Abstract
The fibroblast growth factor (FGF) regulatory axis is phylogenetically ancient, evolving into a large mammalian/human gene family of 22 ligands that bind to four receptor tyrosine kinases for a complex physiologic system controlling cell growth, differentiation, and metabolism. The tissue targets for the primary FGF function are mainly in cartilage and in bone for morphogenesis, mineralization, and metabolism. A multitude of complexities in the FGF ligand-receptor signaling pathways have made translation into therapies for FGF-related bone disorders such as osteomalacia, osteoarthritis, and osteoporosis difficult but not impossible.
Keywords: bone, FGF2, FGF receptors
The article reviews the role of FGF2 and its receptors in bone biology and disease, focusing on our current understanding, gaps in knowledge, and future studies that are needed.
1. Fibroblast Growth Factor 2: A Molecular Prototype and Enigma
“Basic fibroblast growth factor (FGF),” later renamed FGF2, was one of two prototypical growth factors discovered through experiments nearly 50 years ago. First, new cell culture techniques that are now common provided for transfer of conditioned medium from cultured fibroblasts to newly seeded cells, resulting in stimulation of growth [1, 2]. Second, tumor extracts were analyzed for growth factors that regulated tumor angiogenesis [3]. Third, heparin affinity purification and fractionation of homogenized brain pituitary proteins likewise stimulated growth of cultured cells [2]. The growth-stimulating proteinaceous factors were isolated through heparin affinity chromatography and fractionation, leading to a family of heparin-binding growth factors that includes the FGFs [4].
Acidic and basic FGF dissociated from heparin depending on pH and their respective isoelectric points, hence their original names that were later changed to FGF1 for acidic FGF and FGF2 for basic FGF [5]. The FGF family then expanded with isolation of other heparin-binding growth factor proteins through FGF5 [6]. Rapid development of recombinant nucleic acid biochemistry technology then resulted in isolation of FGF family sequence homology clones from cDNA expression and genomic libraries, and the FGF family rapidly expanded to 23 FGFs; the number was later reduced to 22 FGFs when FGF15 and FGF19 were considered orthologs [7–9]. The Human Gene Nomenclature Committee has used bioinformatic analysis to characterize six other FGFs (FGFs 4, 5, 6, 7, 8, and 9) as structurally and functionally related to the prototypes FGF1 and FGF2 [10, 11], and other subclasses of FGFs within the larger family have been constructed [12].
Immunohistochemistry and antisense probes have been used to develop FGF2 expression profiles for tissues and various cells lines that presumed a functional significance [13]. FGF2 expression, gene function, and in vitro activity has been demonstrated in brain [14, 15], peripheral nerves [16], skeletal muscle [17], smooth muscle [18], heart [19], angiogenesis [3, 6, 8, 20], hematopoietic cells [21], chondrocytes [22–25], and osteoblasts [26–34], in both adult and developmental stages. FGF2 was presumed to be a major regulator of tissue growth and differentiation in development and for several diseases, principally cancer and cardiovascular disease.
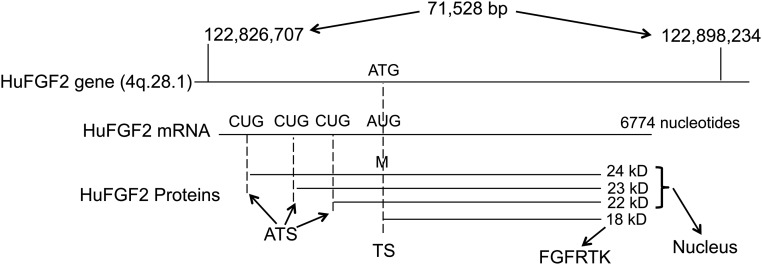
Continuously expanding nucleic acid technology has allowed sequencing and dissection of the FGF2 gene structure and function. Most interesting has been the discovery and analysis of alternative translation CUG start sites from the 5′ end of the FGF2 mRNA [35–38]. This is a rare feature in the mammalian genome that allows translation and expression of multiple protein isoforms from a single FGF2 mRNA, an evolutionary primitive device that is different from the more common alternative splicing of mRNAs. Novel site-directed mutagenesis experiments in COS cells revealed that the different FGF2 protein isoforms (four in humans, three in mice) are differentially trafficked in cells [35, 38]. The high molecular weight (HMW) FGF2 isoforms (24, 23, and 22 kD) are localized to the nucleus, whereas the low molecular weight (LMW) FGF2 (18 kD) is cytoplasmic and membrane associated as the predominant fibroblast growth factor receptor (FGFR) ligand (Fig. 1). Intracellular trafficking of the HMW FGF2 isoforms has been elucidated, but the precise molecular mechanism in the nucleus for regulation of cell growth and differentiation remains unclear [39, 40].
Figure 1.
Representation of the human FGF2 gene, the mRNA transcript with alternative CUG translation start sites, with the LMWFGF2 and human N-terminal expansion HMWFGF2 protein products. The HMWFGF2 proteins (22, 23, and 24 kD) traffic to the nucleus, whereas the 18-kD LMWFGF2 species binds to the FGFRTK for signal transduction. The LMWFGF2 protein is anabolic, whereas the HMWFGF2 proteins cause decreased BMD and osteopenia.
Pursuit of the functional biological, physiologic, and pathological significance for FGF2 as a major target for drug development and therapeutics continued with mapping of FGF2 receptor expression, contiguous with FGF2 ligand expression. FGF2 agonists and antagonists alike were presumed to hold the most potential for cancer, cardiovascular, and neuroscience therapies [41]. Interestingly, FGF2 expression had been detected in bone and cartilage, and a small subset of the FGF2 literature proposed that FGF2 regulated bone growth and mineralization [25, 26, 28, 30, 32]. However, the intense focus of the bone research community was on calcification, and the priority for FGF2 drug development was cancer and cardiovascular disease, and both somewhat obscured the bone results and significance. Development of animal models would change that.
2. Biology of FGFRs
Shortly after the discovery of the FGF2 ligand, the search began for FGFRs. Several different experimental approaches were taken, with FGF2 protein affinity and cross-linking experiments leading the way. A variety of FGF2 ligands were found, but the most fruitful results came from relating the epidermal growth factor receptor tyrosine kinase (RTK) family to the pursuit of similar FGF2-binding RTKs. Through both protein and nucleic acid biochemical approaches, the four-member FGFRTK family has been well defined [42].
The FGFRTKs are part of the immunoglobulin superfamily with three immunoglobulin extracellular domains, a hydrophobic transmembrane domain, and the tyrosine kinase cytoplasmic/enzymatic region [43]. There are other FGF-binding proteins that lack the kinase domain with putative FGF-related functions (including bone metabolism), such as FGFRL1 [44]. However, the majority of the physiologic and genetic data for FGF-related functional significance lies with the 22 FGF ligands and the four principal FGFRTKs.
Expression data for the FGFRTKs map to a wide variety of cells, offering little clue to tissue-specific physiologic function of FGFRTKs or their ligands [45]. When the FGF2 ligand binds its RTK, it generates dimerization generating either homodimers or heterodimers between the FGFRTK family members. The four FGFRs have different affinities for the multiple FGF family members. Ligand binding studies in vitro have shown that FGFRTKs 1, 3, and 4 had the greatest affinity, but FGFRTK 2 also shows biological activity from binding FGF2 [46]. The four FGFRTKs combined with 22 FGF ligands present a complex biological paradigm for FGF function as a peptide growth factor. Then the FGF2 ligand itself further complicates the picture with its four translational protein products. Moreover, the FGFRTK genes are each capable of generating alternatively spliced transcripts, resulting in numerous FGFRTK protein isoforms with differential functions [12, 42].
After discovery of the FGFRTKs came elucidation of the signal transduction pathways. This effort has proven complicated because they vary between cell types. A multitude of reports describe various enzymes downstream of the FGFRTKs, but they follow three principal signaling pathways: an inositol phosphate pathway that activates protein kinase C for calcium regulation [47], a STAT pathway that regulates bone growth [23, 29], and the GRB2-SOS-Ras-Raf-MapK pathway that intersects with Wnt for osteoblast-mediated control of bone growth and mineralization [42, 45, 48, 49]. Signaling bridges between these pathways by molecules such as SHP2 may integrate the net effect of FGF2 on the target cell [50]. The balance of signaling between these pathways [11, 45, 46] could vary depending on the cell, the ligand, and the receptor depending on the splice isoform (of any variant for the four FGFRTKs) and whether it forms a heterodimer or homodimer (Fig. 2).
