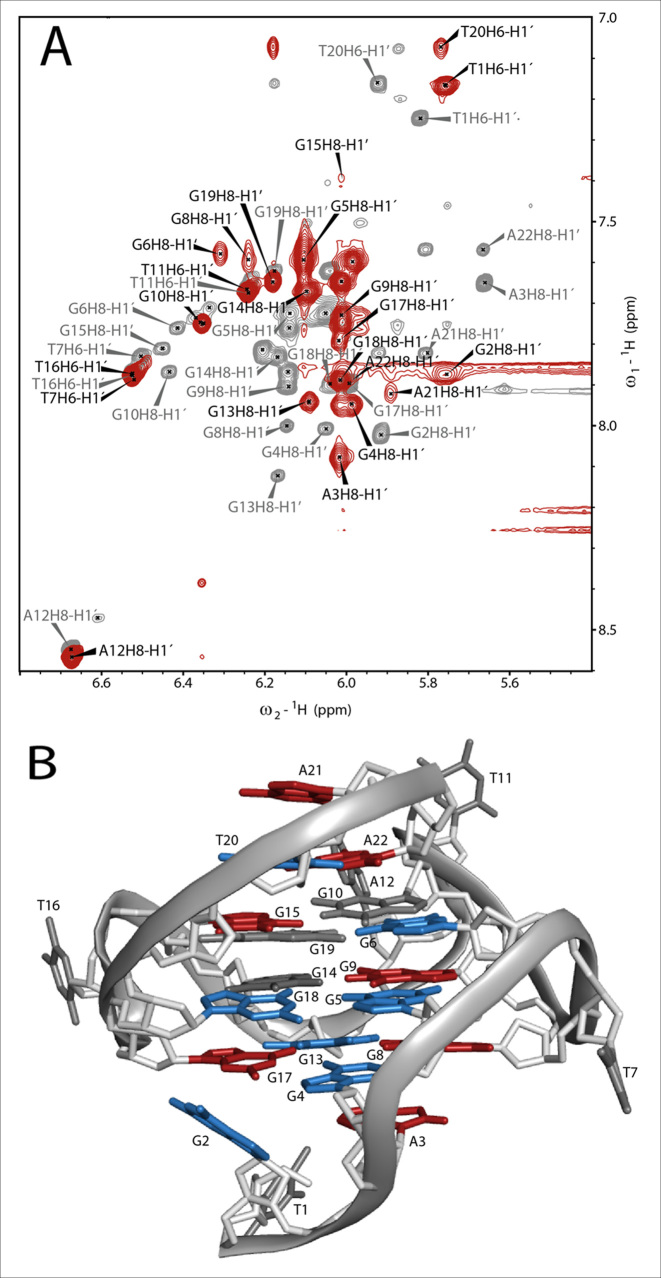
The interaction of TH3 with c-MYC14/23 (45) and the c-MYCPu27 (45) (Supplementary Figure S13) (Figure 4) was investigated by recording 1D 1H NMR spectra at different [TH3]:[DNA] ratios. The titrations were performed in 10% d6-DMSO/90% H2O due to low solubility of the peptide TH3 in NMR buffer (Supplementary Figures S14 and S15). The sequence c-MYCPu27 is known to form a mixture of G-quadruplex conformations with different loops (45) and the imino pattern in the 1D 1H NMR spectrum is very broad and not suitable for a detailed NMR assignment. Only minor changes could be observed on the 1D 1H NMR spectrum of c-MYCPu27 DNA after ligand addition and none of the various conformations seemed to be stabilized by TH3 (Supplementary Figure S13). The c-MYC14/23 G-quadruplex mutant forms a defined G-quadruplex structure that has been characterized by Yang et al. (PDB: 1XAV) (10). The effect of DMSO on the G-quadruplex formed by the sequence c-MYC14/23 G-quadruplex has been previously discussed (46) and the assignment for the target DNA has been transferred from Yang et al. (10,11). Clear changes in the 1D 1H spectrum upon addition of the ligand to the G-quadruplex formed by the sequence c-MYC14/23 were detected. Upon addition of 0.25 eq. of TH3, strong line broadening was observed for all the imino signals (Figure 4A) as well as for most aromatic signals (Figure 4B). At a [TH3]:[DNA] ratio of 2:1, a new set of imino proton signals appeared (Figure 4A), suggesting that the ligand is binding to the DNA with a 2:1 stoichiometry in slow exchange on the NMR time scale. With further addition of ligand, the imino signals became sharper.
