Abstract
The metabolic properties of cells are formed under the influence of environmental factors such as nutrients and hormones. Although such a metabolic program is likely initiated through epigenetic mechanisms, the direct links between metabolic cues and activities of chromatin modifiers remain largely unknown. In this study, we show that lysine-specific demethylase-1 (LSD1) controls the metabolic program in myogenic differentiation, under the action of catabolic hormone, glucocorticoids. By using transcriptomic and epigenomic approaches, we revealed that LSD1 bound to oxidative metabolism and slow-twitch myosin genes, and repressed their expression. Consistent with this, loss of LSD1 activity during differentiation enhanced the oxidative capacity of myotubes. By testing the effects of various hormones, we found that LSD1 levels were decreased by treatment with the glucocorticoid dexamethasone (Dex) in cultured myoblasts and in skeletal muscle from mice. Mechanistically, glucocorticoid signaling induced expression of a ubiquitin E3 ligase, JADE-2, which was responsible for proteasomal degradation of LSD1. Consequently, in differentiating myoblasts, chemical inhibition of LSD1, in combination with Dex treatment, synergistically de-repressed oxidative metabolism genes, concomitant with increased histone H3 lysine 4 methylation at these loci. These findings demonstrated that LSD1 serves as an epigenetic regulator linking glucocorticoid action to metabolic programming during myogenic differentiation.
INTRODUCTION
Environmental factors exert profound influences on the epigenome, leading to phenotypic variations in cells and organisms (1). In particular, nutritional conditions, such as malnutrition and high fat feeding, induce genome-scale rearrangement of epigenomic signatures, including DNA methylation and histone modifications (2,3). However, direct mechanistic links between environmental cues and the activities of individual chromatin modifiers are poorly understood.
Lysine-specific demethylase-1 (LSD1 or KDM1A) is a member of the amine oxidase family that catalyzes demethylation of methylated lysine side chains within proteins, including histone H3 lysine 4 (H3K4) (4,5). LSD1 contributes to a variety of cellular processes, such as stem cell maintenance, development and carcinogenesis (6–8). We previously demonstrated that LSD1 epigenetically suppressed oxidative phosphorylation (OXPHOS) through H3K4 demethylation in adipogenic and cancer cells (9,10). Notably, metabolic regulation by LSD1 depended on environmental conditions such as energetic state, hormones and neurological stimuli (9,11), indicating that LSD1 reacts to the nutritional and metabolic conditions of cells and modulates their metabolic properties.
In mammals, skeletal muscle is composed of mechanically distinct fiber types, each having unique metabolic properties (12). Slow-twitch (type I) fibers participate in persistent mechanical activities, such as posture, through their slow contractile activity, supported by a high capacity for OXPHOS. In contrast, fast-twitch (type II) fibers, with their shift toward anaerobic energy production, are capable of fast contraction, contributing to strong and dynamic movements. Skeletal muscle can flexibly alter its metabolic character in response to environmental factors, contributing to energy homeostasis and physical fitness. Anabolic hormones, such as insulin, activate the glycolytic program under nutritionally rich conditions. On the other hand, when energy sources are scarce, catabolic hormones, such as glucocorticoids, activate oxidative metabolism and suppress formation of the fast fibers (13). Previous studies established that dynamic epigenomic remodeling, including histone modifications and DNA methylation, contribute to highly ordered expression of myogenesis-associated genes (14,15). However, it is unknown how these environmental contexts can be translated into epigenetic remodeling in myogenic cells.
In this study, we demonstrated that LSD1 directly repressed expression of oxidative metabolism and slow myosin genes in differentiating myoblasts, resulting in enhanced OXPHOS capacity in myotubes. We also found that glucocorticoid signaling induced expression of an E3 ligase, JADE-2, leading to proteasomal degradation of LSD1. LSD1 inhibition, in combination with Dex treatment, significantly increased expression of oxidative genes and H3K4 methylation levels, indicating that LSD1 counteracted the glucocorticoid-mediated gene regulation. Our study demonstrated a novel epigenetic mechanism linking hormonal signaling to metabolic reprogramming during myogenic differentiation.
MATERIALS AND METHODS
Cell culture
C2C12 mouse myoblasts and Hepa1-6 mouse hepatoma cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) in a 37°C incubator equilibrated with 95% air, 5% CO2. For myogenic induction, subconfluent C2C12 myoblasts were cultured in DMEM with 2% (v/v) horse serum. The medium was changed every other day. For protein assays, Hepa1-6 cells were cultured for 48 h in DMEM supplemented with 10% (v/v) dextran-coated charcoal (DCC)-treated FBS followed by treatment with 1 μM dexamethasone (Sigma-Aldrich) for 48 h. For immunofluorescence, Hepa1-6 cells were cultured for 24 h in DMEM supplemented with 2% (v/v) DCC-treated FBS followed by treatment with 1 μM Dex for 24 h.
For shRNA expression, SureSilencing Plasmids harboring shRNA against LSD1 (shLSD1, #336312 KM27305H, Qiagen) and non-targeting control (shControl, Qiagen #336312) were used. C2C12 cells were transfected with shLSD1 and shControl vectors using FuGENE6 Transfection Reagent (Promega). Stable transfectants were selected in the presence of 750 μg/ml hygromycin B (Wako). For transient siRNA introduction, specific siRNAs were introduced to the Hepa1-6 cells using RNAiMAX reagent (Invitrogen) when they were ∼50% confluent. The effective siRNA target sequences are as follows: siGL3 (control) CTTACGCTGAGTACTTCGA, siJade2 CGTTAGAGCGTGTTCTAGA.
Reagents and antibodies
Tranylcypromine hydrochloride, dexamethasone, insulin (bovine pancreas), testosterone, 3,3′,5-triiodo-L-thyronine, MG132 and cycloheximide were from Sigma-Aldrich. S2101 was from Millipore. 17-β-Estradiol was from Wako. The antibodies used were anti-LSD1 (Abcam, ab17721), anti-mono-methylated histone H3K4 (Abcam, ab8895), anti-di-methylated histone H3K4 (Millipore, 07-030), anti-tri-methylated histone H3K4 (Millipore, 07-473), anti-pan histone H3 (Abcam, ab1791), anti-myosin heavy chain (Santa Cruz, sc-20641), anti-slow-myosin (Sigma-Aldrich, M8421), anti-glucocorticoid receptor (Santa Cruz, sc-8992) and anti-PHF15 (JADE-2) (Sigma-Aldrich, HPA025959).
Animal studies
Animal experiments were conducted in accordance with the guidelines of the Animal Care and Use Committee of Kumamoto University and the Animal Ethics Committee of the Institute of Medical Science, The University of Tokyo. For gene expression analysis in gastrocnemius and soleus muscle, 8-week-old male C57BL/6J mice were fasted for 4 h, then sacrificed. Intraperitoneal (i.p.) administration of dexamethasone (Dex) to mice harboring skeletal muscle-specific deletion of glucocorticoid receptor (GRmKO) was performed as previously described (16). Briefly, 11-week-old male GRflox/flox (control) and GRmKO mice were injected with Dex at 1 mg/kg or vehicle (saline), daily for 7 days, then were sacrificed. Adrenalectomy was performed as previously described (16). Briefly, 19-week-old male C57BL/6J mice were adrenalectomized and maintained with 0.9% sodium chloride in the drinking water for 7 days, then sacrificed.
Myoblast isolation and culture
Mouse myoblasts were isolated as described previously with some modifications (17). Briefly, hindlimbs of 2-day-old C57BL/6J mice were removed and muscle was separated from the skin and bone with sterile forceps. The cells were dispersed with 0.2% collagenase I (Gibco) and 0.2% Dispase II (Sigma-Aldrich) for 30 min, and then filtered through a 40 μm cell strainer. To remove non-myogenic cells, cells were plated on normal culture dish and incubated for 30 min. Subsequently, floating cells were re-plated on a collagen coated culture dish. Myoblasts were initially cultured in Ham's F10 nutrient mixture (Gibco) and then the medium was switched to DMEM/Ham's F10 (1:1). Both growth media were supplemented with 20% FBS, 2.5 ng/ml bFGF and 1% penicillin/streptomycin. To induce differentiation, myoblasts were cultured in DMEM with 5% HS and penicillin/streptomycin. Medium was changed every other day.
Quantitative RT-PCR
Total RNA from cells was extracted using a Trizol reagent (Invitrogen). Complementary DNA was synthesized using a ReverTra Ace qPCR RT Kit (Toyobo). Quantitative real-time PCR was performed using THUNDERBIRD SYBR qPCR Mix (Toyobo) and an ABI 7300 Real-Time PCR System (Applied Biosciences). Relative RNA quantities were calculated by ΔΔCt method. Primers are listed in Supplementary Table S1.