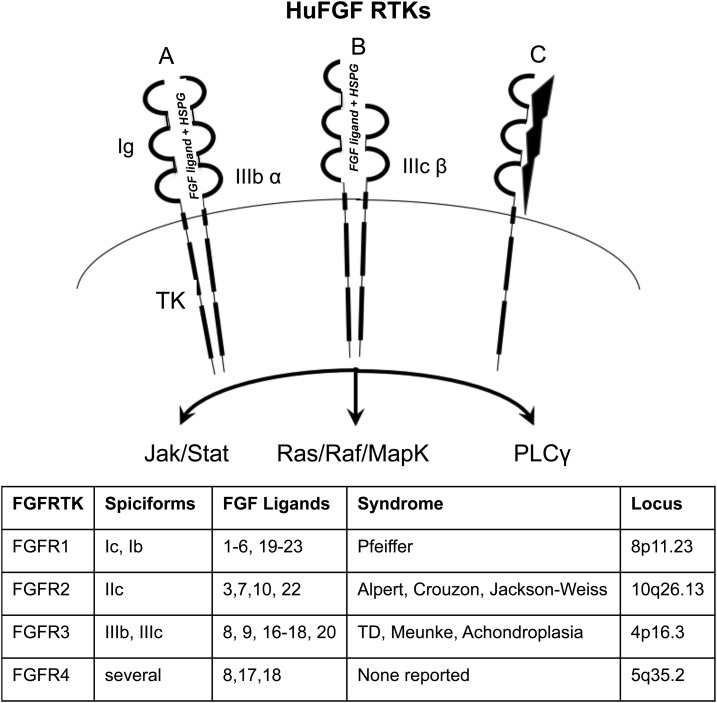
Figure 2.
Representative configurations for FGFRTKs. The canonical FGFRTK contains three immunoglobulin domains (Ig) and a two-domain cytoplasmic tyrosine kinase (TK) that phosphorylate downstream substrates. (A) The four FGFRTK genes are capable of forming homodimers or heterodimers that bind the FGF ligands, with heparin sulfate proteoglycan (HSGP) as a cofactor. The FGFRTKs putatively dimerize (B) between spliciforms or (C) with non-FGFR/ligands such as Klotho or NCAM. The system becomes more complex with alternative splicing that can generate variable Ig spliciforms, such as FGFR1 IIIC with (A) three Ig or (B) two Ig. Different combinations of dimers and spliciforms dictate variable affinities among 18 FGF ligands and also modulate downstream signaling among different pathways (JAK/STAT, PI3K, RAS/RAF/MAPK, or PLCγ) to regulate cell (osteoblast) proliferation or differentiation and calcium metabolism. Polymorphism or sporadic point mutations among the FGFR family can result in skeletal syndromes that are autosomal dominant as gain of function for the FGF system in skeletal physiology and carcinogenesis.
Perhaps the most important scientific results for the FGFRTKs came from genetic mapping of human dwarfisms and chondrodysplasias to the FGFRTK loci. Pfeiffer syndrome [fibroblast growth factor receptor 1 (FGFR1)], Crouzon syndrome (FGFR2), achondroplasia (FGFR3), and thanatophoric dysplasia (FGFR3) all mapped to the fully functional FGFRTKs [51, 52]. These results set the foundation for the FGFs as major regulatory genes in skeletal development and bone physiology. Moreover, the human genetic data were independently confirmed by initial production of animal models with a human FGF2 cDNA that unexpectedly generated chondrodysplasia as the principle phenotype [53]. Together these initial reports set the stage for a series of experiments that used animal models for the human syndromes that defined the genetic and molecular mechanisms for regulation of bone growth and mineralization. Although the FGFRTKs mapped to a major human skeletal disorder, the accompanying FGF2 ligand has not been mapped to a genetic syndrome. That may speak to redundancy in the FGF ligand family that is evident in the murine gene targeting studies [54].
3. Animal Models for FGF2 and FGFRTKs
Consistent with most gene families, elucidation of FGF physiologic function was dramatically advanced by animal modeling, including transgenesis and gene targeting. Basically, transgenesis is used for gain of function, whereas gene targeting is used for loss of function. Initial production of an FGF2 transgenic mouse using the full-length human cDNA unexpectedly resulted in full chondrodysplasia as the principal phenotype [30, 55]. The FGF2 null mouse, in another unexpected result, showed modest vascular effects but no obvious bone phenotype [54]. However, aging the FGF2 null mouse revealed osteoporosis as a major skeletal phenotype [26, 27, 31, 34, 56]. The results of the initial FGF2 animal modeling experiments showed chondrodysplasia for gain of function and osteoporosis for loss of function. Both results were consistent with the human genetic data showing FGF2 (and the FGFRTKs) as a negative regulator for bone growth [57]. Generation of transgenic mice with FGFRTK constructs that contained the human mutations generated chondrodysplasias as dominant negatives, similar to the human condition and consistent with the ligand overexpression transgenic models [53, 55]. Clearly, the FGF2 regulatory system lent itself well to production of animal models to test physiologic function and model human pathologies.
Those principles continued to be reliable for elucidating the function of the FGFRTKs and uncovering the differential function of the FGF2 alternative protein isoforms in bone development and growth. Production of separate transgenic lines for overexpression of the specific FGF2 isoforms proved to be consistent with the previous results. Expression of the HMW (24, 23, and 21 kD) FGF2 isoforms in mice recapitulated the chondrodysplasia phenotype [58–60]. However, overexpression or singular expression of the LMW (18 kD) isoform did not result in chondrodysplasia but cortical thickening with increased bone mass [32, 61, 62]. Overall, animal models generated with human FGF2 transcripts showed that negative regulation of bone growth as chondrodysplasia was caused by the HMW isoforms, whereas the LMW isoform was responsible for increasing bone mass in a positive regulatory mode.
4. Function of FGF2 in Bone Growth and Metabolism
Multiple studies have established the importance of FGF2 in bone development, maintenance, and fracture healing. Further understanding of the molecular mechanisms and pathways involved in these processes can allow therapeutic manipulation and improved patient outcomes in the future. FGF2 expression occurs in stromal cells and osteoblasts in bone, with storage in the extracellular matrix [63]. The differential function of the nuclear high molecular weight (HMWFGF2) and secreted low molecular weight (LMWFGF2) isoforms has been established; the HMW has inhibitory effects on mineralization and the LMW promotes bone formation [30, 32, 55, 59, 62, 64]. The main contributors to the expressed phenotypes are Wnt signaling [32, 61, 65], bone morphogenetic protein 2 (BMP2) [34, 66, 67], FGF23 [49, 59], and phosphate homeostasis via the bone-kidney axis [58, 62, 68].
The LMWFGF2 is exported and functions locally in the bone matrix by binding to FGFR1 and FGFR2, forming TRK dimers, and initiating the Wnt cascade through PI3K in osteoblasts and mesenchymal stem cells [58, 65, 69]. Wnt activation also occurs through inhibition of the sFRP1, a Wnt antagonist [58, 70], allowing Wnt ligands to bind Frizzled receptor and low-density lipoprotein receptor-related protein 5 (LRP5) complex [71, 72]. Another secreted protein involved in Wnt signaling is Dkk1, which can sequester to LRP5 and LRP6, allowing the degradation of β-catenin and inhibiting Wnt signaling [73, 74]. The Frizzled/LRP5 complex activates Disheveled, which in turn inhibits the Axin/APC/GSK3β destruction complex from ubiquitinating β-catenin [73, 75]. This process allows β-catenin to accumulate in the cytoplasm and translocate into the nucleus, where it stimulates activation of TCF/LEF1, a transcription factor [72, 76]. TCF/LEF promotes the expression of Runx2, osterix, and OCN in osteoblasts [61, 62, 77]. Runx2 functions as a transcription factor in early osteoblast development and maturation [77, 78]. Osterix expression is promoted by a Runx2-binding element [79] and functions as a zinc-finger transcription factor promoting collagen IA and also inhibits TCF binding to DNA and therefore causes feedback inhibition of Wnt signaling [80, 81]. These elements initiate the process of osteoblast proliferation, differentiation, and subsequent bone mineralization.
