Abstract
We present a real-time fitter for 3D single-molecule localization microscopy using experimental point spread functions (PSFs) that achieves optimal 3D resolution on any microscope and is compatible with any PSF engineering approach. This allowed us to image cellular structures with a 3D resolution unprecedented for astigmatic PSFs. The fitter compensates for most optical aberrations and makes accurate 3D superresolution microscopy broadly accessible, even on standard microscopes without dedicated 3D optics.
As most biological structures have a three-dimensional (3D) organization, it is desirable to achieve not only a high lateral, but also a high axial resolution in superresolution microscopy. Therefore, several methods have been developed to extend single molecule localization microscopy (SMLM) to 3D. Most commonly, the z-position is extracted from the shape of an astigmatic PSF1. Other approaches use more complex engineered PSFs (double-helix2, phase-ramp3 or tetrapod4), or a bi-plane configuration5. The vast majority of such 3D SMLM data is analysed by fitting a Gaussian PSF model due to its computational simplicity6. However, real PSFs are poorly approximated by a Gaussian function (Supplementary Fig. 1), and they often show aberrations due to imperfect microscope optics. As a result, current 3D fitting routines do not reach the optimal 3D resolution, produce distortions, and are limited to a thin slice around the focal plane. Thus, they cannot realize the full potential of 3D SMLM for biological discovery.
As an alternative to simple Gaussian PSF models, fitting methods using experimentally acquired PSF have been developed that can in theory achieve a higher precision, such as PSF correlation7, phase retrieval8,9, or interpolated PSFs3,10–13. In practice however, at the moment these methods are limited in their usability due to either a) low accuracy and robustness, b) a challenging process to generate an accurate PSF model14, c) slow speeds preventing online fitting during data acquisition or d) lack of camera-specific noise models, limiting the use of increasingly popular sCMOS cameras. Additionally, non-intuitive interfaces, restrictive licenses, and dependencies on specific programming languages and libraries fundamentally complicate their use, especially for users without an expert programming background. Thus, simple Gaussian, and not experimental PSF models are still generally used in 3D SMLM. Particularly for the majority of labs that have microscopes without perfect optics, this leads to a resolution that is very much worse in z than in x and y such that little meaningful 3D information is obtained.
Here, we present a software that overcomes these limitations and makes experimental PSF fitting generally accessible and practically useable, and thereby enables 3D SMLM with optimal z-resolution on any microscope (Supplementary Software 1,2). It contains an intuitive tool to robustly calibrate the experimental PSF and a fitter for cubic spline (cspline) interpolated PSF models that reaches the necessary fitting speeds for real-time localization (>105 fits/second). It also achieves the highest possible localization precision, the Cramér-Rao lower bound (CRLB) on simulated (Supplementary Fig. 2) and experimental (Supplementary Fig. 3) data.
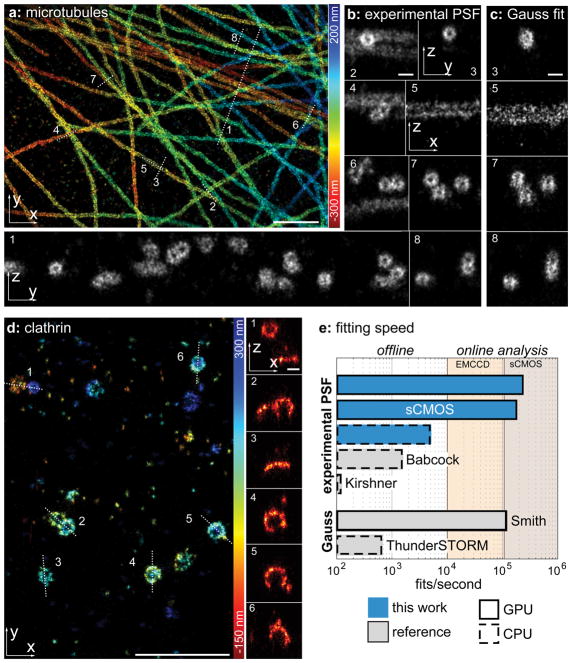
With this new fitter, we were able to resolve very fine structural details on biological structures (Fig. 1, Supplementary Fig. 4), which were previously accessible only by extremely complex interferometric microscopes (Supplementary Fig. 5). We were able to resolve in 3D the hollow cylinder of immunolabeled microtubules both with DNA-PAINT15 (Fig. 1a–b) and dSTORM16 (Supplementary Fig. 6) using the simple astigmatic 3D method. In comparison to the commonly used Gaussian fit17, our new fitter achieved a higher precision and avoided distortions (Fig. 1b, c). Furthermore, we could visualize the spherical geometry of clathrin-coated pits without distortions and found that almost all localizations were in the clathrin coat, highlighting the high localization accuracy (Fig 1d).
Figure 1.