Microarray analysis
Genome-wide expression analysis was performed using a GeneChip Mouse Genome 430 2.0 Array in combination with a GeneChip Hybridization, Wash and Stain Kit (Affymetrix). TC- and S2101-treated or shLSD1 stably expressing C2C12 cells were induced to differentiate for 48 h, followed by RNA extraction and a quality check using a Bioanalyzer RNA 6000 Nano Assay (Agilent). Data annotation analysis was performed using GeneSpring GX software (Agilent). Gene set enrichment analysis was performed using GSEA version 2.0 from the Broad Institute of MIT and Harvard (Cambridge, MA, http://www.broadinstitute.org/gsea/). Significantly enriched gene sets were identified by using the criterion of nominal P-value < 0.05 and FDR q-value < 0.25 according the software instructions (http://software.broadinstitute.org/gsea/index.jsp).
Chromatin immunoprecipitation (ChIP)
ChIP experiments for detecting modified histones and glucocorticoid receptor were performed as previously described (10). Briefly, cells were crosslinked with 1% formaldehyde, then sonicated for chromatin fragmentation. ChIP experiments for detecting LSD1 enrichment were performed according to the Epigenesys protocol by Porro and Perini (Prot 29, http://www.epigenesys.eu/), with some modifications. Briefly, dual crosslinking using 1% formaldehyde and disuccinimidyl glutarate (DSG) (ThermoFisher Scientific) was performed to increase stability of protein-DNA complexes. All chromatin fragmentation was done in a RIPA buffer (50 mM Tris–HCl, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS and 0.5% sodium deoxycholate) by two-step sonication, first with a probe-type sonifier (Branson) and then with a water bath sonifier, Bioruptor (Cosmo Bio). Chromatin fragments were incubated at 4°C overnight with the appropriate antibodies, followed by a pulldown using protein A/G-conjugated agarose beads (Millipore) or Dynabeads protein A/G (Life Technologies). Purified DNA was subjected to quantitative PCR (qPCR).
ChIP-seq analysis
For ChIP-seq analyses to detect LSD1–DNA interactions, 5 × 107 C2C12 cells, differentiated for 2 days or 5 days, were collected. For analyses to detect methylated H3K4, 6.4 × 106 C2C12 cells, differentiated for 48 h with or without S2101, were collected. Crosslinking, fragmentation and immunoprecipitation were performed as described for ChIP. Each protein-bound chromatin fraction was collected using Dynabeads protein A/G (Life Technologies) and DNA was purified using an Agencourt AMPure XP (Beckman Coulter). A DNA fragment library for sequencing was constructed using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs). Adapter-ligated DNA fragments were purified using Agencourt AMPure XP. High-throughput sequencing was performed using a NextSeq 500 Desktop Sequencer (Illumina) according to the manufacturer's instructions. The qualified reads were aligned onto the mouse reference genome mm9 using the Bowtie 2 algorithm (18). Duplicate reads and reads with low overall quality or low mapping quality were excluded. The final numbers of mapped reads are listed in Supplementary Table S2. We identified 44,039 in day 2 and 17,446 in day 5 LSD1-enriched peaks across the whole genome using a model-based analysis of ChIP-Seq data (MACS) algorithm (19). LSD1 binding sites were detected based on the LSD1 peaks significantly enriched over input peaks at a P-cutoff value of 10−5. Detected LSD1 peaks were linked to neighboring genes with ChIPpeakAnno algorithm, measuring the distance from the TSS. The ChIP-seq data for Rest (GSM915175), Max (GSM915174), Six4 (GSM1633919), MyoD (GSM915165), and myogenin (GSM915164) were imported from the National Center for Biotechnology Information (NCBI). Data visualizations were performed with Strand NGS software (Strand Genomics). The prediction of co-occupancy of LSD1 with other proteins was done using ChIP-Atlas (http://chip-atlas.org/). Subsequent data analyses were performed on the Galaxy platform (http://usegalaxy.org/). The number of reads in LSD1 and pan- or modified-histone data were normalized to that of input data with the bamCompare tool. Enrichment values of LSD1 around histone modifications or Pol II peaks were calculated with the computeMatrix tool and visualized with the plotHeatmap tool. Our ChIP-seq data (H3K4me1, H3K4me2 and H3K4me3 in control cells) or the ones obtained from ChIP-Atlas with Q-value of 1.0 × 10−10 (pol II: SRX062103, H3K9me3: SRX862738 and H3K27ac: SRX103218) were used as references. Enrichment values of LSD1 and modified histones at LSD1 peaks were calculated with the multiBigwig tool.
Immunofluorescence and quantitative analysis
Cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min at room temperature. The cells were washed 3 times with PBS for 5 min, then permeabilized with PBS containing 0.5% Triton X-100 for 5 min on ice. The cells were washed 3 times with PBS for 5 min then incubated with the primary antibodies for 60 min, followed by the Alexa 488-conjugated secondary antibody for 60 min. The cells were washed with PBS 3 times for 5 min each, after each incubation. DNA was counterstained with 1 μg/ml 5,6-diamidino-2-phenylindole (DAPI).
Quantitative analysis was performed with HCS Studio Analysis Software (ThermoFisher Scientific). To count nuclei in MHC-expressing myotubes, we used the ‘Colocalization’ command in the ‘Bioapplication’ tool of the HCS software. First, we identified a DAPI-stained nucleus as the main object, then defined regions of interest (ROIs) as 2-pixel rings around each nucleus. When an ROI was co-localized with the fluorescent signal of MHC, the nucleus was identified as an MHC-expressing nucleus. To quantify the area and intensity of the MHC signal, we used the ‘Spot Detector’ command in the ‘Bioapplication’ tool in HCS software, in which MHC-stained myofibers were identified as pixels with MHC signals.
Real-time measurement of OXPHOS activity
Real-time monitoring of cellular OXPHOS activity was performed using the XF24 Extracellular Flux Analyzer (Seahorse Bioscience), according to the manufacturer's instructions. C2C12 cells were seeded on the Seahorse assay plate and allowed to differentiate under LSD1 inhibition for 5 days before the assay. Maximum OXPHOS capacity was determined as previously described (9).
Determination of intracellular ATP
To determine intracellular ATP concentrations, C2C12 cells were allowed to differentiate under LSD1 inhibition for 4 days. After differentiation, cells were incubated in glucose-free DMEM (Gibco) with 1 mM sodium pyruvate and 2% HS for 3 h, and then, in the presence of 1g/l glucose for 15 min. Cells were collected and ATP concentrations were determined with an ATP Bioluminescence Assay Kit CLS II (Roche), according to the manufacturer's instructions. ATP concentration values were normalized to DNA contents, measured in the same samples.
Statistical analyses
Data are presented as means ± standard deviation (s.d.). All statistical analyses, except for microarray analyses, were performed to compare two groups. We first used F-test for examining equality of variances. Where variances were equal, two-tailed Student's t-test was used, and if not, two-tailed Welch's t-test was used. For microarray analyses, Pearson's χ2 test was used.