Another important factor in the LMWFGF2-Wnt cascade is BMP2. LMWFGF2 and BMP2 play synergistic roles in osteoblast activation and differentiation by activating the expression of each other [34, 66, 82]. The effect of LMWFGF2 on BMP2 expression, and vice versa, make BMP2 a vital signaling molecule in FGF2 signaling. BMP2 can further activate Wnt signaling through the Smad pathway [34]. BMP2 binds its receptor BMPR1/2, which stimulates the signaling cascade by phosphorylating Smad 1/5/8 [83–85]. These proteins can bind to Smad4, translocate to the nucleus, and activate a Smad-binding element [83, 86]. Binding to this element promotes expression of Dlx5, a promoter of Runx2, and mediates Osx expression [87, 88]. The resulting gene expression causes osteoblast proliferation and differentiation similar to the Wnt signaling activated by LMWFGF2. Phosphorylated Smad proteins are also able to bind to the CpG island of the Sost promoter, inhibiting expression; Sost is a locally secreted protein and potent Wnt inhibitor [89, 90]. Inhibition occurs through Sost binding LRP4/5/6 and preventing dimerization with the Frizzled receptor and subsequent activation of the Wnt cascade [91, 92].
Sost expression is promoted by the nuclear HMWFGF2 [59, 62]. The HMW isoforms are expressed through a nontraditional CUG start codon upstream of the LMWFGF2 AUG start site [93, 94]. HMWFGF2 proteins contain a nuclear localization sequence that allows them to function in an intracrine manner and affect gene expression [95]. HMWFGF2 gains this ability to function intracellularly by binding FGFRs; specifically, FGFR1 plays an extensive role in this mechanism [96, 97]. The complex of HMWFGF2 and FGFR1 uses importin-β–mediated transport to enter the nucleus and function as a transcriptional activator [96, 97]. Gene expression occurs through the transcriptional activator CREB-binding protein; the FGFR1 complex promotes release of CREB-binding protein from the RSK inactivation complex [98, 99]. The resulting gene expression is probably dependent on the cell type. In bone metabolism, HMWFGF2 nuclear signaling through the described mechanism has been shown to promote a number of genes inhibitory to mineralization. Sost has already been described, but another potent inhibitor of calcification is Matrix GLA protein through sequestration of BMP2 [100]. HMWFGF2 was shown to greatly increase expression in bone when present. Physiologically this could function as feedback mechanism during bone growth and fracture repair but could also be overly expressed in bone disease, such as hypophosphatemic rickets [59]. In addition to Sost and Matrix GLA protein, the most established and influential factor promoted by HMWFGF2 is FGF23 [49, 59].
FGF23 plays a substantial role in modulating the bone-renal axis by regulating phosphate homeostasis [68, 101]. FGF23 expression is promoted by HMWFGF2 nuclear translocation and transcriptional activity [49, 59, 62], resulting in secretion of FGF23 into circulation and subsequent action on kidney, increasing phosphate wasting and modulating vitamin D homeostasis [102, 103]. FGF23 induces its effects on the kidney by binding to Klotho on the cell surface, which can then interact with FGFRs to form an FGF23-specific receptor [104–106]. FGFRs function as tyrosine kinases and activate the RAS-MAPK-ERK pathway intracellularly [12, 59]. Activation of this pathway in the kidney results in decreased expression of type II sodium-dependent phosphate cotransporter in the renal proximal tubule and consequently loss of phosphate reabsorption [107]. Type II sodium-dependent phosphate cotransporter expression is positively regulated by all-trans-retinoic acid, retinoic acid receptor, and 1,25(OH)vitamin D; FGF23 inhibits this activity [108, 109]. FGF23 has not been proven to modulate the retinoic acid pathway but does modulate vitamin D metabolism. FGF23 modulates Cyp proteins in the kidney, specifically decreasing Cyp27b1 (activates vitamin D) and stimulating Cyp24 (degrades vitamin D), resulting in lower circulating vitamin D [109]. HMWFGF2 has been shown to increase parathyroid hormone (PTH) expression [60], and PTH in turn promotes FGF23 expression, resulting in a negative feedback loop on PTH expression [110]. Therefore, HMWFGF2 can manipulate phosphate homeostasis by altering PTH and FGF23 expression.
FGF23 regulation in the bone-renal axis is controlled by a family of short integrin-binding ligand-interacting glycoproteins, specifically Dmp1 and Phex. These proteins interact through an acidic serine aspartate–rich MEPE-associated (ASARM) motif [111]. Dmp1 activates Phex through the ASARM motif, and together they downregulate FGF23 expression, leading to improved bone mineralization [112]. Dmp1 also improves mineralization through its ability to nucleate hydroxyapatite crystals in the bone matrix during bone formation and turnover [91]. HMWFGF2 opposes the action of Dmp1 and Phex through the upregulation of FGF23 and Sost, which upregulates activity of Mepe-ASARM protein [99]. Mepe-ASARM competitively binds the ASARM motif of Phex, inhibiting activation via Dmp1 and the resulting FGF23 downregulation [111].
In summary, LMWFGF2 promotes bone mineralization by acting locally to increase osteoblast proliferation and differentiation, mainly through the canonical Wnt signaling pathway and the similarly functioning BMP2/Smad pathway. LMWFGF2 initiates these signaling cascades after being secreted locally in the extracellular bone matrix via binding and activating cellular surface FGFR1 tyrosine kinase activity. The action of LMWFGF2 is opposed by the HMWFGF2 isoforms that function in the nucleus as transcriptional activators in complex with FGFR1. The main effects of HMWFGF2 are through gene expression of mineralization inhibitors: FGF23 and Sost. Sost inhibits Wnt signaling locally, whereas FGF23 functions, systemically affecting vitamin D metabolism and phosphate homeostasis, resulting in overall inhibition of bone mineralization.
5. Human FGF-Related Bone Disorders
FGF2 and FGFRs have been linked to a number of bone diseases and pathological conditions. Therapeutic manipulation of the involved pathways could prove to be of substantial benefit for people with painful, deforming, and crippling conditions. Impairment of genetic expression of FGFs and their receptors has been linked to a number of bone-related diseases, most significantly osteoporosis or osteomalacia [90, 113], hypophosphatemic rickets [114], and pathological bone fractures [115]. Discovering the molecular roles of FGFs and FGFRs in the development of pathological conditions can provide insights into possible curative treatments through manipulation of the signaling pathways. This manipulation can be accomplished by introducing exogenous FGFs [116], monoclonal antibody inhibitors [117], or possibly direct genetic manipulation [118]. This section will delve into the pathologic disease states related to FGFs and current research into potential treatments.
FGF2 has been demonstrated as an important part of bone development and has been linked to a number of pathologic states, most notably childhood rickets. The FGF-related pathways manipulating phosphate homeostasis have been used to model hypophosphatemic rickets in mice and could lead to potential avenues for treatment of this deforming disease. Much of the involvement of FGF2 in the development of the rickets phenotype has been demonstrated through mouse models with differential expression of the HMWFGF2 isoform, which promotes FGF23 expression and control of the bone-renal axis, leading to systemic hypophosphatemia [59, 62]. The resulting low level of phosphate decreases the potential for formation of hydroxyapatite crystals and results in poor mineralization. In rickets, the phenotype presents as low bone mineral density, bowing of the long bones, and overall poor bone development [114]. In addition to the murine models of disease, a number of human genetic studies reveal mutations in FGF-related pathways. Gene sequencing of families that have been affected by autosomal dominant hypophosphatemic rickets displayed gain of function mutations in FGF23, leading to the characteristic phenotype [119–121]. These studies displayed alterations in the FGF23 cleavage site, allowing subsequent overexpression, hypophosphatemia, and diminished active vitamin D, resulting in the observed phenotype.
The most common initial therapy is dietary supplementation of phosphate, vitamin D, and calcium along with physical therapy and surgery depending on the affected person’s needs. Diet therapy has shown to have positive results initially, with long-term resistance to treatment caused by rising FGF23 levels in response to therapy [122, 123]. This resistance demonstrates a need for improved therapy options. There has been some evidence in both mouse models [124] and a human trial [125] for benefits of iron supplementation providing improvement of symptoms and phenotype. A more definitive long-acting treatment could come in the form of an FGF23 antibody [60, 115] or FGFR-specific antibody [58, 126], which have demonstrated promising results in mouse models. These antibody treatments may be promising but will need to undergo further animal and clinical trials to become viable options in a clinical setting.
