Abstract
Protein depalmitoylation describes the removal of thioester-linked long chain fatty acids from cysteine residues in proteins. For many S-palmitoylated proteins, this process is promoted by acyl protein thioesterase enzymes, which catalyze thioester hydrolysis to solubilize and displace substrate proteins from membranes. The closely related enzymes acyl protein thioesterase 1 (APT1; LYPLA1) and acyl protein thioesterase 2 (APT2; LYPLA2) were initially identified from biochemical assays as G protein depalmitoylases, yet later were shown to accept a number of S-palmitoylated protein and phospholipid substrates. Leveraging the development of isoform-selective APT inhibitors, several studies report distinct roles for APT enzymes in growth factor and hormonal signaling. Recent crystal structures of APT1 and APT2 reveal convergent acyl binding channels, suggesting additional factors beyond acyl chain recognition mediate substrate selection. In addition to APT enzymes, the ABHD17 family of hydrolases contribute to the depalmitoylation of Ras-family GTPases and synaptic proteins. Overall, enzymatic depalmitoylation ensures efficient membrane targeting by balancing the palmitoylation cycle, and may play additional roles in signaling, growth, and cell organization. In this review, we provide a perspective on the biochemical, structural, and cellular analysis of protein depalmitoylases, and outline opportunities for future studies of systems-wide analysis of protein depalmitoylation.
Keywords: Palmitoylation, thioesterase, inhibitor, serine hydrolase, post-translational modification
Introduction
S-palmitoylation describes the addition of a long-chain fatty acid to a cysteine residue via a thioester linkage (Tom and Martin 2013). This hydrophobic post-translational modification promotes the membrane tethering, trafficking, and localization of a significant fraction of membrane-associated proteins. While the term “S-palmitoylation” implies exclusive modification by 16:0 fatty acids, the cellular profile of fatty acylated cysteine residues likely includes a variety of long chain fatty acids with differing unsaturation (Liang et al. 2001; Thinon et al. 2016; Greaves et al. 2017), and is interchangeably used with the term “S-acylation”. Protein S-palmitoylation is catalyzed by a family of protein acyl transferases, termed zDHHC enzymes due to their predicted Zn2+-binding and conserved Asp-His-His-Cys motif (Lobo et al. 2002; Fukata et al. 2004; Gottlieb et al. 2015). These multipass transmembrane proteins catalyze acyl transfer from long chain fatty acyl-CoAs to cysteine residues in proteins. Humans express 23 zDHHC enzymes that influence many diverse cellular pathways. For example, deletion of zDHHC5 blocks neuronal stem cell differentiation (Li et al. 2012). In addition, zDHHC5 and its close homologue zDHHC8 localize to synapses in neurons (Thomas et al. 2012) where they influence activity-dependent endocytic trafficking, excitability, and synaptic connectivity (Brigidi et al. 2015). In addition, zDHHC2 palmitoylates Src-family kinases (Zeidman et al. 2011), while inhibition of close homologue zDHHC20 prevents EGFR S-palmitoylation and enhances the action of EGFR inhibitors (Runkle et al. 2016). zDHHC9 is reported to S-palmitoylate Ras (Swarthout et al. 2005; Chai et al. 2013), and is genetically linked to X-linked mental retardation (Raymond et al. 2007). Across dozens of publications, zDHHC enzymes have emerged as critical regulators of protein S-palmitoylation and membrane targeting (Tom and Martin 2013; Lemonidis et al. 2015).
Perhaps the most intriguing aspect S-palmitoylation is its reversibility, since thioester hydrolysis eliminates the fatty acyl anchor to modulate membrane localization and/or function. For example, cycles of acylation and deacylation are required for the plasma membrane association of Ras-family small GTPases and G proteins (Buss and Sefton 1986; Hancock et al. 1989; Linder et al. 1993), establishing a distinct mode of peripheral membrane protein directionality dependent on a continuous palmitoylation cycle (Rocks et al. 2005; Rocks et al. 2010). Indeed, pharmacological inhibition with promiscuous inhibitors of either zDHHC PATs or depalmitoylases block the plasma membrane targeting of Ras and G proteins (Webb et al. 2000; Rocks et al. 2010; Martin and Lambert 2016). Microinjection of semi-synthetic fluorescently-labeled S-palmitoylated Ras rapidly redistributes to the Golgi, which is established as the destination of depalmitoylated Ras (Rocks et al. 2005). Based on these findings, peripheral membrane protein S-palmitoylation requires a balanced cycle of both zDHHC enzymes and depalmitoylating enzymes for efficient transport along the flux of the secretory pathway towards the plasma membrane (Rocks et al. 2010). Furthermore, both Ras and G proteins are rapidly depalmitoylated in their GTP-bound state (Mumby et al. 1994; Wedegaertner and Bourne 1994; Willumsen et al. 1996), which coincides, but is not required for agonist dependent-internalization to internal membranes (Martin and Lambert 2016). Based on these examples, constitutive protein S-palmitoylation is required for proper trafficking of many peripheral membrane proteins, but may also function as a reversible signal analogous to protein phosphorylation (Figure 1). For example, serum, growth factor, or phorbol ester addition triggers deacylation of a 64 kD protein in [3H]-palmitate labeled mouse BH3C1 cells (James and Olson 1989), which in light of more recent proteomics studies (Martin et al. 2011), likely corresponds to depalmitoylation of metadherin, a major driver of cancer and metastasis (Wan L and Kang 2013). Therefore, external stimuli trigger either direct activation of depalmitoylases, or facilitate conformational changes that enhance depalmitoylase accessibility, which then tip the balance of the palmitoylation cycle to reorganize membrane distribution and signaling outputs.
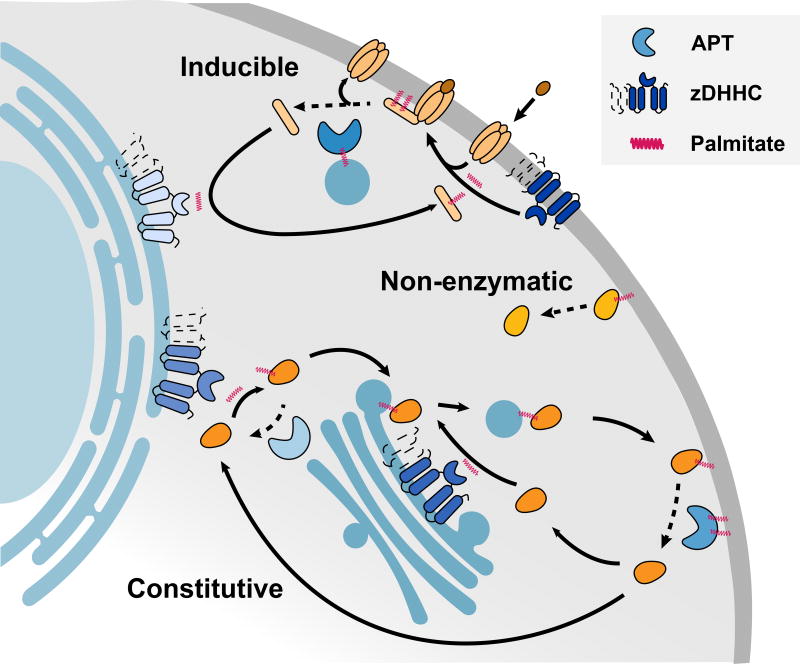
Figure 1. Cellular pathways of protein depalmitoylation.
Protein depalmitoylation can proceed either by non-enzymatic hydrolysis, inducible depalmitoylation, or constitutive cycles of palmitoylation and depalmitoylation during trafficking. zDHHC enzymes are shown as either 4 or 6 transmembrane domains. A color version of the figure is available online.
While this model is appealing, a number of fundamental questions remain, such as 1) what enzymes catalyze depalmitoylation of different S-palmitoylated proteins, 2) what, if anything, triggers their activation, and 3) is there a coincident dysregulation of protein acyl transferase enzymes to block re-palmitoylation? In addition to peripheral membrane proteins, many integral membrane proteins such as channels, receptors, and adhesion proteins also require S-palmitoylation for proper activation, sensitization, or microdomain localization. Given the ever-expanding number of bona fide S-palmitoylated proteins (Blanc et al. 2015), how broad is the role for depalmitoylases in regulating S-palmitoylation stability? What is the scope of enzymatic depalmitoylation compared with the intrinsic hydrolysis of the high-energy thioester linkage? Through the development of new methods and model systems, these questions are central to understanding the role of dynamic S-palmitoylation in physiological systems.
Assays for depalmitoylation
Protein S-palmitoylation was first reported nearly 40 years ago (Schlesinger et al. 1980), but even the most basic experiments were hindered by the limitations of [3H]-palmitate labeling. The low radioactivity and incorporation efficiency typically required weeks to return results for a single experiment (Tsai et al. 2014). Furthermore, assaying [3H]-palmitate incorporation requires immunoprecipitation of a single protein, limiting the scope of the analysis to only specific hypotheses. Nonetheless, [3H]-palmitate metabolic labeling is the most accepted method for studying S-palmitoylation, particularly for measuring stability in pulse-chase assays. In this approach, cells are labeled with [3H]-palmitate for a fixed amount of time, followed by washes and incubation in media supplemented with excess unlabeled palmitic acid (Tsai et al. 2014). After immunoprecipitation and SDS-PAGE, the rate of palmitate turnover on a specific protein can be quantified by autoradiography. Through such analysis, the rate of protein S-palmitoylation turnover is generally faster than the rate of protein turnover, establishing that multiple S-palmitoylation and depalmitoylation events occur throughout the lifetime of an S-palmitoylated protein (Martin et al. 2011). Thus, different PATs and APT enzymes may function at different stages of protein trafficking to promote plasma membrane delivery and residency.