RESULTS
LSD1 inhibition de-represses oxidative metabolism genes in differentiating myoblasts
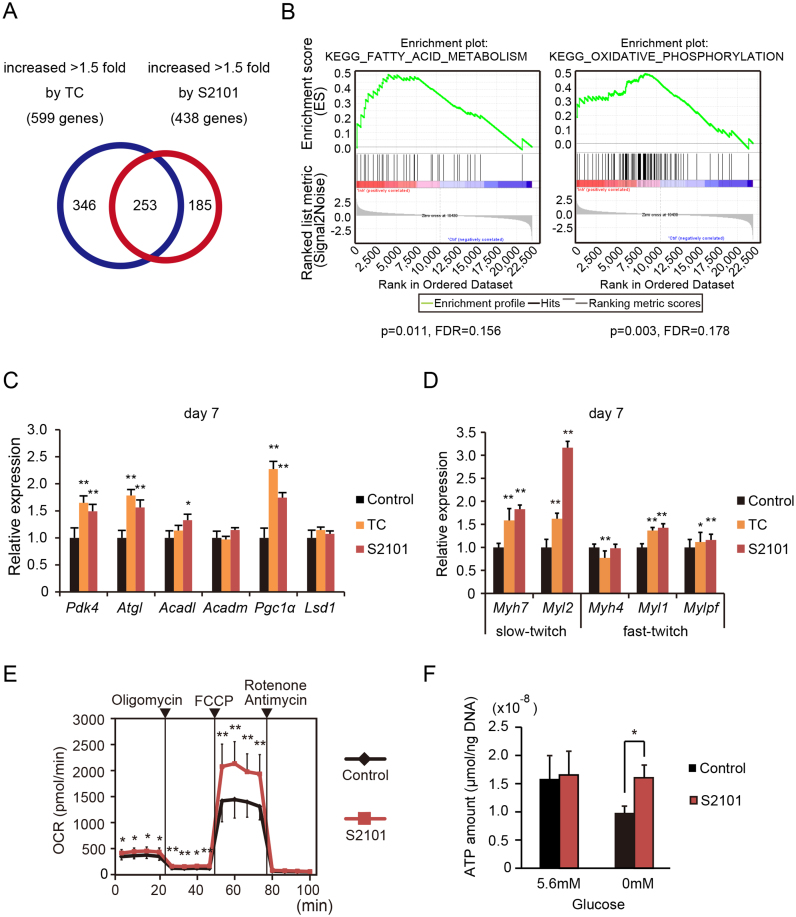
To examine the role of LSD1 in transcriptional regulation during myogenesis, we treated differentiating C2C12 myoblasts with the enzymatic LSD1 inhibitors tranylcypromine (TC) or S2101 (Supplementary Figure S1A). TC is the first reported potent inhibitor of LSD1 (20), and S2101 is highly selective against LSD1 over structurally related enzymes including monoamine oxidases and LSD2 (21,22). Under our experimental conditions, LSD1 inhibition did not interfere with differentiation, as assessed by expression of myogenic markers, Myod1 and Myog, and by myotube formation (days 5 to 7) (Supplementary Figure S1B–D) (23). We performed microarray-based expression analysis of differentiating C2C12 cells under LSD1 inhibition (day 2). We identified 253 genes whose signal intensities were increased by 1.5-fold or more in cells treated with TC and S2101 (Figure 1A). To test whether co-regulated genes were involved in specific biological processes, we performed a Gene Set Enrichment Analysis (GSEA) (24). GSEA identified 14 gene categories that were significantly upregulated by LSD1 inhibition, many of which were related to lipid and oxidative metabolism (Figure 1B and Supplementary Figure S2A). Conversely, gene categories such as ‘cell cycle’ and ‘DNA replication’ were downregulated by LSD1 inhibition (Supplementary Figure S2B). After differentiation (day 7), LSD1-inhibited cells maintained elevated expression of oxidative metabolism genes, compared with in controls (Figure 1C) (25). Because C2C12 myoblasts undergo fast twitch-like skeletal myogenesis upon induction (26), we next tested whether muscle structural genes were co-regulated with metabolic genes. We found that expression of slow fiber-associated genes, myosin heavy chain-7 (Myh7) and light chain-2 (Myl2), was significantly upregulated 2- to 3-fold in LSD1-inhibited myotubes (day 7), while that of fast-twitch genes, Myh4, Myl1 and Mylpf, was less affected (Figure 1D). To further validate the effects of LSD1 inhibition on metabolic gene expression, we characterized the metabolic phenotype of myotubes formed under LSD1 inhibition. Maximum OXPHOS capacity of the cells was enhanced by LSD1 inhibition, while glycolytic activity was unaffected (Figure 1E, Supplementary Figure S3A and B). In addition, while intracellular ATP levels were decreased by glucose deprivation in control myotubes, LSD1-inhibited cells maintained constant ATP levels under glucose deprivation, indicating that LSD1 inhibition augmented OXPHOS-dependent energy production by using alternative fuels such as fatty acids (Figure 1F). We previously reported that LSD1 promoted expression of glycolytic genes through stabilization of HIF-1α protein in human hepatocellular carcinoma cells (10). In C2C12 myoblasts, glycolytic genes were mostly unaffected by LSD1 inhibition (Supplementary Figure S3C). These results show that inhibition of LSD1 demethylase activity during myogenesis activates oxidative metabolism in the resulting myotubes.
Figure 1.
LSD1 inhibition during myogenic differentiation enhances expression of oxidative metabolism genes. (A) Venn diagram of genes increased more than 1.5-fold by treatment with LSD1 inhibitors, TC (10−4 M) or S2101 (10−5 M). C2C12 cells were analyzed at 48 h after myogenic induction. Vehicle was administered to the control samples. (B) Gene set enrichment analysis (GSEA) of genes upregulated by LSD1 inhibition. Nominal p values and false discovery rates (FDRs) are indicated. (C and D) Expression levels of LSD1 target genes (C) and slow and fast muscle myosin genes (D) in TC and S2101 treated cells (n = 4). Cells were cultured in differentiation medium for 7 days in the presence of LSD1 inhibitors. qRT-PCR values were normalized to values for the 36B4 gene and are shown as fold differences compared with vehicle-treated controls. Full names of the genes examined are listed in Supplementary Table S1. (E) Effects of LSD1 inhibition on the OXPHOS capacity of differentiated C2C12 cells. Oxygen consumption rate (OCR) is shown (n = 10). During real-time measurements, respiratory chain inhibitors were added to the medium at the indicated time points. Maximum OXPHOS capacity was evaluated as the maximized OCR levels under FCCP treatment. Values are means ± s.d. (F) Effects of LSD1 inhibition on glucose dependent ATP production. Intracellular ATP concentrations in differentiated cells in the presence or absence of glucose (n = 3). ATP concentration was normalized to DNA content. Detailed procedures are described in MATERIALS AND METHODS. *P < 0.05, **P < 0.01.
LSD1 is associated with transcriptionally competent chromatin
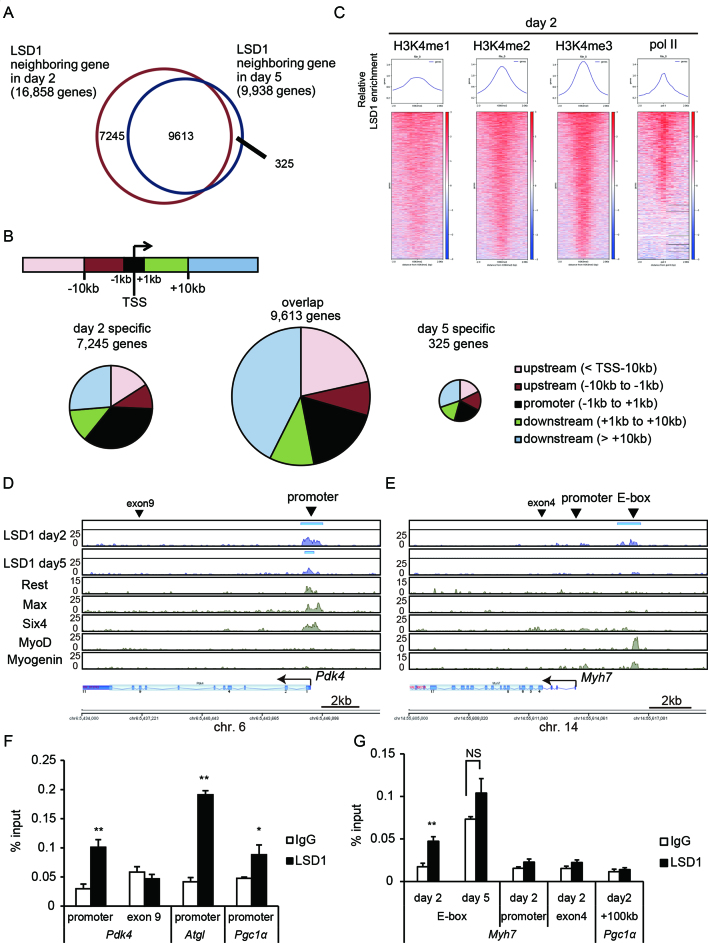
To identify LSD1-bound genes, we performed ChIP-seq analyses in differentiating (day 2) and differentiated (day 5) C2C12 cells. The number of LSD1-bound genes declined from 16,858 at day 2 to 9938 at day 5, of which 9613 were maintained throughout differentiation (Figure 2A). Substantial fraction of LSD1- bound regions was found within ±10 kb of the TSS (Figure 2B). To characterize the chromatin signature associated with LSD1 binding, we merged our LSD1-ChIP-seq results with publicly available ChIP-seq data from C2C12 cells. LSD1-bound sites prominently coincided with active chromatin marks, such as H3K4 methylation and H3K27 acetylation, but not with a repressive mark, H3K9me3 (Figure 2C, Supplementary Figure S4A and B). Furthermore, LSD1-bound regions were also coincident with RNA polymerase II (pol II), suggesting that LSD1 was enriched at transcriptionally competent chromatin. To identify transcription factors (TFs) and/or chromatin modifiers exhibiting distributions similar to that of LSD1, we compared our ChIP-seq data with published datasets using ChIP-Atlas (http://chip-atlas.org/). LSD1-bound promoters (<TSS ± 1 kb) were highly enriched by repressive factors such as Sin3A and Rest in C2C12 cells (Table 1). On the other hand, in genomic regions outside of promoters (>TSS ± 1 kb), myogenic TFs, MyoD (MYOD1) and myogenin (MYOG), were significantly co-localized with LSD1 (Table 1).
