Abstract
Kidney cancer, or renal cell carcinoma (RCC), is a disease of increasing incidence which is commonly seen in the general practice of nephrology. Despite this state of affairs, this fascinating and highly morbid disease is frequently underrepresented, or even absent, from the curriculum of nephrologists in training and is generally underemphasized in national nephrology meetings, both scientific as well as clinical. While classical concepts in cancer research in general had led to the concept that cancer is a disease resulting from mutations in the control of growth-regulating pathways, reinforced by the discovery of oncogenes, more contemporary research particularly in kidney cancer has uncovered changes in metabolic pathways mediated by those same genes which control tumor energetics and biosynthesis. This adaptation of classical biochemical pathways to the tumor’s advantage has been labeled “metabolic reprogramming”. For example, in the case of kidney cancer there exists near universal presence of pVHL inactivation in the most common form, clear cell RCC (ccRCC), leading to activation of hypoxia-relevant and other metabolic pathways. Studies of this and other pathways in clear cell RCC (ccRCC) have been particularly revealing, leading to the concept that ccRCC can itself be considered a “metabolic disease”. For this reason, the relatively new method of metabolomics has become a useful technique in the study of ccRCC to tease out those pathways which have been reprogrammed by the tumor to its maximum survival advantage. Furthermore, identification of the “nodes” of such pathways can lead to novel areas for drug intervention in a disease for which such targets are seriously lacking. Further research and dissemination of these concepts, likely using omics techniques, will lead to clinical trials of therapeutics specifically targeted to tumour metabolism, rather than those generally toxic to all proliferating cells. Such novel agents are highly likely to be more effective than existing drugs and to have far fewer adverse effects. This review provides a general overview of the technique of metabolomics and then discusses how it and other omics techniques have been used to further our understanding of the basic biology of kidney cancer as well as identify new therapeutic approaches.
Keywords: kidney cancer, metabolomics, therapeutics, reprogramming
Utility of metabolomics in kidney cancer
With the discovery of clonal proliferation and subsequently of oncogenes, cancer had long been considered a disease of uncontrolled cell proliferation mediated by such phenomena as constitutively activated growth factor receptors. However, long after the initially puzzling observation of enhanced glycolysis — an energetically inefficient process — occurring in aggressive tumors despite normoxia1, a steady stream of more recent discoveries has linked cancers with a variety of metabolic changes that are only indirectly related to the more obvious phenotype of cell proliferation. Many of these metabolic derangements, especially in cancer of the kidney2,3, have now been linked directly to oncogenic mutations (Table 1, permission obtained from 4)5–7. This research has led to the concept that the evolution of altered metabolism, so-called metabolic reprogramming, is associated with a kidney cancer that is most likely to thrive within the body. When evaluated critically, such reprogramming makes obvious sense because in order to enable uncontrolled proliferation of cancer cells, a tumor has basic needs for augmented levels of cellular building components, such as DNA, nucleotides, and membrane constituents, as well as high levels of molecules that regulate the enhanced tumour energetics.
Table 1.
Selected oncogenes and tumour suppressor genes that regulate metabolism4 (with permission)
| Gene | Metabolic pathway | Relevance to RCC | Refs |
|---|---|---|---|
| PTEN | Inhibition of glycolysis through inactivation of Akt | 2.6% of patients have biallelic loss and 16.6% of patients have monoallelic loss of PTEN Loss of PTEN is associated with high stage and grade of RCC | 49,50 |
| TSC1/2 | Deficiency leads to the Warburg effect and glutamine addiction through activation of mTOR | Mutation is a risk factor for RCC | 51,52 |
| AKT | Upregulation of glycolysis through activation of enzymes including HK | Akt mutation is rare in RCC; activation is through PTEN loss. Akt inhibitors are being tested in clinical trials for RCC |
53,54,55,56 |
| VHL | Inhibition of the Warburg effect through deactivation of HIF | Loss-of-function mutation found in >90% of patients | 57,58 |
| p53 | Downregulation of glycolysis by deactivation of GLUT1/4 and upregulation of TIGAR Upregulation of glutamine metabolism via increased transcription of GLS2 |
Mutation is rare in RCC | 59,60,61,62 |
| LKB1 | Upregulation of glycolysis and β-oxidation and downregulation of lipid synthesis through activation of AMPK | Activity compromised in vitro, in vivo, and in patients | 63,64,65 |
| Myc | Upregulation of the Warburg effect through activation of HK, LDHA, and PDK1 Upregulation of glutamine metabolism through GLS1 activation Upregulation of lipid synthesis through activation of FAS and SCD1 |
Often mutated and overexpressed in RCC Activated by HIF-2α Overexpression induces RCC in mice |
66,67,68,69 70 |
mTOR, mammalian target of rapamycin; HK, hexokinase; RCC, renal cell carcinoma; HIF, hypoxia-inducible factor; GLUT, glucose transporter; TIGAR, p53-inducible glycolysis and apoptosis regulator; GLS, glutaminase; APMK, AMP-activated protein kinase; LDHA, lactate dehydrogenase A; PDK1, pyruvate dehydrogenase kinase 1; FAS, fatty acid synthase; SCD1, Stearoyl-CoA desaturase 1.