The recent development of non-radioactive detection methods has rejuvenated interest in S-palmitoylation, providing simple methods that accelerate the identification and quantitation of S-palmitoylated proteins (Hang et al. 2007; Martin and Cravatt 2009). The primary method for biochemical analysis of S-palmitoylation uses hydroxylamine to hydrolyze all thioesters across the proteome, followed by capture of the newly liberated thiols (Drisdel and Green 2004; Wan J et al. 2007; Kang et al. 2008). These hydroxylamine-switch methods begin with reduction of disulfides with tris(2-carboxyethyl)phosphine (TCEP), followed by treatment with maleimide to alkylate all reduced thiols. Next, neutral hydroxylamine is added to hydrolyze thioesters, which coincidentally hydrolyzes other mechanistic protein-linked thioesters including ubiquitin ligases and lipoamide-dependent dehydrogenases. Any hydroxylamine sensitive thiols are then captured using a pyridyl-disulfide biotin conjugate for streptavidin-resin enrichment (biotin-exchange) or captured directly with pyridyl-disulfide resin (Acyl-resin assisted capture; Acyl-RAC) for further analysis (Forrester et al. 2011). Pyridyl-disulfide resin capture has at least three advantages. First, it limits capture to hydroxylamine-sensitive thioesters, and does not capture products from other ester or other functional groups. Second, the captured disulfide adduct is easily reversible by TCEP, allowing simple elution for gel-based or proteomic analysis of site-specific S-palmitoylation. Finally, since these methods avoid metabolic labeling, primary tissues can be readily analyzed to provide a snapshot of the steady state levels of S-palmitoylation. Most importantly, biotin-exchange and acyl-RAC have enabled mass spectrometry-based discovery and analysis across hundreds of S-palmitoylated proteins, allowing correlated annotation of the exact sites of S-palmitoylation (Forrester et al. 2011). Such large-scale profiling efforts provide long lists of putative S-palmitoylated proteins, which warrant additional biochemical validation. This is somewhat mitigated by the S-palmitoylation database SwissPalm (Blanc et al. 2015), which provides a searchable platform to qualify candidates across published datasets and compare data from distinct experimental systems, enrichment methods, and laboratories.
The commercially available alkynyl fatty acid derivative 17-octadecynoic acid (17-ODYA) provides a second non-radioactive approach for detecting S-palmitoylation (Charron et al. 2009; Martin and Cravatt 2009; Hernandez et al. 2013). After addition to the cell culture media, cells process the free alkynyl fatty acid to form the coenzyme A (CoA) conjugate, which then serves as a substrate for enzymatic transfer to endogenous sites of S-palmitoylation. Cells collected at different time points are then lysed and conjugated by copper-catalyzed azide alkyne cycloaddition (CuAAC) to reporter-linked azides (fluorophores or biotin) for gel-based or mass spectrometry analysis. Saturated incorporation typically occurs in a few hours, although shorter time points can probe more rapid acylation events. For mass spectrometry-based analysis, lysates are conjugated to biotin-azide for streptavidin enrichment, and comparative mass spectrometry is performed using hydroxylamine-treated or palmitic acid treated samples as controls. Additional precautions are necessary to prevent thioesterase hydrolysis during sample preparation, including addition of generic esterase inhibitors, limiting total sample processing time, and careful monitoring of the pH of all reagents (Martin 2013). Several proteomic studies report bioorthogonal alkynyl-fatty acid labeling for profiling S-palmitoylation, including analyses in immune cells (Yount et al. 2010; Martin et al. 2011; Li et al. 2012), epithelial cells (Hernandez et al. 2016), and protozoa (Jones et al. 2012; Foe et al. 2015). Importantly, alkynyl-fatty acid labeling provides a natural complement to hydroxylamine-switch methods, establishing two distinct enrichment methods for more confident analysis.
Alkynyl-fatty acid metabolic labeling has opened new opportunities for non-radioactive profiling of dynamic changes in S-palmitoylation. For example, Lck S-palmitoylation turnover in Jurkat T-cells was assayed using alkynyl-fatty acid pulse-chase methods and quantified by in-gel fluorescence (Zhang et al. 2010). Surprisingly, Lck depalmitoylation was accelerated after treatment with the phosphatase inhibitor pervanadate, yet stabilized by incubation with the non-selective hydrolase inhibitor methyl arachidonyl fluorophosphonate (MAFP). Thus, activated T-cells increase Lck S-palmitoylation cycling, which is dependent on one or more MAFP-sensitive thioesterase enzymes.
Using similar pulse-chase methods, proteome-wide S-palmitoylation dynamics were profiled by quantitative proteomics in a mouse B-cell line (Martin et al. 2011). Using stable isotope labeling with amino acids in cell culture (SILAC) methods, matched “light” and “heavy” replicates were labeled with 17-ODYA over a 2 hr pulse. One of the samples was then further incubated with 10-fold excess palmitic acid in the presence of the translation inhibitor cycloheximide. After combining the two samples together, the pooled lysates were conjugated to biotin-azide for enrichment and quantitative proteomic analysis. While most S-palmitoylated proteins cycled palmitate at a uniform rate in the pulse-chase assay, a subset of proteins demonstrated accelerated turnover, including G proteins, Ras-family GTPases, and a number of cell polarity proteins. Furthermore, treatment with the non-selective lipase inhibitor hexadecylfluorophosphonate (HDFP) stabilized the same subset of dynamically S-palmitoylated proteins, confirming direct enzymatic contribution to S-palmitoylation dynamics on key signaling and polarity scaffolding proteins. Based on these results, the majority of S-palmitoylated proteins undergo basal hydrolysis, while a small subset of S-palmitoylated proteins are rapidly depalmitoylated by HDFP-sensitive lipases.
Alkynyl fatty acid labeling has several inherent limitations, primarily the direct incorporation of the labeled fatty acid across a variety of phospholipids. Longer incubation times lead to more extensive alkynyl-fatty acid incorporation into phospholipid pools (Thiele et al. 2012), reducing the effectiveness of any later chase with free palmitate. This pulse-chase suppression potentially excludes a number of proteins from dynamic analyses. Furthermore, addition of palmitate has major effects on cellular metabolism. For instance, addition of 500 µM palmitate to human primary melanocytes expressing the variants of the melanocortin 1 receptor (MC1R) increased cAMP levels by almost 3-fold, potentially through direct enhancement of GPCR S-palmitoylation (Chen et al. 2017). Exogenous fatty acids also increase cellular acyl-CoA levels (Greaves et al. 2017), which may enhance S-palmitoylation levels by driving zDHHC activity or by non-enzymatic thioester exchange with accessible cysteine residues. For example, 17-ODYA in the presence of 2-bromopalmitate (a non-selective palmitoylation inhibitor) only partially reduces incorporation of the 17-ODYA (Davda et al. 2013). Therefore, non-enzymatic palmitoylation levels may be enhanced by 17-ODYA incubation, elevating acyl-CoA pools to driving non-physiological thioester exchange to enhance levels of S-acylation.
Another major caveat is metabolic labeling can only occur at accessible, reduced cysteines. Thus, stable S-palmitoylated cysteines are essentially invisible, since they do not turnover to present a free thiol substrate for zDHHC PAT enzymes. Accordingly, the more stable S-palmitoylation site, the more challenging it is to assay by metabolic labeling. In addition, high palmitate levels can induce oxidative stress (Xu S et al. 2015), which may trap depalmitoylated thiols as oxidized disulfides or glutathione adducts. Different zDHHC enzymes have also been shown to prefer different fatty acyl-CoAs (Greaves et al. 2017), suggesting an additional level of diversity that could select against a single alkynyl fatty acid species to bias probe incorporation. Varying the acyl length of the alkynyl reporter revealed different acyl preferences, particularly since terminal azide conjugates surprisingly have little influence on zDHHC acyl specificity (Greaves et al. 2017). Moreover, any differential turnover rates between distinct sites in the same protein are effectively averaged in shotgun proteomics experiments, since there are no robust mass spectrometry methods to broadly profile fatty S-acylated peptides. [3H]-palmitate can be elongated or shortened by ß-oxidation, further diversifying the labeled acyl species. This is particularly problematic when 17-ODYA is shortened to 13-tridecynoic acid or 15-pentadecynoic acid, which can be enzymatically incorporated into N-myristoylation sites (Hang et al. 2007; Martin and Cravatt 2009). Despite these pitfalls, metabolic labeling remains the only approach to profile S-palmitoylation dynamics, on select proteins or across the proteome.
Palmitoyl protein thioesterase 1
Several early reports demonstrated enhanced depalmitoylation upon hormonal stimulation, suggesting activation of specific depalmitoylases might regulate intracellular signaling pathways (Wedegaertner and Bourne 1994; Willumsen et al. 1996; Wang et al. 2015). For example, both Ras and G proteins are more rapidly depalmitoylated after activation, which was believed to contribute to more efficient internalization. Several candidate depalmitoylating enzymes were later identified by activity-guided fractionation and purification from soluble tissue homogenates (Camp and Hofmann 1993; Duncan and Gilman 1998). While technically feasible, this approach excluded analysis of any membrane-bound depalmitoylating enzymes.
Nonetheless, the lysosomal hydrolase PPT1 was identified as the most robust H-Ras depalmitoylase in vitro (Camp and Hofmann 1993). PPT1 was later found to localize exclusively in lysosomes and late endosomes (Verkruyse and Hofmann 1996), functionally separating PPT1 from depalmitoylase activity in the cytosol or on plasma membrane. PPT1 is one of 14 genes genetically linked to the family of human neuronal ceroid lipofuscinosis (NCL) lysosomal storage diseases, characterized by accumulation of lysosomal autofluorescent storage material, neurodegeneration, and childhood mortality (Vesa et al. 1995). Interestingly, while not localized in the lysosome, mutations in cysteine string protein alpha gene (CSPα) also lead to NCL (Greaves et al. 2012). CSPα is multiply S-palmitoylated, and functions as a HSC70 interacting J-protein important for protein folding and synaptic vesicle function. In CSPα mutant cells, PPT1 is highly over-expressed (Henderson et al. 2016). Furthermore, PPT1 can depalmitoylate CSPα in vitro, which is reported to lead to the formation of insoluble aggregates. Acyl-RAC analysis of CSPα mutant cells revealed decreased S-palmitoylation of several synaptic proteins and neuronal signaling proteins. Overall, PPT1 is likely not contributing to plasma membrane depalmitoylation, but could play a role in vesicular depalmitoylation and lysosomal degradation of S-palmitoylated proteins.
Acyl protein thioesterases
The reported lysophospholipase LYPLA1 was identified by screening soluble tissue homogenates for G protein depalmitoylase activity (Duncan and Gilman 1998), and subsequently renamed acyl protein thioesterase 1 (APT1). In addition to its depalmitoylase activity, APT1 hydrolyzes a broad profile of lysophospholipids and other long-chain mono-acyl glycerol esters, albeit at a lower catalytic efficiency than high-energy S-palmitoylated substrates (Sugimoto et al. 1996; Hedberg et al. 2011). Since APT1 was first reported before the development of RNAi methods, much of the first decade of depalmitoylase research relied on in vitro biochemistry or over-expression studies. Over-expression of APT1 enhances the depalmitoylation of small GTPases (Dekker et al. 2010; Kong et al. 2013), endothelial nitric oxide synthase (Yeh et al. 1999), and a number of other peripheral membrane proteins in transfected cells. Such over-expression could lead to a number of potential artifacts, either by disruptive fusion of epitope tags or fluorescent proteins, or through saturation of binding partners at non-physiological expression levels. If the enzyme concentration increases much beyond physiological levels, the rate of substrate hydrolysis will also increase, and potentially promote depalmitoylation of non-physiological substrates. In addition, over-expression could also impart non-physiological depalmitoylase activity to otherwise dedicated lipid modifying enzymes, particularly since thioesters are high energy bonds readily hydrolyzed by even weak nucleophiles. Over-expressing candidate S-palmitoylated substrates is also problematic, since high expression levels can approach the enzyme Km, yielding measureable APT1-dependent depalmitoylation of non-native substrates. Even so, APT1 can depalmitoylate a number of proteins in cells. In fact, the neuronal microRNA mi138 modulates APT1 levels in synaptic spines, which decreases Gα13 S-palmitoylation to reduce spine volume (Siegel et al. 2009).