Figure 4.
(A) Imino and (B) aromatic region of 1D 1H NMR spectrum of the c-MYC14/23 G-quadruplex DNA with increasing [TH3]:[DNA] ratio and ligand alone. The spectra were recorded at 298 K, 700 MHz. Experimental conditions: 100 μM DNA in 25 mM Tris•HCl (pH 7.4) buffer containing 100 mM KCl in 10% d6-DMSO/90% H2O. (C) Sequence of c-MYC14/23 with the numbering used for the partial assignment of DNA signals. Signals marked with a star are arising from ligand self-aggregation (∼10%, Supplementary Figure S16) after transferring in buffer during the titration. These aggregated species do not interact with the DNA (Supplementary Figure S17).
In the literature, different binding modes to G-quadruplex have been proposed for distamycin A, structurally related to TH3. Maizels and co-workers observed a 4:1 stoichiometry with two molecules binding to each of the two external tetrads (44). Randazzo and co-workers reported that distamycin A binds as a dimer to the grooves of a tetramolecular G-quadruplex with a 4:1 stoichiometry (47,48).
CSP analysis
In order to gain an insight into the binding mode of TH3, 2D 1H, 1H-NOESY and 2D 1H, 13C-HSQC have been recorded for the assignment of the G-quadruplex in complex with TH3 (Supplementary Figures S18–S19). However, the signals of the ligand in the bound form could not be detected and no NOE intermolecular crosspeaks could be observed under the experimental conditions used in our studies. Therefore, the chemical shift perturbations (CSPs) after ligand binding have been analyzed in detail. The assignment of the complex used for CSP calculation (Supplementary Equation S2) is shown in Figure 5A with black labels, while the assignment of the G-quadruplex alone is in gray labels. The combined CSPs (Supplementary Table S4) of the imino protons (H1, Supplementary Figure S19, Supplementary Table S2) and of the anomeric (C1′H1′, Supplementary Figure S18A, Supplementary Table S3) and aromatic (C8/C6, H8/H6, Supplementary Figure S18B, Supplementary Table S3) signals are displayed as bar chart in Supplementary Figure S20 and on the structure of c-MYC14/23 (Figure 5B and Supplementary Figure S21A), where the nucleobases were colored according to three different categories: non or hardly shifted (CSP ≤ 0.1, gray), moderately shifted (CSP = 0.1–0.2, blue) and strongly shifted (CSP > 0.2, red).
Figure 5.
(A) Overlay of the H1′H6/H1′H8 region of the 1H,1H-NOESYs of c-MYC14/23 (gray) and the complex (red). The assignment of c-MYC14/23 (gray labels) has been transferred from Yang et al. (10). The attempt of assignment of the signals for the complex (black labels) is based on the analysis of 1H,13C-HSQC (Supplementary Figure S18) and the 1H, 1H-NOESY spectra. In the overlay it is already possible to estimate qualitatively the strength of chemical shift perturbation (CSP) of the anomeric (H1′) and aromatic (H6/H8) protons after ligand binding. Experimental conditions: 298 K, 25 mM potassium phosphate pH 7.0, 70 mM KCl, 10% d6-DMSO,1 mM DNA (gray spectrum) and 1 mM DNA with 3 eq. of TH3. (B) Structure of c-MYC14/23 (PDB: 1XAV) with colored nucleobases according to their combined CSP after ligand binding with residue numbering according to Figure 4C. Gray: non shifted (CSP ≤ 0.1), blue: moderately shifted (CSP = 0.1 – 0.2) and red: strongly shifted (CSP > 0.2).
The nucleobases with the strongest CSP (red color code) are located at the 3′- and 5′-ends (A3, G8, G15, G17, A21 and A22) of the G-quadruplex structure. The lack of ligand signals in the bound state does not allow us to determine whether the ligand interacts with the 3′- and 5′-capping structures, with the terminal G-quartets or it is sandwiched between both of them. According to the CSP analysis together with the titration monitored by 1D 1H spectra pointing at a complex with a 2:1 [TH3]:[DNA] ratio, we propose that one ligand molecule is binding at the 3′-end and the other one at the 5′-end. This binding mode is comparable with the one observed by Maizels and co-workers (44), although they reported a different binding stoichiometry. We did not observe induced CD (circular dichroism) signals upon titrating c-MYC14/23 with TH3, which suggests that ligand does not bind to the G-quadruplex grooves as observed in case of distamycin A by Randazzo and coworkers (Supplementary Figure S22).