With the recent advances in high-throughput technologies, the pace of parallel advances in the “omics” fields that are relevant to clinical medicine, including nephrology, has markedly accelerated. If used cautiously and with full knowledge of their inherent limitations, these techniques have the potential to alter how we diagnose and treat human diseases, and their use in nephrology, while lagging as compared to other fields, has now begun to bear fruit most notably in kidney cancer. The advent of metabolomics, in which all of the metabolites in a given tissue or biofluid are examined (with the caveat that some metabolites will be below the detection limit of the instrumentation used and others will not be readily chemically identified), is one of the latest advances in the field of omics. Although not necessarily superior to the other, older omics technologies, metabolomics does have several advantages, in that there are comparatively fewer metabolites compared with genes, transcripts and proteins, which means that there is less data to manipulate. In addition, changes in metabolites are the most accurate predictors of the signature of the actual processes that are occurring within the body, rather than changes in compounds (such as untranscribed DNA or proteins modified before or after translation) that might be superfluous to these processes8. In light of this advantage and the fact that metabolomics analysis can readily be performed on biofluids such as blood and urine as well as more difficultly obtained tissue, it is becoming increasingly clear that metabolomics has great potential for biomarker discovery9. However, serious limitations to this technique exist, mostly connected to consistency of sample collection, potential confounding effects, and limitations in detection of low-abundance or unidentifiable metabolites9.
Metabolomics, in its non-targeted, hypothesis-generating, and thus most useful form, is defined as the non-targeted measurement of all metabolites produced by the body. Now recognized as an “established” omics10, metabolomics has shown utility in biomarker discovery and, more successfully (at least in our hands), in the identification of new therapeutic targets and paradigms. The utility of metabolomics, perhaps more so than other established omics, lies in the fact that the metabolites measured in tissue or biofluids represent actual biochemical processes occurring in the body, such that when such processes are altered in disease states, the set of metabolites measured generated will change accordingly. This may not be the case in other omics techniques since the quantitation of genes and proteins may not correlate with their activities. Indeed, combining metabolomics with proteomics11 or with transcriptomics12 can greatly increase its utility in helping to understand RCC biology.
Metabolic reprogramming is to the cancer’s advantage
Many classical metabolic pathways are increased, decreased, or bypassed entirely in cancer cells; kidney cancer is one of the most studied, and could therefore be considered the “poster child” of malignancies characterized by metabolic reprogramming11,12. For example, in addition to the well-known and continually validated Warburg effect, reductive carboxylation occurs in many clear cell renal cell carcinoma (ccRCC) cells13,14. This glutamine-dependent pathway involves “backwards flow” of the tricarboxylic acid (TCA) cycle, and in addition increases an intrinsic oxidative stress buffer pathway both of which help the tumor grow in a hostile environment15. Such unexpected aberrations of metabolism in RCC have provided new opportunities for both imaging and for the development of new targets for therapeutic intervention15.
Metabolic reprogramming in ccRCC
The labelling of ccRCC as a metabolic disease arises from the fact that many metabolic pathways in this disease are reprogrammed as discussed above. The most studied gene abnormality is in VHL, the mutation of which results in upregulation of several key hypoxia pathways; this results in the abundant angiogenesis, as a result of vascular endothelial growth factor (VEGF), and accounts for many of the paraneoplastic effects, for example increased erythropoietin, seen in this disease. Indeed, due to these protean clinical manifestations, many endocrine related (Table 2), RCC has often been labelled the “internist’s tumor”.
Table 2.