In addition to APT1, vertebrates express the highly similar depalmitoylase APT2, (68% identical, 81% similar). APT2 also hydrolyzes lysophospholipids (Toyoda et al. 1999), but in contrast to APT1, it also hydrolyzes prostaglandin glycerol esters (Manna et al. 2014). Both APT1 and APT2 act as efficient depalmitoylases in vitro (Hedberg et al. 2011), and are presumed to broadly regulate S-palmitoylation and trafficking of peripheral membrane proteins in cells. Unfortunately, there are no high quality commercial sources of APT1 and APT2 antibodies, since they are likely poorly immunogenic due to nearly exact conservation across most mammals, including mice, rabbits, and humans. Thus, most studies on APT1 and APT2 localization have used GFP or epitope fusions for subcellular analysis. The literature consensus reveals both enzymes are primarily cytoplasmic, although partially localized on internal membranes (Vartak et al. 2014) or in some instances on the plasma membrane (Kong et al. 2013; Adachi et al. 2016). Both APT1 and APT2 share a cysteine at the second position immediately following the initiator methionine, and are reportedly both S-palmitoylated (Yang et al. 2010; Kong et al. 2013; Vartak et al. 2014). After removal of the initiator methionine, if the N-terminus is not acetylated, it may readily undergo N-acyl transfer to form a stable amide linkage. Interestingly, APT1 S-palmitoylation has been reported in only one proteomics experiments using hydroxylamine switch methods, but never by alkynyl fatty acid labeling (Yang et al. 2010; Blanc et al. 2015). APT1 and APT2 are expressed and active across nearly all tissues (Bachovchin et al. 2010; Abrami et al. 2017), yet their absence in most large-scale profiling efforts suggests they may not be stoichiometrically S-palmitoylated. This low-level S-palmitoylation may be linked to an alternative translational start site (potentially Met-6) or possibly through auto-depalmitoylation. Nonetheless, knockdown of either APT1 or APT2 promotes plasma membrane association of the other APT enzyme, suggesting an interdependent S-palmitoylation cycle (Kong et al. 2013).
It also remains unclear if APT1 and APT2 are functionally redundant or hydrolyze distinct substrates. While APT1 over-expression has no effect on GAP-43 S-palmitoylation, APT2 over-expression promotes more rapid GAP-43 depalmitoylation in cells (Tomatis et al. 2010). In another example, APT2 (but not APT1) is essential for depalmitoylation and stabilization of zDHHC6 (Abrami et al. 2017), which is important for efficient S-palmitoylation of calnexin cytoplasmic domains, as well as a number of other S-palmitoylated ER proteins. Additionally, agonist-dependent S-palmitoylation of the β2-adrenergic receptor is reversed by APT1, but not APT2 (Adachi et al. 2016). Clearly, APT1 and APT2 either access a different pool of substrates or are mechanistically directed toward select S-acylated proteins.
Based on their predicted role in regulating peripheral membrane protein localization, peptidomimetic inhibitors of APT1 were synthesized that upon microinjection could mislocalize N-Ras away from the plasma membrane (Deck et al. 2005). Based on these studies, derivatives of the generic natural product lipase inhibitor tetrahydrolipstatin were identified that potently inhibited both APT1 and APT2 (Dekker et al. 2010). The β-lactone mechanism-based inhibitors Palmostatin B and the choline-derivative Palmostatin M (Hedberg et al. 2011) were developed as dual inhibitors of both APT enzymes. Palmostatin B treatment led to accumulation of N-Ras on internal membranes, as well as partial rescue of E-Cadherin membrane localization (Dekker et al. 2010). Interestingly, knockdown of APT1 was not sufficient to reproduce the pharmacological effects to statistical significance, demonstrating possible compensatory mechanisms, either by APT2 or by other unannotated depalmitoylating enzymes. Single cell studies revealed altered trafficking dynamics of over-expressed N-Ras fluorescent protein fusions and microinjection of semi-synthetic, fluorescently labeled N-Ras. Upon growth factor stimulation, Palmostatin B treated cells showed reduced Ras activation on the Golgi. Later studies demonstrated selective growth inhibition of N-Ras, but not K-Ras driven growth of myeloid progenitors (Xu J et al. 2012). Accordingly, Palmostatin B directly modulates N-Ras localization and activity by disrupting the Ras palmitoylation cycle (Grecco et al. 2011).
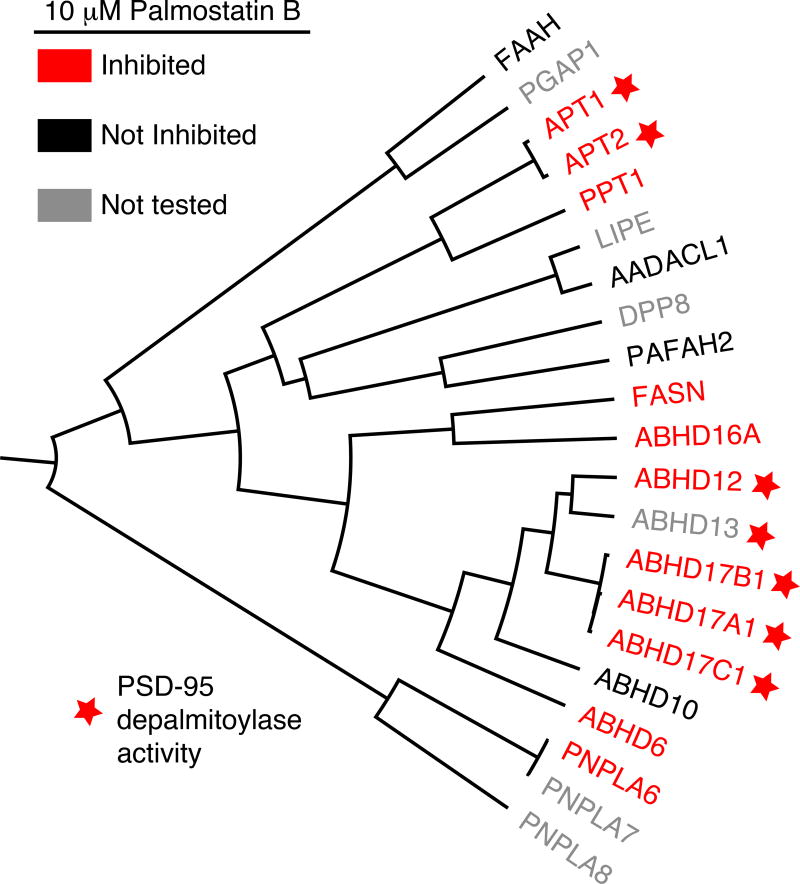
While Palmostatin B demonstrates high potency against both APTs (IC50 < 5 nM) (Hedberg et al. 2011), phenotypic effects often require > 20 µM (Dekker et al. 2010; Yokoi et al. 2016; Hernandez et al. 2017). Early studies suggested that Palmostatin derivatives are highly unstable in serum, requiring repeated cycles of inhibitor addition to maintain inactivation (Siegel et al. 2009). Nevertheless, this compound has been used in numerous studies to profile APT1 and APT2 function. For instance, Palmostatin B addition accelerated the incorporation of the 17-ODYA in Lck in unstimulated T-cells (Akimzhanov and Boehning 2015). However, the compound failed to block depalmitoylation once the cells were stimulated with Fas ligand. The authors suggested that there may be distinct pools of depalmitoylases once the receptors are activated. In addition, the neuronal RGS-binding protein R7BP accelerates the deactivation of Gi/o proteins. Inhibition with Palmostatin B or HDFP both redistributed R7BP from the plasma membrane to endomembranes, disrupted association with G protein-regulated inwardly rectifying potassium (GIRK) channels, and slowed GIRK channel closure (Jia et al. 2014). Importantly, knockdown of both APT1 and APT2 had no effect on GIRK inactivation, suggesting the presence of additional depalmitoylating enzymes. Chronic lymphocytic leukemia (CLL) B-cells express nearly 10-fold more APT2 and 2-fold more APT1. Palmostatin B treatment or APT knockdown increased CD95-mediated apoptosis (Berg et al. 2015). Activity-based protein profiling (ABPP) studies later demonstrated that Palmostatin B is not exclusive for APT1/APT2, and inactivates a number of other lipid processing serine hydrolases with somewhat weaker potency, including ABHD6, ABHD16A, ABHD17A-C, PNPLA6, and FASN (Lin and Conibear 2015). Accordingly, Palmostatin B should be used only as a generic depalmitoylase inhibitor, with an understanding that multiple enzymes are blocked at concentrations required for sustainable depalmitoylase inhibition, and the biological outcomes may reflect polypharmacology summed across many enzymes.
Despite these issues, Palmostatin B remains a popular tool for blocking depalmitoylase activity, and even attenuates depalmitoylation in vivo after intraperitoneal injection (Chen et al. 2017). The MC1R G-protein-coupled receptor triggers melanin production and enhances DNA repair after ultraviolet irradiation. Mice with red hair MC1R variants have reduced MC1R S-palmitoylation, weaker cAMP stimulation, and are more susceptible to developing melanoma. Remarkably, intraperitoneal injection of Palmostatin B (10 mg kg−1) prior to UV irradiation increased MC1R S-palmitoylation and clearance of DNA photoproducts while reducing tumor growth. While Palmostatin B is promiscuous, at this relatively low-dose, APTs are likely to be the primary targets. APT inhibition also prevents melanoma tumor cell invasion by regulating the S-palmitoylation of the polarity organizing protein MCAM (Wang et al. 2015). Wnt5a stimulation promotes APT1-dependent depalmitoylation of MCAM, inducing asymmetric MCAM localization to promote a more invasive state. Taken together, APT inhibitors may be valuable prophylactics in topical sunscreens to prevent melanoma. Nonetheless, the APT1 and APT2 inhibitors ML348 and ML349 had no effect on MAPK signaling or growth across several N-Ras-dependent melanoma cell lines (Vujic et al. 2016), demonstrating APT activity is not essential across all cancers stages or cell types.
