Abstract
Novel halogenated purines and pseudopurines with diverse aryl-substituted 1,2,3-triazoles were prepared. While p-(trifluoromethyl)-substituted 1,2,3-triazole in N-9 alkylated purine and 3-deazapurine was critical for strong albeit unselective activity on pancreatic adenocarcinoma cells CFPAC-1,1-(p-fluorophenyl)-1,2,3-triazole derivative of 7-deazapurine showed selective cytostatic effect on metastatic colon cancer cells SW620. Importantly, 1-(p-chlorophenyl)-1,2,3-triazole-tagged benzimidazole displayed the most pronounced and highly selective inhibitory effect in nM range on non-small cell lung cancer A549. This compound revealed to target molecular processes at the extracellular side and inside the plasma membrane regulated by GPLD1 and growth factor receptors PDGFR and IGF-1R leading to the inhibition of cell proliferation and induction of apoptosis mediated by p38 MAP kinase and NF-κB, respectively. Further optimisation of this compound as to reduce its toxicity in normal cells may lead to the development of novel agent effective against lung cancer.
Keywords: Purine; 1,2,3-triazole; purinomimetic; non-small cell lung cancer A549; p38 MAPK
Introduction
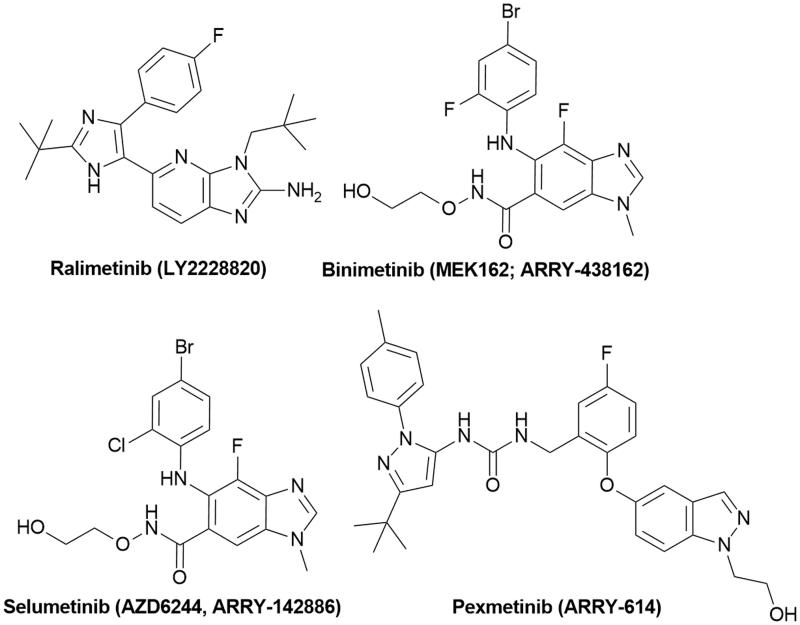
Mitogen-activated protein kinases (MAPKs) are a family of kinases of different lineages that participate in intracellular signalling during proliferation, differentiation, cellular stress responses and apoptosis 1 , 2 . Over the recent years, a large body of evidence has revealed that overexpression and activation of MAPKs play a crucial role in the development and progression of cancer indicating the promising prospect of MAPK signalling as a valuable target in tumour therapy 3 . In search of MAPK pathway inhibitors, purine and purine-like scaffolds have been recognised as an important constituent of small molecules acting as highly potent MAPK inhibitors. Among them, ralimetinib (p38 MAPK inhibitor) 4 , binimetinib 5–7 , selumetinib (MEK1/2 inhibitors) 8 , 9 and pexmetinib (Tie-2/p38 MAPK inhibitor) 10 , 11 have reached the clinical trial stage for therapeutic application as anticancer drugs (Figure 1). Prevalence of halogenated drugs and the introduction of halogen atoms in pseudopurines presented in Figure 1 shows that halogenated compounds have been widely exploited in drug discovery indicating the importance of halogens in biological activity. In support of this finding, it was demonstrated that the formation of halogen bonds has been recognised to contribute to the stability of formed protein–ligand complexes 12–14 .
Figure 1.
Roscivitine and purine isosteres as small-molecule inhibitors of CDK under clinical evaluations for the treatment of cancer.
Over the recent years, p38 MAPK, as a member of the MAPK superfamily, has emerged as an important regulator of cancer progression 15 . Increased p38 MAPK phosphorylation has been reported in human lung tumours compared with normal tissue 16 , suggesting that this pathway contributes to lung tumour progression. In addition, p38 MAPK cascade has been demonstrated to be involved in the pathogenesis of other cancer types, such as hepatocellular carcinoma (HCC) 17 . More recently, antiproliferative effect of imidazoline and N-isopropylamidine benzimidazoles on non-small cell lung cancer cells A549 was ascribed to downregulation of p38 MAPK activity 18 . Considering the aforementioned and as a continuation of our research project on development of new cytostatic agents, herein we have reported the synthesis of N-alkylated halogenated purine and purine isosteres with 1,4-disubstituted 1,2,3-triazole unit. Different contribution of diverse purine and purinomimetics, such as 6-chloropurine, 6-chloro-3-deazapurine, 6-chloro-7-deazapurine, 6-chloro-7-bromo-7-deazapurine, 5-fluoroindole, indole and benzimidazole on cytostatic effect was anticipated. Furthermore, the influence of the type and position of halogen atoms at both heterocycle and phenyl moiety on their antiproliferative activities was investigated. Finally, apoptosis induction, in silico prediction and structural analysis of biological targets and the evaluation of p38 MAPK/NF-κB signalling pathways, as potential mediators of cytostatic effects in non-small cell lung cancer cells (A549) were carried out.
Materials and methods
General
All the solvents and chemicals including starting purinomimetics (6-chloropurine, 1a, 4-chloro-1 H-imidazo[4,5-c]pyridine, 1b, 6-chloro-7-deazapurine, 1c, 7-bromo-6-chloro-7-deazapurine, 1d, 5-fluoroindole, 1e, indole, 1f and benzimidazole, 1g), as well as halogen-substituted phenyl azides (1-azido-4-fluorobenzene, 1-azido-4-chlorobenzene, 1-azido-2-fluorobenzene and 1-azido-4-(trifluoromethyl)benzene) were purchased from Aldrich (St. Louis, MO), Acros (Geel, Belgium) and Ark Pharm Inc. (Arlington Heights, IL) thin layer chromatography was performed on pre-coated Merck silica gel 60F-254 plates, while glass column slurry-packed under gravity with silica gel (Fluka, 0.063–0.2 mm) was employed for column chromatography. Melting points of compounds were determined using Kofler micro hot-stage (Reichert, Wien). One- (1D) and two-dimensional (2D) homonuclear and heteronuclear NMR spectra were recorded on a Varian Gemini 300 (300 and 75 MHz) or Varian Gemini 600 (600 and 150 MHz) as well as Agilent Technologies DD2 NMR (300 and 600 MHz) spectrometers. All data were recorded in dimethyl sulfoxide (DMSO)-d6 at 298 K. NMR chemical shifts were referenced to the residual solvent signal of DMSO at б 2.50 ppm for 1H and б 39.50 ppm for 13C. Individual resonances were assigned on the basis of their chemical shifts, signal intensities, multiplicity of resonances, H–H coupling constants and with the use of a set of 2D experiments: correlation spectroscopy (1H–1H COSY), heteronuclear single-quantum coherence (1H–13C HSQC) and heteronuclear multiple-bond correlation (1H–13C HMBC). Microwave-assisted syntheses were performed in a Milestone start S microwave oven using quartz cuvettes.