Paraneoplastic manifestations are present in up to 10–40% of patients with renal cell carcinoma
| ENDOCRINE | NON-ENDOCRINE |
|---|---|
| Hypertension | Kidney failure |
| Polycythemia | Anemia |
| Hepatic dysfunction (not due to metastasis) | Coagulopathy |
| Hypercalcemia | Neuropathy and myopathy |
| Cushing’s syndrome | Vasculopathy |
| Glucose metabolism alterations | Amyloidosis |
| Galactorrhea |
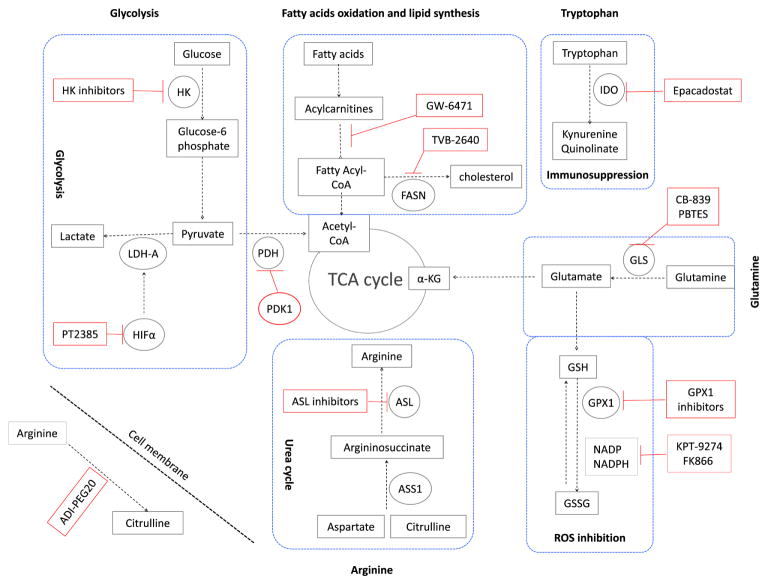
Several recent studies have evaluated tumor tissues from ccRCC patients using combined metabolomic, transcriptomic, and proteomic methods. These investigations have shown expected reprogramming, such as in the glycolysis pathway (Warburg metabolism) as well as relatively unexpected, such as glutamine reprogramming into antioxidant pathways15. A summary of the reprogrammed pathways discovered from combining metabolomics and proteomics appears in Fig. 1 (Reproduced with permission from Abu Aboud O, Weiss RH: Translating Metabolic Reprogramming into New Targets for Kidney Cancer, Kidney Cancer, in press). Close inspection of the altered metabolic pathways invariably leads (1) to identification of “nodes” or metabolic crossroads common to several pathways where a given pathway branches into other pathways, or (2) to the recognition of compounds (primarily enzymes) which can be impacted by drugs or other compounds to specifically attenuate the pathway(s) of interest. Overlaid on Fig. 1 are such pathways and proposed target therapies based thereupon. The distinct advantage of targeting reprogrammed pathways is that, since they primarily occur in the tumor and not in normal tissue, they are less likely than traditional chemotherapy to result in toxic effects in normal tissue.
Fig. 1. Targeting metabolic reprogramming in ccRCC (from Abu Aboud O, Weiss RH: Translating Metabolic Reprogramming into New Targets for Kidney Cancer, Kidney Cancer, in press, with permission).
Glycolysis can be inhibited using inhibitors for hexokinase (HK), the hypoxia-inducible factor α (HIF-α) such as PT2385, Lactate dehydrogenase (LDH-A). Glycolysis can also be inhibited by activators of pyruvate dehydrogenase kinase-1 (PDK-1), which inhibits pyruvate dehydrogenase (PDH). Fatty acid oxidation can be inhibited using peroxisome proliferator-activated receptor (PPARα) antagonist (GW-6471). Also fatty acid synthesis can be inhibited using the fatty acid synthase (FASN) inhibitor TVB-2640. In ccRCC, upregulation of tryptophan metabolism results in increased production of the immunosuppressive metabolites kynuernine and quinolinate. This pathway can be inhibited using the indoleamine 2,3-dioxygenase (IDO) inhibitor epacadostat. Glutamine metabolism can be inhibited using the glutaminase (GLS) inhibitor CB-839. α–KG is α–ketoglutarate. Glutamine is also feeding glutathione (GSH) pathway. Inhibitors of glutathione peroxidase (GPX1) and inhibitors of NADPH such as KPT-9274 and FK866 will stop changing GSH to GSSG. This reaction is responsible for scavenging reactive oxygen species (ROS) in the cell. Arginine is synthesized from citrulline in two steps of the urea cycle using the enzymes argininosuccinate synthase-1 (ASS1), and argininosuccinate lyase (ASL). As ASS1 levels are markedly decreased in all grades of ccRCC, the tumour cells are dependent on extracellular sources of arginine for their survival. Extracellular arginine can be depleted using the pegylated form of arginine deaminase enzyme (ADI-PEG20). ASL inhibitors can also be used for targeted therapy of ccRCC.