Structure-activity relationship
The differences in 3D structures of peptides TH 1-3 may provide the rationale behind their differential affinity towards different G-quadruplexes. Out of these three peptides, only TH3 exist in planar crescent structure due to extended π-electron conjugation between three thiazole moieties (Supplementary Figure S3). Owing to the lack of extended π-electron conjugation in TH1 (only two thiazole moieties present) and TH2 (additional –CH2 present between thiazole and triazole moieties), they do not exhibit the planar crescent structure. As a result, their recognition sites in a G-quadruplex structure is expected to be different.
Although the G-quadruplex structures (c-MYC14/23, c-KIT1, c-KIT2 and BCL-2), used in this study exist in parallel topology, the intermediate loop sequences and flanking regions (capping structures) are quite different (Supplementary Figure S21). A close inspection of these structures indicates that c-MYC14/23 G-quadruplex contains AT-rich capping structures at both 5′ and 3′ ends, which are absent or truncated in other G-quadruplexes. The presence of extended planar structure of TH3 promotes higher stacking interaction with the terminal G-quartets compared to TH1 and TH2. In addition, the NMR analysis shows that the peptide TH3 interacts with the AT-rich 5′ and 3′ ends of c-MYC G-quadruplex structure. These capping structures are unique in c-MYC G-quadruplex structure, which may be the reason for the observed selectivity of peptide TH3 towards c-MYC G-quadruplex over other G-quadruplexes.
Ligand dependent c-MYC expression in cancer cells
Since TH3 was found to be a selective ligand for targeting c-MYC G-quadruplex structure over other promoter G-quadruplexes, the influence of TH3 on the c-MYC gene expression was evaluated using qRT-PCR analysis, western blot and dual-luciferase assay. For a comparison, the influence of a less potent G-quadruplex binder, TH2 on the c-MYC gene expression was also investigated.
qRT-PCR analysis
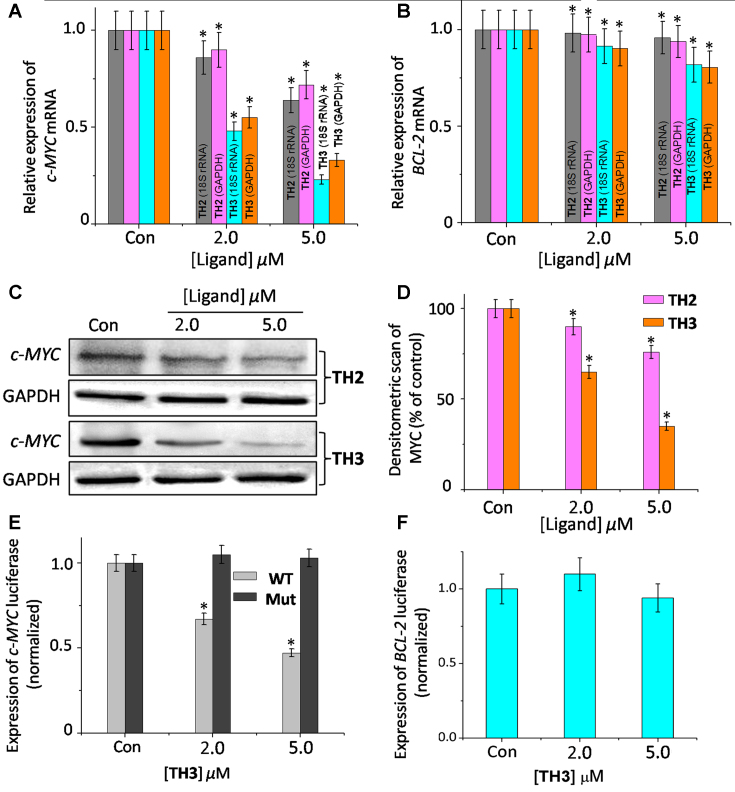
The effect of ligands TH2 and TH3 on c-MYC transcription was investigated by monitoring the mRNA expression profiles in human cervical cancer cells (HeLa) cells (Figure 6A, B and Supplementary Figure S23). Cells were treated with various concentrations (2.0 and 5.0 μM) of TH2 and TH3 for 24 h, and the expression of c-MYC mRNA was normalized against the constitutively expressed housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). TH3 reduced the c-MYC mRNA level to 0.3-fold (decreased by 70% relative to control cells, treated with 0.1% DMSO), whereas TH2 exhibited a small change in the c-MYC mRNA expression (0.72 fold, i.e. 28% decrease) relative to the control cells (treated with 0.1% DMSO). When 18S rRNA was used to normalize gene expression, similar results were obtained; the c-MYC mRNA levels were reduced to 0.7 fold and 0.3 fold by TH2 and TH3, respectively. We have also monitored the c-MYC mRNA profiles upon treatment with peptides TH2 and TH3 in human alveolar basal epithelial cancer cells (A549) with respect to control GAPDH and 18S rRNA. The results show that TH2 and TH3 could reduce the c-MYC mRNA levels to ∼0.8- and ∼0.25-fold (i.e. decrease by 20% for TH2 and decrease by 75% for TH3), respectively relative to the control cells (Supplementary Figure S24). In addition, the effect of peptides TH2 and TH3 was also studied on the expression of another G-quadruplex forming BCL-2 oncogene. Interestingly, TH2 and TH3 did not induce any significant change in BCL-2 mRNA (<25%) expression (Figure 6B). Furthermore, TH2 and TH3 did not essentially alter the mRNA levels of GAPDH and 18S rRNA. These results suggest that TH3 inhibits the c-MYC transcription with three fold higher efficiency than TH2 and further, both these peptides do not alter the expression of housekeeping genes.