Experimental procedures for the synthesis of compounds
6-Chloro-9-(prop-2-yn-1-yl)-9H-purine (2a) 19 , 6-chloro-9-(prop-2-yn-1-yl)-9H-purine (3a) 19 , 4-chloro-7-(prop-2-yn-1-yl)-7H-pyrrolo[2,3-d]pyrimidine (2c) 20 , 5-fluoro-1-(prop-2-yn-1-yl)-1H-indole (2e) 21 , 1-(prop-2-yn-1-yl)-1H-indole (2f) 22 , 1-(prop-2-yn-1-yl)-1H-benzo[d]imidazole (2g) 21 , 1-{[1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl]met-hyl}-1H-benzo[d]imidazole (12a) 21 , 1-{[1-(4-chlorophenyl)-1H-1,2, 3-triazol-4-yl]methyl}-1H-benzo[d]imidazole (12b) 21 , 1-{[1–(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-benzo[d]imidazole (12c) 21 , 1-{[1–(2-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-benzo[d]imidazole (12d) 21 and 4-{4-[((1H-benzo[d]imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-4a,8a-dihydro-2H-chromen-2-on (12e) 21 and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one 23 were prepared according to known procedures.
General procedure for N-alkylation of compounds 2b, 3b and 2d
To a solution of the corresponding heterocyclic base in dry dimethylformamide (DMF), NaH (1.2 eq) was added and stirred for 30 min under argon atmosphere. Propargyl bromide (1.2 eq) was added and the reaction mixture was heated at 60 °C and stirred overnight. Solvent was evaporated and the residue was purified by column chromatography (CH2Cl2:CH3OH =60:1).
4-Chloro-1-(prop-2-yn-1-yl)-1 H-imidazo[4,5-c]pyridine (2 b)
Compound 2b was prepared using the above-mentioned procedure using 4-chloro-imidazo[4,5-c]pyridine (1b) (100 mg, 0.65 mmol) to obtain 2b as white powder (44.3 mg, 67%, m.p. = 101–103 °C). 1H (300 MHz, DMSO-d6): δ 8.52 (1H, s, H8), 8.21 (1H, d, J = 5.6 Hz, H2), 7.76 (1H, d, J = 5.6 Hz, H3), 5.29 (2H, d, J = 2.5 Hz, CH2), 3.60 (1H, t, J = 2.5 Hz, CCH).13 C NMR (75 MHz, DMSO-d6) δ 152.4 (C6), 145.9 (2), 141.3 (C8), 141.2 (C4), 139.8 (C5), 107.0 (C3), 77.4 (CCH), 77.1 (CCH), 34.7 (CH2).
5-Bromo-4-chloro-7-(prop-2-yn-1-yl)-7 H-pyrrolo[2,3-d]pyrimidine (2d)
Compound 2d was prepared using the above-mentioned procedure using 5-bromo-4-chloro-7H-pyrrolo[2,3-d]pyrimidine (1d) (400 mg, 1.72 mmol) to obtain 2d as white powder (348.9 mg, 70%, m.p. > 250 °C). 1H (300 MHz, DMSO-d6): δ 8.73 (1H, s, 1H, H2), 8.08 (1H, s, H8), 5.15 (2H, d, J = 2.5 Hz, CH2), 3.50 (1H, t, J = 2.5 Hz, CCH). 13C (151 MHz, DMSO-d6): δ 151.3 (C2), 150.7 (C6), 149.6 (C4), 130.6 (C8), 114.1 (C5), 86.3 (C7), 78.0 (CCH), 76.3 (CCH), 34.2 (CH2).
4-Chloro-3-(prop-2-yn-1-yl)-3 H-imidazo[4,5-c]pyridine (3b)
Compound was prepared using the above-mentioned procedure using 4-chloro-imidazo[4,5-c]pyridine (1b)(100 mg, 0.65 mmol) to obtain 3b as crude oil (24.1 mg, 36%). 1H NMR (600 MHz, DMSO-d6) δ 8.62 (1H, s, H8), 8.18 (1H, d, J = 5.5 Hz, H2), 7.74 (1H, d, J = 5.5 Hz, H3), 5.40 (2H, d, J = 2.4 Hz, CH2), 3.58 (1H, t, J = 2.5 Hz, CCH). 13C (75 MHz, DMSO-d6): δ 151.1 (C6), 148.7 (C8), 141.3 (C2), 133.2 (C4), 127.5 (C5), 115.1 (C3), 78.8 (CCH), 77.4 (CH2), 36.2 (CH2).
General procedure for the synthesis of N-1 substituted 1,2,3-triazolyl purinomimetics
The corresponding N-propargylated heterocyclic base (2a–f, 3a or 3 b) was dissolved in a mixture of t-BuOH: H2O = 1: 1 and DMF. 1 M CuSO4 (0.3 eq), Cu(0) (0.8 eq) and the corresponding terminal azide (1.2 eq) were added. The reaction mixture was stirred under microwave irradiation (300 W) at 80 °C for 45 min. The solvent was removed under reduced pressure and the residue was purified by column chromatography using CH2Cl2: CH3OH= 60: 1, as an eluent.
6-Chloro-9-{[1–(4-fluorophenyl)-1 H-1,2,3-triazol-4-yl]methyl}-9 H-purine (4a)
Compound 4a was prepared using the above-mentioned procedure using compound 2a (50 mg, 0.28 mmol) and 1-azido-4-fluorobenzene to obtain 4a as white powder (43.4 mg, 47%, m.p. = 170–171 C). 1H (600 MHz, DMSO-d6): δ 8.83 (1H, s, H5'), 8.80 (1H, s, H2), 8.79 (1H, s, H8), 7.91–7.88 (2H, m, Ph''), 7.43 (2H, t, J = 8.8 Hz, Ph''), 5.72 (2H, s, CH2). 13C (151 MHz, DMSO-d6): δ 162.6; 161.0 (d, JCF = 246.0 Hz, Ph-q''), 151.9 (C6), 151.8 (C2), 149.2 (C4), 143.1 (C4'), 133.05; 133.0 (d, J CF = 2.8 Hz, Ph-q''), 130.9 (C5), 122.6; 122.6 (d, JCF = 8.8 Hz, Ph''), 122.3 (C5'), 116.9; 116.7 (d, JCF = 23.3 Hz, Ph''), 39.0 (CH2), 38.9 (CH2). Anal. calcd. for C14H9ClFN7: C, 51.00; H, 2.75; N, 29.74. Found: C, 51.23; H, 2.56; N, 29.68.