In addition to biomarker and therapeutic target discovery, it is possible to capitalize on cancer’s altered metabolism for purposes of diagnostic as well as real-time therapeutic imaging. The current standard staging procedure for RCC employs computed tomography (CT) scanning. The high uptake of glucose by cancer cells (a signature of the Warburg effect; discussed below) can be evaluated in vivo using positron emission tomography (PET) imaging with the glucose analogue 18F-fluorodeoxyglucose (18F-FDG)16. This technique has revolutionized imaging for some malignancies17–19, yet is problematic in RCC due to variable tumour uptake of 18F-FDG20 and its secretion into the urinary system21, so is not commonly used for routine staging of kidney tumors.
The recent identification of glutamine reprogramming (discussed below and in reference 11) and the consequent requirement for this amino acid for ccRCC survival has opened up a new area of diagnostic imaging using 18F-glutamine PET scanning15. While not yet evaluated in human disease, pre-clinical models show fairly convincing data that this technique will ultimately be applicable to diagnosis as well as to stratification of patients with glutamine-avid tumors and the progress of therapy with glutaminase inhibition. Given the increasing rate of advances in imaging techniques as well as radiotracers, it is likely that other reprogrammed metabolic pathways could also be exploited in this manner; this is a prime example of the clinical exploitation of metabolic reprogramming.
Metabolomic Reprogramming in RCC
The extent of reprogrammed biochemical pathways in any cancer is unlikely to ever be fully worked out, but the fact that many malignancies have common pathways5 has led to the investigation of those pathways in particular. In the following sections, I discuss a few aspects of metabolic reprogramming which are, at least in the current state of knowledge, most germane to ccRCC (Fig. 1).
Glycolysis
Warburg metabolism, or aerobic glycolysis, was the first historical description of metabolic reprogramming, although it is now fairly clear that Otto Warburg did not fully understand the ramifications of his discovery at the beginning of the 20th century1. Contemporary investigations of ccRCC cells and tissues from xenografted mice and patients, using metabolomics22,23, proteomics11,23,24, and transcriptomics12, have unambiguously confirmed that this phenomenon is a key component of ccRCC metabolic reprogramming. As an example of Warburg metabolism, levels of enzymes involved in glycolysis such as hexokinase-1 (HK-1), pyruvate kinase (PK-2), and lactate dehydrogenase A (LDH-A) were significantly increased in ccRCC cells and tissues24. While the chemically modified glucose analog 2-deoxy-D-glucose (2-DG) acts as substrate to inhibit hexokinase25, 3-bromopyruvate (3-BrPa) is believed to interfere directly with hexokinase enzymatic activity and bind to hexokinase 226. In ccRCC, HIFα plays a major role in activating LDH-A, a critical enzyme in glycolysis. Recent studies targeting HIFα using PT2385 resulted in inhibition of tumor growth in the different RCC cell lines22 and now is being tested in clinical trials27.
Fatty acid oxidation and lipid synthesis
Non-targeted metabolomics studies in ccRCC have demonstrated an increase in utilization of fatty acids in these cells28. Consistent with this finding, inhibitors of fatty acids oxidation such as the Peroxisome proliferator-activated receptor (PPAR) α antagonist GW6471 was tested in several ccRCC models and shown to inhibit xenograft ccRCC tumor growth29,30. In addition, fatty acid synthase (FASN), which is involved in building long chain fatty acids and cholesterol, was found to be overexpressed in ccRCC and associated with tumor aggressiveness and poor prognosis in this disease31. Inhibition of FASN by cerulin or its derivative C75 induced a rapid increase in malonyl-coA with a marked reduction in lipogenesis in multiple cancer models, including RCC cells32,33.