Figure 2.
LSD1 is enriched at transcriptionally competent chromatin in differentiating myoblasts. (A) Venn diagram of LSD1 neighboring genes in differentiating (day 2) and differentiated (day 5) C2C12 cells. (B) Pie charts showing genomic distribution of LSD1 peaks. The regions were classified into five classes based on the distance from transcription starting site (TSS) of each neighboring gene. (C) LSD1 enrichment relative to methylated H3K4 and RNA polymerase II at day 2. Normalized enrichment values of LSD1 from –2 kb to +2 kb of indicated protein peaks are shown. (D and E) Enrichment of LSD1 at the Pdk4 (D) and slow-associated Myh7 (E) gene loci. Y axis indicates the read counts for each data. MACS LSD1 peaks are indicated by blue bars above the enrichment data. Data were visualized using Strand NGS software (Strand Genomics). Publicly available ChIP-seq data for Rest, Max, Six4, MyoD and myogenin in C2C12 cells are also shown (Data IDs are listed in MATERIALS AND METHODS). Arrowheads indicate the target regions for ChIP-qPCR. (F and G) ChIP-qPCR analyses of LSD1 occupancy at metabolic gene promoters (F) and Myh7 gene loci (G). ChIP analyses were performed using differentiating (day 2) and differentiated (day 5) C2C12 cells (n = 3). *P < 0.05, **P < 0.01. NS: no significant difference.
Table 1. (A and B) Transcription factors (TFs) exhibiting similar genomic distributions to that of LSD1. LSD1 peaks were divided into two groups, LSD1-bound promoters (<TSS ± 1 kb) (A) and outside of promoters (>TSS ± 1 kb) (B). An extensive comparison with publicly available datasets was conducted for each group, using the ChIP-Atlas (http://chip-atlas.org/). Top 10 rankings are shown.
| Rank | ID | Antigen | Cell | Num of peaks | Overlaps / LSD1 | Overlaps / random | Fold enrichment | P-value | Q-value |
|---|---|---|---|---|---|---|---|---|---|
| (A) TFs similarly distributed with LSD1 (<TSS ± 1 kb) | |||||||||
| 1 | SRX3206550 | Brd4 | Muscle, Skeletal | 5236 | 4424/11268 | 27/11268 | 163.852 | <1.0 × 10−324 | <1.0 × 10−324 |
| 2 | SRX3206547 | Brd4 | Muscle, Skeletal | 6926 | 5690/11268 | 39/11268 | 145.897 | <1.0 × 10−324 | <1.0 × 10−324 |
| 3 | SRX3206545 | Brd4 | Muscle, Skeletal | 9561 | 7152/11268 | 50/11268 | 143.04 | <1.0 × 10−324 | <1.0 × 10−324 |
| 4 | SRX142538 | E2f4 | C2C12 | 2379 | 1628/11268 | 13/11268 | 125.231 | <1.0 × 10−324 | <1.0 × 10−324 |
| 5 | SRX522661 | Sin3a | C2C12 | 2230 | 1845/11268 | 15/11268 | 123 | <1.0 × 10−324 | <1.0 × 10−324 |
| 6 | SRX956815 | Six4 | C2C12 | 4850 | 3510/11268 | 29/11268 | 121.034 | <1.0 × 10−324 | <1.0 × 10−324 |
| 7 | SRX344972 | Sin3a | C2C12 | 1797 | 1405/11268 | 12/11268 | 117.083 | <1.0 × 10−324 | <1.0 × 10−324 |
| 8 | SRX150192 | Hdac1 | Myoblasts | 2823 | 1217/11268 | 11/11268 | 110.636 | <1.0 × 10−324 | <1.0 × 10−324 |
| 9 | SRX142525 | Max | C2C12 | 6364 | 2661/11268 | 30/11268 | 88.7 | <1.0 × 10−324 | <1.0 × 10−324 |
| 10 | SRX142526 | Rest | C2C12 | 5494 | 1466/11268 | 17/11268 | 86.2353 | <1.0 × 10−324 | <1.0 × 10−324 |
| (B) TFs similarly distributed with LSD1 (>TSS ± 1 kb) | |||||||||
| 1 | SRX770038 | Myod1 | C2C12 | 4298 | 3786/40746 | 60/40746 | 63.1 | <1.0 × 10−324 | <1.0 × 10−324 |
| 2 | SRX142533 | Fosl1 | C2C12 | 3696 | 2664/40746 | 45/40746 | 59.2 | <1.0 × 10−324 | <1.0 × 10−324 |
| 3 | SRX328690 | Myod1 | C2C12 | 10980 | 8705/40746 | 153/40746 | 56.8954 | <1.0 × 10−324 | <1.0 × 10−324 |
| 4 | SRX142529 | Tcf12 | C2C12 | 7817 | 6126/40746 | 112/40746 | 54.6964 | <1.0 × 10−324 | <1.0 × 10−324 |
| 5 | SRX142537 | Myod1 | C2C12 | 12521 | 8722/40746 | 166/40746 | 52.5422 | <1.0 × 10−324 | <1.0 × 10−324 |
| 6 | SRX142515 | Myog | C2C12 | 5718 | 4341/40746 | 84/40746 | 51.6786 | <1.0 × 10−324 | <1.0 × 10−324 |
| 7 | SRX142516 | Myod1 | C2C12 | 3446 | 2568/40746 | 50/40746 | 51.36 | <1.0 × 10−324 | <1.0 × 10−324 |
| 8 | SRX1817465 | Tead4 | C2C12 | 1697 | 1478/40746 | 29/40746 | 50.9655 | <1.0 × 10−324 | <1.0 × 10−324 |
| 9 | SRX039346 | Epitope tags | C2C12 | 3133 | 1462/40746 | 29/40746 | 50.4138 | <1.0 × 10−324 | <1.0 × 10−324 |
| 10 | SRX213538 | Mef2d | C2C12 | 3551 | 1840/40746 | 37/40746 | 49.7297 | <1.0 × 10−324 | <1.0 × 10−324 |
We observed that LSD1 was enriched at the promoter regions of oxidative metabolism genes at both days 2 and 5 (Figure 2D and F, Supplementary Figure S5A and B). In addition, LSD1 peaks were also detected in glycolytic genes such as Gapdh (Supplementary Figure S5C). LSD1 was bound at about 3 kb upstream of the transcription start site of Myh7 (Figure 2E and G). LSD1 enrichment was also observed downstream of another slow fiber gene, Tnnc1 (Supplementary Figure S5D), but not near the fast fiber gene Myh4 (Supplementary Figure S5E). Interestingly, enrichment of LSD1 at the Myh7 and Tnnc1 gene loci was detected at day 2 but not on day 5. In agreement with the ChIP-Atlas results, in the Myh7 and Tnnc1 loci, the LSD1 peaks were located at the MyoD- and myogenin-bound sites, which harbored the enhancer box (E-box) sequence, a consensus binding sequence for myogenic TFs (Figure 2E and Supplementary Figure S5D) (27). These results indicate that, in differentiating myoblasts, LSD1 preferentially binds to transcriptionally competent chromatin regions in oxidative fiber-associated genes.