6-Chloro-9-{[1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-9H-purine (4c)
Compound 4c was prepared using the above-mentioned procedure using compound 2a (50 mg, 0.28 mmol) and 1-azido-4-(trifluoromethyl)benzene to obtain 4c as colourless crystals (72.6 mg, 72%, m.p. = 174–177 °C). 1H (300 MHz, DMSO-d6): δ 8.95 (1H, s, H5'), 8.85 (1H, s, H2), 8.81 (1H, s, H8), 8.12 (2H, d, J = 8.5 Hz, Ph''), 7.97 (2H, d, J = 8.6 Hz, Ph''), 5.75 (2H, s, CH2). 13C (151 MHz, DMSO-d6): δ 151.9 (C6), 151.8 (C2), 149.2 (C4), 143.5 (C4'), 139.3 (Ph-q''), 130.9 (C5), 129.2; 129.0; 128.8; 128.6 (q, JCF = 32.4 Hz, CCF3), 127.3; 127.3; 127.3; 127.2 (q, JCF = 3.6 Hz, Ph''), 124.7; 122.4 (d, JCF = 272.4 Hz, CCF 3), 122.4 (C5'), 120.7 (C8), 39.0 (CH2). Anal. calcd. for C15H9ClF3N7: C, 47.44; H, 2.39; N, 25.82. Found: C, 47.38; H, 2.14; N, 26.09.
6-Chloro-9-{[1-(2-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-9H-purine (4d)
Compound 4d was prepared using the above-mentioned procedure using compound 2a (50 mg, 0.28 mmol) and 1-azido-2-fluorobenzene to obtain 4d as white powder (36.1 mg, 39%, m.p. = 143–145 °C). 1H (600 MHz, DMSO-d6): δ 8.85 (1H, s, H5'), 8.81 (1H, s, H2), 8.67 (1H, d, J = 1.8 Hz, H8), 7.80 (1H, td, J = 7.8, 1.5 Hz, Ph''), 7.64–7.50 (2 H, m, Ph''), 7.42 (1H, td, J = 8.0, 0.9 Hz, Ph''), 5.75 (2H, s, CH2). 13C (151 MHz, DMSO-d6): δ 154.7; 153.1 (d, JCF = 250.6 Hz, Ph-q''), 151.9 (C6), 151.9 (C2), 149.3 (C4), 142.6 (C4'), 131.6; 130.9 (d, JCF = 7.9 Hz, Ph''), 130.9 (C5), 126.1 (C8), 125.7; 125,7 (d, JCF = 3.8 Hz, Ph''), 125.5; 125.4 (d, JCF = 4.3 Hz, C5'), 124.7; 124.4 (d, JCF = 10.7 Hz, Ph-q''), 117.3; 117.2 (d, JCF = 19.5 Hz, Ph''), 38.9 (CH2). Anal. calcd. for C14H9ClFN7: C, 51.00; H, 2.75; N, 29.74. Found: C, 51.17; H, 2.82; N, 29.71.
4-{[4-((6-Chloro-9H-purin-9-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (4e)
Compound 4e was prepared using the above-mentioned procedure using compound 2a (50 mg, 0.28 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (73.8 mg, 0.34 mmol) to obtain 4e as yellow powder (37.4 mg, 33%, m.p. = 178–77 °C). 1H (600 MHz, DMSO-d6): δ 10.69 (1H, bs, OH''), 8.81 (1H, s, H8), 8.77 (1H, s, H2), 8.31 (1H, s, H5'), 7.65 (1H, d, J = 8.8 Hz, H6''), 6.81 (1H, dd, J = 8.8, 2.2 Hz, C7''), 6.75 (1H, s, H-8''), 5.87 (2H, s, CH2), 5.66 (2H, s, CH2), 5.55 (1H, s, H3''). 13C (75 MHz, DMSO-d6): δ 161.8 (C7''), 160.1 (C2''), 155.2 (C8a''), 154.9 (C4), 151.9 (C5), 151.8 (C2), 150.5 (C6), 149.3 (C4''), 142.4 (C4'), 126.2 (C5''), 125.1 (C5'), 113.3 (C6''), 109.5 (C4a''), 109.4 (C3''), 102.6 (C8''), 49.4 (CH2), 38.9 (CH2). Anal. calcd. for C18H12ClN7O3: C, 52.76; H, 2.95; N, 23.93. Found: C, 52.99; H, 3.06; N, 24.25.
4-Chloro-1-{[1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-imidazo[4,5-c]pyridine (5c)
Compound 5c was prepared using the above-mentioned procedure using compound 2b (25 mg, 0.13 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.31 ml, 0.16 mmol) to obtain 5c as white powder (39.5 mg, 80%, m.p. = 151–154 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.99 (1H, s, H5'), 8.63 (1H, s, H8), 8.15 (3H, m, H2; Ph''), 7.97 (2H, d, J = 8.7 Hz, Ph''), 7.79 (1H, d, J = 5.6 Hz, H3), 5.78 (2H, s, CH2).13C (75 MHz, DMSO-d6): δ 146.3 (C8), 143.4 (C4'), 141.1 (C6), 140.9 (C2), 140.0 (C4), 139.2 (C5), 128.7 (m, Ph-q''), 127.2 (q, J = 3.5 Hz, Ph''), 122.4 (C5'), 122.1 (m, CF3), 120.6 (Ph''), 106.9 (CH-3), 40.0 (CH2). Anal. calcd. for C16H10ClF3N6: C, 50.74; H, 2.66; N, 22.19. Found: C, 50.58; H, 2.34; N, 22.37.
4-Chloro-1-{[1–(2-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-imidazo[4,5-c]pyridine (5d)
Compound 5d was prepared using the above-mentioned procedure using compound 2b (25 mg, 0.13 mmol) and 1-azido-2-fluorobenzene (0.31 ml, 0.16 mmol) to obtain 5d as white crystals (30.9 mg, 72%, m.p. = 124–127 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.72 (1H, d, J = 2.2 Hz, H5'), 8.63 (1H, s, H8), 8.17 (1H, d, J = 5.6 Hz, H2), 7.85–7.77 (2H, m, H3; Ph''), 7.65–7.50 (2H, m, Ph''), 7.46–7.38 (1H, m, Ph''), 5.77 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 155.5; 152.1 (d, JCF = 250.4 Hz, Ph-q''), 146.4 (C8), 142.6 (C4'), 141.0 (C2), 140.1 (C6), 137.2 (C4), 136.3 (C5), 131.6; 131.5 (d, JCF = 8.0 Hz, Ph''), 126.0 (C5'), 125.7; 125.6 (d, JCF = 3.8 Hz, Ph''), 125.5; 125.5 (d, JCF = 4.7 Hz, Ph''), 117.6; 117.4 (d, JCF = 15.7 Hz, Ph-q''), 117.6 (d, J = 19.5 Hz, Ph''), 107.1 (C3), 38.9 (CH2). Anal. calcd. for C15H10ClFN6: C, 54.80; H, 3.07; N, 25.46. Found: C, 54.63; H, 3.28; N, 25.57.
4-{[4-((4-Chloro-1H-imidazo[4,5-c]pyridin-1-yl)methyl-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (5e)
Compound 5e was prepared using the above-mentioned procedure using compound 2b (25 mg, 0.13 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (34.7 mg, 0.16 mmol) to obtain 5e as yellow powder (53.1 mg, 60%, m.p. = 228–231 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.58 (1H, s, H8), 8.31 (1H, s, H5'), 8.14 (1H, d, J = 5.6 Hz, H2), 7.70 (1H, d, J = 5.6 Hz, H3), 7.61 (1H, d, J = 8.7 Hz, H5''), 6.78 (1H, dd, J = 8.8, 1.8 Hz, H6''), 6.73 (1H, d, J = 1.6 Hz, H8''), 5.86 (2H, s, CH2), 5.69 (2H, s, CH2), 5.55 (1H, s, H3'')0.13 C NMR (151 MHz, DMSO-d6) δ 161.8 (C7''), 159.9 (C2''), 155.1 (C8a''), 150.2 (C4''), 146.3 (C8), 142.4 (C4'), 141.0 (C6), 140.8 (C2), 134.0 (C4), 137.1 (C5), 126.0 (C5''), 124.9 (5'), 113.2 (C6''), 109.4 (C3''), 109.2 (C4a''), 106.9 (C3), 102.5 (C8''), 49.3 (CH2), 40.1 (CH2). Anal. calcd. for C15H10ClFN6: C, 55.82; H, 3.20; N, 20.56. Found: C, 55.96; H, 3.22; N, 20.78.