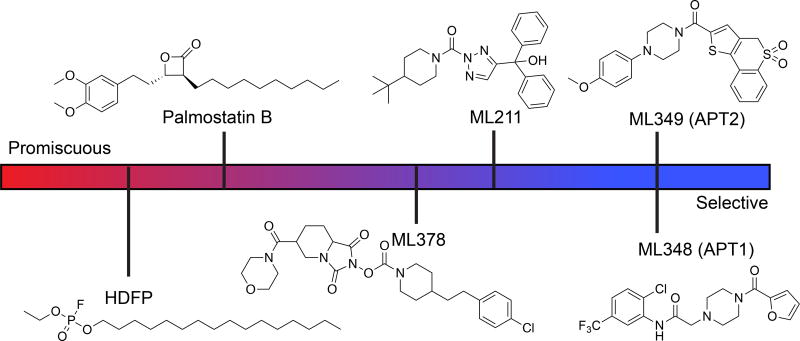
A number of more selective APT inhibitors have been developed with more drug-like properties. Isoform-selective inhibitors of APT1 (ML348, Ki = 280 nM) and APT2 (ML349, Ki = 120 nM) were identified by high throughput screening using a competitive fluorescence polarization assay (Adibekian, Martin, et al. 2010b, 2010a; Adibekian et al. 2012). These inhibitors share a common piperazine-amide scaffold, but diverge through additional modifications to impart isoform selectivity. For example, APT1 inhibitor, ML348 has piperazine amide adjacent to a furanyl group but APT2 inhibitor, ML349 has a thiophene conjugated to thiochromane 1,1-dioxide. These isoform-selective inhibitors are active in vivo following intraperitoneal injection, and engage their respective targets across all major tissues (Adibekian et al. 2012). Furthermore, chemical proteomics analysis of biotin-conjugated ML349 confirmed largely selective inhibition across the proteome at low micromolar concentrations (Won et al. 2017). Other covalent APT1/APT2 dual inhibitors have been reported, including the commercially available triazole urea ML211 (Adibekian, Martin, Speers, et al. 2010; Adibekian et al. 2011) and the N-hydroxyhydantoin carbamate ML378 (Hulce et al. 2010; Cognetta et al. 2015). ML211 is more potent, yet the higher reactivity reflects a smaller selectivity window limiting in vivo applications. ML378 also inhibits ABHD6 and FAAH in mouse brain homogenates, yet selective inhibitors to ABHD6 and FAAH can be used as anti-target controls. Based on our studies, we highly recommend ML348 and ML349 as probes for exploring APT function in cells and model organisms (Figure 2).
Figure 2. APT inhibitor selectivity index.
All listed inhibitors are commercially available, except the promiscuous lipase inhibitor HDFP. HDFP (fluorophosphonate), Palmostatin B (b-lactone), ML378 (N-hydroxyhydantoin carbamate), and ML211 (triazole urea) are covalent inhibitors, although the covalent Palmostatin B adduct is slowly reversible. ML211 inhibits ABHD11 (Adibekian, Martin, Speers, et al. 2010) and PPT1 (unpublished results). ML378 inhibits ABHD6, FAAH, and PPT1 at higher doses (Cognetta et al. 2015). A color version of the figure is available online.
Arguably, the development of APT inhibitors has outpaced detailed biochemical analysis of APT function in cells. According to pulse-chase S-palmitoylation proteomics studies, the tumor suppressor Scribble (Scrib) demonstrated the fastest S-palmitoylated turnover in aggressive B-cell hybridoma cells (Martin et al. 2011). Importantly, this turnover was completely blocked by treatment with the non-selective lipase inhibitor HDFP. Originally identified in Drosophila, the cell polarity tumor suppressor Scrib has emerged as a central regulator of cell growth, invasion, and malignancy (Bilder and Perrimon 2000; Dow et al. 2003; Feigin et al. 2014). While Scrib is not widely mutated in cancers, its mislocalization and amplification are clinically correlated with high-grade cervical carcinomas (Nakagawa and Huibregtse 2000; Nakagawa et al. 2004), as well as malignant colon (Gardiol et al. 2006), prostate (Pearson et al. 2011), breast cancers (Zhan et al. 2008; Pearson et al. 2011), and other epithelial cancers (Vaira et al. 2011) (Figure 3A). In addition, Scrib+/− heterozygous male mice develop widespread epithelial hyperplasia (Pearson et al. 2011), and MMTV-driven conditional knockout of Scrib in female mice induces breast hyperplasia and widespread tumor formation (Feigin et al. 2014).
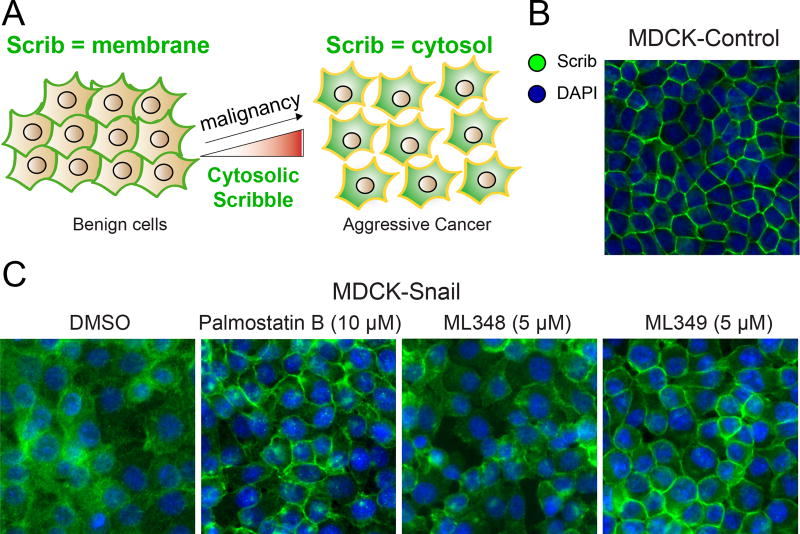
Figure 3. APT2 inhibition restores Scribble plasma membrane localization in Snail-expressing cells.
(A) Scribble (Scrib) moves from the plasma membrane to the cytosol in more malignant cells. (B) Scrib is localized at the plasma membrane in MDCK polarized epithelial cells. (C) APT2 inhibition significantly rescues Scrib plasma membrane localization in Snail expressing MDCK cells. Adapted with permission from (Hernandez et al. 2017). A color version of the figure is available online.
In order to explore the mechanism of Scrib membrane mislocalization in cancer, cells were engineered cells to over-express the epithelial to mesenchymal transition transcription factor (EMT-TF) Snail. Snail reprograms cells to transform to a more stem cell-like state, which eliminates basolateral polarity and leads to Scrib translocation from membrane to the cytosol (Cordenonsi et al. 2011) (Figure 3B). Importantly, Scrib fusion to a C-terminal CAAX prenylation motif is reported to rescue plasma membrane localization and reduces levels of activated TAZ (Cordenonsi et al. 2011), the central transducer of the Hippo signaling pathway. Without its plasma membrane localization, Scrib no longer represses Ras/MAPK-driven cell invasion and EMT phenotypes, yet retains the ability to suppress anchorage-independent growth (Elsum and Humbert 2013). Interestingly, Snail expression in either MDCK or MCF10A cells led to the repression of select zDHHC protein acyl transferases and increased expression of the depalmitoylase APT2, suggesting Snail initiates a transcriptional program that directly targets the S-palmitoylation cycle (Hernandez et al. 2016; Hernandez et al. 2017). When Snail over-expressing cells are treated with the APT2 inhibitor ML349 or APT2 siRNAs, Scrib localization is largely restored to the plasma membrane, rescuing markers of cell polarity and suppressing MAPK signaling (Hernandez et al. 2017) (Figure 3C). ML349 had no effect on Ras-GTP levels, but markedly reduced Raf and MEK activation, suggesting S-palmitoylated Scrib organizes a signaling complex that regulates Raf activation downstream of Ras. Importantly, overnight incubation with Palmostatin B also rescued Scrib membrane localization, but required 100-times higher concentrations than ML349. Surprisingly, the APT1 inhibitor ML348 had no effect on Scrib localization, confirming isoform-selective depalmitoylase regulation of S-palmitoylation. Altogether, this data demonstrates that APT2, but not APT1, participates in an altered S-palmitoylation cycle that becomes imbalanced to promote pathways that drive malignancy. Furthermore, this work established a unique biochemical pathway that reverts certain malignant phenotypes in cancer through pharmacological perturbation of the Scrib palmitoylation cycle, without directly affecting Ras activation or signaling.
Beyond their role as protein depalmitoylases, APT enzymes can also hydrolyze a variety of other esters and thioester metabolites (Davda and Martin 2014). APT1 and APT2 were originally characterized as lysophospholipase enzymes (Sugimoto et al. 1996; Toyoda et al. 1999), yet later studies demonstrated enhanced catalytic efficiency towards acyl-thioester-linked proteins (Duncan and Gilman 1998). Interestingly, APT2 was identified as the primary prostaglandin glycerol esterase, while APT1 had no significant activity (Manna et al. 2014). Both APT1 and APT2 accelerate the deacylation of octanoyl-ghrelin in serum, suggesting these enzymes may also be secreted from cells (Satou et al. 2010). Lipopolysaccharide (LPS) treatment increased in APT1 serum levels, potentially through release from liver. Furthermore, the intracellular APT1 mRNA and protein levels were reduced after LPS stimulation, yet APT2 levels were unaffected. Overall, APTs likely hydrolyze a number of more soluble acyl-esters and thioesters, and may have broader functional roles in vivo beyond depalmitoylation.
To add spatial and temporal resolution to depalmitoylation enzymes, a series of mechanism-based octanoyl-thioester fluorogenic probes were developed for live cell imaging (Kathayat et al. 2017). Approximately half of the probe fluorescence produced in HEK293T cells was blocked by ML348 or Palmostatin B, and APT1 siRNA knockdown reduced probe fluorescence produced by 25%. Interestingly, EGF stimulation reduced the probe fluorescence produced by about 10% in live cells, suggesting some growth factor regulation of deacylase activity. While intriguing, further evidence is needed to establish if such reductions affect steady-state cellular S-palmitoylation levels, particularly of S-palmitoylated growth signaling mediators. Given the large number of candidate octanoyl-thioesterases, these probes provide a useful cell-based platform to explore deacylase activity in live cells, and measure contributions across different candidate enzymes.
Structural insights to APT enzymes
In order to explain the functional differences between APT1 and APT2, the first high-resolution structures bound to their respective inhibitors were recently solved (Won et al. 2016). While the structure of apo-APT1 was reported (Devedjiev et al. 2000), there were no additional studies to analyze the structure of APT1 or any structures of APT2 for comparison. Before solving the structures of each enzyme, we profiled the thermostability of APT1 and APT2 bound to different ligands. APT1 was more thermostable than APT2 by 5°C, suggesting previous efforts to crystalize APT2 were likely stymied by additional flexibility or disorder. Compared to free enzyme, APT1 binding to ML348 or APT2 binding to ML349 increased the thermostability by 4°C, which confirms thermodynamically favorable inhibitor engagement. Furthermore, incubation with HDFP led to a surprising 10°C stabilization. Accordingly, APT enzymes were predicted to harbor an acyl-binding pocket to stabilize the enzyme upon substrate binding. Based on these observations, we identified co-crystallization conditions and solved the structures of both APT1•ML348 (1.55 Å; 5SYM) and APT2•ML349 (1.64 Å; 5SYN) by x-ray crystallography (Figure 4).