LSD1 inhibition increases H3K4 methylation at target regions of oxidative metabolism genes
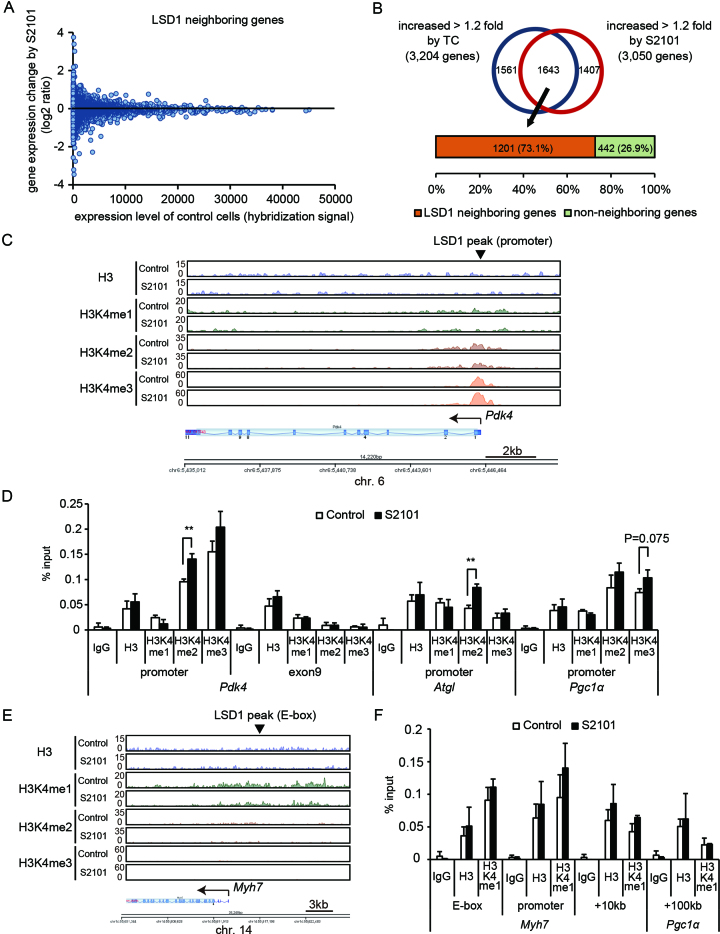
To gain mechanistic insights into the role of LSD1 in gene regulation, we analyzed the effects of LSD1 inhibition on expression of LSD1-bound genes. The overall expression profile of LSD1-bound genes was not greatly altered by LSD1 inhibition in C2C12 cells on day 2 (Figure 3A). However, a large percentage of LSD1 inhibition-induced genes (73.1%) was bound to LSD1 (Figure 3B), suggesting that LSD1 was involved in activation of these genes. To evaluate effects of LSD1 inhibition on H3K4 methylation status, we performed ChIP-seq using antibodies against H3K4me1, H3K4me2 and H3K4me3. Most of the genomic regions with high LSD1 enrichment had decreased H3K4me1 levels in the presence of LSD1 inhibition, while H3K4me3 levels were increased (Supplementary Figure S6), suggesting a link between LSD1 activity and H3K4 methylation equilibrium. In the Pdk4 gene locus, H3K4me2 and H3K4me3 were condensed near the promoter region, while H3K4me1 showed a dispersed distribution, both in control and LSD1-inhibited cells (Figure 3C). By quantitative ChIP-PCR, we observed elevated H3K4me2 levels at the promoters of Pdk4 and Atgl and a relatively high H3K4me3 level at the Pgc1a promoter in LSD1-inhibited cells (Figure 3D). In the Myh7 gene locus, there was widespread distribution of H3K4me1, while H3K4me2 and H3K4me3 were nearly absent (Figure 3E). A region harboring an E-box in Myh7 did not have detectable redistribution of H3K4 methylation (Figure 3F).
Figure 3.
LSD1 fine-tunes expression of actively transcribed genes related to oxidative metabolism. (A) Scatterplot showing correlation between gene expression levels and changes in expression under LSD1 inhibition. LSD1-neighboring genes in C2C12 cells on day 2 (16,858 genes) are shown. (B) Percentage of LSD1 inhibition-induced genes associated with neighboring LSD1-bound regions. (C) H3K4 methylation at the Pdk4 gene locus. Control or S2101 (10−5 M) treated C2C12 cells were analyzed at day 2 after myogenic induction. ChIP-seq data are presented in the same manner as in Figure 2D. (D) ChIP-qPCR analyses of H3K4 methylation at metabolic gene promoters. ChIP analyses were performed using differentiating (day 2) C2C12 cells (n = 3). (E) Histone modifications at the Myh7 gene locus. (F) ChIP-qPCR analyses of H3K4me1 enrichment at the Myh7 gene locus (n = 3). **P < 0.01.
To further confirm regulation of oxidative metabolism genes by LSD1 in myogenic cells, we generated stable LSD1 knockdown (KD) myoblasts by introducing a shRNA expression vector into C2C12 cells (Supplementary Figure S7A). Consistent with previous reports (28–30), LSD1-KD cells failed to differentiate into myotubes upon induction (Supplementary Figure S7B). We performed a microarray analysis of control and LSD1-KD C2C12 cells at differentiation day 2, identifying 2104 genes upregulated by LSD1-KD (Supplementary Figure S7C). Of these, 435 were also upregulated by LSD1 inhibitors and genes in this group were enriched for oxidative genes (Supplementary Figure S7C and D, Supplementary Table S3). By ChIP-qPCR analyses, we found increased H3K4 methylation at a number of oxidative gene promoters and the Myh7-E-box region (Supplementary Figure S7E and F). Taken together, these results suggest that loss of LSD1 caused remodeling of local H3K4 dynamics to increase expression of oxidative metabolism genes during myogenesis.
Glucocorticoid induces JADE2-mediated LSD1 degradation
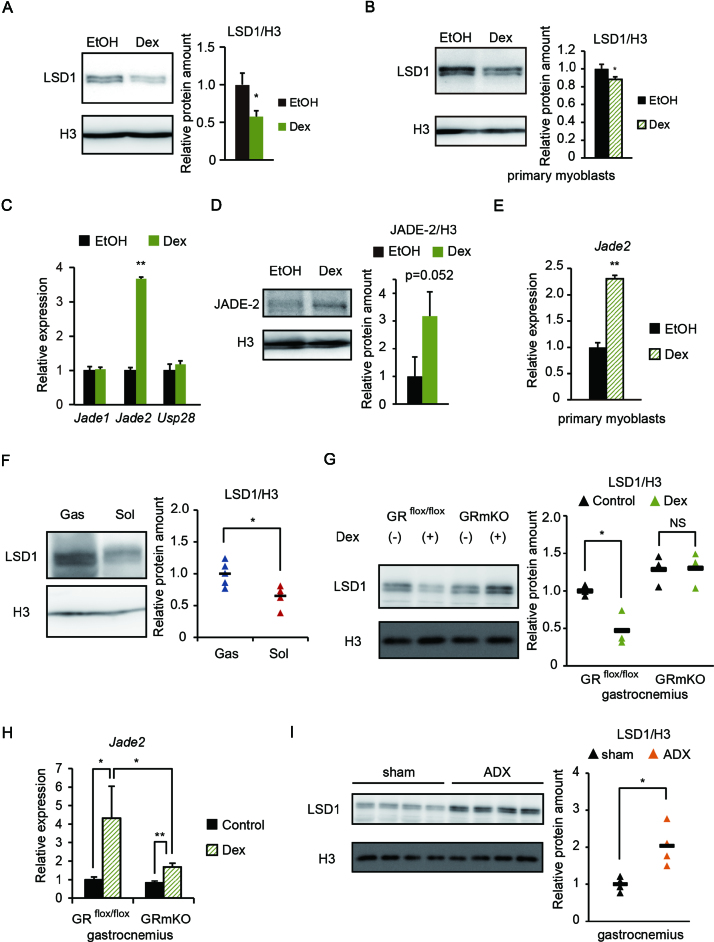
To investigate effects of the endocrine environment on LSD1 activity, we treated differentiated C2C12 myotubes with hormones associated with either anabolic or catabolic signaling. Interestingly, the glucocorticoid analog dexamethasone (Dex) caused a marked decrease in levels of LSD1 protein (Supplementary Figure S8A and B). These results were confirmed by showing a significant decrease in LSD1 levels after Dex treatment in both C2C12 and mouse primary myotubes (Figure 4A and B). Despite the decreased levels of LSD1 protein in the presence of Dex, LSD1 mRNA expression was unaffected, suggesting post-transcriptional regulation (Supplementary Figure S8C and D). It was reported that LSD1 was actively degraded by the ubiquitin-proteasome system (UPS), depending on the cellular context (31,32). To assess the contribution of UPS in Dex-induced LSD1 regulation, cells were treated with MG132, a proteasome inhibitor (Supplementary Figure S9A). To eliminate de novo synthesis of LSD1, cycloheximide (CHX) was also added. Combined treatment with MG132 and CHX prevented the suppressive effect of Dex on LSD1 protein levels, indicating that the glucocorticoid triggered proteasomal LSD1 degradation. Several factors were implicated in regulation of LSD1 protein levels. The plant homeodomain finger-containing protein, JADE-2 (also known as PHF15), was reported to act as an E3 ubiquitin ligase for LSD1 in neural cells (31), while USP28 acted as a deubiquitinase for LSD1 in cancer cells (32). Intriguingly, JADE-2 expression was markedly increased in C2C12 and primary myoblasts treated with Dex (Figure 4C–E). A glucocorticoid response element is located at approximately 17 kb upstream of the transcription start site of the Jade2 gene (33) (Supplementary Figure S9B). By ChIP analysis, we determined that the glucocorticoid receptor (GR) was prominently targeted to this region in response to Dex (Supplementary Figure S9B). To confirm involvement of JADE-2 in Dex-induced LSD1 degradation, we used mouse hepatoma Hepa1-6 cells showing nuclear translocation of abundantly expressed GR (Supplementary Figure S9C). We found that JADE-2 levels were similarly increased by Dex treatment in these cells, and that the Dex-induced decrease in LSD1 levels was attenuated by Jade2 KD (Supplementary Figure S9D). These data show that glucocorticoid signaling induces JADE-2 expression, resulting in degradation of LSD1 protein.
Figure 4.