Other conditions related to FGF expression are involved with bone aging and fragility. Osteomalacia and osteoporosis are part of a spectrum of poor bone mineralization related to aging and commonly resulting in fragility fractures. The role of FGF2 in bone mineralization and osteoporosis has been long demonstrated in mouse models [26, 114] and more recent genetic studies in human populations [127, 128]. The human models demonstrated a strong correlation of FGF2 polymorphisms causing decreased LMWFGF2 expression and significantly increased risk for developing osteoporosis. Recently, treatment of osteoporosis has included increased weight-bearing exercise, diet modification, smoking and glucocorticoid cessation, PTH, denosumab, selective estrogen receptor modulators, and bisphosphonates. Bisphosphonate are well tolerated and can be beneficial [117]. PTH can be used in patients with risk of fracture because of its anabolic action in bone but can be used only for ≤2 years because of its potential for bone catabolism in the long term [118].
FGF2 has been shown to play a role in current treatment through the response to PTH. PTH has been demonstrated to increase FGF2 levels in patients being treated for glucocorticoid-induced osteoporosis [129]. The anabolic PTH response has also been shown to be FGF2 dependent in mouse models [31]. The FGF2 and BMP2 pathways have been long established as promoters of bone mineralization and risk factors for the development of osteoporosis but have not been widely used in the treatment setting. A recent study of femoral head osteonecrosis used an adenovirus vector to promote expression of FGF2 and BMP2 in beagle dogs, with promising results [130]. Perhaps viral vector delivery or exogenous BMP2/FGF2 treatment could provide additional benefit to patients with severe osteoporosis.
With the development of decreased bone density in the case of osteomalacia and osteoporosis comes the increased risk of pathological fractures. Patients with previously diminished ability to support proper bone metabolism are at increased risk of poor healing after undergoing a fracture. Intervention into improving fracture healing has begun, and FGF2 is a viable candidate in the cascade because it has been shown to initiate bone repair [131]. The viability of FGF2 expression has displayed benefits in a number of animal fracture models including rabbits [131], transgenic mice [132], and nonhuman primates [133]. The improvement in fracture healing appears to involve activation of the mesenchymal stem cell population derived from the periosteum, leading to rapid collagenous callous formation and a shorter timeline for full fracture repair. Exogenous BMP2 has also been used clinically to improve fracture repair, but high doses are needed to achieve benefits in older adults [134]. BMP2 expression is promoted by LMWFGF2, as described earlier, and can have synergistic functions during activation of osteoblastic cell lineage. One study in mice used a two-phase biomaterial scaffold to administer low doses of FGF2 along with low-dose BMP2 and displayed improved bone repair in calvarial defects of older mice [135]. The application of FGF2 in the form of biodegradable hydrogels has shown improvement of bone union and decreased healing time in a number of studies including patients undergoing tibial osteotomy [136], long bone repair [137], and numerous animal studies using FGF-loaded gels and nanoparticles to repair bone defects [138, 139]. The improvements in tissue engineering and further clinical trials into the efficacy of biomaterials and FGF/BMP-loaded hydrogels could prove extremely beneficial in fracture repair, especially in an osteoporotic, aging population.
FGFR-related genetic mutations have been shown to play a role in disease etiologies related to skeletal development, such as craniosynostosis syndromes including Pfeiffer, Jackson-Weiss, Crouzon, Apert, and Muenke [140–142]. These diseases can cause deformities and deficits starting at a very young age. The only available intervention is a series of surgical procedures to correct the premature fusion of the cranial suture lines. Currently animal models are being used to explore possible treatment through genetic manipulation of the involved pathways, and this method could also open the possibility to targeted antibody therapies in the future, such as tyrosine kinase inhibitors and small molecular inhibitors [143]. All nonsurgical interventions are at very early stages of development and require further testing and investigation before becoming clinically viable options in the treatment of craniosynostosis syndromes.
Another strong correlation to FGFRs is that of FGFR3 mutations in the development of disease phenotypes related to bone growth and development. Achondroplasia, which causes skeletal dwarfism, has been demonstrated to have a causal mutation in FGFR3 [144]. A number of mutations in FGFR3 are correlated with phenotypes of different severities of achondroplasia ranging from mild to lethal forms [145, 146]. FGFR3 appears to regulate bone growth. Mouse models have demonstrated that inactivation of FGFR3 results in an overgrowth phenotype [147], whereas activating mutations display a phenotype of diminished growth potential similar to achondroplasia [148]. Possible therapies for treatment remain in the early stages, with some promise in murine models. One treatment relies on introducing a soluble form of FGFR3 to function as a decoy for ligands and theoretically decrease intracellular signaling [149]. Another potential target is abrogating downstream signaling from the mutated receptor, which has been accomplished in mice through C-type natriuretic peptide that inhibits the MAP kinase pathway [150, 151]. These are both viable theoretical therapies, but like many potential treatments they need further animal testing and clinical trials.
6. Discussion and Future Directions
Differential expression of FGFs and FGFRs through various mechanisms can play a substantial role in the overall health of an individual. These conditions can manifest early in life, such as the craniosynostosis syndromes, skeletal growth abnormalities, and variations of rickets due to genetic polymorphisms resulting in altered gene expression. A person’s FGF expression levels could alter his or her risk of developing osteoporotic disease and subsequent pathological fracture risk. FGF profile can also affect the rate of bone repair and response to certain treatments. The multitude of FGF-related pathways involved in bone maintenance, metabolism, and disease progression present targets for therapy, but many are still in the experimental stages. There are promising results in numerous animal models for improving disease and fracture management, especially in frail older adults.
Acknowledgments
Financial Support: Supported by National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant AR072986.05 to M.M.H.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ASARM
acidic serine- and aspartic acid-rich motif
- BMP
bone morphogenetic protein
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- HMW
high molecular weight
- LMW
low molecular weight
- LRP
low-density lipoprotein receptor-related protein
- PTH
parathyroid hormone
- RTK
receptor tyrosine kinase
References and Notes
- 1. Gospodarowicz D. Localisation of a fibroblast growth factor and its effect alone and with hydrocortisone on 3T3 cell growth. Nature. 1974;249(453):123–127. [DOI] [PubMed] [Google Scholar]
- 2. Gospodarowicz D, Handley HH. Stimulation of division of Y1 adrenal cells by a growth factor isolated from bovine pituitary glands. Endocrinology. 1975;97(1):102–107. [DOI] [PubMed] [Google Scholar]
- 3. Shing Y, Folkman J, Sullivan R, Butterfield C, Murray J, Klagsbrun M. Heparin affinity: purification of a tumor-derived capillary endothelial cell growth factor. Science. 1984;223(4642):1296–1299. [DOI] [PubMed] [Google Scholar]
- 4. Gospodarowicz D, Cheng J, Lui GM, Baird A, Böhlent P. Isolation of brain fibroblast growth factor by heparin-Sepharose affinity chromatography: identity with pituitary fibroblast growth factor. Proc Natl Acad Sci USA. 1984;81(22):6963–6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Böhlen P, Esch F, Baird A, Gospodarowicz D. Acidic fibroblast growth factor (FGF) from bovine brain: amino-terminal sequence and comparison with basic FGF. EMBO J. 1985;4(8):1951–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klagsbrun M. The fibroblast growth factor family: structural and biological properties. Prog Growth Factor Res. 1989;1(4):207–235. [DOI] [PubMed] [Google Scholar]
- 7. Abraham JA, Whang JL, Tumolo A, Mergia A, Friedman J, Gospodarowicz D, Fiddes JC. Human basic fibroblast growth factor: nucleotide sequence and genomic organization. EMBO J. 1986;5(10):2523–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abraham JA, Mergia A, Whang JL, Tumolo A, Friedman J, Hjerrild KA, Gospodarowicz D, Fiddes JC. Nucleotide sequence of a bovine clone encoding the angiogenic protein, basic fibroblast growth factor. Science. 1986;233(4763):545–548. [DOI] [PubMed] [Google Scholar]
- 9. Gospodarowicz D, Neufeld G, Schweigerer L. Fibroblast growth factor: structural and biological properties. J Cell Physiol Suppl. 1987;133(suppl 5)15–26. [DOI] [PubMed] [Google Scholar]
- 10. InterPro.uk InterPro protein sequence analysis & classification. http://www.ebi.ac.uk/interpro/entry/IPR002209. Accessed 26 March 2018.