Figure 6.
Determination of transcriptional regulation of (A) c-MYC mRNA, (B) BCL-2 mRNA in the presence of TH2 and TH3 in cancer cells (HeLa) by qRT-PCR and quantified by double delta Ct analysis using 18S rRNA and GAPDH as reference genes. Data is presented in terms of fold change. Data shown as mean ± SD. *P < 0.05, versus untreated cancer cells; (C) immunoreactive bands of c-MYC protein were analyzed by Western blot in HeLa cells. Data shown as mean ± SD. *P < 0.05, versus untreated cancer cells; (D) the protein expression of c-MYC protein in the presence of TH2 and TH3 in HeLa cells. Relative luciferase expression in (E) Del4 c-MYC/mut-c-MYC and (F) LB322 BCL-2 promoter containing firefly plasmid normalized with pRL-TK Renilla plasmid (FF/RL) upon treatment of TH3 in HeLa cells, Data shown as mean ± SD. *P < 0.05, versus untreated cancer cells.
Western blot
Western blot analysis was carried out to examine the effect of peptides TH2 and TH3 on the translation of c-MYC in HeLa cells (Figure 6C). Consistent with the qRT-PCR results, the densitometric analysis showed that TH2 and TH3, at 5.0 μM concentration, reduced the c-MYC protein levels by ∼27% and ∼56%, respectively (Figure 6D), compared to the control cells. Importantly, the treatment of peptides TH2 and TH3 did not significantly alter the BCL-2 gene expression at protein level (<25%) (Supplementary Figure S25). Furthermore, the protein levels of housekeeping GAPDH gene remained unchanged upon treatment with TH2 and TH3. These results along with the qRT-PCR analysis indicate that the tripetide, TH3 is more potent in reducing the c-MYC expression compared to the dipeptide TH2.
Dual-luciferase assay
In order to investigate the effect of these peptides (TH2 and TH3) on the c-MYC gene expression, the dual-luciferase reporter assay was carried out (Figure 6E). The assay was performed using two reporter vectors containing (i) wild-type c-MYC G-quadruplex forming sequence in the upstream region of firefly luciferase coding gene (Del 4) and (ii) non G-quadruplex promoter sequence in the upstream region of renilla luciferase gene (pRL-TK). These two vectors were co-transfected into HeLa cells with subsequent treatment of 5 μM TH2 or TH3. Owing to the absence of G-rich sequence in pRL-TK, the Renilla luciferase expression was unaffected by the peptides. The expression of c-MYC firefly luciferase was normalized relative to the Renilla luciferase expression. Our results suggest that peptide TH3 decreased the wild-type (WT) c-MYC promoter-linked luciferase expression by 52%, relative to the untreated sample (Figure 6E). However, only a small change in the WT-c-MYC luciferase expression was observed upon treatment with TH2 (Supplementary Figure S26).