(a) Immunolabeled microtubules imaged using the DNA-PAINT15 approach. Localizations are color-coded according to their z-position. Corresponding localization precisions and profiles are shown in Supplementary Fig. 4. (b) Side-view cross-sections along the lines denoted in (a) clearly reveal the hollow, cylinder-shaped structure of microtubules. (c) Side-view reconstructions of the same area as in (b) analysed with ThunderSTORM17 using an elliptical Gaussian MLE fit. (d) Immunolabeled clathrin imaged using dSTORM16. Side-view cross-sections clearly show the geometry of clathrin-coated pits with low and high curvatures. (e) Fitting speed of the fitter presented in this work, compared to previous implementation of fitters for experimental PSF models (Babcock et al.13 and Kirshner et al.12) and Gaussian PSF models (Smith et al.25 and ThunderSTORM17). Fits/second were measured on a i7-5930 CPU and a GTX1070 consumer graphics card. Width of the cross-sections: 150 nm (b1,4,6), 200 nm (b2,3,7,8), 30 nm (b5), 50 nm (d). Scale bars: 1 μm (a, d) and 100 nm (b, c, and x-z reconstructions in d).
We achieved this high performance with our fitter by a systematic optimization in which we overcame previous bottlenecks in the use of experimental PSFs. Firstly, we optimized the precision by developing a robust implementation of maximum likelihood estimation (MLE) for spline-interpolated PSF models (Supplementary Fig. 7). Compared to the simple Gaussian PSF models, our fitter reaches substantially higher localization accuracies on both simulated (Supplementary Fig. 8) and experimental (Supplementary Fig. 9 and Fig. 1b, c) data. Moreover, our fitter avoids the systematic error of Gaussian PSF models in estimating the number of photons per localization18 (Supplementary Fig. 10). We note that the quality of the PSF model is vital to avoid artifacts commonly observed when using experimental PSFs (Supplementary Fig. 11). To avoid these artifacts, we developed a simple and user-friendly tool to robustly create accurate experimental PSF models from several bead stacks (Supplementary Software 1,2).
Secondly, we optimized the speed of our fitter (Fig. 1e, Supplementary Fig. 12). High speeds are essential to enable fitting during data acquisition. This allows monitoring the image quality in real time, and to stop acquisitions as soon as a sufficient number of molecules have been localized19 or if the sample is deemed unsatisfactory. By implementing our fitter on the Graphical Processing Unit (GPU), we obtained fitting speeds more than a hundred times faster than for the fastest previously available implementation13. This now enables online analysis, even of dense structures and large fields of view (Fig. 1e).
Thirdly, we extended our fitter to sCMOS cameras (Fig. 1e), which offer fast imaging speeds and large fields of view, making them exquisitely suitable for SMLM20. For this, we included an sCMOS-specific noise model, which was previously limited to simple Gaussian PSF models20, in our fitter for experimental PSFs. We then validated that our fitter avoids camera noise-induced localization errors (Supplementary Fig. 13) and is fast enough for online analysis also for sCMOS cameras (Fig. 1e).
Taken together, our software retains the ease-of-use and accessibility of astigmatic 3D SMLM while approaching a 3D image quality that has to date only been achieved with highly complex 4Pi microscopes21.
However, astigmatism, as any other PSF engineering approach, requires dedicated 3D optics. Many users do not have access to these microscopes and are thereby limited to acquiring two-dimensional data. But even unmodified PSFs contain information on the z-position of the fluorophore22, which can be estimated from the PSF size, or by using the recently published photometry approach18. However, the z-resolution or axial range remain limited and these methods cannot distinguish fluorophores above and below the focus because of the high symmetry of the PSF.
Here we overcome these limitations and extract accurate and precise z-positions by fitting 2D SMLM data with an experimental model of the unmodified PSF. This allows us to exploit subtle differences between the upper and lower halves of the PSF to correctly localize the fluorophore. To this end, we developed a bi-directional fitting approach, in which we fit once with a z starting parameter above the focus and a second time below the focus and choose the solution with the maximum likelihood.
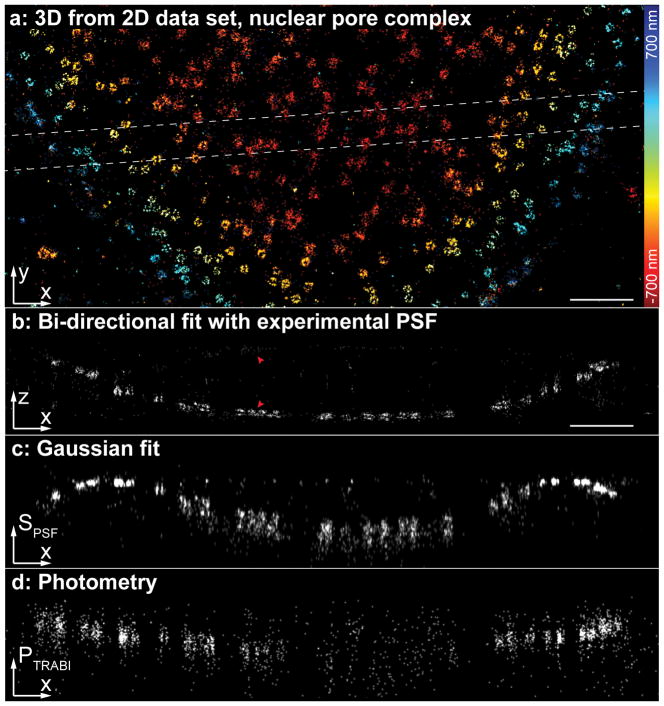
We achieved a z-resolution almost as good as that of astigmatic PSFs (Fig. 2a, b, Supplementary Fig. 4, Supplementary Fig. 8, Supplementary Fig. 14). We then directly compared our approach with existing methods based on PSF size (Fig. 2c) or photometry (TRABI)18 (Fig. 2d). We found that only our software could resolve the nucleoplasmic and cytoplasmic rings of the nuclear pore complex, which are axially spaced apart by 53 nm23. While close to the focal plane the z resolution was slightly decreased, and 5% of miss-assignments lead to a faint mirror image (Fig. 2b), our fitter enabled high-resolution 3D imaging directly on standard microscopes without any 3D optics.