- 11. HUGO.org HGNC FGF2 symbol report.org. 2018. https://www.genenames.org/cgi-bin/gene_symbol_report?match=FGF2. Accessed 26 March 2018.
- 12. Ornitz DM, Itoh N. The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol. 2015;4(3):215–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. EMBL-EBI.uk Expression atlas.org. https://www.ebi.ac.uk/gxa/genes/ENSG00000138685?bs=%7B%22homo%20sapiens%22%3A%5B%22ORGANISM_PART%22%2C%22CELL_LINE%22%5D%7D&ds=%7B%22kingdom%22%3A%5B%22animals%22%5D%7D#baseline. Accessed 26 March 2018.
- 14. Carlson GA, Borchelt DR, Dake A, Turner S, Danielson V, Coffin JD, Eckman C, Meiners J, Nilsen SP, Younkin SG, Hsiao KK. Genetic modification of the phenotypes produced by amyloid precursor protein overexpression in transgenic mice. Hum Mol Genet. 1997;6(11):1951–1959. [DOI] [PubMed] [Google Scholar]
- 15. Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, Hung YT. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis [published corrections appear in Nat Neurosci 1999;2:848 and Nat Neurosci 1999;2:485] Nat Neurosci. 1999;2(3):246–253. [DOI] [PubMed] [Google Scholar]
- 16. Kuzis K, Coffin JD, Eckenstein FP. Time course and age dependence of motor neuron death following facial nerve crush injury: role of fibroblast growth factor. Exp Neurol. 1999;157(1):77–87. [DOI] [PubMed] [Google Scholar]
- 17. Pawlikowski B, Vogler TO, Gadek K, Olwin BB. Regulation of skeletal muscle stem cells by fibroblast growth factors. Dev Dyn. 2017;246(5):359–367. [DOI] [PubMed] [Google Scholar]
- 18. Davis MG, Zhou M, Ali S, Coffin JD, Doetschman T, Dorn GW II. Intracrine and autocrine effects of basic fibroblast growth factor in vascular smooth muscle cells. J Mol Cell Cardiol. 1997;29(4):1061–1072. [DOI] [PubMed] [Google Scholar]
- 19. Liao S, Bodmer J, Pietras D, Azhar M, Doetschman T, Schultz JJ. Biological functions of the low and high molecular weight protein isoforms of fibroblast growth factor-2 in cardiovascular development and disease. Dev Dyn. 2009;238(2):249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fulgham DL, Widhalm SR, Martin S, Coffin JD. FGF-2 dependent angiogenesis is a latent phenotype in basic fibroblast growth factor transgenic mice. Endothelium. 1999;6(3):185–195. [DOI] [PubMed] [Google Scholar]
- 21. Itkin T, Ludin A, Gradus B, Gur-Cohen S, Kalinkovich A, Schajnovitz A, Ovadya Y, Kollet O, Canaani J, Shezen E, Coffin DJ, Enikolopov GN, Berg T, Piacibello W, Hornstein E, Lapidot T. FGF-2 expands murine hematopoietic stem and progenitor cells via proliferation of stromal cells, c-Kit activation, and CXCL12 down-regulation. Blood. 2012;120(9):1843–1855. [DOI] [PubMed] [Google Scholar]
- 22. Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13(11):1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sahni M, Raz R, Coffin JD, Levy D, Basilico C. STAT1 mediates the increased apoptosis and reduced chondrocyte proliferation in mice overexpressing FGF2. Development. 2001;128(11):2119–2129. [DOI] [PubMed] [Google Scholar]
- 24. Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18(3):290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weksler NB, Lunstrum GP, Reid ES, Horton WA. Differential effects of fibroblast growth factor (FGF) 9 and FGF2 on proliferation, differentiation and terminal differentiation of chondrocytic cells in vitro. Biochem J. 1999;342(Pt 3):677–682. [PMC free article] [PubMed] [Google Scholar]
- 26. Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105(8):1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Okada Y, Montero A, Zhang X, Sobue T, Lorenzo J, Doetschman T, Coffin JD, Hurley MM. Impaired osteoclast formation in bone marrow cultures of Fgf2 null mice in response to parathyroid hormone. J Biol Chem. 2003;278(23):21258–21266. [DOI] [PubMed] [Google Scholar]
- 28. Xiao L, Liu P, Sobue T, Lichtler A, Coffin JD, Hurley MM. Effect of overexpressing fibroblast growth factor 2 protein isoforms in osteoblastic ROS 17/2.8 cells. J Cell Biochem. 2003;89(6):1291–1301. [DOI] [PubMed] [Google Scholar]
- 29. Xiao L, Naganawa T, Obugunde E, Gronowicz G, Ornitz DM, Coffin JD, Hurley MM. Stat1 controls postnatal bone formation by regulating fibroblast growth factor signaling in osteoblasts. J Biol Chem. 2004;279(26):27743–27752. [DOI] [PubMed] [Google Scholar]
- 30. Sobue T, Naganawa T, Xiao L, Okada Y, Tanaka Y, Ito M, Okimoto N, Nakamura T, Coffin JD, Hurley MM. Over-expression of fibroblast growth factor-2 causes defective bone mineralization and osteopenia in transgenic mice. J Cell Biochem. 2005;95(1):83–94. [DOI] [PubMed] [Google Scholar]
- 31. Hurley MM, Okada Y, Xiao L, Tanaka Y, Ito M, Okimoto N, Nakamura T, Rosen CJ, Doetschman T, Coffin JD. Impaired bone anabolic response to parathyroid hormone in Fgf2−/− and Fgf2+/− mice. Biochem Biophys Res Commun. 2006;341(4):989–994. [DOI] [PubMed] [Google Scholar]
- 32. Xiao L, Liu P, Li X, Doetschman T, Coffin JD, Drissi H, Hurley MM. Exported 18-kDa isoform of fibroblast growth factor-2 is a critical determinant of bone mass in mice. J Biol Chem. 2009;284(5):3170–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sabbieti MG, Agas D, Marchetti L, Coffin JD, Xiao L, Hurley MM. BMP-2 differentially modulates FGF-2 isoform effects in osteoblasts from newborn transgenic mice. Endocrinology. 2013;154(8):2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Naganawa T, Xiao L, Coffin JD, Doetschman T, Sabbieti MG, Agas D, Hurley MM. Reduced expression and function of bone morphogenetic protein-2 in bones of Fgf2 null mice. J Cell Biochem. 2008;103(6):1975–1988. [DOI] [PubMed] [Google Scholar]
- 35. Florkiewicz RZ, Sommer A. Human basic fibroblast growth factor gene encodes four polypeptides: three initiate translation from non-AUG codons. Proc Natl Acad Sci USA. 1989;86(11):3978–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prats AC, Vagner S, Prats H, Amalric F. cis-Acting elements involved in the alternative translation initiation process of human basic fibroblast growth factor mRNA. Mol Cell Biol. 1992;12(10):4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vagner S, Gensac MC, Maret A, Bayard F, Amalric F, Prats H, Prats AC. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Arese M, Chen Y, Florkiewicz RZ, Gualandris A, Shen B, Rifkin DB. Nuclear activities of basic fibroblast growth factor: potentiation of low-serum growth mediated by natural or chimeric nuclear localization signals. Mol Biol Cell. 1999;10(5):1429–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bonnal S, Schaeffer C, Créancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem. 2003;278(41):39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Audigier S, Guiramand J, Prado-Lourenco L, Conte C, Gonzalez-Herrera IG, Cohen-Solal C, Récasens M, Prats AC. Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA. 2008;14(9):1852–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou K, Fan YD, Duysenbi S, Wu PF, Feng ZH, Qian Z, Zhang TR. siRNA-mediated silencing of bFGF gene inhibits the proliferation, migration, and invasion of human pituitary adenoma cells. Tumour Biol. 2017;39(6):1010428317704805. [DOI] [PubMed] [Google Scholar]
- 42. Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29(14):1463–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uniprot.org Uniprot FGFR.org. http://www.uniprot.org/uniprot/P11362. Accessed 26 March 2018.
- 44. Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68(6):951–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.NCBI.gov. FGFR1 fibroblast growth factor receptor 1 [Homo sapiens (human)]. NCBI FGFR https://www.ncbi.nlm.nih.gov/gene/2260. Accessed 26 March 2018.