Tryptophan
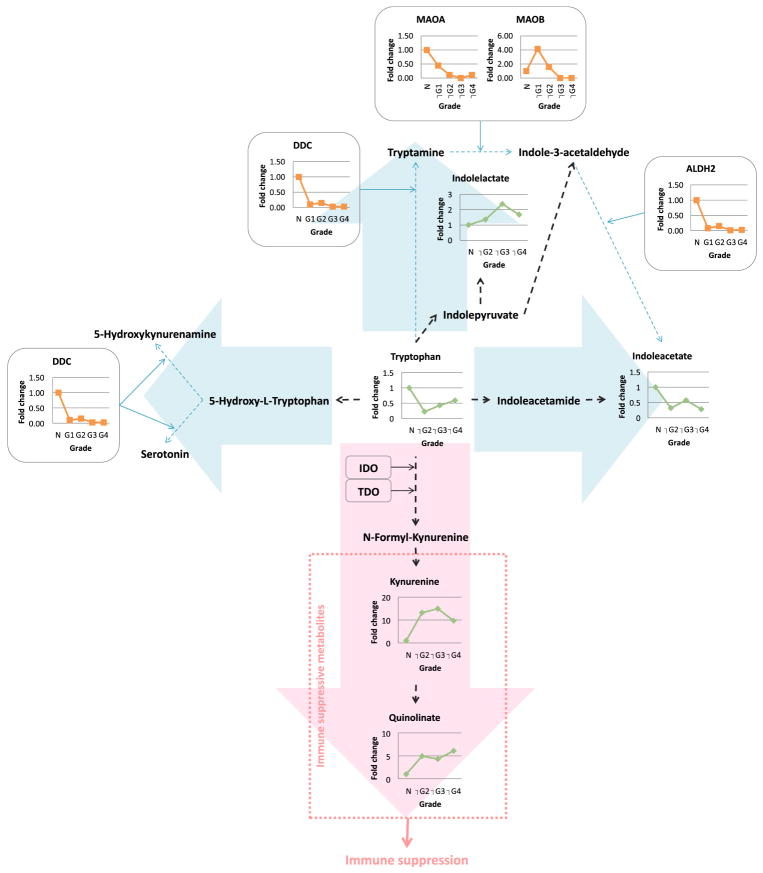
Tryptophan is an essential amino acid and is catabolized through the kynurenine pathway via the rate-limited enzyme activity of the indoleamine 2,3-dioxygenase (IDO)34. Important although underreported consequences of tryptophan metabolism are (1) the formation of metabolites in the kynurenine pathway which have immunosuppressive properties35, and (2) that tryptophan depletion, itself immunosuppressive, obviously follows its catabolism36. In a classic example of the translational power of metabolomics, the level of tryptophan in ccRCC was shown to be reduced and the pathways of its metabolism, as analyzed by metabolomics and proteomics techniques, lead strikingly into the kynurenine pathway (Fig. 2; permission obtained from11), suggesting upregulation of this pathway in this disease leading to immunosuppression11. Indeed, the historical failure of IFNα therapy for human RCC is likely due to its inability to overcome the immunosuppressive environment created by increased IDO1. Consequently, inhibition of IDO prevents the purported immunosuppressive effect and enables T-cell activation in a murine immune-competent RCC model37. The selective IDO inhibitor epacadostat was tested in ccRCC and found to enhance the lytic ability of tumor-antigen-specific T cells in preclinical models38.
Fig. 2. Tryptophan metabolism favors a grade-dependent increase in immune suppressive metabolites (11 with permission).
Combined proteomics and metabolomics data of human RCC tissue was overlayed onto a stylized KEGG-based pathway diagram. Green, metabolite; orange, enzyme; black dot arrow, metabolism; red arrow, upregulated pathway; blue arrow, downregulated pathway; MAO, monoamine oxidase; DDC, dopa decarboxylase; ALDH, aldehyde dehydrogenase; IDO, indoleamine 2,3-dioxygenase; TDO, tryptophan 2,3-dioxygenase
As far as translational work relevant to this paradigm, there are several ongoing clinical studies using IDO inhibitors, either singly or in combination, in refractory RCC. Two combination IDO inhibitor and checkpoint inhibitor antibody studies (clinicaltrials.gov NCT02178722, NCT02318277), a combination IDO inhibitor and JAK inhibitor study in advanced solid tumors (NCT02559492), and an IDO inhibitor alone study on refractory solid tumors (NCT02048709) are all recruiting. However, none of these studies is complete and none have utilized interferon in combination with an IDO inhibitor; however, in light of new data showing that interferon (IFN) α increases IDO levels37, IFNα therapy with an IDO inhibitor should be revisited for RCC.
Glutamine
Several independent studies have shown that glutamine utilization is increased in ccRCC as compared to normal kidney tissues11,12,39. Based on this data, the targeting of enzymes involved in glutamine metabolism, such as glutaminase (GLS) with the drug CB83940, is being tested both in pre-clinical as well as clinical trials. In pre-clinical work, glutamine in ccRCC was shown to be feeding the glutathione/oxidized glutathione pathway (GSH/GSSG), an important pathway for neutralizing ROS in cells11,12,15; one study even demonstrated the use of this phenomenon for real-time PET imaging of RCC which could be used not only for stratification of patients who might respond to glutaminase inhibitors, but also to be able to follow the response of these patients to such therapies15. In a clinical trial, the small molecule inhibitor of glutaminase, CB-839, either alone or in combination with the mTOR inhibitor everolimus, showed favorable results in patients with ccRCC41. Furthermore Inhibitors of NADPH synthesis such as KPT-9274 have showed decreased tumor burden in RCC mouse model42.