Figure 2. Our spline fitter extracts accurate 3D positions from a simple 2D dataset with an unmodified PSF.
(a) Nup107-SNAP-AlexaFluor647 was imaged using dSTORM on a standard microscope without 3D optics. (b) Side-view reconstruction of the region denoted in (a). The nucleoplasmic and cytoplasmic rings of the nuclear pore complex, spaced 53 nm apart, can be easily resolved. The arrows denote nuclear pore complexes and their mirror images caused by misassignments. From the respective number of localizations, we estimated the fraction of misassignments to be ~5%. (c) Side view reconstruction of the same region using the size of the PSF SPSF from a fit with a symmetric Gaussian PSF model as a measure for the z-position. (d) Side view reconstruction of the same region using the photometry-based intensity ratio18 PTRABI as a measure for the z-position. Corresponding localization precisions and profiles can be found in Supplementary Fig. 4. Scale bars: 1 μm.
Besides astigmatism, a variety of sophisticated PSF engineering approaches have been developed (double helix2, self-bending24, tetrapod4, phase-ramp3 etc.), which increase the depth of field beyond ~1 μm, but require complex data analysis. As our fitter is directly applicable to all those PSFs (Supplementary Fig. 14), it will allow many more labs to exploit the advantages of advanced PSF engineering for large sample volumes. Moreover, for the first time it allows the use of advanced PSF engineering with sCMOS cameras while accurately accounting for the camera noise.
To summarize, we presented a fast, robust and precise single-molecule fitter for arbitrary PSF models. This allowed us to achieve a substantially improved 3D resolution and image quality using engineered astigmatic PSFs or unmodified PSFs from a standard microscope. As deformations of the PSF are included in the experimental PSF model, our fitter is robust with respect to field-independent aberrations, leading to a high accuracy even for objectives with a mediocre PSF or imperfect alignment of the microscope (Supplementary Fig. 14, 15). The presented framework is not restricted to bead-stack based PSFs, but can be used in the same way to obtain and rapidly fit a spline-interpolation of an arbitrary analytical or phase retrieved PSF model, for which aberrations can be calculated and added computationally.
To enable the broad community to profit from these innovations, we developed an easy-to-use fitting software that allows anybody to use the fitter directly on their own data. Additionally, we provide our CPU-based C-code and the GPU-based CUDA-code with extensive example code as open-source (Supplementary Software 1,2, github.com/jries/fit3Dcspline.git). It can be easily incorporated in any programming language as it is not library dependent, and thus will greatly improve speed and accuracy of any existing single-molecule fitting software.
With this, we hope that our software will transform 3D SMLM from an experts-only technique into a high-resolution imaging methodology that is broadly accessible.
ONLINE METHODS
Robust averaging of experimental bead stacks
Stacks of beads, immobilized on a coverslip, were acquired in a range of ±1000 nm with respect to the coverslip. A spacing in z between 10 nm and 50 nm works well. Beads in each stack were segmented in a maximum intensity projected image by maximum finding and thresholding. Sub regions around each bead location were cropped. Next, we aligned all beads with sub-pixel accuracy by 3D cross-correlation using a single bead as reference. To gain precision, another round of 3D alignment of the central part of each stack was performed using the average of the aligned bead stacks as the reference. We scaled up the central part of the cross-correlation by a factor of 20 by cubic spline interpolation and determined the x, y, and z shifts from the position of the maximum26. The bead stacks were shifted using cubic spline interpolation. Iteratively, bead stacks which showed a large dissimilarity from the average were identified based on the maximum value of the cross-correlation and the mean square error and excluded from the average. To eliminate the background, the minimum value of the bead stack was subtracted and the amplitude was normalized by the total (summed up) intensity of the central slice. We further regularized the bead stack by smoothing it in the z-direction with a smoothing B-spline27. In the presence of field-dependent aberrations, systematic fitting errors can be corrected in a post-processing step based on a precise calibration28. In presence of strong field-dependent aberrations, we suggest to perform the calibration and fitting only locally on small sub-regions of near uniform aberrations.