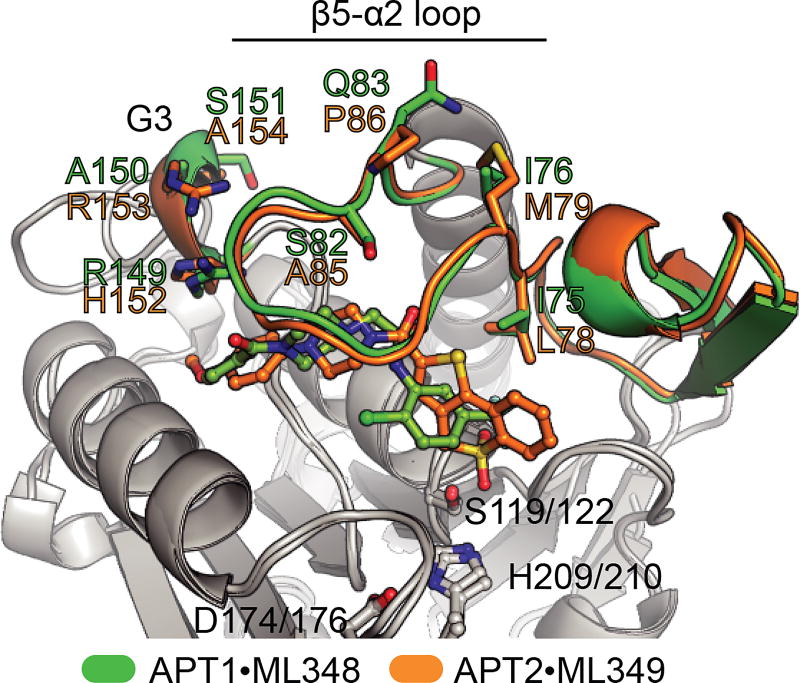
Figure 4. Divergence in APT1•ML348 and APT2•ML349 structures.
Blue regions signify divergence between APT1 and APT2. The inhibitor selectivity filter APT1-L176M and APT2-M178L is shown in grey sticks in the β8/α4 region. Adapted with permission from (Won et al. 2016). A color version of the figure is available online.
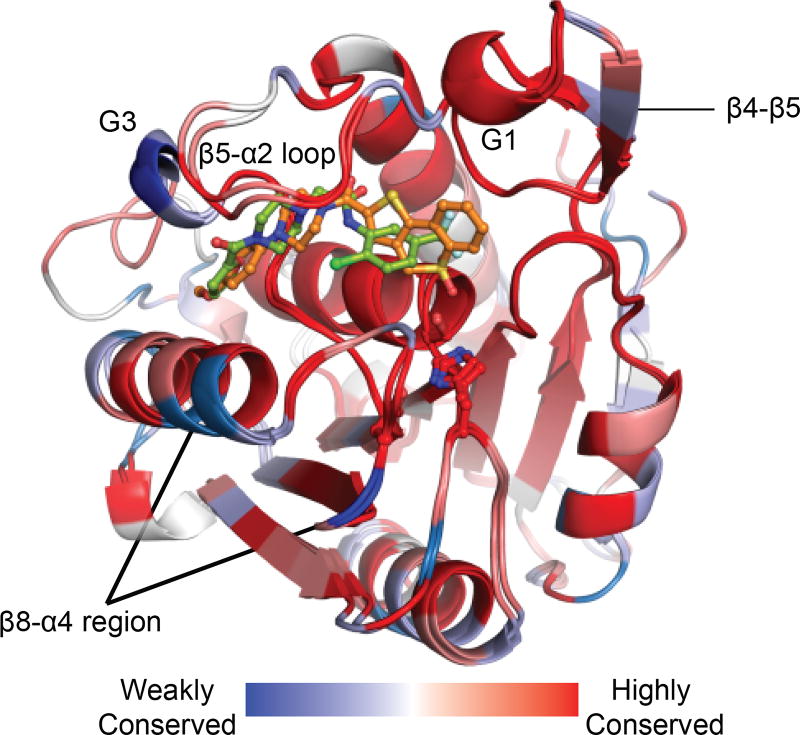
APT1 and APT2 feature unusual α/β serine hydrolase fold where the first β1 strand in the ‘canonical’ fold is missing and the fourth α-helix is replaced by a short α-helical segment termed G3. These enzymes include unique insertions, which add the short β4-β5 sheet and the β5-α2 loop. This loop contributes to forming a relatively long channel flanking the catalytic triad of both enzymes. Comparison of apoAPT1, APT1•ML348, and APT2•ML349 revealed this loop has varying degrees of openness, suggesting dynamic movement may engage various substrates. This feature functionally replaces the ‘cap’ or ‘lid’ domain found in many lipases, contributing to substrate engagement and providing thermodynamically stable environment for the catalysis (Khan et al. 2017). Both ML348 and ML349 make unique interactions with the divergent residues along this channel (Figure 5). However, these residues are either protruding away from the core of the protein (I76/M79, Q83/P86) or are similar in characteristics (eg. I75/L78). Importantly, mutational analysis identified a single residue I75/L78 (APT1/APT2) that establishes steric constraints to impart isoform selective inhibition. Similarly, the residues within the loop of the catalytic D174/176 does not show obvious differences. Other divergent residues in the G3 helix show marginal contributions to inhibitor selectivity, potentially by constraining the dynamics of the β5-α2 loop.
Figure 5. Isoform-selective inhibitor binding in APT1•ML348 and APT2•ML349.
Alignment demonstrates inhibitor engagement spanning along a hydrophobic channel towards the catalytic triad. Select residues are highlighted with sequence divergence between APT1 and APT2. Adapted from (Won et al. 2016). A color version of the figure is available online.
The β5-α2 loop and the surrounding residues around the active site form a hydrophobic channel, that may enhance the entry and engagement of hydrophobic substrates. While it is tempting to think that peptide component of substrates might occupy the channel, docking simulations further support exclusive acyl chain occupancy. As further evidence, ML349-fluorescein binding to APT2 is blocked by pre-treatment with palmitic acid. Together with the observed 10°C thermal stabilization upon HDFP engagement, we hypothesize long chain fatty acyl substrates bind in the channel, locking the substrate to promote ester hydrolysis. The channel and residues in proximity are probably not involved in the selectivity of the enzymes towards palmitoylated substrates, but rather select for acyl chain length through stabilization of the β5-α2 loop. Of note, the structures of both APT1•ML348, and APT2•ML349 lack the first few N-terminal residues. Since APT enzymes may be S-palmitoylated themselves near the N-terminus, it remains unclear how this may influence hydrophobic channel orientation and engagement of palmitoylated substrates. However, APT enzymes are largely soluble present in the cytosol, so we predict APTs may only be fractionally S-palmitoylated.
Our structural analysis of APT1 and APT2 reveal strikingly similar substrate binding regions and surface polarity, and only conservative mutations across the enzyme active site (Figure 6). ML348 and ML349 are isoform selective apparently by coincidence and not by evolutionary selection in substrate discrimination. Therefore, any specific biological roles for each enzyme may be due to distinct protein-protein interactions, post-translational processing, differential expression, or distinct subcellular localizations. This raises several questions, such as why would evolution keep two isoforms of APT enzymes? What imparts the functional divergence between APT enzymes? Although these enzymes share nearly 70% identity, overexpression and knockdown studies clearly show preferences for specific palmitoylated substrates. One explanation could be the large difference in melting temperatures between isoforms, which suggest APT2 is more flexible and may be able to accept more diverse substrates than APT1. Furthermore, the single acyl binding channel explains the reduced activity towards diacylated substrates, and emphasizes a potential role for APT enzymes as general esterases for long-chain monoacylated substrates, including lysophospholipids, prostaglandin glycerol esters, and S-palmitoylated proteins. Overall, structural analysis of APT1 and APT2 does not provide a definitive explanation of how these enzymes are functionally divergent.
Figure 6. Surface polarity of APT1•ML348 and APT2•ML349.
Both APT1 and APT2 have similar surface polarity, with similar distributions of polar and hydrophobic surfaces. A color version of the figure is available online.
ABHD17 family thioesterases
Until recently, APT1 was recognized as the primary depalmitoylase in cells, yet mutants in yeast, Drosophila, and C. elegans have no reported phenotypes. Given that APT1 was initially described as the primary depalmitoylase regulating N-Ras activity, this was quite puzzling. Cracks in this model began to appear throughout a number of publications, most strikingly from evidence that H-Ras depalmitoylase activity was exclusively in insoluble cell fraction (P100), and not with APT1 in the soluble fraction (S100) (Ahearn et al. 2011). Later, APT1/APT2 knockdown was shown to have no effect on R7BP depalmitoylation, while treatment with Palmostatin B or HDFP both blocked activity (Jia et al. 2014). This was followed by detailed activity-based profiling of Palmostatin B targets, and the realization that Palmostatin B is broadly reactive across a number of candidate depalmitoylases (Lin and Conibear 2015). Clearly other depalmitoylase activities beyond APT1/APT2 are present in cells, and likely contribute to the depalmitoylation of plasma membrane localized targets, including Ras.