4-{[4-((6-Chloro-7H-purin-7-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (6e)
Compound 6e was prepared using the above-mentioned procedure using compound 3a (20 mg, 0.11 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (29.3 mg, 0.14 mmol) to obtain 6e as white powder (14.9 mg, 33%, m.p. > 250 °C). 1H (600 MHz, DMSO-d6): 8.96 (1H, s, H8), 8.82 (1H, s, H2), 8.30 (1H, s, H5'), 7.63 (1H, d, J = 8.8 Hz, H5''), 6.80 (1H, dd, J = 8.8, 2.3 Hz, H6''), 6.76 (1H, d, J = 2.3 Hz, H8''), 5.87 (2H, s, CH2''), 5.85 (2H, s, CH2), 5.55 (1H, s, H3''). 13C (150 MHz, DMSO-d6): 161.8 (C7''), 161.6 (C4), 160.1 (C2''), 155.1 (C8a''), 151.8 (C2), 151.1 (C8), 150.3 (C4''), 143.3 (C4'), 126.2 (C5''), 124.4 (C5'), 122.1 (C5), 113.2 (C6''), 109.5 (C4a''), 109.2 (C3''), 102.6 (C8''), 49.3 (CH2''), 42.0 (CH2). Anal. calcd. for C18H12ClN7O3: C, 52.76; H, 2.95; N, 23.93. Found: C, 52.84; H, 3.75; N, 23.90.
4-Chloro-3-{[1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-3H-imidazo[4,5-c]pyridine (7c)
Compound 7c was prepared using the above-mentioned procedure using compound 3b (20 mg, 0.10 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.24 ml, 0.12 mmol) to obtain 7c as white powder (30.1 mg, 79%, m.p. = 183–185 °C). 1H (300 MHz, DMSO-d6): δ 8.98 (1H, s, H5'), 8.76 (s, 1H, H8), 8.16 (1H, d, 1H, J = 5.4 Hz, H2), 8.14 (2H, m, Ph''), 7.97 (2H, m, Ph), 7.76 (1H, d, J = 5.4 Hz, H3), 5.97 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 151.0 (C4), 149.3 (C8), 144.8 (C4'), 140.7 (C2), 139.2 (Ph-q''), 132.9 (C6), 128.7 (q, JCF = 32.6 Hz, CCF3''), 127.2 (C5), 127.1 (q, JCF = 3.8 Hz, Ph''), 122.0 (m, CCF 3), 121.8 (C5'), 120.6 (Ph''), 114.9 (C3), 41.3 (CH2). Anal. calcd. for C16H10ClF3N6: C, 50.74; H, 2.66; N, 22.19. Found: C, 50.76; H, 2.58; N, 22.17.
4-{[4-((4-Chloro-3H-imidazo[4,5-c]pyridin-3-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (7e)
Compound 7e was prepared using the above-mentioned procedure using compound 3b (15 mg, 0.08 mmol) and 4-(azidomethyl)-7-hydroxy-2 H-chromen-2-one (19.5 mg, 0.09 mmol) to obtain 7e as white powder (23.8 mg, 74%, m.p. > 250 °C). 1H (300 MHz, DMSO-d6): 8.73 (1H, s, H8), 8.25 (1H, s, H5'), 8.15 (1H, d, J = 5.4 Hz, H2), 7.74 (1H, d, J = 5.4 Hz, H3), 7.63 (1H, d, J = 8.8 Hz, H5''), 6.79 (1H, dd, J = 8.8, 2.1 Hz, H6''), 6.74 (1H, d, J = 2.1 Hz, H8''), 5.88 (2H, s, CH2''), 5.86 (2H, s, CH2), 5.51 (1H, s, H3''). 13C (75 MHz, DMSO-d6): 161.7 (C7''), 159.9 (C2''), 155.1 (C8a''), 150.4 (C4), 150.2 (C4''), 143.8 (C4'), 140.7 (C2), 133.2 (C6), 127.5 (C5), 126.0 (C5''), 124.2 (C5'), 114.9 (C3), 113.2 (C6''), 109.1 (C4a''), 109.1 (C3''), 102.5 (C8''), 49.2 (CH2''), 41.5 (CH2). Anal. calcd. for C15H10ClFN6: C, 55.82; H, 3.20; N, 20.56. Found: C, 55.72; H, 3.16; N, 20.85.
4-Chloro-7-{[1-(4-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-7H-pyrrolo[2,3-d]pyrimidine (8a)
Compound 8a was prepared using the above-mentioned procedure using compound 2c (100 mg, 0.56 mmol) and 1-azido-4-fluorobenzene (1.34 ml, 0.67 mmol) to obtain 8a as white powder (131.6 mg, 74%, m.p. = 176–178 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.74 (1H, s, H5'), 8.68 (1H, s, H2), 7.93–7.84 (3H, m, H6; Ph''), 7.43 (2H, t, J = 8.8 Hz, Ph''), 6.71 (1H, d, J = 3.6 Hz, H5), 5.68 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 163.4; 160.1 (d, JCF = 245.8 Hz, Ph-q'') 152.1 (C4), 150.8 (C7a), 150.6 (C2), 143.9 (C4'), 133.1; 133.1 (d, JCF = 2.8 Hz, Ph-q''), 131.4 (C6), 122.7; 122.6 (d, JCF = 8.9 Hz, Ph''), 122.2 (C5'), 117.0; 116.6 (d, JCF = 23.3 Hz, Ph''), 113.1 (C4a), 99.1 (C5), 39.6 (CH2). Anal. calcd. for C15H10ClFN6: C, 54.80; H, 3.07; N, 25.56. Found: C, 54.98; H, 2.94; N, 25.36.
4-Chloro-7-{[1-(4-chlorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-7H-pyrrolo[2,3-d]pyrimidine (8b)
Compound 8b was prepared using the above-mentioned procedure using compound 2c (100 mg, 0.56 mmol) and 1-azido-4-chlorobenzene (1.34 ml, 0.67 mmol) to obtain 8b as white powder (107.8 mg, 56%, m.p. = 221–223 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.79 (1H, s, H5'), 8.68 (1H, s, H2), 7.90 (2H, d, J = 8.9 Hz, Ph''), 7.86 (1H, d, J = 3.6 Hz, H6), 7.65 (2H, d, J = 8.9 Hz, Ph''), 6.71 (1H, d, J = 3.6 Hz, H5), 5.68 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 150.7 (C4), 150.5 (C7a), 150.5 (C2), 144.0 (C4'), 135.3 (Ph-q''), 133.0 (Ph-q''), 131.3 (C6), 129.8 (Ph''), 121.9 (C5'), 121.8 (Ph''), 116.9 (C4a), 99.0 (C5), CH2 in DMSO. Anal. calcd. for C15H10Cl2N6: C, 52.19; H, 2.92; N, 24.35. Found: C, 52.12; H, 2.94; N, 24.29.