Calculation of cspline-interpolated PSFs
Spline functions are piecewise polynomials for which high order derivatives are continuous at the knots, where the pieces connect. Cubic splines are the most commonly used splines, e.g. in computer graphics, geometric modeling, etc. Recently, this type of approximation theory has also been used for single molecule localization10,12,13. We implemented the cspline interpolation both in terms of cubic splines and cubic B-splines. A B-spline interpolation is generally less memory intensive since only one B-spline coefficient is needed in each spline interval. In comparison, (d + 1)n coefficients are required in each spline interval for spline polynomials, where d is the spline degree and n is the dimension. However, our implementation of a 3D fit based on cubic splines is about 2.5 times faster than the cubic B-spline form due to the fact that cubic splines are more explicit and less calculations are needed to calculate spline values and derivatives. Therefore, the software used in this work is based on cubic splines with 64 coefficients in each voxel of the 3D PSF stack.
Similar to Ref. 13, the 3D PSF is described by a three dimensional cubic spline for voxel (i, j, k) as follows:
where Δx and Δy is the pixel size of the PSF in the object space in x and y directions, respectively. Δz is the step size in the objective space in z direction. xi, yj and zk are the start positions of voxel (i, j, k) in x, y and z directions, respectively.
In order to calculate the cspline coefficients, the 3D PSF stack was firstly built by averaging the bead stacks from different fields of view by 3D cross correlation and by regularization, as described above. The spline coefficients were built based on the averaged and smoothed 3D PSF stack. As 64 cspline coefficients are required to describe each voxel, we up sampled (cubic spline interpolation) each voxel 3 times in x, y and z directions, respectively. The 64 up sampled coordinates (including boundary of neighboring voxels) were used to calculate the 64 cspline coefficients. The code to calculate the spline coefficients from bead stacks can be found in the Supplementary Software 1.
z-calibration of astigmatic Gaussian PSF models
Our PSF calibration tool also allows extracting z-positions using two widely used algorithms: a) calculate the z-positions directly from the calibrated σx(z) and σy(z) returned by the elliptical Gaussian fit; b) determine the z-positions by directly fitting the single molecules with the calibrated astigmatic Gaussian PSF model29.
For both calibrations, the bead stacks are fitted with an elliptical Gaussian PSF model and shifted in z according to their true z-positions where σx(z) == σy(z). The outliers are removed based on the root mean error of σx(z) and σy(z) with respect to the average curves.
For algorithm a), we calculate dσ2(z) = σx(z)2 − σy(z)2 and interpolate the functional relationship z(dσ)2 by a smoothing cubic B-spline. This B-spline interpolation is then used to directly read out z from dσ2.
For algorithm b), σx(z) and σy(z) are fitted with a polynomial approximation for the astigmatic Gaussian model:
The parameters σ0x, Ax, Bx, σ0y, Ay, By, γ and d are input parameters for the Gaussian fitter, which directly returns the z-coordinates of the fluorophores. We follow the formula in Ref. 25 to calculate the derivatives of the parameters. However, the iterative process was re-implemented using the Levenberg-Marquardt (L-M) algorithm.
Newton and Levenberg-Marquardt iterative schemes for MLE
Maximum likelihood estimation is the method of choice for fitting data with Poisson statistics30. The objective function for MLE is given by31:
where μk is the expected number of photons in pixel k from the model PSF function, xk is the measured number of photons. By minimizing , we obtain the maximum likelihood for the Poisson process.
Methods for nonlinear optimization are usually iterative. For Newton iterative schemes, the search direction Δθi of each iteration is given by25
where θi is the i -th free fit parameter. However, computing the second derivatives is often quite difficult and can be destabilizing when the model fits badly or is contaminated by outlier points30.
An alternative method is the L-M algorithm. The L-M algorithm is often used for least squares fitting as it is quick and robust. With relatively simple modifications31, the L-M algorithm has also been used to minimize . In the L-M algorithm, the second derivatives term is neglected and only the first derivatives are used. In the L-M algorithm, the update Δθi is given by
where, Hi,j is the Hessian matrix without the second partial derivatives term, defined as , Jj is the Jacobian matrix defined as , λ is the damping factor, I is a diagonal matrix equal to the diagonal elements of the Hessian matrix. This method is more robust since a damping factor is introduced and the second derivatives do not contribute. This damping factor is increased (multiplied by 10 in this work) if an iteration step does not decrease or Hi,j is not positive definite.
GPU Implementation
This GPU implementation of the iterative method follows the framework developed for fitting a Gaussian PSF model using a GPU25, however using the L-M algorithm. We note that a general framework for L-M fitting on the GPU has been recently published32. Unlike previous work for EMCCD and sCMOS noise models20,25, where the shared memory was used to store the molecule candidate data and readout noise map, we kept the data in the GPU global memory. Each thread is pointed to each molecule candidate and performs all the computations for each molecule candidate. No thread synchronization is required. 64 threads per block were used. The overall speed is about 1.9 times (small window size) to 47.9 times (large window size) faster (Supplementary Fig. 12) than for the original code where shared memory was employed for the sCMOS noise model. We assume that this is due to the compiler optimization where more registers are used and the time for copying data from the global to the shared memory is saved. Both, the CPU based C-code and the GPU based CUDA-code were compiled using Microsoft Visual Studio 2010. The software was called via Matlab (Mathworks) mex files. It was run on a personal computer using an Intel(R) Core(TM) i7-5930 processor clocked at 3.50 GHz with 64 GB memory. An NVIDIA GeForce GTX 1070 graphics card with 8.0 GB memory was used for GPU based computation.