Downregulation of LSD1 by a glucocorticoid in cultured myotubes and in mouse skeletal muscle in vivo. (A and B) Expression of LSD1 protein in C2C12 myotubes (A) and primary myoblasts (B) under Dex (10−6 M) treatment (n = 3). Following myogenic induction for 5 days in C2C12 and 4 days in primary cells, cells were treated with Dex or vehicle (EtOH) for 24 h. Band densities were quantified by densitometry and values, normalized to those for histone H3, are shown. (C) Expression of genes related to LSD1 ubiquitination (n = 3). Cells were subjected to myogenic induction for 4 days and then treated with Dex for 24 h. Values are shown as fold differences against those for EtOH-treated controls. (D) Effects of Dex on JADE-2 protein levels in C2C12 myotubes (n = 3). Values are shown as fold differences against those for EtOH-treated controls. (E) Expression of the Jade2 gene in myotubes derived from primary myoblasts treated with Dex for 24 h (n = 3). (F) Expression of LSD1 protein in gastrocnemius (Gas) and soleus (Sol) muscle in 8-week-old male C57BL/6J mice (n = 5). Band densities were quantified by densitometry, normalized to those for histone H3 and plotted as triangles. Values are shown as fold differences against the average for Gas. Black bars show the means. (G) Effects of Dex administration on LSD1 protein levels in mouse skeletal muscle (n = 6). Protein was extracted from Gas muscle from GRflox/flox and GRmKO mice treated with PBS (controls) or Dex for 7 days before sacrifice. Band densities were quantified by densitometry, normalized to those for histone H3 and plotted as triangles. Values are shown as fold differences against the average of vehicle (PBS)-treated GRflox/flox samples. (H) Effects of Dex administration on Jade2 expression in mouse skeletal muscle (n = 5). RNA was extracted from Gas muscle from GRflox/flox and GRmKO mice treated with PBS (controls) or Dex for 7 days before sacrifice. Values are shown as fold differences compared with those for PBS-treated GRflox/flox samples. (I) Effects of adrenalectomy on LSD1 protein expression in mouse skeletal muscle (n = 4). Protein was extracted from Gas muscle from C57BL/6J mice at 7 days after adrenalectomy (ADX) or sham surgery. Band densities were quantified by densitometry and normalized to those for histone H3. Values are shown as fold differences against the averages of sham control samples. *P < 0.05, **P < 0.01. NS: no significant difference.
LSD1 downregulation is mediated by GR in mouse skeletal muscle
Among skeletal muscle tissues in mice, the fast fiber enriched gastrocnemius had higher LSD1 expression than did the slow fiber enriched soleus (Figure 4F, Supplementary Figure S9E). To test glucocorticoid-induced LSD1 suppression in vivo, we used muscle-specific glucocorticoid receptor knockout (GRmKO) mice, which exhibited fast fiber enrichment (16). In control mice, continuous administration of Dex for one week induced a moderate decline in LSD1 levels in the skeletal muscle (Figure 4G). In contrast, Dex treatment did not affect LSD1 levels in GRmKO mice (Figure 4G). In agreement with these observations, Jade2 expression was markedly elevated after continuous Dex treatment, while this effect was attenuated in Dex treated GRmKO mice (Figure 4H). Furthermore, in muscles of adrenalectomized mice, thus devoid of corticosterone production (16), LSD1 levels were significantly elevated (Figure 4I). We next analyzed publicly available transcriptome datasets (34), finding that Jade2 expression was significantly upregulated in metabolic tissues, such as adipose tissue, liver and skeletal muscle, in fasting mice (Supplementary Figure S9F). These results indicate that LSD1 protein level is negatively regulated by GR-mediated signaling in skeletal muscle.
LSD1 inhibition facilitates glucocorticoid-induced expression of oxidative metabolism genes
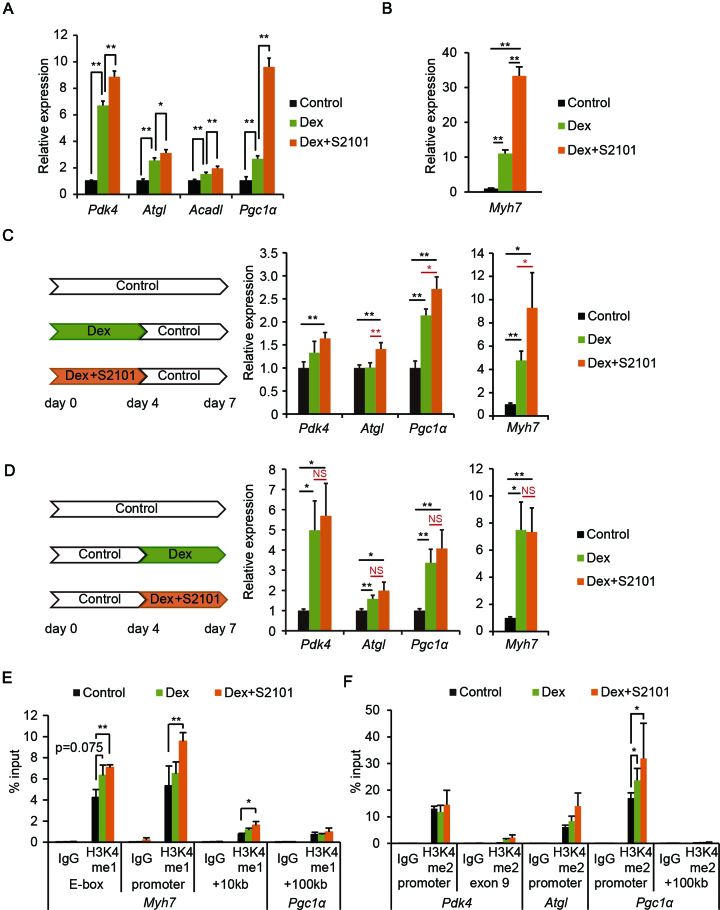
Glucocorticoid signaling was shown to enhance OXPHOS and promote the selective wasting of fast muscle fibers (35,36). Having observed that LSD1 repressed expression of oxidative genes (Figure 1), we hypothesized that loss of LSD1 function would enhance the hormonal actions of glucocorticoids. Indeed, Dex treatment during differentiation increased expression of oxidative metabolism genes, an effect further augmented by S2101 (Figure 5A). In addition, LSD1 inhibition and Dex synergistically enhanced expression of the slow fiber gene Myh7 (Figure 5B and Supplementary Figure S10A). By immunofluorescence, we confirmed that staining with anti-slow-myosin heavy chain (slow-MHC) antibody was enhanced in myotubes treated with Dex and S2101 (Supplementary Figure S10B and C). These data suggest that LSD1 counteracts glucocorticoid-induced expression of oxidative fiber genes. To gain further insights into functional interactions between LSD1 and glucocorticoid signaling, we treated myoblasts with Dex and S2101 during either the early or late phase of differentiation. LSD1 inhibition during the early phase, but not the late phase, markedly augmented Dex-induced expression of both oxidative and Myh7 genes (Figure 5C and D). These results emphasized that LSD1 contributes to differentiation-coupled metabolic programming in myoblasts.
Figure 5.
LSD1 inhibition facilitates Dex-induced expression of oxidative genes. (A) Effects of Dex and S2101 on expression of oxidative metabolism genes (n = 3). C2C12 cells were cultured in differentiation medium for 7 days in the presence of Dex (10−6 M) alone or with S2101 (10−5 M). Values shown are fold differences against vehicle-treated controls. (B) Expression of the Myh7 gene in Dex- and S2101-treated cells. Values shown are fold differences against vehicle-treated controls. (C and D) Expression of oxidative metabolism and Myh7 genes in C2C12 myotubes treated with Dex and S2101 during the early phase (C) or late phase (D) of differentiation (n = 4). RNA was extracted from C2C12 cells on day 7 after differentiation with the indicated treatments. (E) Enrichment of H3K4me1 at the Myh7 gene locus. ChIP analyses were performed using differentiating C2C12 cells (48 h) treated with Dex and S2101 (n = 3). Values are each shown as % input. (F) Enrichment of H3K4me2 at the promoter regions of oxidative metabolism genes. *P < 0.05, **P < 0.01. NS: no significant difference.
To explore how LSD1 counteracted glucocorticoid-induced gene expression, we evaluated changes in H3K4 methylation in cells treated with Dex and S2101. H3K4me1 levels were analyzed at the E-box region of the Myh7 locus, because ChIP-seq data indicated an enrichment of the enhancer signature, including H3K4me1 and acetylated H3K27, at this region (Figure 3E and data not shown) (37). Treatment with S2101, together with Dex, significantly increased H3K4me1 levels at the E-box and at additional regions in the Myh7 locus (Figure 5E). We also found increased levels of H3K4me2 at the promoter regions of oxidative metabolism genes in cells treated with Dex and S2101, especially at the promoter of Pgc1α (Figure 5F). Collectively, our results suggest that LSD1 maintains transcriptionally competent chromatin by H3K4 demethylation, priming these sites for glucocorticoid-induced gene activation.