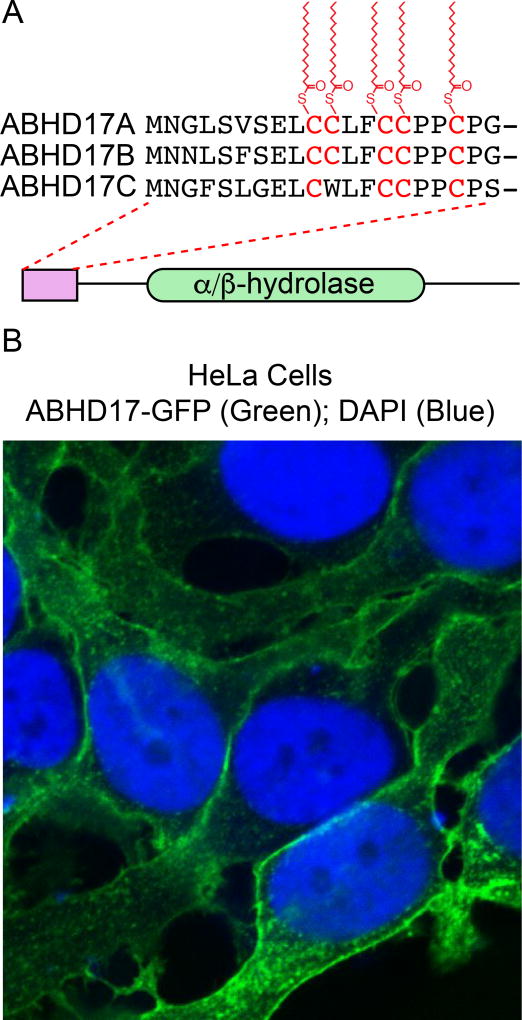
In order to identify these additional depalmitoylases, Palmostatin B targeted hydrolases were over-expressed and assayed for S-palmitoylation turnover using 17-ODYA pulse-chase methods. This led to the identification of ABHD17 enzymes as candidate N-Ras depalmitoylases in COS-7 cells (Lin and Conibear 2015). Over-expression of catalytic dead enzyme or N-terminal truncated mutants did not affect the N-Ras palmitoylation cycle, confirming ABHD17 enzymes are functional depalmitoylases in cells. ABHD17 hydrolases (ABHD17A, ABHD17B, and ABHD17C) are broadly expressed in all vertebrates, and harbor multiple conserved cysteines near their N-termini (Figure 7A). The N-terminal cysteine rich domain is essential for both S-palmitoylation and plasma membrane association (Martin and Cravatt 2009). Thus, ABHD17 enzymes are themselves S-palmitoylated, which is necessary for plasma membrane association and proximity to other potential S-palmitoylated protein substrastes (Figure 7B). Importantly, deletion of the N-terminal cysteine rich domain has no effect on ABHD17 reactivity with fluorophosphonate activity-based probes, suggesting the palmitoylation motif is primarily responsible for directing membrane localization and not for enzyme activity. ABHD17A over-expression shifted N-Ras localization from the plasma membrane to internal membranes, indicative of more rapid S-palmitoylation turnover. Triple ABHD17 (A-C) knockdown stabilized N-Ras 17-ODYA labeling, while APT1/APT2 inhibition with ML348 and ML348 had no effect. Accordingly, ABHD17 enzymes (but not APT1/APT2) contribute to N-Ras depalmitoylation in cells.
Figure 7. ABHD17 enzymes are S-palmitoylated and localize to the plasma membrane.
(A) ABHD17A-C are S-palmitoylated in a conserved N-terminal cysteine-rich motif separate from the α/β-hydrolase domain. (B) ABHD17A-GFP localization in HeLa cells demonstrates predominant plasma membrane localization. A color version of the figure is available online.
The synaptic protein PSD-95 is rapidly de-palmitoylated after depolarization. In order to identify PSD-95 depalmitoylase(s), a panel of serine hydrolases were individually co-expressed with PSD-95 in 293T and COS-7 cells (Yokoi et al. 2016; Tortosa et al. 2017). Over-expressed ABHD17 enzymes efficiently depalmitoylated PSD-95 in 293T, COS-7, and primary neuronal cultures, while other Palmostatin B targets ABHD12, ABHD13, APT1, and APT2 had only fractional reductions in PSD-95 S-palmitoylation (Figure 8). ABHD17 over-expression reduced the synaptic clusters of PSD-95 and AMPA receptors in neurons, while ABHD17A/ABHD17B/ABHD17C knockdown stabilized palmitoylated PSD-95 and prevented the decrease in synaptic PSD-95 clustering. In addition, ABHD17 overexpression in primary neurons reduces the S-palmitoylation of microtubule-associated protein 6 (MAP6), causing MAP6 retention in axons and microtubule stability (Tortosa et al. 2017). Recently the international knockout mouse consortium identified ABHD17A as one of the top suppressors of distal cancer metastasis in mice (van der Weyden et al. 2017), suggesting it may play a role in immune cell activation or surveillance.
Figure 8. Homology, depalmitoylase activity, and Palmostatin B inhibition of HDFP-sensitive serine hydrolases.
HDFP-sensitive seine hydrolases reported from mouse B-cell hybidoma cells (Martin et al. 2011) are shown based on active-site anchored homology. Palmostatin B targets are highlighted in red (Savinainen et al. 2014; Lin and Conibear 2015). Enzymes that reduced PSD-95 S-palmitoylation after over-expression are labeled with a red star (Yokoi et al. 2016). A color version of the figure is available online.
Despite this growing number of cell-based ABHD17 studies, there is no biochemical evidence demonstrating in vitro depalmitoylation, enzyme kinetics, substrate specificity, or other functional properties of each ABHD17 enzyme. ABHD17 enzymes may hydrolyze other lipids, particularly since their plasma membrane association delivers them in proximity to a number of potential substrates. A reported biochemical high throughput screen identified moderately potent inhibitors of an N-terminal truncated ABHD17B protein, yet these inhibitors failed to inhibit full-length, mammalian cell expressed protein (National Center for Biotechnology Information. PubChem BioAssay Database; AID=2200). Structural studies are clearly necessary to understand if the N-terminal 100 amino acids before the conserved α/β-hydrolase domain function solely as a platform for S-palmitoylation and membrane tethering, or if they impart additional structural features important for substrate engagement. Clearly future efforts are needed to explore the substrate profile, redundancy between isoforms, and physiological roles for each ABHD17 enzymes.
Conclusions
Cycles of S-palmitoylation and depalmitoylation provide a mechanism for membrane sampling and trafficking of peripheral membrane proteins to the plasma membrane (Rocks et al. 2010). Such directional palmitoylation cycles are demonstrated for G proteins, Ras, Scrib, and other central regulators of cell growth and organization. G protein internalization occurs even in the presence of generic depalmitoylation inhibitors, demonstrating that while depalmitoylation may be necessary for plasma membrane trafficking, agonist-dependent internalization occurs independent of depalmitoylation (Martin and Lambert 2016). Based on these findings, many peripheral membrane proteins require cycles of S-palmitoylation and depalmitoylation for plasma membrane delivery, but there are no definitive examples of post-translational regulation of depalmitoylation activity. Rather, depalmitoylation appears to depend more on sub-cellular localization and regulated expression. ABHD17 enzymes localize to the plasma membrane, where they have direct access to neuronal scaffolding proteins and growth regulators. APT enzymes are soluble, and largely cytoplasmic, and play active roles in maintaining the directional palmitoylation cycle important for peripheral membrane protein trafficking.
Future efforts to understand regulation of depalmitoylation will continue to leverage emerging chemical and genetic technologies. While Palmostatin B and HDFP are useful generic inhibitors of depalmitoylases, selective inhibitors of APT enzymes should now be routinely used to individually perturb each APT isoform. These reagents will be valuable for parsing how APT1 and APT2 function either independently or in conjunction to influence distinct cellular outcomes. Future efforts are needed to validate APT1 and APT2 localization, interaction partners, and modifications to fully understand the divergent phenotypes. This would be greatly accelerated by knockout animal models, allowing detailed biochemical and phenotypic analysis for each APT enzyme in their native physiological environments.
Similarly, ABHD17 enzymes are largely unannotated, and without biochemical analysis, it is not clear if each of the three enzymes are bona fide depalmitoylases or if each isoform carries out unique cellular functions. Clearly, ABHD17A plays a major role in suppressing growth of distal metastases, which likely reflects a role in immune regulation. Surely there are additional depalmitoylating enzymes, whether as their primary biochemical role or through non-specific acyl-thioester hydrolysis. Furthermore, Palmostatin B and HDFP likely act though polypharmacology, inhibiting a number of depalmitoylase (or lipase) enzyme activities to affect global depalmitoylation dynamics. Further functional studies of other hydrolases are likely to add to the growing number of factors contributing depalmitoylation dynamics in cells.
Finally, with the emergence of novel depalmitoylating enzymes, it is important to annotate the substrate profile and functional regulation for each enzyme. For example, cells could be treated with ML348 or ML349 to profile differential enrichment by mass spectrometry. Such experiments may be complicated by compensatory mechanisms, requiring simultaneous inhibition across a panel of depalmitoylases. Current enrichment and mass spectrometry detection methods are either indirect (Acyl-RAC) or require non-physiological metabolic labeling (17-ODYA). Since current liquid chromatography methods are optimized for polar peptides, future efforts are needed to extend quantitative proteomics to more hydrophobic peptides, allowing direct analysis of the modified peptide and the specific acyl chain. This could readily be accomplished by changing the column resin or adapting the mobile phase composition. Investment in sample preparation methods to optimize acyl-thioester stability, separation, ionization, and fragmentation have not been explored outside of single peptide measurements (Ji et al. 2013). In addition, multiplexed quantitative proteomics has emerged as a promising approach for time-dependent analyses, providing multiple measurements in a single combined proteomics experiment. We anticipate that ongoing advances in sample preparation and analysis will ultimately resolve many unanswered questions, providing a direct approach to profile the hydrophobic proteome and enzyme regulation.
Acknowledgments
This work was supported by the National Institutes of Health under Grant DP2 GM114848 and the University of Michigan.
Footnotes
Disclosure statement
The authors disclose no conflict of interest.