- 46. Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271(25):15292–15297. [DOI] [PubMed] [Google Scholar]
- 47. Lemonnier J, Delannoy P, Hott M, Lomri A, Modrowski D, Marie PJ. The Ser252Trp fibroblast growth factor receptor-2 (FGFR-2) mutation induces PKC-independent downregulation of FGFR-2 associated with premature calvaria osteoblast differentiation. Exp Cell Res. 2000;256(1):158–167. [DOI] [PubMed] [Google Scholar]
- 48. Hurley MM, Tetradis S, Huang YF, Hock J, Kream BE, Raisz LG, Sabbieti MG. Parathyroid hormone regulates the expression of fibroblast growth factor-2 mRNA and fibroblast growth factor receptor mRNA in osteoblastic cells. J Bone Miner Res. 1999;14(5):776–783. [DOI] [PubMed] [Google Scholar]
- 49. Xiao L, Esliger A, Hurley MM. Nuclear fibroblast growth factor 2 (FGF2) isoforms inhibit bone marrow stromal cell mineralization through FGF23/FGFR/MAPK in vitro. J Bone Miner Res. 2013;28(1):35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krejci P, Masri B, Salazar L, Farrington-Rock C, Prats H, Thompson LM, Wilcox WR. Bisindolylmaleimide I suppresses fibroblast growth factor-mediated activation of Erk MAP kinase in chondrocytes by preventing Shp2 association with the Frs2 and Gab1 adaptor proteins. J Biol Chem. 2007;282(5):2929–2936. [DOI] [PubMed] [Google Scholar]
- 51. Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, et al. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet. 1994;8(3):269–274. [DOI] [PubMed] [Google Scholar]
- 52. Muenke M, Schell U. Fibroblast-growth-factor receptor mutations in human skeletal disorders. Trends Genet. 1995;11(8):308–313. [DOI] [PubMed] [Google Scholar]
- 53. Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125(24):4977–4988. [DOI] [PubMed] [Google Scholar]
- 54. Zhou M, Sutliff RL, Paul RJ, Lorenz JN, Hoying JB, Haudenschild CC, Yin M, Coffin JD, Kong L, Kranias EG, Luo W, Boivin GP, Duffy JJ, Pawlowski SA, Doetschman T. Fibroblast growth factor 2 control of vascular tone. Nat Med. 1998;4(2):201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coffin JD, Florkiewicz RZ, Neumann J, Mort-Hopkins T, Dorn GW II, Lightfoot P, German R, Howles PN, Kier A, O’Toole BA, et al. Abnormal bone growth and selective translational regulation in basic fibroblast growth factor (FGF-2) transgenic mice. Mol Biol Cell. 1995;6(12):1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fei Y, Xiao L, Hurley MM. The impaired bone anabolic effect of PTH in the absence of endogenous FGF2 is partially due to reduced ATF4 expression. Biochem Biophys Res Commun. 2011;412(1):160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ornitz DM, Legeai-Mallet L. Achondroplasia: development, pathogenesis, and therapy. Dev Dyn. 2017;246(4):291–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xiao L, Du E, Homer-Bouthiette C, Hurley MM. Inhibition of FGFR signaling partially rescues hypophosphatemic rickets in HMWFGF2 Tg male mice. Endocrinology. 2017;158(10):3629–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiao L, Naganawa T, Lorenzo J, Carpenter TO, Coffin JD, Hurley MM. Nuclear isoforms of fibroblast growth factor 2 are novel inducers of hypophosphatemia via modulation of FGF23 and KLOTHO. J Biol Chem. 2010;285(4):2834–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Du E, Xiao L, Hurley MM. FGF23 neutralizing antibody ameliorates hypophosphatemia and impaired FGF receptor signaling in kidneys of HMWFGF2 transgenic mice. J Cell Physiol. 2017;232(3):610–616. [DOI] [PubMed] [Google Scholar]
- 61. Xiao L, Ueno D, Catros S, Homer-Bouthiette C, Charles L, Kuhn L, Hurley MM. Fibroblast growth factor-2 isoform (low molecular weight/18 kDa) overexpression in preosteoblast cells promotes bone regeneration in critical size calvarial defects in male mice. Endocrinology. 2014;155(3):965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Homer-Bouthiette C, Doetschman T, Xiao L, Hurley MM. Knockout of nuclear high molecular weight FGF2 isoforms in mice modulates bone and phosphate homeostasis. J Biol Chem. 2014;289(52):36303–36314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gospodarowicz D. Fibroblast growth factor. Chemical structure and biologic function. Clin Orthop Relat Res. 1990;(257):231–248. [PubMed]
- 64. Sørensen V, Nilsen T, Wiedłocha A. Functional diversity of FGF-2 isoforms by intracellular sorting. BioEssays. 2006;28(5):504–514. [DOI] [PubMed] [Google Scholar]
- 65. Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116(5):1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fakhry A, Ratisoontorn C, Vedhachalam C, Salhab I, Koyama E, Leboy P, Pacifici M, Kirschner RE, Nah HD. Effects of FGF-2/-9 in calvarial bone cell cultures: differentiation stage-dependent mitogenic effect, inverse regulation of BMP-2 and noggin, and enhancement of osteogenic potential. Bone. 2005;36(2):254–266. [DOI] [PubMed] [Google Scholar]
- 67. Sato MM, Nakashima A, Nashimoto M, Yawaka Y, Tamura M. Bone morphogenetic protein-2 enhances Wnt/beta-catenin signaling-induced osteoprotegerin expression. Genes Cells. 2009;14(2):141–153. [DOI] [PubMed] [Google Scholar]
- 68. Bergwitz C, Jüppner H. Regulation of phosphate homeostasis by PTH, vitamin D, and FGF23. Annu Rev Med. 2010;61(1):91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ding VMY, Ling L, Natarajan S, Yap MGS, Cool SM, Choo ABH. FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3beta signaling. J Cell Physiol. 2010;225(2):417–428. [DOI] [PubMed] [Google Scholar]
- 70. Bodine PVN, Zhao W, Kharode YP, Bex FJ, Lambert AJ, Goad MB, Gaur T, Stein GS, Lian JB, Komm BS. The Wnt antagonist secreted frizzled-related protein-1 is a negative regulator of trabecular bone formation in adult mice. Mol Endocrinol. 2004;18(5):1222–1237. [DOI] [PubMed] [Google Scholar]
- 71. Urano T. [Wnt-beta-catenin signaling in bone metabolism]. Clin Calcium. 2006;16(1):54–60. [PubMed] [Google Scholar]
- 72. Levasseur R, Lacombe D, de Vernejoul MC. LRP5 mutations in osteoporosis-pseudoglioma syndrome and high-bone-mass disorders. Joint Bone Spine. 2005;72(3):207–214. [DOI] [PubMed] [Google Scholar]
- 73. Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. [DOI] [PubMed] [Google Scholar]
- 74. Butler JS, Murray DW, Hurson CJ, O’Brien J, Doran PP, O’Byrne JM. The role of Dkk1 in bone mass regulation: correlating serum Dkk1 expression with bone mineral density. J Orthop Res. 2011;29(3):414–418. [DOI] [PubMed] [Google Scholar]
- 75. Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem. 1996;237(1):295–302. [DOI] [PubMed] [Google Scholar]
- 76. Armstrong VJ, Muzylak M, Sunters A, Zaman G, Saxon LK, Price JS, Lanyon LE. Wnt/beta-catenin signaling is a component of osteoblastic bone cell early responses to load-bearing and requires estrogen receptor alpha. J Biol Chem. 2007;282(28):20715–20727. [DOI] [PubMed] [Google Scholar]
- 77. Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–49. [DOI] [PubMed] [Google Scholar]
- 79. Nishio Y, Dong Y, Paris M, O’Keefe RJ, Schwarz EM, Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. [DOI] [PubMed] [Google Scholar]
- 80. Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K, Takayanagi H. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11(8):880–885. [DOI] [PubMed] [Google Scholar]
- 81. Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD, de Crombrugghe B. Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc Natl Acad Sci USA. 2008;105(19):6936–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van Gastel N, Stegen S, Stockmans I, Moermans K, Schrooten J, Graf D, Luyten FP, Carmeliet G. Expansion of murine periosteal progenitor cells with fibroblast growth factor 2 reveals an intrinsic endochondral ossification program mediated by bone morphogenetic protein 2. Stem Cells. 2014;32(9):2407–2418. [DOI] [PubMed] [Google Scholar]