Arginine
Arginosuccinate synthase 1 (ASS1) is the rate-limiting enzyme for the conversion of citrulline to arginine for ammonia detoxification through the urea cycle in kidney43. In ccRCC, ASS1 has been shown to be either not expressed or is downregulated in tumors44, causing these cancers to be auxotrophic for arginine. This finding was confirmed in all grades of ccRCC in a recent proteomic study24. Consistent with this model, arginine deprivation inhibited tumor growth in the RENCA mouse model of RCC44. The driver of the evolution of dependence of some ccRCCs on extracellular amino acids, such as arginine, is not obvious but may also relate to enhanced synthesis of essential building blocks45 in parallel with Warburg metabolism. The pegylated form of arginine deaminase (ADI-PEG20) can be used to deplete arginine through deamination of arginine to citrulline44,46.
Summary and future directions
Despite the fact that this disease is commonly diagnosed in the renal clinic, kidney cancer has woefully been missing from the curriculum, or at best underrepresented, in many if not most nephrology training programs. However, this “internists’ tumor” is associated with fascinating changes in metabolism as compared to other cancers and has thus been aptly labelled a “metabolic disease” characterized by a high degree of “metabolic reprogramming” in multiple biochemical pathways. For these reasons, it surely merits the attention of, and study by, the intellectually-inclined nephrologist. With the advent of omics and other high throughput research techniques, kidney cancer is now recognized to be more than a disease of aberrant cell cycle progression, as was previously been considered the mechanism of all malignancies, but also critically involves reprogramming of classical metabolic pathways which are important in energy and cellular synthetic component production as well as control of immune surveillance. Thus, many aspects of diagnosis and management, from imaging to pathology to therapeutics, can capitalize on these reprogrammed pathways in ways which will lead to new insights in disease management. We foresee in the future an era in which malignancies, including RCC, will be classified on the basis of those metabolic pathways which are enhanced or otherwise altered, in addition to site of occurrence or microscopic pathology47. However, a word of caution is in order regarding tumor heterogeneity48 which has shown to be especially relevant for RCC: it is likely that separate regions of the tumor have distinct reprogramming, and this will need to be worked out in the future before optimally successful treatments are developed. Based on the knowledge to date and the rapidity by which metabolic reprogramming has been translated to clinical advances, it is highly likely that additional therapies will soon be developed which target the aberrant metabolic pathways.
Acknowledgments
Financial support for this work:
NIH/NCI grant 1R01CA135401
NIH/NCI grant 1R03CA181837-01
NIH/NIDDK grant 1R01DK082690-01A1
NIH/NCI Cancer Center Support Grant P30 CA093373
Jerry and Susan Knapp cancer research fund
Dialysis Clinics Incorporated
Footnotes
Financial disclosure and conflict of interest statements: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warburg O. On the origin of cancer cells. Science (New York, NY) 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Weiss RH, Lin P-Y. Kidney Cancer: Identification of Novel Targets for Therapy. Kidney Int. 2006;69:224–32. doi: 10.1038/sj.ki.5000065. [DOI] [PubMed] [Google Scholar]
- 3.Hu SL, Chang A, Perazella MA, Okusa MD, Jaimes EA, Weiss RH. The Nephrologist’s Tumor: Basic Biology and Management of Renal Cell Carcinoma. J AmSoc Nephrol. 2016 doi: 10.1681/ASN.2015121335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wettersten HI, Aboud OA, Lara PN, Jr, Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nature reviews Nephrology. 2017 doi: 10.1038/nrneph.2017.59. [DOI] [PubMed] [Google Scholar]
- 5.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–98. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.van der Mijn JC, Panka DJ, Geissler AK, Verheul HM, Mier JW. Novel drugs that target the metabolic reprogramming in renal cell cancer. Cancer & metabolism. 2016;4:14. doi: 10.1186/s40170-016-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. UrolOncol. 2011;29:551–7. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. NatRevNephrology. 2011;8:22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 10.Baker M. Big biology: The ‘omes puzzle. Nature. 2013;494:416–9. doi: 10.1038/494416a. [DOI] [PubMed] [Google Scholar]