References
- Abrami L, Dallavilla T, Sandoz PA, Demir M, Kunz B, Savoglidis G, Hatzimanikatis V, van der Goot FG. Identification and dynamics of the human ZDHHC16-ZDHHC6 palmitoylation cascade. Elife. 2017;6 doi: 10.7554/eLife.27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi N, Hess DT, McLaughlin P, Stamler JS. S-Palmitoylation of a Novel Site in the beta2-Adrenergic Receptor Associated with a Novel Intracellular Itinerary. J Biol Chem. 2016;291(38):20232–20246. doi: 10.1074/jbc.M116.725762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010a. Characterization of a Selective, Reversible Inhibitor of Lysophospholipase 1 (LYPLA1) [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010b. Characterization of a Selective, Reversible Inhibitor of Lysophospholipase 2 (LYPLA2) [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, et al. Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J Am Chem Soc. 2012;134(25):10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. Optimization and characterization of a triazole urea dual inhibitor for lysophospholipase 1 (LYPLA1) and lysophospholipase 2 (LYPLA2) [PubMed] [Google Scholar]
- Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7(7):469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, Philips MR. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Mol Cell. 2011;41(2):173–185. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimzhanov AM, Boehning D. Rapid and transient palmitoylation of the tyrosine kinase Lck mediates Fas signaling. Proc Natl Acad Sci U S A. 2015;112(38):11876–11880. doi: 10.1073/pnas.1509929112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening [Research Support, NIH., Extramural Research Support, Non-U.S.Gov't] Proc Natl Acad Sci U S A. 2010;107(49):20941–20946. doi: 10.1073/pnas.1011663107. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg V, Rusch M, Vartak N, Jungst C, Schauss A, Waldmann H, Hedberg C, Pallasch CP, Bastiaens PI, Hallek M, et al. miRs-138 and -424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in CLL. Blood. 2015;125(19):2948–2957. doi: 10.1182/blood-2014-07-586511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D, Perrimon N. Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature. 2000;403(6770):676–680. doi: 10.1038/35001108. eng. [DOI] [PubMed] [Google Scholar]
- Blanc M, David F, Abrami L, Migliozzi D, Armand F, B¸rgi J, van der Goot F. SwissPalm: Protein Palmitoylation database. 2015;4 doi: 10.12688/f1000research.6464.1. : (F1000Research; 261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigidi GS, Santyr B, Shimell J, Jovellar B, Bamji SX. Activity-regulated trafficking of the palmitoyl-acyl transferase DHHC5. Nat Commun. 2015;6:8200. doi: 10.1038/ncomms9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss JE, Sefton BM. Direct Identification of Palmitic Acid as the Lipid Attached to P21ras. Molecular and Cellular Biology. 1986;6(1):116–122. doi: 10.1128/mcb.6.1.116. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem. 1993;268(30):22566–22574. [PubMed] [Google Scholar]
- Chai S, Cambronne XA, Eichhorn SW, Goodman RH. MicroRNA-134 activity in somatostatin interneurons regulates H-Ras localization by repressing the palmitoylation enzyme, DHHC9. Proc Natl Acad Sci U S A. 2013;110(44):17898–17903. doi: 10.1073/pnas.1317528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131(13):4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhu B, Yin C, Liu W, Han C, Chen B, Liu T, Li X, Chen X, Li C, et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature. 2017;549(7672):399–403. doi: 10.1038/nature23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognetta AB, 3rd, Niphakis MJ, Lee HC, Martini ML, Hulce JJ, Cravatt BF. Selective N-Hydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases. Chem Biol. 2015;22(7):928–937. doi: 10.1016/j.chembiol.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. eng. [DOI] [PubMed] [Google Scholar]
- Davda D, El Azzouny MA, Tom CT, Hernandez JL, Majmudar JD, Kennedy RT, Martin BR. Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem Biol. 2013;8(9):1912–1917. doi: 10.1021/cb400380s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davda D, Martin BR. Acyl protein thioesterase inhibitors as probes of dynamic S-palmitoylation. Medchemcomm. 2014;5(3):268–276. doi: 10.1039/C3MD00333G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deck P, Pendzialek D, Biel M, Wagner M, Popkirova B, Ludolph B, Kragol G, Kuhlmann J, Giannis A, Waldmann H. Development and biological evaluation of acyl protein thioesterase 1 (APT1) inhibitors [In Vitro Research Support, Non-U.S. Gov't] Angew Chem Int Ed Engl. 2005;44(31):4975–4980. doi: 10.1002/anie.200462625. eng. [DOI] [PubMed] [Google Scholar]
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6(6):449–456. doi: 10.1038/nchembio.362. eng. [DOI] [PubMed] [Google Scholar]
- Devedjiev Y, Dauter Z, Kuznetsov SR, Jones TL, Derewenda ZS. Crystal structure of the human acyl protein thioesterase I from a single X-ray data set to 1.5 A. Structure. 2000;8(11):1137–1146. doi: 10.1016/s0969-2126(00)00529-3. [DOI] [PubMed] [Google Scholar]
- Dow LE, Brumby AM, Muratore R, Coombe ML, Sedelies KA, Trapani JA, Russell SM, Richardson HE, Humbert PO. hScrib is a functional homologue of the Drosophila tumour suppressor Scribble. Oncogene. 2003;22(58):9225–9230. doi: 10.1038/sj.onc.1207154. eng. [DOI] [PubMed] [Google Scholar]
- Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36(2):276–285. doi: 10.2144/04362RR02. eng. [DOI] [PubMed] [Google Scholar]
- Duncan JA, Gilman AG. A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate from G Protein α Subunits and p21RAS. Journal of Biological Chemistry. 1998;273(25):15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- Elsum IA, Humbert PO. Localization, not important in all tumor-suppressing properties: a lesson learnt from scribble. Cells Tissues Organs. 2013;198(1):1–11. doi: 10.1159/000348423. [DOI] [PubMed] [Google Scholar]
- Feigin ME, Akshinthala SD, Araki K, Rosenberg AZ, Muthuswamy LB, Martin B, Lehmann BD, Berman HK, Pietenpol JA, Cardiff RD, et al. Mislocalization of the cell polarity protein scribble promotes mammary tumorigenesis and is associated with basal breast cancer. Cancer research. 2014;74(11):3180–3194. doi: 10.1158/0008-5472.CAN-13-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe IT, Child MA, Majmudar JD, Krishnamurthy S, van der Linden WA, Ward GE, Martin BR, Bogyo M. Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe. 2015;18(4):501–511. doi: 10.1016/j.chom.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester MT, Hess DT, Thompson JW, Hultman R, Moseley MA, Stamler JS, Casey PJ. Site-specific analysis of protein S-acylation by resin-assisted capture. J Lipid Res. 2011;52(2):393–398. doi: 10.1194/jlr.D011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44(6):987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Gardiol D, Zacchi A, Petrera F, Stanta G, Banks L. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int J Cancer. 2006;119(6):1285–1290. doi: 10.1002/ijc.21982. eng. [DOI] [PubMed] [Google Scholar]
- Gottlieb CD, Zhang S, Linder ME. The Cysteine-rich Domain of the DHHC3 Palmitoyltransferase Is Palmitoylated and Contains Tightly Bound Zinc. J Biol Chem. 2015;290(49):29259–29269. doi: 10.1074/jbc.M115.691147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Lemonidis K, Gorleku OA, Cruchaga C, Grefen C, Chamberlain LH. Palmitoylation-induced aggregation of cysteine-string protein mutants that cause neuronal ceroid lipofuscinosis. J Biol Chem. 2012;287(44):37330–37339. doi: 10.1074/jbc.M112.389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NC, Chamberlain LH. Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci U S A. 2017;114(8):E1365–E1374. doi: 10.1073/pnas.1612254114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grecco HE, Schmick M, Bastiaens PI. Signaling from the living plasma membrane. Cell. 2011;144(6):897–909. doi: 10.1016/j.cell.2011.01.029. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, Marshall CJ. All Ras Proteins Are Polyisoprenylated but Only Some Are Palmitoylated. Cell. 1989;57(7):1167–1177. doi: 10.1016/0092-8674(89)90054-8. English. [DOI] [PubMed] [Google Scholar]
- Hang HC, Geutjes EJ, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL. Chemical probes for the rapid detection of Fatty-acylated proteins in Mammalian cells. J Am Chem Soc. 2007;129(10):2744–2745. doi: 10.1021/ja0685001. eng. [DOI] [PubMed] [Google Scholar]
- Hedberg C, Dekker FJ, Rusch M, Renner S, Wetzel S, Vartak N, Gerding-Reimers C, Bon RS, Bastiaens PI, Waldmann H. Development of highly potent inhibitors of the Ras-targeting human acyl protein thioesterases based on substrate similarity design. Angew Chem Int Ed Engl. 2011;50(42):9832–9837. doi: 10.1002/anie.201102965. Eng. [DOI] [PubMed] [Google Scholar]
- Henderson MX, Wirak GS, Zhang YQ, Dai F, Ginsberg SD, Dolzhanskaya N, Staropoli JF, Nijssen PC, Lam TT, Roth AF, et al. Neuronal ceroid lipofuscinosis with DNAJC5/CSPalpha mutation has PPT1 pathology and exhibit aberrant protein palmitoylation. Acta Neuropathol. 2016;131(4):621–637. doi: 10.1007/s00401-015-1512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JL, Davda D, Cheung See Kit M, Majmudar JD, Won SJ, Gang M, Pasupuleti SC, Choi AI, Bartkowiak CM, Martin BR. APT2 Inhibition Restores Scribble Localization and S-Palmitoylation in Snail-Transformed Cells. Cell Chem Biol. 2017;24(1):87–97. doi: 10.1016/j.chembiol.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JL, Davda D, Majmudar JD, Won SJ, Prakash A, Choi AI, Martin BR. Correlated S-palmitoylation profiling of Snail-induced epithelial to mesenchymal transition. Mol Biosyst. 2016;12(6):1799–1808. doi: 10.1039/c6mb00019c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JL, Majmudar JD, Martin BR. Profiling and inhibiting reversible palmitoylation. Curr Opin Chem Biol. 2013;17(1):20–26. doi: 10.1016/j.cbpa.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce JJ, Joslyn C, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. An in Vivo Active Carbamate-based Dual Inhibitor of Lysophospholipase 1 (LYPLA1) and Lysophospholipase 2 (LYPLA2) [PubMed] [Google Scholar]
- James G, Olson EN. Identification of a novel fatty acylated protein that partitions between the plasma membrane and cytosol and is deacylated in response to serum and growth factor stimulation. J Biol Chem. 1989;264(35):20998–21006. [PubMed] [Google Scholar]
- Ji Y, Leymarie N, Haeussler DJ, Bachschmid MM, Costello CE, Lin C. Direct detection of S-palmitoylation by mass spectrometry. Anal Chem. 2013;85(24):11952–11959. doi: 10.1021/ac402850s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Chisari M, Maktabi MH, Sobieski C, Zhou H, Konopko AM, Martin BR, Mennerick SJ, Blumer KJ. A mechanism regulating G protein-coupled receptor signaling that requires cycles of protein palmitoylation and depalmitoylation. J Biol Chem. 2014;289(9):6249–6257. doi: 10.1074/jbc.M113.531475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Analysis of protein palmitoylation reveals a pervasive role in Plasmodium development and pathogenesis. Cell Host Microbe. 2012;12(2):246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456(7224):904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathayat RS, Elvira PD, Dickinson BC. A fluorescent probe for cysteine depalmitoylation reveals dynamic APT signaling. Nat Chem Biol. 2017;13(2):150–152. doi: 10.1038/nchembio.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan FI, Lan D, Durrani R, Huan W, Zhao Z, Wang Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front Bioeng Biotechnol. 2017;5:16. doi: 10.3389/fbioe.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB. Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J Biol Chem. 2013;288(13):9112–9125. doi: 10.1074/jbc.M112.421073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonidis K, Werno MW, Greaves J, Diez-Ardanuy C, Sanchez-Perez MC, Salaun C, Thomson DM, Chamberlain LH. The zDHHC family of S-acyltransferases. Biochem Soc Trans. 2015;43(2):217–221. doi: 10.1042/BST20140270. [DOI] [PubMed] [Google Scholar]