To further validate our results, the effect of TH2 and TH3 on c-MYC firefly luciferase vector containing a mutated G-quadruplex promoter sequence (Mut-c-MYC) was also investigated (Supplementary Figure S26). Interestingly, we did not observe any notable change in Mut-c-MYC firefly luciferase expession upon treatment of TH2 and TH3 compared to the untreated control cells. In addition, TH3 did not show any significant changes in the expression of reporter vector containing other promoter G-quadruplex forming sequences such as BCL-2 (LB322 plasmid, Figure 6F). These results indicate that TH3 downregulates c-MYC expression by directly targeting the c-MYC promoter quadruplex in cancer cells.
Antiproliferative activity of thiazole peptides in cancer cells
Cell cytotoxicity assay
As thiazole derivatives exhibit anticancer activities (49,50), we have evaluated the ability of thiazole peptides TH2 and TH3 to inhibit the growth of cancer cells. The growth inhibitory activities of peptides TH2 and TH3 for HeLa and A549 cancer cells as well as for normal human kidney epithelial cells (NKE) were investigated using XTT assay (Thermo scientific) (Supplementary Figures S27 and S28). Cells treated with 0.1% DMSO was used as control. TH3 exhibited an IC50 value of 3.8 ± 0.6 μM for HeLa cells and 3.2 ± 0.4 μM for A549 cells, whereas TH2 showed significantly higher IC50 values of 17.6 ± 2.8 μM and 15.8 ± 2.2 μM for HeLa and A549 cells, respectively. Comparatively, both ligands TH2 and TH3 exhibited negligible cytotoxicity for normal cells (IC50 ≥ 50 μM).
Cell cycle analysis
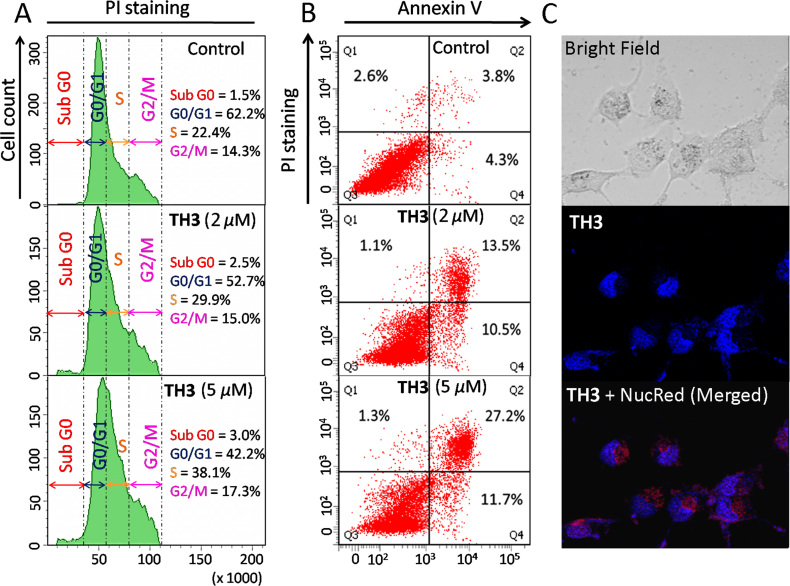
Flow cytometric analysis was conducted to examine whether the inhibitory effect of ligand TH3 in HeLa cell proliferation was associated with cell cycle arrest (Figure 7A). The analysis of cell cycle histograms revealed an arrest in S phase population (22.4–38.1%) upon treatment with 5.0 μM TH3. The observed growth arrest in the S phase (Synthesis phase) by TH3 indicates that it induces growth inhibition in HeLa cells.
Figure 7.
(A) Flow cytometric analysis of cell cycle parameters after incubation of HeLa cancer cells with TH3 (2.0 and 5.0 μM). (B) Flow cytometric analysis of the mode of cancer cell death after treatment with TH3 (2.0 and 5.0 μM) in HeLa cancer cells; Lower left (Q3), lower right (Q4), upper right (Q2) and upper left (Q1) quadrants indicate healthy cells, early, late apoptotic and necrotic cells, respectively. (C) Fluorescence microscopic image of localization of TH3 (5.0 μM) in HeLa cell.
Annexin V-FITC/PI apoptosis assay
The mode of cell death induced by peptide TH3 was investigated by flow cytometry using Annexin V and PI dual staining assay (Figure 7B). HeLa cells were treated with various concentration of TH3 (2.0 and 5.0 μM) for 24 h. The analyses of dot-plots show a dose dependent increase in the population of late apoptotic cells from ∼3.8% to ∼27.2% at 5.0 μM concentration of TH3. However, the population of necrotic cells decreases from ∼2.8% to ∼1.3% relative to the control cells (treated with 0.1% DMSO). These results indicate that G-quadruplex binding peptide TH3 causes cancer cell death by inducing apoptosis.