- 83. Chen G, Deng C, Li Y-P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8(2):272–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhou Z, Xie J, Lee D, Liu Y, Jung J, Zhou L, Xiong S, Mei L, Xiong WC. Neogenin regulation of BMP-induced canonical Smad signaling and endochondral bone formation. Dev Cell. 2010;19(1):90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Podkowa M, Attisano L. A skeleton in the closet: neogenin guides bone development. Dev Cell. 2010;19(1):1–2. [DOI] [PubMed] [Google Scholar]
- 86. Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, Lan Y, Cheng X, Hou N, Liu H, Ding J, Lin F, Yang R, Gao X, Chen D, Yang X. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci. 2007;120(Pt 13):2162–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lee M-H, Kwon T-G, Park H-S, Wozney JM, Ryoo H-M. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309(3):689–694. [DOI] [PubMed] [Google Scholar]
- 88. Holleville N, Matéos S, Bontoux M, Bollerot K, Monsoro-Burq A-H. Dlx5 drives Runx2 expression and osteogenic differentiation in developing cranial suture mesenchyme. Dev Biol. 2007;304(2):860–874. [DOI] [PubMed] [Google Scholar]
- 89. Papathanasiou I, Kostopoulou F, Malizos KN, Tsezou A. DNA methylation regulates sclerostin (SOST) expression in osteoarthritic chondrocytes by bone morphogenetic protein 2 (BMP-2) induced changes in Smads binding affinity to the CpG region of SOST promoter. Arthritis Res Ther. 2015;17(1):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Atkins GJ, Rowe PS, Lim HP, Welldon KJ, Ormsby R, Wijenayaka AR, Zelenchuk L, Evdokiou A, Findlay DM. Sclerostin is a locally acting regulator of late-osteoblast/preosteocyte differentiation and regulates mineralization through a MEPE-ASARM-dependent mechanism. J Bone Miner Res. 2011;26(7):1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. He G, Dahl T, Veis A, George A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein 1. Nat Mater. 2003;2(8):552–558. [DOI] [PubMed] [Google Scholar]
- 92. Kumar J, Swanberg M, McGuigan F, Callreus M, Gerdhem P, Akesson K. LRP4 association to bone properties and fracture and interaction with genes in the Wnt- and BMP signaling pathways. Bone. 2011;49(3):343–348. [DOI] [PubMed] [Google Scholar]
- 93. Okada-Ban M, Thiery JP, Jouanneau J. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32(3):263–267. [DOI] [PubMed] [Google Scholar]
- 94. Prats H, Kaghad M, Prats AC, Klagsbrun M, Lélias JM, Liauzun P, Chalon P, Tauber JP, Amalric F, Smith JA, et al. High molecular mass forms of basic fibroblast growth factor are initiated by alternative CUG codons. Proc Natl Acad Sci USA. 1989;86(6):1836–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Delrieu I. The high molecular weight isoforms of basic fibroblast growth factor (FGF-2): an insight into an intracrine mechanism. FEBS Lett. 2000;468(1):6–10. [DOI] [PubMed] [Google Scholar]
- 96. Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90(4):662–691. [DOI] [PubMed] [Google Scholar]
- 97. Dunham-Ems SM, Lee Y-W, Stachowiak EK, Pudavar H, Claus P, Prasad PN, Stachowiak MK. Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol Biol Cell. 2009;20(9):2401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Myers JM, Martins GG, Ostrowski J, Stachowiak MK. Nuclear trafficking of FGFR1: a role for the transmembrane domain. J Cell Biochem. 2003;88(6):1273–1291. [DOI] [PubMed] [Google Scholar]
- 99. Fang X, Stachowiak EK, Dunham-Ems SM, Klejbor I, Stachowiak MK. Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1: a novel mechanism of gene regulation. J Biol Chem. 2005;280(31):28451–28462. [DOI] [PubMed] [Google Scholar]
- 100. Zebboudj AF, Imura M, Boström K. Matrix GLA protein, a regulatory protein for bone morphogenetic protein-2. J Biol Chem. 2002;277(6):4388–4394. [DOI] [PubMed] [Google Scholar]
- 101. Quarles LD. FGF23, PHEX, and MEPE regulation of phosphate homeostasis and skeletal mineralization. Am J Physiol Endocrinol Metab. 2003;285(1):E1–E9. [DOI] [PubMed] [Google Scholar]
- 102. Yu X, White KE. FGF23 and disorders of phosphate homeostasis. Cytokine Growth Factor Rev. 2005;16(2):221–232. [DOI] [PubMed] [Google Scholar]
- 103. Lu Y, Feng JQ. FGF23 in skeletal modeling and remodeling. Curr Osteoporos Rep. 2011;9(2):103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Quarles LD. Skeletal secretion of FGF-23 regulates phosphate and vitamin D metabolism. Nat Rev Endocrinol. 2012;8(5):276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem. 2006;281(10):6120–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, Fujita T, Fukumoto S, Yamashita T. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444(7120):770–774. [DOI] [PubMed] [Google Scholar]
- 107. Jüppner H. Phosphate and FGF-23. Kidney Int Suppl. 2011;79(121):S24–S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Masuda M, Yamamoto H, Kozai M, Tanaka S, Ishiguro M, Takei Y, Nakahashi O, Ikeda S, Uebanso T, Taketani Y, Segawa H, Miyamoto K, Takeda E. Regulation of renal sodium-dependent phosphate co-transporter genes (Npt2a and Npt2c) by all-trans-retinoic acid and its receptors. Biochem J. 2010;429(3):583–592. [DOI] [PubMed] [Google Scholar]
- 109. Quarles LD. Role of FGF23 in vitamin D and phosphate metabolism: implications in chronic kidney disease. Exp Cell Res. 2012;318(9):1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lanske B, Razzaque MS. Molecular interactions of FGF23 and PTH in phosphate regulation. Kidney Int. 2014;86(6):1072–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rowe PSN. Regulation of bone-renal mineral and energy metabolism: the PHEX, FGF23, DMP1, MEPE ASARM pathway. Crit Rev Eukaryot Gene Expr. 2012;22(1):61–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Martin A, Liu S, David V, Li H, Karydis A, Feng JQ, Quarles LD. Bone proteins PHEX and DMP1 regulate fibroblastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling. FASEB J. 2011;25(8):2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Fromigué O, Modrowski D, Marie PJ. Growth factors and bone formation in osteoporosis: roles for fibroblast growth factor and transforming growth factor beta. Curr Pharm Des. 2004;10(21):2593–2603. [DOI] [PubMed] [Google Scholar]
- 114. ADHR Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26(3):345–348. [DOI] [PubMed] [Google Scholar]
- 115. Aono Y, Yamazaki Y, Yasutake J, Kawata T, Hasegawa H, Urakawa I, Fujita T, Wada M, Yamashita T, Fukumoto S, Shimada T. Therapeutic effects of anti-FGF23 antibodies in hypophosphatemic rickets/osteomalacia. J Bone Miner Res. 2009;24(11):1879–1888. [DOI] [PubMed] [Google Scholar]
- 116. Franceschi RT. Biological approaches to bone regeneration by gene therapy. J Dent Res. 2005;84(12):1093–1103. [DOI] [PubMed] [Google Scholar]
- 117. Kennel KA, Drake MT. Adverse effects of bisphosphonates: implications for osteoporosis management. Mayo Clin Proc. 2009;84(7):632–637, quiz 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Morley P, Whitfield JF, Willick GE. Parathyroid hormone: an anabolic treatment for osteoporosis. Curr Pharm Des. 2001;7(8):671–687. [DOI] [PubMed] [Google Scholar]
- 119. Gribaa M, Younes M, Bouyacoub Y, Korbaa W, Ben Charfeddine I, Touzi M, Adala L, Mamay O, Bergaoui N, Saad A. An autosomal dominant hypophosphatemic rickets phenotype in a Tunisian family caused by a new FGF23 missense mutation. J Bone Miner Metab. 2010;28(1):111–115. [DOI] [PubMed] [Google Scholar]