- Li Y, Martin BR, Cravatt BF, Hofmann SL. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem. 2012;287(1):523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Nazarian A, Erdjument-Bromage H, Bornmann W, Tempst P, Resh MD. Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J Biol Chem. 2001;276(33):30987–30994. doi: 10.1074/jbc.M104018200. [DOI] [PubMed] [Google Scholar]
- Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. Elife. 2015;4:e11306. doi: 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Middleton P, Hepler JR, Taussig R, Gilman AG, Mumby SM. Lipid modifications of G proteins: alpha subunits are palmitoylated. Proc Natl Acad Sci U S A. 1993;90(8):3675–3679. doi: 10.1073/pnas.90.8.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo S, Greentree WK, Linder ME, Deschenes RJ. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J Biol Chem. 2002;277(43):41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- Manna JD, Wepy JA, Hsu KL, Chang JW, Cravatt BF, Marnett LJ. Identification of the major prostaglandin glycerol ester hydrolase in human cancer cells. J Biol Chem. 2014;289(49):33741–33753. doi: 10.1074/jbc.M114.582353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR. Nonradioactive analysis of dynamic protein palmitoylation. Curr Protoc Protein Sci. 2013;73 doi: 10.1002/0471140864.ps1415s73. Unit 14 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6(2):135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Lambert NA. Activated G Protein Galphas Samples Multiple Endomembrane Compartments. J Biol Chem. 2016;291(39):20295–20302. doi: 10.1074/jbc.M116.729731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2011;9(1):84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby SM, Kleuss C, Gilman AG. Receptor regulation of G-protein palmitoylation. Proc Natl Acad Sci U S A. 1994;91(7):2800–2804. doi: 10.1073/pnas.91.7.2800. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Huibregtse JM. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20(21):8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Yano T, Nakagawa K, Takizawa S, Suzuki Y, Yasugi T, Huibregtse JM, Taketani Y. Analysis of the expression and localisation of a LAP protein, human scribble, in the normal and neoplastic epithelium of uterine cervix. Br J Cancer. 2004;90(1):194–199. doi: 10.1038/sj.bjc.6601465. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson HB, Perez-Mancera PA, Dow LE, Ryan A, Tennstedt P, Bogani D, Elsum I, Greenfield A, Tuveson DA, Simon R, et al. SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. Journal of Clinical Investigation. 2011;121(11):4257–4267. doi: 10.1172/JCI58509. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond FL, Tarpey PS, Edkins S, Tofts C, O'Meara S, Teague J, Butler A, Stevens C, Barthorpe S, Buck G, et al. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am J Hum Genet. 2007;80(5):982–987. doi: 10.1086/513609. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 2010;141(3):458–471. doi: 10.1016/j.cell.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- Runkle KB, Kharbanda A, Stypulkowski E, Cao XJ, Wang W, Garcia BA, Witze ES. Inhibition of DHHC20-Mediated EGFR Palmitoylation Creates a Dependence on EGFR Signaling. Mol Cell. 2016;62(3):385–396. doi: 10.1016/j.molcel.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou M, Nishi Y, Yoh J, Hattori Y, Sugimoto H. Identification and characterization of acyl-protein thioesterase 1/lysophospholipase I as a ghrelin deacylation/lysophospholipid hydrolyzing enzyme in fetal bovine serum and conditioned medium. Endocrinology. 2010;151(10):4765–4775. doi: 10.1210/en.2010-0412. [DOI] [PubMed] [Google Scholar]
- Savinainen JR, Patel JZ, Parkkari T, Navia-Paldanius D, Marjamaa JJ, Laitinen T, Nevalainen T, Laitinen JT. Biochemical and pharmacological characterization of the human lymphocyte antigen B-associated transcript 5 (BAT5/ABHD16A) PLoS One. 2014;9(10):e109869. doi: 10.1371/journal.pone.0109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger MJ, Magee AI, Schmidt MF. Fatty acid acylation of proteins in cultured cells. J Biol Chem. 1980;255(21):10021–10024. [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11(6):705–716. doi: 10.1038/ncb1876. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, Hayashi H, Yamashita S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J Biol Chem. 1996;271(13):7705–7711. doi: 10.1074/jbc.271.13.7705. eng. [DOI] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280(35):31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- Thiele C, Papan C, Hoelper D, Kusserow K, Gaebler A, Schoene M, Piotrowitz K, Lohmann D, Spandl J, Stevanovic A, et al. Tracing fatty acid metabolism by click chemistry. ACS Chem Biol. 2012;7(12):2004–2011. doi: 10.1021/cb300414v. [DOI] [PubMed] [Google Scholar]
- Thinon E, Percher A, Hang HC. Bioorthogonal Chemical Reporters for Monitoring Unsaturated Fatty-Acylated Proteins. Chembiochem. 2016;17(19):1800–1803. doi: 10.1002/cbic.201600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GM, Hayashi T, Chiu SL, Chen CM, Huganir RL. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron. 2012;73(3):482–496. doi: 10.1016/j.neuron.2011.11.021. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom CT, Martin BR. Fat chance! Getting a grip on a slippery modification. ACS Chem Biol. 2013;8(1):46–57. doi: 10.1021/cb300607e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One. 2010;5(11):e15045. doi: 10.1371/journal.pone.0015045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa E, Adolfs Y, Fukata M, Pasterkamp RJ, Kapitein LC, Hoogenraad CC. Dynamic Palmitoylation Targets MAP6 to the Axon to Promote Microtubule Stabilization during Neuronal Polarization. Neuron. 2017;94(4):809–825. e807. doi: 10.1016/j.neuron.2017.04.042. [DOI] [PubMed] [Google Scholar]
- Toyoda T, Sugimoto H, Yamashita S. Sequence, expression in Escherichia coli, and characterization of lysophospholipase II [doi: DOI: 10.1016/S1388-1981(99)00007-4] Biochim Biophys Acta. 1999;1437(2):182–193. doi: 10.1016/s1388-1981(99)00007-4. [DOI] [PubMed] [Google Scholar]
- Tsai FD, Wynne JP, Ahearn IM, Philips MR. Metabolic labeling of Ras with tritiated palmitate to monitor palmitoylation and depalmitoylation. Methods Mol Biol. 2014;1120:33–41. doi: 10.1007/978-1-62703-791-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaira V, Faversani A, Dohi T, Maggioni M, Nosotti M, Tosi D, Altieri DC, Bosari S. Aberrant overexpression of the cell polarity module scribble in human cancer. Am J Pathol. 2011;178(6):2478–2483. doi: 10.1016/j.ajpath.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Arends MJ, Campbell AD, Bald T, Wardle-Jones H, Griggs N, Velasco-Herrera MD, Tuting T, Sansom OJ, Karp NA, et al. Genome-wide in vivo screen identifies novel host regulators of metastatic colonization [Letter] Nature. 2017;541(7636):233–236. doi: 10.1038/nature20792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartak N, Papke B, Grecco HE, Rossmannek L, Waldmann H, Hedberg C, Bastiaens PI. The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys J. 2014;106(1):93–105. doi: 10.1016/j.bpj.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkruyse LA, Hofmann SL. Lysosomal targeting of palmitoyl-protein thioesterase. J Biol Chem. 1996;271(26):15831–15836. doi: 10.1074/jbc.271.26.15831. [DOI] [PubMed] [Google Scholar]
- Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuori P, Hofmann SL, Peltonen L. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376(6541):584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- Vujic I, Sanlorenzo M, Esteve-Puig R, Vujic M, Kwong A, Tsumura A, Murphy R, Moy A, Posch C, Monshi B, et al. Acyl protein thioesterase 1 and 2 (APT-1, APT-2) inhibitors palmostatin B, ML348 and ML349 have different effects on NRAS mutant melanoma cells. Oncotarget. 2016;7(6):7297–7306. doi: 10.18632/oncotarget.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Roth AF, Bailey AO, Davis NG. Palmitoylated proteins: purification and identification [10.1038/nprot.2007.225] Nat Protoc. 2007;2(7):1573–1584. doi: 10.1038/nprot.2007.225. [DOI] [PubMed] [Google Scholar]
- Wan L, Kang Y. Pleiotropic roles of AEG-1/MTDH/LYRIC in breast cancer. Adv Cancer Res. 2013;120:113–134. doi: 10.1016/B978-0-12-401676-7.00004-8. [DOI] [PubMed] [Google Scholar]
- Wang W, Runkle KB, Terkowski SM, Ekaireb RI, Witze ES. Protein Depalmitoylation Is Induced by Wnt5a and Promotes Polarized Cell Behavior. J Biol Chem. 2015;290(25):15707–15716. doi: 10.1074/jbc.M115.639609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275(1):261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- Wedegaertner PB, Bourne HR. Activation and depalmitoylation of Gs alpha. Cell. 1994;77(7):1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- Willumsen BM, Cox AD, Solski PA, Der CJ, Buss JE. Novel determinants of H-Ras plasma membrane localization and transformation. Oncogene. 1996;13(9):1901–1909. eng. [PubMed] [Google Scholar]
- Won SJ, Davda D, Labby KJ, Hwang SY, Pricer R, Majmudar JD, Armacost KA, Rodriguez LA, Rodriguez CL, Chong FS, et al. Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2) ACS Chem Biol. 2016;11(12):3374–3382. doi: 10.1021/acschembio.6b00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won SJ, Eschweiler JD, Majmudar JD, Chong FS, Hwang SY, Ruotolo BT, Martin BR. Affinity-Based Selectivity Profiling of an In-Class Selective Competitive Inhibitor of Acyl Protein Thioesterase 2. ACS Med Chem Lett. 2017;8(2):215–220. doi: 10.1021/acsmedchemlett.6b00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. Inhibiting the palmitoylation/depalmitoylation cycle selectively reduces the growth of hematopoietic cells expressing oncogenic Nras. Blood. 2012;119(4):1032–1035. doi: 10.1182/blood-2011-06-358960. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Nam SM, Kim JH, Das R, Choi SK, Nguyen TT, Quan X, Choi SJ, Chung CH, Lee EY, et al. Palmitate induces ER calcium depletion and apoptosis in mouse podocytes subsequent to mitochondrial oxidative stress [Original Article] Cell Death Dis. 2015;6:e1976. doi: 10.1038/cddis.2015.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol Cell Proteomics. 2010;9(1):54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh DC, Duncan JA, Yamashita S, Michel T. Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca(2+)-calmodulin. J Biol Chem. 1999;274(46):33148–33154. doi: 10.1074/jbc.274.46.33148. [DOI] [PubMed] [Google Scholar]
- Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M. Identification of PSD-95 Depalmitoylating Enzymes. J Neurosci. 2016;36(24):6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount JS, Moltedo B, Yang YY, Charron G, Moran TM, Lopez CB, Hang HC. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6(8):610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidman R, Buckland G, Cebecauer M, Eissmann P, Davis DM, Magee AI. DHHC2 is a protein S-acyltransferase for Lck. Mol Membr Biol. 2011;28(7–8):473–486. doi: 10.3109/09687688.2011.630682. [DOI] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135(5):865–878. doi: 10.1016/j.cell.2008.09.045. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci U S A. 2010;107(19):8627–8632. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]