4-Chloro-7-{[1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-7H-pyrrolo[2,3-d]pyrimidine (8c)
Compound 8c was prepared using the above-mentioned procedure using compound 2c (50 mg, 0.28 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.67 ml, 0.34 mmol) to obtain 8c as white powder (84.2 mg, 80%, m.p. = 202–204 °C). 1H (300 MHz, DMSO-d6): δ 8.91 (1H, s, C5'), 8.68 (1H, s, H2), 8.13 (2H, d, J = 8.5 Hz, Ph''), 7.96 (2H, d, J = 8.6 Hz, Ph''), 7.87 (1H, d, J = 3.6 Hz, H6), 6.72 (1H, d, J = 3.6 Hz, H5), 5.71 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 150.9 (C4), 150.7 (C7a), 150.7 (C2), 144.4 (C4'), 139.4 (Ph-q''), 131.5 (C6), 129.3; 128.8 (d, JCF = 32.3 Hz, CCF3), 127.4; 127.4; 127.3; 127.3 (q, JCF = 3.6 Hz, Ph''), 122.8; 122.1 (d, JCF = 272.3 Hz, CCF 3), 122.3 (C5'), 120.9 (Ph''), 117.1 (C4a), 99.3 (C5), 39.6 (CH2). Anal. calcd. for C16H10ClF3N6: C, 54.74; H, 2.66; N, 22.19. Found: C, 54.54; H, 2.73; N, 22.00.
4-Chloro-7-{[1-(2-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-7H-pyrrolo[2,3-d]pyrimidine (8d)
Compound 8d was prepared using the above-mentioned procedure using compound 2c (100 mg, 0.56 mmol) and 1-azido-2-fluorobenzene (1.34 ml, 0.67 mmol) to obtain 8d as white crystals (53.3 mg, 29%, m.p. = 112–114 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.68 (1H, s, H2), 8.61 (1H, d, J = 2.0 Hz, H5'), 7.88 (1H, d, J = 3.6 Hz, H6), 7.84–7.75 (1H, m, Ph''), 7.64–7.52 (2H, m, Ph''), 7.45–7.38 (1H, m, Ph''), 6.71 (1H, d, J = 3.6 Hz, H5), 5.70 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 163.9; 160.3 (d, JCF = 246.0 Hz, Ph-q''), 152.1 (C4), 150.4 (C2), 150.1 (C7a), 143.2 (C4'), 132.1; 131.9 (d, JCF = 19.5 Hz, Ph-q''), 131.4 (Ph''), 131.3 (C6), 127.6 (C5'), 125.9 (Ph''), 125.5; 125.5 (d, JCF = 3.8 Hz, Ph''), 117.2; 116.9 (d, JCF = 19.6 Hz, Ph''), 117.0 (C4a), 99.0 (C5), 39.3 (CH2). Anal. calcd. for C15H10ClFN6: C, 54.80; H, 3.07; N, 25.56. Found: C, 54.81; H, 3.25; N, 25.69.
4-{[4-((4-Chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (8e)
Compound 8e was prepared using the above-mentioned procedure using compound 2c (50 mg, 0.28 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (274.0 mg, 0.34 mmol) in DMF (0.5 ml) to obtain 8e as white powder (15.1 mg, 40%, m.p. = 202–204 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.65 (1H, s, H2), 8.22 (1H, s, H5'), 7.84 (1H, d, J = 3.6 Hz, H6), 7.63 (1H, d, J = 8.8 Hz, H5''), 6.78 (1H, dd, J = 9.0, 1.7 Hz, H6''), 6.71 (1H, d, J = 1.8 Hz, H8''), 6.69 (1H, d, J = 3.6 Hz, H5), 5.84 (2H, s, CH2), 5.62 (2H, s, CH2), 5.49 (1H, s, H3''). 13C NMR (75 MHz, DMSO-d6) δ 161.8 (C7''), 160.2 (C2''), 156.6 (C4), 155.2 (C8a''), 150.7 (C2), 150.6 (C4''), 145.0 (C7a), 143.4 (C4'), 131.6 (C6), 126.2 (C5''), 124.9 (C5'), 117.0 (C4a), 113.4 (C6''), 109.5 (C4a''), 109.4 (C3''), 102.7 (C8''), 99.1 (C5), 49.4 (CH2). Anal. calcd. for C19H13ClN6O3: C, 55.82; H, 3.20; N, 25.56. Found: C, 55.53; H, 3.16; N, 25.70.
5-Bromo-4-chloro-7-{[1-(4-(trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-7H-pyrrolo[2,3-d]pyrimidine (9c)
Compound 9c was prepared using the above-mentioned procedure using compound 2d (50 mg, 0.18 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.43 ml, 0.22 mmol) to obtain 9c as white powder (21.2 mg, 26%, m.p. = 220–221 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.91 (1H, s, H5'), 8.72 (1H, s, H2), 8.16–8.10 (3H, m, H6; Ph''), 7.97 (2H, d, J = 8.7 Hz, Ph''), 5.69 (2H, s, CH2). 13C (75 MHz, DMSO-d6): δ 151.3 (C2), 150.1 (C7a), 147.6 (C4), 144.0 (C4'), 139.4 (Ph-q''), 131.3 (C6), 128.8 (Ph-q''), 127.4; 127.4; 127.3; 127.3 (q, JCF = 3.8 Hz, Ph''), 125.7; 122.2 (d, JCF = 281.9 Hz, CCF 3), 122.4 (C5'), 120.8 (Ph''), 119.1 (C4a), 86.3 (C5), 38.9 (CH2). Anal. calcd. for C16H9BrClF3N6: C, 41.99; H, 1.98; N, 18.36. Found: C, 42.11; H, 1.92; N, 18.07.
5-Bromo-4-chloro-7-{[1–(2-fluorophenyl)-1 H-1,2,3-triazol-4-yl]methyl}-7 H-pyrrolo[2,3-d]pyrimidine (9d)
Compound 9d was prepared using the above-mentioned procedure using compound 2d (50 mg, 0.18 mmol) and 1-azido-2-fluorobenzene (0.43 ml, 0.22 mmol) to obtain 9d as white powder (18.4 mg, 25%, m.p. = 145–147 °C). 1H NMR (600 MHz, DMSO-d6) δ 8.72 (1 H, s, H2), 8.62 (1 H, d, J = 1.4 Hz, H5'), 8.12 (1 H, s, H6), 7.82–7.78 (1 H, m, Ph''), 7.63–7.53 (2 H, m, Ph''), 7.42 (1 H, t, J = 7.6 Hz, Ph''), 5.69 (2 H, s, CH2). 13 C NMR (151 MHz, DMSO-d6) δ 162.2; 160.4 (d, JCF = 265.4 Hz, Ph-q''), 151.3 (C2), 150.7 (C4), 150.0 (C7a), 143.0 (C4'), 131.6; 131.6 (d, JCF = 8.0 Hz, Ph''), 131.3 (C6), 126.1 (C5'), 125.7; 125.7 (d, JCF = 3.6 Hz, Ph''), 125.5; 125.4 (d, JCF = 4.4 Hz, Ph''), 124.7; 124.6 (d, JCF = 10.9 Hz, Ph-q''), 117.3; 117.2 (d, JCF = 19.6 Hz, Ph''), 114.2 (C4a), 86.3 (C5), 40.0 (CH2). Anal. calcd. for C15H9BrClFN6: C, 44.20; H, 2.22; N, 20.62. Found: C, 43.99; H, 2.25; N, 20.48.