DISCUSSION
Environmental conditions, such as nutritional state, exert profound influences on epigenome, but little is known about the underlying mechanisms. Our study demonstrated that LSD1 transcriptionally repressed oxidative metabolism and slow fiber myosin genes via the H3K4 demethylation, with a concomitant decrease of oxygen consumption. A catabolic glucocorticoid signal decreased LSD1 protein levels by the glucocorticoid receptor-mediated expression of the E3 ubiquitin ligase, JADE-2. In agreement with these observations, the combination of glucocorticoid stimulation and LSD1 inhibition synergistically enhanced oxidative fiber gene expression with increased levels of H3K4 methylation at LSD1-bound promoters and enhancers. These results indicated that LSD1 serves as an epigenetic hub to translate environmental information into epigenetic remodeling (Supplementary Figure S11).
Previous reports showed that LSD1 was required for differentiation of cultured myoblasts (28,29). Those findings were based on LSD1 loss-of-function experiments using stable shRNA expression or CRSPR-Cas9-based genome editing, which required extended culture of undifferentiated myoblasts after LSD1 inactivation. This extended culture might have significantly affected the myogenic potential of the cells. More recently, Scionti et al. reported that LSD1 was required for MyoD expression at the initial step of myogenesis in myogenic stem cells (30). Notably, they demonstrated that H3K9 methylation level at an enhancer of Myod gene was increased in LSD1-KD cells before differentiation induction, suggesting that those cells had lost myogenic identity. Consistent with these previous reports, we observed a loss of differentiation potential after establishment of LSD1-KD in C2C12 cells, accompanied by decreased MyoD expression (Supplementary Figure S7 and data not shown). In our study, we demonstrated that LSD1 restricted the metabolic properties of differentiating myoblasts by suppressing expression of oxidative genes without affecting the differentiation itself. Of note, we added LSD1 inhibiting drugs only during the differentiation period, when MyoD expression had already been established. Scionti et al. tested the effects of LSD1 inhibitors other than the ones we used, and demonstrated that inhibitor treatment upon differentiation did not affect Myod expression (30). Thus, both our study and previous reports demonstrated that LSD1 function during myogenesis depends on the differentiation state. In particular, our data show that LSD1 has context-dependent activities as an environment-responsive epigenetic factor.
LSD1 contributes to repressive chromatin structure through H3K4 demethylation, but in some cases, activates transcription through H3K9 demethylation (8,38). Our ChIP-seq data revealed that LSD1 was highly coincident with active chromatin marks such as methylated H3K4, H3K27ac and pol II, suggesting that LSD1 preferentially binds to transcriptionally competent chromatin. At genomic regions with high LSD1 occupancy, loss of LSD1 activity was associated with reduced H3K4me1 and increased H3K4me3 levels. At oxidative metabolism gene loci, loss of LSD1 led to upregulated expression with moderate increase in H3K4 methylation. By contrast, LSD1 did not show a prominent engagement with H3K9me3 peaks. We previously reported using adipogenic cells that LSD1 inhibition did not affect H3K9 methylation at the promoters of oxidative metabolism genes (9). Collectively, the line of evidences suggests that, for the regulation of metabolic plasticity, LSD1 contribute to the balancing of H3K4 methylation state at transcriptionally competent genes, rather than affecting tightly repressive H3K9 methylation marks. LSD2, the other flavin-dependent lysine demethylase, is also expressed in myogenic cells (data not shown). We previously reported that LSD2 repressed lipid transport genes including CD36 to limit lipid influx and metabolism in human hepatic cells (39). Previous report demonstrated that CD36 was preferentially expressed in slow-twitch rather than fast-twitch fiber of human skeletal muscle (40). Thus, LSD2 together with LSD1 might contribute to the suppression of slow type metabolism in the muscle.
A glucocorticoid was shown to enhance oxidative metabolism and contractility in fetal cardiomyocytes by inducing expression of the PGC-1α gene (41). In addition, glucocorticoid signaling promotes fast fiber-selective muscle wasting, suggesting that this class of hormones favored oxidative muscle activity (36,42). Here, we demonstrated that Dex treatment led to increased expression of oxidative and slow-type myosin genes, effects further augmented by LSD1 inhibition. Our results suggest that the catabolic action of the glucocorticoid in muscle was counteracted by LSD1-mediated chromatin modification. Also of interest, a glucocorticoid surge in late gestation was reported to contribute to maturation of cardiac muscle, which shows a high bias toward oxidative energy metabolism (43). Thus, it is possible that crosstalk between glucocorticoids and LSD1 may be involved in early programming of oxidative capacity in muscle lineages.
Epidemiologic and animal studies indicated that malnutrition or over-nutrition in early life is linked to an increased risk of metabolic disorders such as obesity, diabetes and cardiovascular diseases in adulthood (44,45). Although relationships between diet and the epigenome have been very actively studied, it is not yet clear how nutritional conditions can affect health and disease through specific epigenetic mechanisms (3). Previously, we demonstrated that, under anabolic conditions, LSD1 repressed expression of oxidative metabolism and lipid metabolism associated genes in cultured adipose cells and tissues (9,10). Our current study further showed that glucocorticoids, involved in fetal and postnatal growth in mammals, targeted LSD1-mediated metabolic gene regulation. Interestingly, patients with type 2 diabetes or obesity were reported to have more glycolytic and fewer oxidative fibers, compared with healthy individuals (46,47). In addition, the PGC-1α gene was epigenetically silenced in the skeletal muscle of individuals with diabetes (48). Based on these findings, it is tempting to speculate that the nutritional environment could influence LSD1-mediated epigenome formation, which may then affect long-term metabolic outcomes.
In conclusion, our study demonstrated that multiple metabolic signaling processes affect LSD1 activity, which could then influence the trajectory of metabolic reprogramming in differentiating myoblasts. Our findings extend the current understanding of how environmental factors can control the epigenome during transitions of cellular states.
DATA AVAILABILITY
The accession number for the microarray data presented in this paper are GEO: GSE86524 (LSD1 inhibitors) and GSE109839 (LSD1-KD), and that for the ChIP-seq data are GSE109845 (methylated H3K4) and GSE109846 (LSD1).