Cell imaging using thiazole peptides
Fluorescence microscopy was employed to examine the cellular internalization and localization of TH3 (Figure 7C). HeLa cells were treated with TH3 (5.0 μM) for 2 h, and the fluorescence microscopic images were taken. The merged image shows that TH3 localizes inside the nucleus. These results suggest that ligand TH3 is cell membrane permeable and it binds to the cellular DNA.
CONCLUSION
In summary, we have synthesized novel thiazole peptides and demonstrated that a thiazole tripeptide, structurally related to distamycin A, selectively binds to c-MYC quadruplex over other investigated G-quadruplexes and duplex DNA. Intriguingly, NMR analysis suggests that two molecules of this crescent-shaped tripeptide bind to two terminals of c-MYC quadruplex; one molecule binds to the 5′-end and another one at the 3′-end. More importantly, the tripeptide significantly inhibits the transcription of the c-MYC oncogene by a quadruplex dependent mechanism. Since structurally related natural product distamycin derivatives are known as DNA binding ligands, the insights gained from this study would inspire the development of structural mimics of natural products for site-specific targeting of DNA structures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Department of Science and Technology (DST) and CSIR-India for funding. J.D. thanks DST for a SwarnaJayanti fellowship. D.D. thanks CSIR-India for research fellowship. M.D. and R.P. thank DST for INSPIRE fellowships. The authors thank DBT-IPLS unit, University of Calcutta for confocal microscopy. The work was supported by LOEWE program: SYNCHEMBIO and by iNEXT, project number 653706, funded by the Horizon 2020 program of the European Union. H.S. is member of the DFG-funded cluster of excellence: macromolecular complexes (EXC 115). Work at BMRZ is supported by the state of Hesse.
Author Contributions: All authors have given approval to the final version of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Science and Technology [DST/SJF/CSA-01/2015-16]. Funding for open access charge: Institute of Organic Chemistry and Chemical Biology, Goethe University Frankfurt.
Conflict of interest statement. None declared.
REFERENCES
- 1. Collie G.W., Parkinson G.N.. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011; 40:5867–5892. [DOI] [PubMed] [Google Scholar]
- 2. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hänsel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 4. Simonsson T., Kubista M., Pecinka P.. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998; 26:1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fernando H., Reszka A.P., Huppert J., Ladame S., Rankin S., Venkitaraman A.R., Neidle S., Balasubramanian S.. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006; 45:7854–7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dexheimer T.S., Sun D., Hurley L.H.. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006; 128:5404–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brooks T.A., Hurley L.H.. Targeting MYC expression through G-quadruplexes. Genes Cancer. 2010; 1:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Watson J.D., Crick F.H.C.. The structure of DNA. Cold Spring Harbor Symp. Quantum Biol. 1953; 18:123–131. [DOI] [PubMed] [Google Scholar]
- 9. Phan A.T., Modi Y.S., Patel D.J.. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004; 126:8710–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ambrus A., Chen D., Dai J., Jones R.A., Yang D.. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005; 44:2048–2058. [DOI] [PubMed] [Google Scholar]
- 11. Mathad R.I., Hatzakis E., Dai J., Yang D.. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III 1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011; 39:9023–9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alzeer J., Vummidi B.R., Roth P.J., Luedtke N.W.. Guanidinium-Modified phthalocyanines as High-Affinity G-Quadruplex fluorescent probes and transcriptional regulators. Angew. Chem. Int. Ed. 2009; 48:9362–9365. [DOI] [PubMed] [Google Scholar]