Simulation of realistic single molecule data
To simulate single molecule images using a realistic PSF model, we used the cspline-interpolated PSF model generated from experimental bead stacks as described above. We generated the single molecule image from this PSF model at a random 3D position, multiplied it with the number of photons/localization and added a constant background. Finally, we applied Poisson noise to the images. For simulations of sCMOS data, we added a pixel-dependent Gaussian noise. The code to simulate PSFs from the calculated cspline coefficients can be found in the Supplementary Software 1.
MLE fit using an sCMOS camera noise model
sCMOS cameras have become more and more attractive for localization microscopy due to their fast data acquisition even for large fields of view, low readout noise and relatively low price. However, their intrinsic pixel-dependent gain, offset and readout noise can create a localization bias, which has to be corrected when localizing the single molecule20. Gain gk and offset ok in pixel k can be taken into account when converting the camera image in analog digital units (ADU) into photons:
The readout noise, however, has to be taken into account during the fitting in the noise model and can be calculated from many dark camera images as the pixel-wise variance. Here, we use the same model as proposed by Huang, et al.20 which approximates the normal distributed readout noise (vark, in units of photoelectrons) with a Poisson distribution. By adding a pixel-dependent constant, vark, to the measured photoelectrons, one can expect the new value to approximate a Poisson distribution with a mean of uk + vark. Here, uk is the expected photon number in pixel k of the PSF model function. Therefore, in comparison to the conventional MLE fit for EMCCD data, only one more parameter, vark, is required for sCMOS data. Also vark is only kept in the global memory of the GPU. Compared to the EMCCD noise model, the speed performance of the algorithm was only reduced by less than 25% by additionally accounting for the pixel dependent readout noise (Supplementary Fig. 12).
3D Fitting of SMLM data acquired with standard microscopes without 3D optics
As our code includes fitting with arbitrary PSFs, it is directly applicable on 2D data, acquired with an unmodified PSF in a standard microscope. A model for the unmodified PSF can be calculated directly from bead stacks, in an analogous way to engineered PSFs. However, the unmodified PSF has a high symmetry with respect to the focal plane (Supplementary Figure 1), making it difficult for an iterative fitting procedure to converge through the focal plane. To overcome this problem, we fit every localization twice: once with a starting parameter for the z-position 500 nm above the focal plane, and once with a starting parameter 500 nm below the focal plane. The maximum likelihood is then used to select the better fit. As a real PSF is not completely symmetric33, this breaks the degeneracy previously encountered when extracting z-positions in 2D data sets from only a single photometry or PSF size parameter18.
Due to the rather large size of the calibration bead (100 nm) and small inaccuracies during the averaging of many bead stacks, the cspline PSF model is slightly blurred compared to a single-molecule PSF. This had no apparent effect on 3D data, but in 2D data it lead to an accumulation of fitted localizations at the focal plane. To overcome this problem, we filtered the raw images with a Gaussian kernel (standard deviation σ < 0.5 pixels), thus applying the blur in the PSF model to the data. To find the right σ, we fitted a subset of the data with several values for σ = 0, 0.1, …, 0.5 and selected the σ value for which we found neither an accumulation nor a depletion of localizations around the focal plane.
Post processing
As the positions used above are all based on the objective positions, which differ from the true absolute positions due to refractive index mismatch, we further multiply the z-positions with a refractive index mismatch factor of 0.751. Then, x, y, and z-positions were corrected for residual drift by a custom algorithm based on redundant cross-correlation. Localizations persistent in consecutive frames were grouped into one localization, and superresolution images were constructed with every localization rendered as a 2D elliptical Gaussian with a width proportional to the localization precision.
For measurements deep in the sample in combination with oil objectives, aberrations induced by the refractive index mismatch can be corrected for as described in Ref. 34.
Sample preparation of clathrin-coated pits in SK-MEL-2 cells
All samples were imaged on round 24 mm high precision glass coverslips No. 1.5H (117640, Marienfeld, Lauda-Königshofen, Germany). Coverslips were cleaned overnight in a 1:1 mixture of concentrated HCl and methanol, rinsed with millipore water until neutral, dried and UV sterilized in a standard cell culture hood.