Supplementary Material
ACKNOWLEDGEMENTS
We thank our laboratory members for helpful discussions and technical support. We thank Susan R. Doctrow, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Author Contributions: Conceptualization, K.A., S.H. and M.N.; Methodology, K.A. and S.H.; Software, S.O.; Investigation, K.A., S.H., N.S., A.K., K.N., R.T., K.K., H.A., Y.H and S.U.; Writing, K.A., S.H., N.S. and M.N.; Visualization, K.A., S.H. and M.N.; Supervision, H.T., K.N., F.E. and M.N.; Funding acquisition, S.H. and M.N.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JSPS KAKENHI [JP15H04707 to M.N., JP15K15068 to M.N., JP16K07215 to S.H., 25430178 to S.H.]; Takeda Science Foundation (to M.N. and S.H.); Kanae Foundation for the Promotion of Medical Science (to S.H.); Ono Medical Research Foundation (to S.H.); Mochida Memorial Foundation for Medical and Pharmaceutical Research (to S.H.). Funding for open access charge: Takeda Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Feil R., Fraga M.F.. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2011; 13:97–109. [DOI] [PubMed] [Google Scholar]
- 2. Siebel A.L., Fernandez A.Z., El-Osta A.. Glycemic memory associated epigenetic changes. Biochem. Pharmacol. 2010; 80:1853–1859. [DOI] [PubMed] [Google Scholar]
- 3. Jimenez-Chillaron J.C., Diaz R., Martinez D., Pentinat T., Ramon-Krauel M., Ribo S., Plosch T.. The role of nutrition on epigenetic modifications and their implications on health. Biochimie. 2012; 94:2242–2263. [DOI] [PubMed] [Google Scholar]
- 4. Burg J.M., Link J.E., Morgan B.S., Heller F.J., Hargrove A.E., McCafferty D.G.. KDM1 class flavin-dependent protein lysine demethylases. Biopolymers. 2015; 104:213–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y.. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004; 119:941–953. [DOI] [PubMed] [Google Scholar]
- 6. Amente S., Lania L., Majello B.. The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim. Biophys. Acta. 2013; 1829:981–986. [DOI] [PubMed] [Google Scholar]
- 7. Hino S., Nagaoka K., Nakao M.. Metabolism-epigenome crosstalk in physiology and diseases. J. Hum. Genet. 2013; 58:410–415. [DOI] [PubMed] [Google Scholar]
- 8. Hino S., Kohrogi K., Nakao M.. Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci. 2016; 107:1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hino S., Sakamoto A., Nagaoka K., Anan K., Wang Y., Mimasu S., Umehara T., Yokoyama S., Kosai K., Nakao M.. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat. Commun. 2012; 3:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakamoto A., Hino S., Nagaoka K., Anan K., Takase R., Matsumori H., Ojima H., Kanai Y., Arita K., Nakao M.. Lysine Demethylase LSD1 coordinates glycolytic and mitochondrial metabolism in hepatocellular carcinoma cells. Cancer Res. 2015; 75:1445–1456. [DOI] [PubMed] [Google Scholar]
- 11. Duteil D., Metzger E., Willmann D., Karagianni P., Friedrichs N., Greschik H., Gunther T., Buettner R., Talianidis I., Metzger D. et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 2014; 5:4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schiaffino S., Reggiani C.. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011; 91:1447–1531. [DOI] [PubMed] [Google Scholar]
- 13. Hoppeler H. Molecular networks in skeletal muscle plasticity. J. Exp. Biol. 2016; 219:205–213. [DOI] [PubMed] [Google Scholar]
- 14. Moresi V., Marroncelli N., Adamo S.. New insights into the epigenetic control of satellite cells. World J. Stem. Cells. 2015; 7:945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrio E., Suelves M.. DNA methylation dynamics in muscle development and disease. Front Aging Neurosci. 2015; 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shimizu N., Maruyama T., Yoshikawa N., Matsumiya R., Ma Y., Ito N., Tasaka Y., Kuribara-Souta A., Miyata K., Oike Y. et al. A muscle-liver-fat signalling axis is essential for central control of adaptive adipose remodelling. Nat. Commun. 2015; 6:6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rando T.A., Blau H.M.. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994; 125:1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee M.G., Wynder C., Schmidt D.M., McCafferty D.G., Shiekhattar R.. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006; 13:563–567. [DOI] [PubMed] [Google Scholar]
- 21. Mimasu S., Umezawa N., Sato S., Higuchi T., Umehara T., Yokoyama S.. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry. 2010; 49:6494–6503. [DOI] [PubMed] [Google Scholar]
- 22. Niwa H., Umehara T.. Structural insight into inhibitors of flavin adenine dinucleotide-dependent lysine demethylases. Epigenetics. 2017; 12:340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bentzinger C.F., Wang Y.X., Rudnicki M.A.. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012; 4:a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Watt M.J., Hoy A.J.. Lipid metabolism in skeletal muscle: generation of adaptive and maladaptive intracellular signals for cellular function. Am. J. Physiol. Endocrinol. Metab. 2012; 302:E1315–E1328. [DOI] [PubMed] [Google Scholar]
- 26. Yuan Y., Shi X.E., Liu Y.G., Yang G.S.. FoxO1 regulates muscle fiber-type specification and inhibits calcineurin signaling during C2C12 myoblast differentiation. Mol. Cell. Biochem. 2011; 348:77–87. [DOI] [PubMed] [Google Scholar]
- 27. Moncaut N., Rigby P.W., Carvajal J.J.. Dial M(RF) for myogenesis. FEBS J. 2013; 280:3980–3990. [DOI] [PubMed] [Google Scholar]
- 28. Choi J., Jang H., Kim H., Kim S.T., Cho E.J., Youn H.D.. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem. Biophys. Res. Commun. 2010; 401:327–332. [DOI] [PubMed] [Google Scholar]
- 29. Munehira Y., Yang Z., Gozani O.. Systematic analysis of known and candidate lysine demethylases in the regulation of myoblast differentiation. J. Mol. Biol. 2017; 429:2055–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scionti I., Hayashi S., Mouradian S., Girard E., Esteves de Lima J., Morel V., Simonet T., Wurmser M., Maire P., Ancelin K. et al. LSD1 controls timely MyoD expression via MyoD core enhancer transcription. Cell Rep. 2017; 18:1996–2006. [DOI] [PubMed] [Google Scholar]
- 31. Han X., Gui B., Xiong C., Zhao L., Liang J., Sun L., Yang X., Yu W., Si W., Yan R. et al. Destabilizing LSD1 by Jade-2 promotes neurogenesis: an antibraking system in neural development. Mol. Cell. 2014; 55:482–494. [DOI] [PubMed] [Google Scholar]
- 32. Wu Y., Wang Y., Yang X.H., Kang T., Zhao Y., Wang C., Evers B.M., Zhou B.P.. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013; 5:224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuo T., Lew M.J., Mayba O., Harris C.A., Speed T.P., Wang J.C.. Genome-wide analysis of glucocorticoid receptor-binding sites in myotubes identifies gene networks modulating insulin signaling. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:11160–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schupp M., Chen F., Briggs E.R., Rao S., Pelzmann H.J., Pessentheiner A.R., Bogner-Strauss J.G., Lazar M.A., Baldwin D., Prokesch A.. Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genomics. 2013; 14:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee S.R., Kim H.K., Song I.S., Youm J., Dizon L.A., Jeong S.H., Ko T.H., Heo H.J., Ko K.S., Rhee B.D. et al. Glucocorticoids and their receptors: insights into specific roles in mitochondria. Prog. Biophys. Mol. Biol. 2013; 112:44–54. [DOI] [PubMed] [Google Scholar]
- 36. Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S., Nakae J., Tagata Y., Nishitani S., Takehana K. et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011; 13:170–182. [DOI] [PubMed] [Google Scholar]
- 37. Zhou V.W., Goren A., Bernstein B.E.. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 2011; 12:7–18. [DOI] [PubMed] [Google Scholar]
- 38. Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R.. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005; 437:436–439. [DOI] [PubMed] [Google Scholar]
- 39. Nagaoka K., Hino S., Sakamoto A., Anan K., Takase R., Umehara T., Yokoyama S., Sasaki Y., Nakao M.. Lysine-specific demethylase 2 suppresses lipid influx and metabolism in hepatic cells. Mol. Cell. Biol. 2015; 35:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vistisen B., Roepstorff K., Fau-Roepstorff C., Roepstorff C., Fau-Bonen A., Bonen A., Fau-van Deurs B., van Deurs B., Fau-Kiens B., Kiens B.. Sarcolemmal FAT/CD36 in human skeletal muscle colocalizes with caveolin-3 and is more abundant in type 1 than in type 2 fibers. J. Lipid Res. 2004; 45:603–609. [DOI] [PubMed] [Google Scholar]
- 41. Rog-Zielinska E.A., Craig M.A., Manning J.R., Richardson R.V., Gowans G.J., Dunbar D.R., Gharbi K., Kenyon C.J., Holmes M.C., Hardie D.G. et al. Glucocorticoids promote structural and functional maturation of foetal cardiomyocytes: a role for PGC-1alpha. Cell Death Differ. 2015; 22:1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ciciliot S., Rossi A.C., Dyar K.A., Blaauw B., Schiaffino S.. Muscle type and fiber type specificity in muscle wasting. Int. J. Biochem. Cell Biol. 2013; 45:2191–2199. [DOI] [PubMed] [Google Scholar]
- 43. Rog-Zielinska E.A., Richardson R.V., Denvir M.A., Chapman K.E.. Glucocorticoids and foetal heart maturation; implications for prematurity and foetal programming. J. Mol. Endocrinol. 2014; 52:R125–R135. [DOI] [PubMed] [Google Scholar]
- 44. Hales C.N., Barker D.J.. The thrifty phenotype hypothesis. Br. Med. Bull. 2001; 60:5–20. [DOI] [PubMed] [Google Scholar]
- 45. Roseboom T.J., van der Meulen J.H., Ravelli A.C., Osmond C., Barker D.J., Bleker O.P.. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol. Cell. Endocrinol. 2001; 185:93–98. [DOI] [PubMed] [Google Scholar]
- 46. He J., Watkins S., Kelley D.E.. Skeletal muscle lipid content and oxidative enzyme activity in relation to muscle fiber type in type 2 diabetes and obesity. Diabetes. 2001; 50:817–823. [DOI] [PubMed] [Google Scholar]
- 47. Tanner C.J., Barakat H.A., Dohm G.L., Pories W.J., MacDonald K.G., Cunningham P.R., Swanson M.S., Houmard J.A.. Muscle fiber type is associated with obesity and weight loss. Am. J. Physiol. Endocrinol. Metab. 2002; 282:E1191–E1196. [DOI] [PubMed] [Google Scholar]
- 48. Barres R., Osler M.E., Yan J., Rune A., Fritz T., Caidahl K., Krook A., Zierath J.R.. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009; 10:189–198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the microarray data presented in this paper are GEO: GSE86524 (LSD1 inhibitors) and GSE109839 (LSD1-KD), and that for the ChIP-seq data are GSE109845 (methylated H3K4) and GSE109846 (LSD1).