- 14. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Agarwal T., Roy S., Chakraborty T.K., Maiti S.. Selective Targeting of G-Quadruplex Using Furan-Based Cyclic Homooligopeptides: Effect on c-MYC Expression. Biochemistry. 2010; 49:8388–8397. [DOI] [PubMed] [Google Scholar]
- 16. Zeng D.-Y., Kuang G.-T., Wang S.-K., Peng W., Lin S.-L., Zhang Q., Su X.-X., Hu M.-H., Wang H., Tan J.-H.. Discovery of novel 11-Triazole substituted benzofuro [3, 2-b] quinolone derivatives as c-myc G-Quadruplex specific stabilizers via click chemistry. J. Med. Chem. 2017; 60:5407–5423. [DOI] [PubMed] [Google Scholar]
- 17. Debnath M., Ghosh S., Chauhan A., Paul R., Bhattacharyya K., Dash J.. Preferential targeting of i-motifs and G-quadruplexes by small molecules. Chem. Sci. 2017; 8:7448–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kendrick S., Muranyi A., Gokhale V., Hurley L.H., Rimsza L.M.. Simultaneous drug targeting of the promoter MYC G-Quadruplex and BCL2 i-Motif in diffuse large B-Cell lymphoma delays tumor growth. J. Med. Chem. 2017; 60:6587–6597. [DOI] [PubMed] [Google Scholar]
- 19. Wang M., Mao Z., Kang T.-S., Wong C.-Y., Mergny J.-L., Leung C.-H., Ma D.-L.. Conjugating a groove-binding motif to an Ir (III) complex for the enhancement of G-quadruplex probe behavior. Chem. Sci. 2016; 7:2516–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nanjunda R., Musetti C., Kumar A., Ismail M.A., Farahat A.A., Wang S., Sissi C., Palumbo M., Boykin D.W., Wilson W.D.. Heterocyclic dications as a new class of telomeric G-Quadruplex targeting agents. Curr. Pharm. Des. 2012; 18:1934–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma D.-L., Chan D.S.-H., Fu W.-C., He H.-Z., Yang H., Yan S.-C., Leung C.-H.. Discovery of a natural Product-Like c-myc G-Quadruplex DNA Groove-Binder by molecular docking. PLOS ONE. 2012; 7:e43278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang J., Li G., Wu Z., Song Z., Zhou Y., Shuai L., Weng X., Zhou X., Yang G.. Bisbenzimidazole to benzobisimidazole: from binding B-form duplex DNA to recognizing different modes of telomere G-quadruplex. Chem. Commun. 2009; 902–904. [DOI] [PubMed] [Google Scholar]
- 23. Jain A.K., Bhattacharya S.. Interaction of G-Quadruplexes with nonintercalating Duplex-DNA minor groove binding ligands. Bioconjugate Chem. 2011; 22:2355–2368. [DOI] [PubMed] [Google Scholar]
- 24. De Cian A., Guittat L., Kaiser M., Saccà B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M.-P., Lacroix L., Mergny J.-L.. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007; 42:183–195. [DOI] [PubMed] [Google Scholar]
- 25. Liu M., Mao X.-A., Ye C., Huang H., Nicholson J.K., Lindon J.C.. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J. Magn. Reson. 1998; 132:125–129. [Google Scholar]
- 26. Sklenář V., Bax A.. Spin-echo water suppression for the generation of pure-phase two-dimensional NMR spectra. J. Magn. Reson. 1987; 74:469–479. [Google Scholar]
- 27. Palmer A.G. III, Cavanagh J., Wright P.E., Rance M.. Sensitivity improvement in Proton-Detected Two-Dimensional heteronuclear correlation NMR spectroscopy heteronuclear correlation NMR spectroscopy. J. Magn. Reson. 1990; 93:151–170. [Google Scholar]
- 28. Kay L., Keifer P., Saarinen T.. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 1992; 114:10663–10665. [Google Scholar]
- 29. Hwang T.L., Shaka A.J.. Water suppression that works. Excitation sculpting using arbitrary Wave-Forms and Pulsed-Field gradients. J. Magn. Reson. 1995; 112:275–279. [Google Scholar]
- 30. Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J.. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951; 193:265–275. [PubMed] [Google Scholar]
- 31. Roy R.S., Gehring A.M., Milne J.C., Belshaw P.J., Walsh C.T.. Thiazole and oxazole peptides: biosynthesis and molecular machinery. Nat. Prod. Rep. 1999; 16:249–263. [DOI] [PubMed] [Google Scholar]
- 32. Hamada Y., Shioiri T.. Recent progress of the synthetic studies of biologically active marine cyclic peptides and depsipeptides. Chem. Rev. 2005; 105:4441–4482. [DOI] [PubMed] [Google Scholar]
- 33. Bagley M.C., Dale J.W., Merritt E.A., Xiong X.. Thiopeptide antibiotics. Chem. Rev. 2005; 105:685–714. [DOI] [PubMed] [Google Scholar]