SK-MEL-2 cells (kind gift from David Drubin, described in Ref. 35) were cultured under adherent conditions in DMEM/F-12 (Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12) with GlutaMAX and phenol red (ThermoFisher 10565018) supplemented with 10% [v/v] FBS, ZellShield™ (Biochrom AG, Berlin, Germany), and 30 mM HEPES at 37°C, 5% CO2 and 100% humidity. Cells were fixed using 3% [w/v] paraformaldehyde (PFA) in cytoskeleton buffer (CB; 10 mM MES pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM D-glucose, 5 mM MgCl2, described in Ref. 36) for 20 minutes. Fixation was stopped by incubation in 0.1% [w/v] NaBH4 for 7 minutes. The sample was washed with PBS three times, and subsequently permeabilised using 0.01% [w/v] digitonin (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 15 minutes. After washing twice with PBS, the sample was blocked with 2% [w/v] BSA in PBS for 60 minutes, washed again with PBS, and stained for 3–12 hours with anti-clathrin light chain (sc-28276, Santa Cruz Biotechnology, Dallas, TX, USA, diluted 1:300) and anti-clathrin heavy chain rabbit polyclonal antibodies (ab21679, Abcam, Cambridge, UK, diluted 1:500) in 1% [w/v] BSA in PBS. The sample was washed with PBS three times, and incubated with a donkey anti-rabbit secondary antibody (711-005-152, Jackson ImmunoResarch, West Grove, PA, USA), which was previously conjugated with Alexa Fluor 647-NHS at an average degree of labeling of 1.5, for 4 hours. Finally, the sample was washed three times with PBS prior to imaging.
For dSTORM imaging, coverslips were mounted in 500 μL blinking buffer (50 mM Tris pH 8, 10 mM NaCl, 10% [w/v] D-glucose, 35 mM 2-mercaptoethylamine (MEA), 500 μg/mL GLOX, 40 μg/mL catalase, 2 mM COT).
Sample preparation for imaging of the nuclear pore complex and microtubules
Wildtype U-2 OS and genome-edited U-2 OS cells that express Nup107-SNAP (as previously described in Ref. 37) were cultured under adherent conditions in Dulbecco’s Modified Eagle Medium (DMEM, high glucose, w/o phenol red) supplemented with 10% [v/v] FBS, 2 mM L-glutamine, non-essential amino acids, ZellShield™ (Biochrom AG, Berlin, Germany) at 37°C, 5% CO2 and 100% humidity. All incubations were carried out at room temperature. For nuclear pore staining, the coverslips were rinsed twice with PBS and prefixed with 2.4% [w/v] PFA in PBS for 30 seconds. Cells were permeabilized with 0.4% [v/v] Triton X-100 in PBS for 3 minutes and afterwards fixed with 2.4% [w/v] PFA in PBS for 30 minutes. Subsequently, the fixation reaction was quenched by incubation in 100 mM NH4Cl in PBS for 5 minutes. After washing twice with PBS, the samples were blocked with Image-iT™ FX Signal Enhancer (ThermoFisher Scientific, Waltham, MA, USA) for 30 minutes. The coverslips were incubated in staining solution (1 μM benzylguanine Alexa Fluor 647 (S9136S, NEB, Ipswich, MA, USA); 1 mM DTT; 1% [w/v] BSA; in PBS) for 50 minutes in the dark. After rinsing three times with PBS and washing three times with PBS for 5 minutes, the sample was mounted for imaging.
For microtubule staining, wildtype U-2 OS cells were prefixed for 2 minutes with 0.3% [v/v] glutaraldehyde in CB + 0.25% [v/v] Triton X-100 and fixed with 2% [v/v] glutaraldehyde in CB for 10 minutes. Fluorescent background was reduced by incubation with 0.1% [w/v] NaBH4 in PBS for 7 minutes. After 3 washes with PBS, microtubules were stained using anti alpha-tubulin antibody (MS581, NeoMarkers, Fremont, CA, USA) 1:300 in PBS + 2% [w/v] BSA for 2 h and anti-mouse Alexa Fluor 647 (A21236, Invitrogen, Carlsbad, CA, USA) 1:300 in PBS + 2% [w/v] BSA for 2h. After 3 washes with PBS samples were imaged in a blinking buffer as described above, but with pyranose oxidase instead of glucose oxidase.
For DNA-PAINT imaging, microtubules were labelled with anti alpha-tubulin antibodies (MS581, NeoMarkers, and T6074, Sigma-Aldrich, St. Louis, MO, USA) and anti beta-tubulin antibody (T5293, Sigma-Aldrich) each 1:300 diluted in PBS with 2% [w/v] BSA, for 2 hours. After 3 washes with PBS, samples were incubated with a DNA labelled anti-mouse secondary antibody overnight (docking strand sequence: 5′-TT ATA CAT CTA-3′) and imaged after 5 washes with PBS using 50 pM of complementary Atto-655 labelled DNA imager strand (5′-C TAG ATG TAT-3′-Atto655) in PAINT buffer (PBS, 500 mM NaCl, 40 mM Tris, pH 8.0).