4-{[4-((5-Bromo-4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (9e)
Compound 9e was prepared using the above-mentioned procedure using compound 2d (50 mg, 0.18 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (47.7 mg, 0.22 mmol) to obtain 9e as yellow powder (30.7 mg, 35%, m.p. > 250 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.69 (1H, s, H2), 8.25 (1H, s, H5'), 8.09 (1H, s, H6), 7.65 (1H, d, J = 8.7 Hz, H5''), 6.81 (1H, dd, J = 8.7, 2.4 Hz, H6''), 6.75 (1H, d, J = 2.3 Hz, H8''), 5.85 (2H, s, CH2), 5.60 (2H, s, CH2), 5.55 (1H, s, H3''). 13C NMR (75 MHz, DMSO-d6) δ 161.7 (C7''), 160.0 (C2''), 155.1 (C8a''), 150.4 (C4''), 149.2 (C2), 146.1 (C4), 144.1 (C7a), 142.7 (C4'), 131.3 (C6), 126.1 (C5''), 124.9 (C5'), 114.1 (C4a), 113.3 (C6''), 109.6 (C4a''), 109.4 (C3''), 102.6 (C8''), 93.7 (C5), 49.3 (CH2), 40.0 (CH2). Anal. calcd. for C19H12BrClN6O3: C, 46.79; H, 2.48; N, 17.23. Found: C, 46.79; H, 2.57; N, 17.05.
1-{[1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-1H-indole (10a)
Compound 10a was prepared using the above-mentioned procedure using 2e (50 mg, 0.29 mmol) and 1-azido-4-fluorobenzene (0.70 ml, 0.35 mmol) to obtain 10a as yellow oil (19.1 mg, 21%). 1H NMR (300 MHz, DMSO-d6) δ 8.77 (1H, s, H5'), 7.94–7.85 (2H, m, Ph), 7.65–7.60 (1H, m, H7), 7.56 (1H, d, J = 3.2 Hz, H2), 7.43 (2H, t, J = 8.8 Hz, Ph''), 7.31 (1H, dd, J = 9.9, 2.5 Hz, H4), 6.99 (1H, td, J = 9.3, 2.5 Hz, H6), 6.46 (1H, d, J = 3.1 Hz, H3), 5.55 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 163.4; 160.1 (d, JCF = 246.0 Hz, Ph-q''), 155.7 (C5), 144.6 (C4'), 135.5 (Ph-q''), 132.4 (C7a), 130.6 (C2), 124.3 (C3a), 122.7; 122.5 (d, JCF = 8.8 Hz, Ph''), 122.0 (C5'), 116.9, 116.6 (d, JCF = 23.3 Hz, Ph''), 111.3; 111.1 (d, JCF = 9.8 Hz, C7), 109.5; 109.2 (d, JCF = 26.2 Hz, C6), 105.2; 104.9 (d, JCF = 23.2 Hz, C4), 101.3; 101.3 (d, JCF = 4.5 Hz, C3), 41.0 (CH2). Anal. calcd. for C17H12F2N4: C, 65.80; H, 3.90; N, 18.05. Found: C, 65.78; H, 4.26; N, 17.86.
1-{[(1–(4-Chlorophenyl)-1 H-1,2,3-triazol-4-yl]methyl}-5-fluoro-1 H-indole (10b)
Compound 10b was prepared using the above-mentioned procedure using 2e (50 mg, 0.29 mmol) to obtain 10b as white powder (33.3 mg, 35%, m.p. = 117–120 °C). 1 H NMR (300 MHz, DMSO-d6) δ 8.82 (1H, s, H5'), 7.90 (2H, d, J = 9.0 Hz, Ph''), 7.65 (2H, d, J = 9.0 Hz, Ph''), 7.61 (1H, d, J = 4.6 Hz, H7), 7.56 (1H, d, J = 3.1 Hz, H2), 7.31 (1H, dd, J = 9.9, 2.5 Hz, H4), 6.99 (1H, td, J = 9.3, 2.5 Hz, H6), 6.46 (1H, dd, J = 3.1, 0.7 Hz, H3), 5.55 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 158.7 (C5), 144.6 (C4'), 135.3 (Ph-q''), 133.0 (C7a), 132.3 (Ph-q''), 130.6 (C2), 129.9 (C5'), 129.8 (Ph''), 126.2 (C3a), 121.8 (Ph''), 111.2; 111.1 (d, JCF = 9.9 Hz, C7), 109.5; 109.1 (d, JCF = 26.1 Hz, C6), 105.2; 104.9 (d, JCF = 23.3 Hz, C4), 101.2; 101.2 (d, JCF = 4.7 Hz, C3), 40.9 (CH2). Anal. calcd. for C17H12ClFN4: C, 62.48; H, 3.70; N, 17.15. Found: C, 62.33; H, 3.47; N, 17.01.
5-Fluoro-1-{[1–(4-(trifluoromethyl)phenyl)-1 H-1,2,3-triazol-4-yl]methyl}-1 H-indole (10c)
Compound 10c was prepared using the above-mentioned procedure using 2e (50 mg, 0.29 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.70 ml, 0.35 mmol) to obtain 10c as white powder (73.2 mg, 70%, m.p. = 151–154 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.95 (1H, s, H5'), 8.12 (2H, d, J = 8.4 Hz, Ph''), 7.96 (2H, d, J = 8.6 Hz, Ph''), 7.63 (1H, dd, J = 8.9, 4.6 Hz, H7), 7.57 (1H, d, J = 3.1 Hz, H2), 7.31 (1H, dd, J = 9.9, 2.5 Hz, H4), 6.99 (1H, td, J = 9.2, 2.5 Hz, H6), 6.47 (1H, d, J = 3.1 Hz, H3), 5.58 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 158.7 (C5), 144.9 (C4'), 139.3 (Ph-q''), 132.3 (C7a), 130.6 (C2), 128.9; 128.5 (d, JCF = 32.5 Hz, CCF3), 128.6 (C3a) 127.3; 127.2; 127.2; 127.1 (q, JCF = 3.7 Hz, Ph''), 125.3; 121.6 (d, JCF = 285.4 Hz, CCF 3), 122.0 (C5'), 120.6 (Ph''), 111.2; 111.1 (d, JCF = 9.8 Hz, C7), 109.5; 109.1 (d, JCF = 26.1 Hz, C6), 105.2; 104.9 (d, JCF = 23.2 Hz, C4), 101.3; 101.2 (d, JCF = 4.7 Hz, C3), 40.9 (CH2). Anal. calcd. for C18H12F4N4: C, 60.00; H, 3.36; N, 15.55. Found: C, 59.83; H, 3.26; N, 15.78.