- 120. Sun Y, Wang O, Xia W, Jiang Y, Li M, Xing X, Hu Y, Liu H, Meng X, Zhou X. FGF23 analysis of a Chinese family with autosomal dominant hypophosphatemic rickets. J Bone Miner Metab. 2012;30(1):78–84. [DOI] [PubMed] [Google Scholar]
- 121. Seton M, Jüppner H. Autosomal dominant hypophosphatemic rickets in an 85 year old woman: characterization of her disease from infancy through adulthood. Bone. 2013;52(2):640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Imel EA, DiMeglio LA, Hui SL, Carpenter TO, Econs MJ. Treatment of X-linked hypophosphatemia with calcitriol and phosphate increases circulating fibroblast growth factor 23 concentrations. J Clin Endocrinol Metab. 2010;95(4):1846–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Linglart A, Biosse-Duplan M, Briot K, Chaussain C, Esterle L, Guillaume-Czitrom S, Kamenicky P, Nevoux J, Prié D, Rothenbuhler A, Wicart P, Harvengt P. Therapeutic management of hypophosphatemic rickets from infancy to adulthood. Endocr Connect. 2014;3(1):R13–R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Farrow EG, Yu X, Summers LJ, Davis SI, Fleet JC, Allen MR, Robling AG, Stayrook KR, Jideonwo V, Magers MJ, Garringer HJ, Vidal R, Chan RJ, Goodwin CB, Hui SL, Peacock M, White KE. Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice. Proc Natl Acad Sci USA. 2011;108(46):E1146–E1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Kapelari K, Köhle J, Kotzot D, Högler W. Iron supplementation associated with loss of phenotype in autosomal dominant hypophosphatemic rickets. J Clin Endocrinol Metab. 2015;100(9):3388–3392. [DOI] [PubMed] [Google Scholar]
- 126. Wöhrle S, Henninger C, Bonny O, Thuery A, Beluch N, Hynes NE, Guagnano V, Sellers WR, Hofmann F, Kneissel M, Graus Porta D. Pharmacological inhibition of fibroblast growth factor (FGF) receptor signaling ameliorates FGF23-mediated hypophosphatemic rickets. J Bone Miner Res. 2013;28(4):899–911. [DOI] [PubMed] [Google Scholar]
- 127. Lei S-F, Papasian CJ, Deng H-W. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bin X, Lin C, Huang X, Zhou Q, Wang L, Xian CJ. FGF-2 gene polymorphism in osteoporosis among Guangxi’s Zhuang Chinese. Int J Mol Sci. 2017;18(7):E1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hurley M, Yao W, Lane NE. Changes in serum fibroblast growth factor 2 in patients with glucocorticoid-induced osteoporosis treated with human parathyroid hormone (1-34). Osteoporos Int. 2005;16(12):2080–2084. [DOI] [PubMed] [Google Scholar]
- 130. Peng W-X, Wang L. Adenovirus-mediated expression of BMP-2 and BFGF in bone marrow mesenchymal stem cells combined with demineralized bone matrix for repair of femoral head osteonecrosis in beagle dogs. Cell Physiol Biochem. 2017;43(4):1648–1662. [DOI] [PubMed] [Google Scholar]
- 131. Chen W-J, Jingushi S, Aoyama I, Anzai J, Hirata G, Tamura M, Iwamoto Y. Effects of FGF-2 on metaphyseal fracture repair in rabbit tibiae. J Bone Miner Metab. 2004;22(4):303–309. [DOI] [PubMed] [Google Scholar]
- 132. Hurley MM, Adams DJ, Wang L, Jiang X, Burt PM, Du E, Xiao L. Accelerated fracture healing in transgenic mice overexpressing an anabolic isoform of fibroblast growth factor 2. J Cell Biochem. 2016;117(3):599–611. [DOI] [PubMed] [Google Scholar]
- 133. Kawaguchi H, Nakamura K, Tabata Y, Ikada Y, Aoyama I, Anzai J, Nakamura T, Hiyama Y, Tamura M. Acceleration of fracture healing in nonhuman primates by fibroblast growth factor-2. J Clin Endocrinol Metab. 2001;86(2):875–880. [DOI] [PubMed] [Google Scholar]
- 134. Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38(12):1424–1429. [DOI] [PubMed] [Google Scholar]
- 135. Charles LF, Woodman JL, Ueno D, Gronowicz G, Hurley MM, Kuhn LT. Effects of low dose FGF-2 and BMP-2 on healing of calvarial defects in old mice. Exp Gerontol. 2015;64:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kawaguchi H, Jingushi S, Izumi T, Fukunaga M, Matsushita T, Nakamura T, Mizuno K, Nakamura T, Nakamura K. Local application of recombinant human fibroblast growth factor-2 on bone repair: a dose-escalation prospective trial on patients with osteotomy. J Orthop Res. 2007;25(4):480–487. [DOI] [PubMed] [Google Scholar]
- 137. Marcacci M, Kon E, Moukhachev V, Lavroukov A, Kutepov S, Quarto R, Mastrogiacomo M, Cancedda R. Stem cells associated with macroporous bioceramics for long bone repair: 6- to 7-year outcome of a pilot clinical study. Tissue Eng. 2007;13(5):947–955. [DOI] [PubMed] [Google Scholar]
- 138. Yamada K, Tabata Y, Yamamoto K, Miyamoto S, Nagata I, Kikuchi H, Ikada Y. Potential efficacy of basic fibroblast growth factor incorporated in biodegradable hydrogels for skull bone regeneration. J Neurosurg. 1997;86(5):871–875. [DOI] [PubMed] [Google Scholar]
- 139. Kang MS, Kim J-H, Singh RK, Jang J-H, Kim H-W. Therapeutic-designed electrospun bone scaffolds: mesoporous bioactive nanocarriers in hollow fiber composites to sequentially deliver dual growth factors. Acta Biomater. 2015;16:103–116. [DOI] [PubMed] [Google Scholar]
- 140. Roscioli T, Flanagan S, Kumar P, Masel J, Gattas M, Hyland VJ, Glass IA. Clinical findings in a patient with FGFR1 P252R mutation and comparison with the literature. Am J Med Genet. 2000;93(1):22–28. [DOI] [PubMed] [Google Scholar]
- 141. Su N, Jin M, Chen L. Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res. 2014;2(1):14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Robin NH, Falk MJ, Haldeman-Englert CR. FGFR-related craniosynostosis syndromes. In: Adam MP, Ardinger HH, Pagon RA, et al, eds. GeneReviews® Seattle (WA). Seattle, WA: University of Washington; 1993. Accessed 23 February 2018. http://www.ncbi.nlm.nih.gov/books/NBK1455/.
- 143. Melville H, Wang Y, Taub PJ, Jabs EW. Genetic basis of potential therapeutic strategies for craniosynostosis. Am J Med Genet A. 2010;152A(12):3007–3015. [DOI] [PubMed] [Google Scholar]
- 144. Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78(2):335–342. [DOI] [PubMed] [Google Scholar]
- 145. Bellus GA, McIntosh I, Smith EA, Aylsworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet. 1995;10(3):357–359. [DOI] [PubMed] [Google Scholar]
- 146. Brodie SG, Kitoh H, Lachman RS, Nolasco LM, Mekikian PB, Wilcox WR. Platyspondylic lethal skeletal dysplasia, San Diego type, is caused by FGFR3 mutations. Am J Med Genet. 1999;84(5):476–480. [PubMed] [Google Scholar]
- 147. Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84(6):911–921. [DOI] [PubMed] [Google Scholar]
- 148. Wang Y, Spatz MK, Kannan K, Hayk H, Avivi A, Gorivodsky M, Pines M, Yayon A, Lonai P, Givol D. A mouse model for achondroplasia produced by targeting fibroblast growth factor receptor 3. Proc Natl Acad Sci USA. 1999;96(8):4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Garcia S, Dirat B, Tognacci T, Rochet N, Mouska X, Bonnafous S, Patouraux S, Tran A, Gual P, Le Marchand-Brustel Y, Gennero I, Gouze E. Postnatal soluble FGFR3 therapy rescues achondroplasia symptoms and restores bone growth in mice. Sci Transl Med. 2013;5(203):203ra124. [DOI] [PubMed] [Google Scholar]
- 150. Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M, Tamura N, Ogawa Y, Nakao K. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10(1):80–86. [DOI] [PubMed] [Google Scholar]
- 151. Lorget F, Kaci N, Peng J, Benoist-Lasselin C, Mugniery E, Oppeneer T, Wendt DJ, Bell SM, Bullens S, Bunting S, Tsuruda LS, O’Neill CA, Di Rocco F, Munnich A, Legeai-Mallet L. Evaluation of the therapeutic potential of a CNP analog in a Fgfr3 mouse model recapitulating achondroplasia. Am J Hum Genet. 2012;91(6):1108–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]