Microscopy
SMLM image acquisition was performed at room temperature (24 °C) on a customized microscope38 equipped with a high NA oil immersion objective (160x, 1.43-NA oil immersion, Leica, Wetzlar, Germany). We employed a laser combiner (LightHub®, Omicron-Laserage Laserprodukte, Dudenhofen, Germany) with Luxx 405, 488 and 638, Cobolt 561 lasers. The lasers were triggered using a FPGA (Mojo, Embedded Micro, Denver, CO, USA) allowing microsecond pulsing control of lasers. After passing through a speckle reducer (LSR-3005-17S-VIS, Optotune, Dietikon, Switzerland), the laser is then guided through a multimode fiber (M105L02S-A, Thorlabs, Newton, NJ, USA). The output of the fiber is first magnified by an achromatic lens and then imaged into the sample38. A laser clean-up filter (390/482/563/640 HC Quad, AHF, Tübingen, Germany) is placed in the beam path to remove fiber generated fluorescence. A close-loop focus lock system was implemented using the signal of a near infrared laser reflected by the coverslip and its detection by a quadrant photodiode. The focus can be stabilized within ±10 nm over several hours39. The fluorescence emission was filtered by a bandpass filter (700/100, AHF or 676/37, AHF) and recorded by an EMCCD camera (Evolve512D, Photometrics, Tucson, AZ, USA). Typically, we acquire 100,000 – 300,000 frames with 15 ms exposure time (100 ms for DNA-PAINT) and laser power densities of ~15 kW/cm2. The pulse length of the 405 nm laser is automatically adjusted to retain a constant number of localizations per frame.
Controlled induction of optical aberrations was carried out on a similar microscope, albeit with an additional detection path. After the tube-lens, the back-focal plane of the objective is imaged onto a deformable mirror (Mirao 52e, Imagine Optic, Orsay, France) for phase correction. A flip mirror allows guiding the light either towards a Shack-Hartmann wavefront sensor (HASO3, Imagine Optic) or the camera for fluorescence imaging. The PSFs were obtained by correcting the optical aberrations using a closed loop between the mirror and the wavefront sensor in Casao (Imagine Optic) and image-based optimization of a single-bead in Micao (Imagine Optic). Aberrations were then induced based on pure Zernike modes.
Furthermore, SMLM images were acquired on a commercial Leica SR GSD 3D microscope. The setup is equipped with a 500 mW 642 nm laser (MPBC Inc., Montreal, Canada), a 30 mW 405 nm diode laser (Coherent Inc., Santa Clara, CA, USA), a DBP 405/10 642/10 excitation filter, a LP649 dichroic mirror, a BP 710/100 emission filter and a high NA oil immersion objective (160x, 1.43-NA oil immersion, Leica). Astigmatism was induced with a modified tube lens and the image was recorded on an iXon3 897 EMCCD camera (Andor, Belfast, Northern Ireland). Reference images were reconstructed with the Leica Application Suite X (Version 1.9.0.13747) and used as comparison to the cspline fit.
Statistics and reproducibility
All figures show representative data from ≥3 (Figure 1, Supplementary Figures 6, 11) or ≥5 (Figure 2, Supplementary Figures 1) representative experiments, or from single experiments (Supplementary Figure 9, 14, 15). Supplementary Figure 13 shows representative data from 3 similar independent simulations.
Code availability
Source code for the software used in this manuscript is contained in Supplementary Software 1 and updated versions can be freely downloaded at github.com/jries/fit3Dcspline.git.
Data availability statement
The datasets generated and analysed during the current study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank J. Mehl for helping with the sample preparation and data acquisition, S. van der Linde (University of Glasgow) for help with the TRABI analysis, E. Klotzsch (University of New South Wales) for the Zeiss Elyra bead stacks, M. Lampe (ALMF, EMBL Heidelberg) for help with the acquisition on the Leica GSD system and D. Drubin (Berkeley) for the gift of SK-MEL-2 hCLTA(EN)/DNM2(EN) cells. This work was supported by the European Research Council (ERC CoG-724489, M.M., J.R.), the Deutsche Forschungs Gemeinschaft (DFG RI 2380/2, J.R., J.D.), by the EMBL Interdisciplinary Postdoc Programme (EIPOD) under Marie Curie Actions COFUND (Y.L.), the 4D Nucleome/4DN NIH Common Fund (U01 EB021223) (J.E., J.R.) and by the European Molecular Biology Laboratory (Y.L., M.M., P.H., J.D., U.M., B.N., V.J.S., J.E., J.R.).