5-Fluoro-1-{[1-(2-fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-indole (10d)
Compound 10d was prepared using the above-mentioned procedure using 2e (50 mg, 0.29 mmol) and 1-azido-2-fluorobenzene (0.70 ml, 0.35 mmol) in DMF (0.5 ml) and t-BuOH: H2O = 1:1 (4 ml) to obtain 10d as yellow oil (19 mg, 21%). 1H NMR (300 MHz, DMSO-d6) δ 8.61 (1H, d, J = 2.2 Hz, H5'), 7.84–7.77 (1H, m, Ph''), 7.66 (1H, dd, J = 9.0, 4.4 Hz, H7), 7.58 (2H, d, J = 3.1 Hz, H2; Ph''), 7.40 (1H, dd, J = 6.9, 2.5 Hz, Ph''), 7.31 (1H, dd, J = 10.0, 2.5 Hz, H4), 7.03–6.94 (2H, m, Ph''; H6), 6.46 (1H, d, J = 3.1 Hz, H3), 5.57 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 158.3 (C5), 155.6; 152.0 (d, JCF = 271.7 Hz, Ph-q''), 146.7 (C4'), 132.3 (C7a), 131.3; 131.2 (d, JCF = 8.0 Hz, Ph''), 130.5 (C2), 125.8 (C5'), 125.5; 125.5 (d, JCF = 3.7 Hz, Ph''), 125.1 (Ph''), 124.5; 124.5 (d, JCF = 4.7 Hz, Ph-q''), 117.2; 116.9 (d, JCF = 19.4 Hz, Ph''), 111.2; 111.1 (d, JCF = 8.8 Hz, C7), 109.4; 109.1 (d, JCF = 26.4 Hz, C6), 105.1; 104.8 (d, JCF = 23.3 Hz, C4), 101.2; 101.1 (d, JCF = 4.5 Hz, C3), 40.7 (CH2). Anal. calcd. for C17H12F2N4: C, 65.80; H, 3.90; N, 18.05. Found: C, 65.75; H, 3.93; N, 18.16.
4-{[4-((5-Fluoro-1H-indol-1-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2H-chromen-2-one (10e)
Compound 10e was prepared using the above-mentioned procedure using compound 2e (50 mg, 0.29 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (74.3 mg, 0.35 mmol) to obtain 10e as white powder (67.3 mg, 59%, m.p. = 181–184 °C). 1H NMR (300 MHz, DMSO-d6) δ 10.69 (1H, s, OH''), 8.17 (1H, s, H5'), 7.64 (1H, d, J = 8.7 Hz, H5''), 7.60–7.54 (1H, m, H7), 7.53 (1H, d, J = 3.2 Hz, H2), 7.31 (1H, dd, J = 9.9, 2.5 Hz, H4), 6.97 (1H, td, J = 9.2, 2.5 Hz, H6), 6.81 (1H, dd, J = 8.7, 2.4 Hz, H6''), 6.76 (1H, d, J = 2.3 Hz, H8''), 6.45 (1H, d, J = 2.5 Hz, H3), 5.86 (2 H, s, CH2), 5.54 (2H, s, CH2), 5.51 (1H, s, H3''). 13C NMR (75 MHz, DMSO-d6) δ 161.6 (C7''), 159.9 (C2''), 158.7; 155.6 (d, JCF = 231.7 Hz, C5), 155.1 (C8a''), 150.5 (C4''), 144.1 (C4'), 132.3 (C7a), 130.6 (C2), 128.6; 128.4 (d, JCF = 10.2 Hz, C3a), 126.0 (C5''), 124.4 (C5'), 113.2 (C6''), 111.2; 111.0 (d, JCF = 10.0 Hz, C7), 109.4; 109.1 (d, JCF = 25.5 Hz, C6), 109.3 (C4a''), 109.3 (C3''), 105.2; 104.9 (d, JCF = 23.2 Hz, C4), 102.5 (C8''), 101.1; 101.1 (d, JCF = 4.5 Hz, C3), 49.1 (CH2), 41.0 (CH2). Anal. calcd. for C21H15FN4O3: C, 64.61; H, 3.87; N, 14.35. Found: C, 64.43; H, 3.83; N, 14.54.
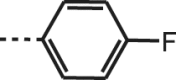
1-{[1-(4-Fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-indole (11a)
Compound 11a was prepared using the above-mentioned procedure using 2f (70 mg, 0.45 mmol) and 1-azido-4-fluorobenzene (0.81 ml, 0.66 mmol) to obtain 11a as white powder (88.5 mg, 67%, m.p. = 141–144 °C). 1H NMR (600 MHz, DMSO-d6) δ 8.77 (1H, s, H5'), 7.90 (2H, dd, J = 9.1, 4.7 Hz, Ph''), 7.62 (1H, d, J = 8.0 Hz, H7), 7.54 (1H, d, J = 7.9 Hz, H4), 7.48 (1H, d, J = 3.1 Hz, H2), 7.42 (2H, t, J = 8.8 Hz, Ph''), 7.16–7.12 (1H, m, H5), 7.040–7.01 (1H, m, H6), 6.47 (1H, dd, J = 3.3, 0.5 Hz, H3), 5.54 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 163.2; 160.0 (d, J CF = 245.7 Hz, Ph-q''), 144.6 (C4'), 135.5 (C7a), 133.1; 133.0 (d, J CF = 2.9 Hz, Ph-q''), 128.6 (C2), 128.2 (C3a), 122.5; 122.4 (d, J CF = 8.8 Hz, Ph''), 121.9 (C5'), 121.1 (C6), 120.4 (C4), 119.2 (C5), 116.8; 116.5 (d, JCF = 23.2 Hz. Ph''), 110.1 (C7), 101.1 (C3), 40.6 (CH2). Anal. calcd. for C17H13FN4: C, 69.85; H, 4.48; N, 19.17. Found: C, 70.13; H, 4.39; N, 19.11.
1-{[1-(4-Chlorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-indole (11b)
Compound 11b was prepared using the above-mentioned procedure using 2f (70 mg, 0.45 mmol) and 1-azido-4-chlorobenzene to obtain 11b as white powder (83.2 mg, 60%, m.p. = 168–171 °C). 1H NMR (300 MHz, DMSO-d6) δ 8.83 (1H, s, H5'), 7.90 (2H, d, J = 8.9 Hz, Ph''), 7.71–7.59 (3H, m, Ph''; H7), 7.54 (1H, d, J = 7.8 Hz, H4), 7.48 (1H, d, J = 3.1 Hz, H2), 7.14 (1H, t, J = 7.5 Hz, H5), 7.02 (1H, t, J = 7.4 Hz, H6), 6.47 (1H, d, J = 3.0 Hz, H3), 5.55 (2H, s, CH2). 13C NMR (151 MHz, DMSO-d6) δ 144.8 (C4'), 135.5 (C7a), 135.3 (Ph-q''), 132.9 (Ph-q''), 129.8 (Ph''), 128.6 (C2), 128.2 (C3a), 121.8 (Ph''), 121.7 (C5'), 121.1 (C6), 120.4 (C4), 119.1 (C5), 110.0 (C7), 101.1 (C3), 40.6 (CH2). Anal. calcd. for C17H13ClN4: C, 66.13; H, 4.24; N, 18.14. Found: C, 66.32; H, 4.32; N, 18.31.