- 11.Wettersten HI, Hakimi AA, Morin D, et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75:2541–52. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakimi AA, Reznik E, Lee CH, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer cell. 2016;29:104–16. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metallo CM, Gameiro PA, Bell EL, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullen AR, Wheaton WW, Jin ES, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu Aboud O, Habib SL, Trott JF, et al. Glutamine addiction in kidney cancer suppresses oxidative stress and can be exploited for real-time imaging. Cancer research. 2017 doi: 10.1158/0008-5472.CAN-17-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber WA, Schwaiger M, Avril N. Quantitative assessment of tumor metabolism using FDG-PET imaging. Nuclear medicine and biology. 2000;27:683–7. doi: 10.1016/s0969-8051(00)00141-4. [DOI] [PubMed] [Google Scholar]
- 17.Shankar LK, Hoffman JM, Bacharach S, et al. Consensus recommendations for the use of 18F-FDG PET as an indicator of therapeutic response in patients in National Cancer Institute Trials. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2006;47:1059–66. [PubMed] [Google Scholar]
- 18.Tunariu N, Kaye SB, Desouza NM. Functional imaging: what evidence is there for its utility in clinical trials of targeted therapies? British journal of cancer. 2012;106:619–28. doi: 10.1038/bjc.2011.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juweid ME, Cheson BD. Positron-emission tomography and assessment of cancer therapy. The New England journal of medicine. 2006;354:496–507. doi: 10.1056/NEJMra050276. [DOI] [PubMed] [Google Scholar]
- 20.Robey IF, Stephen RM, Brown KS, Baggett BK, Gatenby RA, Gillies RJ. Regulation of the Warburg effect in early-passage breast cancer cells. Neoplasia (New York, NY) 2008;10:745–56. doi: 10.1593/neo.07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gofrit ON, Orevi M. Diagnostic Challenges in Kidney Cancer: A Systematic Review of the Role of PET/CT. J Urol. 2016 doi: 10.1016/j.juro.2016.02.2992. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Hill H, Christie A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–7. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perroud B, Lee J, Valkova N, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. MolCancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perroud B, Ishimaru T, Borowsky AD, Weiss RH. Grade-dependent proteomics characterization of kidney cancer. MolCell Proteomics. 2008;8:971–85. doi: 10.1074/mcp.M800252-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raez LE, Papadopoulos K, Ricart AD, et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer chemotherapy and pharmacology. 2013;71:523–30. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Zhang H, Lu W, Huang P. Role of mitochondria-associated hexokinase II in cancer cell death induced by 3-bromopyruvate. Biochimica et biophysica acta. 2009;1787:553–60. doi: 10.1016/j.bbabio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medicine UNLo. 2017 https://clinicaltrials.gov/ct2/show/NCT02293980.
- 28.Ganti S, Taylor SL, Abu AO, et al. Kidney tumor biomarkers revealed by simultaneous multiple matrix metabolomics analysis. Cancer Res. 2012;72:3471–9. doi: 10.1158/0008-5472.CAN-11-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abu Aboud O, Wettersten HI, Weiss RH. Inhibition of PPAR-alpha induces cell cycle arrest and apoptosis and synergizes with glycolysis inhibition in kidney cancer cells. PLoSOne. 2013;8:e71115. doi: 10.1371/journal.pone.0071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abu AO, Donohoe D, Bultman S, et al. PPARalpha inhibition modulates multiple reprogrammed metabolic pathways in kidney cancer and attenuates tumor growth. AmJPhysiol Cell Physiol. 2015;308:C890–C8. doi: 10.1152/ajpcell.00322.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiguchi A, Asano T, Asano T, Ito K, Sumitomo M, Hayakawa M. Fatty acid synthase over expression is an indicator of tumor aggressiveness and poor prognosis in renal cell carcinoma. JUrol. 2008;180:1137–40. doi: 10.1016/j.juro.2008.04.135. [DOI] [PubMed] [Google Scholar]
- 32.Pizer ES, Thupari J, Han WF, et al. Malonyl-coenzyme-A is a potential mediator of cytotoxicity induced by fatty-acid synthase inhibition in human breast cancer cells and xenografts. Cancer research. 2000;60:213–8. [PubMed] [Google Scholar]
- 33.Horiguchi A, Asano T, Asano T, Ito K, Sumitomo M, Hayakawa M. Pharmacological inhibitor of fatty acid synthase suppresses growth and invasiveness of renal cancer cells. The Journal of urology. 2008;180:729–36. doi: 10.1016/j.juro.2008.03.186. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu T, Nomiyama S, Hirata F, Hayaishi O. Indoleamine 2,3-dioxygenase. Purification and some properties. The Journal of biological chemistry. 1978;253:4700–6. [PubMed] [Google Scholar]
- 35.Fallarino F, Grohmann U, Vacca C, et al. T cell apoptosis by tryptophan catabolism. Cell DeathDiffer. 2002;9:1069–77. doi: 10.1038/sj.cdd.4401073. [DOI] [PubMed] [Google Scholar]