Footnotes
AUTHOR CONTRIBUTIONS
Y.L. and J.R. conceived the approach, developed the methods, wrote the software and analyzed the data. M.M. performed the imaging of clathrin coated pits. P.H. performed the imaging of nuclear pore complexes. U.M. performed the DNA-PAINT imaging of microtubules. B.N, V.J.S. and J.E. contributed the Nup107 cell line. I.S. contributed the custom-made DNA-PAINT antibodies. J.D. acquired the aberrated PSFs and helped with the data analysis. Y.L., M.M. and J. R. wrote the manuscript with the input from all other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pavani SRP, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baddeley D, Cannell MB, Soeller C. Three-dimensional sub-100 nm super-resolution imaging of biological samples using a phase ramp in the objective pupil. Nano Res. 2011;4:589–598. [Google Scholar]
- 4.Shechtman Y, Sahl SJ, Backer AS, Moerner WE. Optimal point spread function design for 3D imaging. Phys Rev Lett. 2014;113:1–5. doi: 10.1103/PhysRevLett.113.133902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juette MF, et al. Three-dimensional sub–100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 6.Deschout H, et al. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat Methods. 2014;11:253–266. doi: 10.1038/nmeth.2843. [DOI] [PubMed] [Google Scholar]
- 7.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quirin S, Pavani SRP, Piestun R. Optimal 3D single-molecule localization for superresolution microscopy with aberrations and engineered point spread functions. Proc Natl Acad Sci. 2012;109:675–679. doi: 10.1073/pnas.1109011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Kromann EB, Krueger WD, Bewersdorf J, Lidke KA. Three dimensional single molecule localization using a phase retrieved pupil function. Opt Express. 2013;21:29462. doi: 10.1364/OE.21.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahmasbi A, Ward ES, Ober RJ. Determination of localization accuracy based on experimentally acquired image sets: applications to single molecule microscopy. Opt Express. 2015;23:7630–7652. doi: 10.1364/OE.23.007630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mlodzianoski MJ, Juette MF, Beane GL, Bewersdorf J. Experimental characterization of 3D localization techniques for particle-tracking and super-resolution microscopy. Opt Express. 2009;17:8264–8277. doi: 10.1364/oe.17.008264. [DOI] [PubMed] [Google Scholar]
- 12.Kirshner H, Vonesch C, Unser M. Can localization microscopy benefit from approximation theory?. 2013 IEEE 10th International Symposium on Biomedical Imaging; 2013. pp. 588–591. [Google Scholar]
- 13.Babcock HP, Zhuang X. Analyzing Single Molecule Localization Microscopy Data Using Cubic Splines. Sci Rep. 2017;7:552. doi: 10.1038/s41598-017-00622-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng Y, Shaevitz JW. Effect of aberration on height calibration in three-dimensional localization-based microscopy and particle tracking. Appl Opt. 2009;48:1886. doi: 10.1364/ao.48.001886. [DOI] [PubMed] [Google Scholar]
- 15.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 16.van de Linde S, et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 17.Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franke C, Sauer M, van de Linde S. Photometry unlocks 3D information from 2D localization microscopy data. Nat Methods. 2016;14:41–44. doi: 10.1038/nmeth.4073. [DOI] [PubMed] [Google Scholar]
- 19.Nieuwenhuizen RPJ, et al. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–62. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang F, et al. Video-rate nanoscopy using sCMOS camera–specific single-molecule localization algorithms. Nat Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang F, et al. Ultra-High Resolution 3D Imaging of Whole Cells. Cell. 2016;166:1028–1040. doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aguet F, Van De Ville D, Unser M. A maximum-likelihood formalism for sub-resolution axial localization of fluorescent nanoparticles. Opt Express. 2005;13:10503–10522. doi: 10.1364/opex.13.010503. [DOI] [PubMed] [Google Scholar]
- 23.von Appen A, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia S, Vaughan JC, Zhuang X. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nat Photonics. 2014;8:302–306. doi: 10.1038/nphoton.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CS, Joseph N, Rieger B, Lidke Ka. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat Methods. 2010;7:373–375. doi: 10.1038/nmeth.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33:156. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 27.Unser M, Aldroubi A, Eden M. B-spline signal processing. I Theory. IEEE Trans Signal Process. 1993;41:821–833. [Google Scholar]
- 28.von Diezmann A, Lee MY, Lew MD, Moerner WE. Correcting field-dependent aberrations with nanoscale accuracy in three-dimensional single-molecule localization microscopy. Optica. 2015;2:985. doi: 10.1364/OPTICA.2.000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtzer L, Meckel T, Schmidt T. Nanometric three-dimensional tracking of individual quantum dots in cells. Appl Phys Lett. 2007;90:53902. [Google Scholar]
- 30.Press W, Teukolsky S, Vetterling W, Flannery B. Numerical recipes in C: the art of scientific computing. Cambridge University Press; Cambridge, U.K: 1992. [Google Scholar]
- 31.Laurence TA, Chromy BA. Efficient maximum likelihood estimator fitting of histograms. Nat Methods. 2010;7:338–339. doi: 10.1038/nmeth0510-338. [DOI] [PubMed] [Google Scholar]
- 32.Przybylski A, Thiel B, Keller-Findeisen J, Stock B, Bates M. Gpufit: An open-source toolkit for GPU-accelerated curve fitting. Sci Rep. 2017;7:15722. doi: 10.1038/s41598-017-15313-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirshner H, Aguet F, Sage D, Unser M. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc. 2013;249:13–25. doi: 10.1111/j.1365-2818.2012.03675.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, et al. Fast, robust and precise 3D localization for arbitrary point spread functions. bioRxiv. 2017:172643. [Google Scholar]
- 35.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, Zhong G, Zhuang X. Actin, Spectrin, and Associated Proteins Form a Periodic Cytoskeletal Structure in Axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsuka S, et al. Nuclear pore assembly proceeds by an inside-out extrusion of the nuclear envelope. Elife. 2016;5:1–23. doi: 10.7554/eLife.19071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deschamps J, Rowald A, Ries J. Efficient homogeneous illumination and optical sectioning for quantitative single-molecule localization microscopy. Opt Express. 2016;24:28080–28090. doi: 10.1364/OE.24.028080. [DOI] [PubMed] [Google Scholar]
- 39.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author upon reasonable request.