- 34. Brockmann A., Strittmatter T., May S., Stemmer K., Marx A., Brunner T.. Structure–Function relationship of Thiazolide-Induced apoptosis in colorectal tumor cells. ACS Chem. Biol. 2014; 9:1520–1527. [DOI] [PubMed] [Google Scholar]
- 35. Padroni G., Parkinson J.A., Fox K.R., Burley G.A.. Structural basis of DNA duplex distortion induced by thiazole-containing hairpin polyamides. Nucleic Acids Res. 2018; 46:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. García-Reynaga P., VanNieuwenhze M.S.. A new total synthesis of patellamide A. Org. Lett. 2008; 10:4621–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Banker R., Carmeli S.. Tenuecyclamides A− D, cyclic hexapeptides from the cyanobacterium nostoc spongiaeforme var. tenue. J. Nat. Prod. 1998; 61:1248–1251. [DOI] [PubMed] [Google Scholar]
- 38. Todorova A.K., Juettner F., Linden A., Pluess T., von Philipsborn W.. Nostocyclamide: a new macrocyclic, thiazole-containing allelochemical from Nostoc sp. 31 (cyanobacteria). J. Org. Chem. 1995; 60:7891–7895. [Google Scholar]
- 39. Tera M., Ishizuka H., Takagi M., Suganuma M., Shin-ya K., Nagasawa K.. Macrocyclic hexaoxazoles as Sequence-and Mode-Selective G-Quadruplex binders. Angew. Chem. Int. Ed. 2008; 47:5557–5560. [DOI] [PubMed] [Google Scholar]
- 40. Meunier B. Hybrid molecules with a dual mode of action: dream or reality. Acc. Chem. Res. 2007; 41:69–77. [DOI] [PubMed] [Google Scholar]
- 41. Kim M.-Y., Vankayalapati H., Shin-Ya K., Wierzba K., Hurley L.H.. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 2002; 124:2098–2099. [DOI] [PubMed] [Google Scholar]
- 42. Jantos K., Rodriguez R., Ladame S., Shirude P.S., Balasubramanian S.. Oxazole-based peptide macrocycles: a new class of G-quadruplex binding ligands. J. Am. Chem. Soc. 2006; 128:13662–13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rzuczek S.G., Pilch D.S., Liu A., Liu L., LaVoie E.J., Rice J.E.. Macrocyclic pyridyl polyoxazoles: selective RNA and DNA G-quadruplex ligands as antitumor agents. J. Med. Chem. 2010; 53:3632–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cocco M.J., Hanakahi L.A., Huber M.D., Maizels N.. Specific interactions of distamycin with G-quadruplex DNA. Nucleic Acids Res. 2003; 31:2944–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hatzakis E., Okamoto K., Yang D.. Thermodynamic stability and folding kinetics of the major G-quadruplex and its loop isomers formed in the nuclease hypersensitive element in the human c-Myc promoter: effect of loops and flanking segments on the stability of parallel-stranded intramolecular G-quadruplexes. Biochemistry. 2010; 49:9152–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar Y.P., Bhowmik S., Das R.N., Bessi I., Paladhi S., Ghosh R., Schwalbe H., Dash J.. A fluorescent guanosine dinucleoside as a selective Switch-On sensor for c-myc G-Quadruplex DNA with potent anticancer activities. Chem. Eur. J. 2013; 19:11502–11506. [DOI] [PubMed] [Google Scholar]
- 47. Cosconati S., Marinelli L., Trotta R., Virno A., De Tito S., Romagnoli R., Pagano B., Limongelli V., Giancola C., Baraldi P.G. et al. . Structural and conformational requisites in DNA quadruplex groove binding: another piece to the puzzle. J. Am. Chem. Soc. 2010; 132:6425–6433. [DOI] [PubMed] [Google Scholar]
- 48. Martino L., Virno A., Pagano B., Virgilio A., Di Micco S., Galeone A., Giancola C., Bifulco G., Mayol L., Randazzo A.. Structural and thermodynamic studies of the interaction of distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007; 129:16048–16056. [DOI] [PubMed] [Google Scholar]
- 49. Lombardo L.J., Lee F.Y., Chen P., Norris D., Barrish J.C., Behnia K., Castaneda S., Cornelius L.A., Das J., Doweyko A.M.. Discovery of N-(2-chloro-6-methyl-phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino) thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004; 47:6658–6661. [DOI] [PubMed] [Google Scholar]
- 50. Lu Y., Li C.-M., Wang Z., Chen J., Mohler M.L., Li W., Dalton J.T., Miller D.D.. Design, synthesis, and SAR studies of 4-substituted methoxylbenzoyl-aryl-thiazoles analogues as potent and orally bioavailable anticancer agents. J. Med. Chem. 2011; 54:4678–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.