- 36.Lee GK, Park HJ, Macleod M, Chandler P, Munn DH, Mellor AL. Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology. 2002;107:452–60. doi: 10.1046/j.1365-2567.2002.01526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott JF, Kim J, Aboud OA, et al. Inhibiting tryptophan metabolism enhances interferon therapy in kidney cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jochems C, Fantini M, Fernando RI, et al. The IDO1 selective inhibitor epacadostat enhances dendritic cell immunogenicity and lytic ability of tumor antigen-specific T cells. Oncotarget. 2016;7:37762–72. doi: 10.18632/oncotarget.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gameiro PA, Yang J, Metelo AM, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. MolCancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 41.Meric-Bernstam F, Tannir NMHJJ, Bennett MK. Phase 1 study of CB-839, a small molecule inhibitor of glutaminase (GLS), alone and in combination with everolimus (E) in patients (pts) with renal cell cancer (RCC) JClinOncol. 2016:34. [Google Scholar]
- 42.Abu Aboud O, Chen CH, Senapedis W, Baloglu E, Argueta C, Weiss RH. Dual and specific inhibition of NAMPT and PAK4 by KPT-9274 decreases kidney cancer growth. MolCancer Ther. 2016 doi: 10.1158/1535-7163.MCT-16-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haines RJ, Pendleton LC, Eichler DC. Argininosuccinate synthase: at the center of arginine metabolism. International journal of biochemistry and molecular biology. 2011;2:8–23. [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon CY, Shim YJ, Kim EH, et al. Renal cell carcinoma does not express argininosuccinate synthetase and is highly sensitive to arginine deprivation via arginine deiminase. International journal of cancer. 2007;120:897–905. doi: 10.1002/ijc.22322. [DOI] [PubMed] [Google Scholar]
- 45.Rabinovich S, Adler L, Yizhak K, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature. 2015;527:379–83. doi: 10.1038/nature15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon JK, Frankel AE, Feun LG, Ekmekcioglu S, Kim KB. Arginine deprivation therapy for malignant melanoma. Clinical pharmacology : advances and applications. 2013;5:11–9. doi: 10.2147/CPAA.S37350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peyser ND, Grandis JR. Cancer genomics: Spot the difference. Nature. 2017;541:162–3. doi: 10.1038/nature21112. [DOI] [PubMed] [Google Scholar]
- 48.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. NEnglJMed. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sumitomo M, Iwase A, Zheng R, et al. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- 50.Lee HJ, Lee HY, Lee JH, et al. Prognostic significance of biallelic loss of PTEN in clear cell renal cell carcinoma. J Urol. 2014;192:940–6. doi: 10.1016/j.juro.2014.03.097. [DOI] [PubMed] [Google Scholar]
- 51.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 52.Bjornsson J, Short MP, Kwiatkowski DJ, Henske EP. Tuberous sclerosis-associated renal cell carcinoma. Clinical, pathological, and genetic features. Am J Pathol. 1996;149:1201–8. [PMC free article] [PubMed] [Google Scholar]
- 53.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 54.Miyamoto S, Murphy AN, Brown JH. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 2008;15:521–9. doi: 10.1038/sj.cdd.4402285. [DOI] [PubMed] [Google Scholar]
- 55.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem. 2008;283:34495–9. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park JY, Lin PY, Weiss RH. Targeting the PI3K-Akt pathway in kidney cancer. Expert review of anticancer therapy. 2007;7:863–70. doi: 10.1586/14737140.7.6.863. [DOI] [PubMed] [Google Scholar]
- 57.Ohh M, Park CW, Ivan M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 58.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–33. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 60.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki S, Tanaka T, Poyurovsky MV, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–6. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gurova KV, Hill JE, Razorenova OV, Chumakov PM, Gudkov AV. p53 pathway in renal cell carcinoma is repressed by a dominant mechanism. Cancer Res. 2004;64:1951–8. doi: 10.1158/0008-5472.can-03-1541. [DOI] [PubMed] [Google Scholar]
- 63.Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–35. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yalniz Z, Tigli H, Tigli H, Sanli O, Dalay N, Buyru N. Novel mutations and role of the LKB1 gene as a tumor suppressor in renal cell carcinoma. Tumour Biol. 2014;35:12361–8. doi: 10.1007/s13277-014-2550-4. [DOI] [PubMed] [Google Scholar]
- 65.Duivenvoorden WC, Beatty LK, Lhotak S, et al. Underexpression of tumour suppressor LKB1 in clear cell renal cell carcinoma is common and confers growth advantage in vitro and in vivo. Br J Cancer. 2013;108:327–33. doi: 10.1038/bjc.2012.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shroff EH, Eberlin LS, Dang VM, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci U S A. 2015;112:6539–44. doi: 10.1073/pnas.1507228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gordan JD, Lal P, Dondeti VR, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14:435–46. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]