1-{[1-(4-(Trifluoromethyl)phenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-indole (11c)
Compound 11c was prepared using the above-mentioned procedure using 2f (70 mg, 0.45 mmol) and 1-azido-4-(trifluoromethyl)benzene (0.81 ml, 0.66 mmol) to obtain 11c as white powder (110.9 mg, 72%, m.p. = 185–187 °C). 1H NMR (600 MHz, DMSO-d6) δ 8.95 (1H, s, H5'), 8.12 (2H, d, J = 8.5 Hz, Ph''), 7.96 (2H, d, J = 8.6 Hz, Ph''), 7.63 (1H, d, J = 8.4 Hz, H7), 7.55 (1H, d, J = 7.9 Hz, H4), 7.49 (1H, d, J = 3.2 Hz, H2), 7.17–7.12 (1H, m, H5), 7.04–7.01 (1H, m, H6), 6.47 (1H, dd, J = 3.1, 0.6 Hz, H3), 5.57 (2H, s, CH2). 13C NMR (75 MHz, DMSO-d6) δ 145.0 (C4'), 139.3 (Ph-q''), 135.5 (C7a), 129.2; 128.9; 128.4; 128.0 (q, JCF = 32.4 Hz, CCF3) 128.7 (C2), 128.3 (C3a), 127.2; 127.2; 127.1; 127.1 (q, JCF = 3.8 Hz, Ph''), 125.6; 122.0 (q, JCF = 272.0 Hz, CCF 3), 121.9 (C5'), 121.2 (C6), 120.5 (Ph''), 120.4 (C4), 119.2 (C5), 110.0 (C7), 101.1 (C3), 40.6 (CH2). Anal. calcd. for C18H13F3N4: C, 63.16; H, 3.83; N, 16.37. Found: C, 62.96; H, 4.00; N, 16.30.
1-{[1-(2-Fluorophenyl)-1H-1,2,3-triazol-4-yl]methyl}-1H-indole (11d)
Compound 11d was prepared using the above-mentioned procedure using 2f (70 mg, 0.45 mmol) and 1-azido-2-fluorobenzene (0.81 ml, 0.66 mmol) to obtain 11d as yellow oil (37.4 mg, 28%). 1H NMR (600 MHz, DMSO-d6) δ 8.60 (1H, d, J = 1.8 Hz, H5'), 7.80 (1H, td, J = 7.8, 1.4 Hz, Ph''), 7.65 (1H, d, J = 8.3 Hz, H7), 7.61–7.57 (1 H, m, Ph''), 7.56–7.52 (2H, m, Ph''; H4), 7.50 (1H, d, J = 3.1 Hz, H2), 7.41 (1H, t, J = 7.7 Hz, Ph''), 7.14 (1H, t, J = 7.6 Hz, H5), 7.03 (1H, dt, J = 7.8, 3.9 Hz, H6), 6.46 (1H, d, J = 3.1 Hz, H3), 5.57 (2H, s, CH2). 13C NMR (151 MHz, DMSO-d6) δ 154.5; 152.8 (d, JCF = 250.2 Hz, Ph-q''), 144.1 (C4'), 135.5 (C7a), 131.3; 131.2 (d, JCF = 8.0 Hz, Ph-q''), 128.6 (C2), 128.2 (C3a), 125.8 (C5'), 125.5; 125.5 (d, JCF = 3.7 Hz, Ph''), 124.9; 124.8 (d, JCF = 4.6 Hz, Ph''), 124.7; 124.6 (d, JCF = 10.9 Hz, Ph''), 121.1 (C6), 120.4 (C4), 119.2 (C5), 117.1; 117.0 (d, JCF = 19.5 Hz, Ph''), 110.1 (C7), 101.1 (C3), 40.5 (CH2). Anal. calcd. for C17H13FN4: C, 69.85; H, 4.48; N, 19.17. Found: C, 69.92; H, 4.55; N, 19.17.
4-{[4-((1H-Indol-1-yl)methyl)-1H-1,2,3-triazol-1-yl]methyl}-7-hydroxy-2 H-chromen-2-one (11e)
Compound 11e was prepared using the above-mentioned procedure using compound 2e (70 mg, 0.45 mmol) and 4-(azidomethyl)-7-hydroxy-2H-chromen-2-one (87.9 mg, 0.66 mmol) to obtain 11e as yellow powder (78.8 mg, 47%, m.p. = 155–158 °C). 1H NMR (300 MHz, DMSO-d6) δ 10.69 (1H, s, OH''), 8.15 (1H, s, H7), 7.63 (1H, d, J = 8.7 Hz, H5''), 7.53 (2H, d, J = 8.9 Hz, H4; H5'), 7.44 (1H, d, J = 3.2 Hz, H2), 7.15–7.08 (1H, m, H5), 7.04–6.97 (1H, m, H6), 6.80 (1H, dd, J = 8.7, 2.3 Hz, H6''), 6.75 (1H, d, J = 2.3 Hz, H8''), 6.44 (1H, d, J = 3.1 Hz, H3), 5.84 (2H, s, CH2), 5.53 (1H, s, H3''), 5.50 (2H, s, CH2). 13C NMR (151 MHz, DMSO-d6) δ 161.6 (C7''), 159.8 (C2''), 155.1 (C8a''), 150.4 (C4''), 144.2 (C4'), 135.5 (C7a), 128.6 (C2), 128.2 (C3a), 126.0 (C5''), 124.3 (C5'), 121.1 (C6), 120.4 (C4), 119.1 (C5), 113.1 (C6''), 110.0 (C7), 109.3 (C4a''), 109.2 (C3''), 102.5 (C8''), 101.0 (C3), 49.1 (CH2), 40.8 (CH2). Anal. calcd. for C21H16N4O3: C, 67.73; H, 4.33; N, 15.04. Found: C, 67.62; H, 4.21; N, 14.92.
Cell culturing
Human carcinoma cell lines A549 (lung carcinoma), HeLa (cervical carcinoma), SW620 (colorectal adenocarcinoma, metastatic), and CFPAC-1 (pancreatic cancer, derived from metastatic: liver), normal human lung (WI38) and foreskin (HFF-1) fibroblasts were obtained from the American Type Culture Collection (ATCC). Cells were cultured in humidified atmosphere at 37 °C with 5% CO2. As growth medium, Dulbecco’s modified Eagle medium (DMEM) was used with the addition of foetal bovine serum (10%), L-glutamine (2 mM) and antibiotics: streptomycin (100 µg/ml) and penicillin (100 U/ml).
Proliferation assay
Cells were seeded onto 96-well microtiter plates at a seeding density of 3000 cells/well for carcinoma cell lines, and 5000 cells/well for normal human fibroblasts. The next day, cells were treated with test agents in five different concentrations (0.01–100 µM) and further incubated for 72 h. DMSO (solvent) was tested for potential cytotoxic effect but it did not exceed 0.1%. Following 72 h incubation, the MTT assay was performed and measured absorbances were transformed into percentage of cell growth as described previously 24 . Results were obtained from three independent experiments. IC50 values were calculated using linear regression analysis and results were statistically analysed by ANOVA, Tukey post-hoc test (p < .05).
Apoptosis detection
Cells were seeded into 8-well chambers (Lab-tek II Chmaber Slides) in concentration of 2 × 104 cells per well and treated with 2 × IC50 concentrations of selected compounds for 48 h. Staining of the cells was performed by Annexin-V-FITC Staining kit (Santa Cruz Biotechnology, Dallas, TX) according to the manufacturer’s instructions. Cells were visualised by fluorescent microscope (Olympus) at magnification of 40×.
Western blot analysis
Cells were seeded in 6-well plates in the concentration depending on tested cell line varying from 1 × 105 to 2 × 105 cells/well. Cells were treated for 48 h with 2 × IC50 concentrations of selected compounds. Following treatment, cells were lysed with RIPA buffer supplemented with protease and phosphatase inhibitors (Roche). Total proteins (50 µg) were resolved on 10 or 12% polyacrilamide gels, depending on protein size, and transferred onto PVDF membranes that were blocked for 1 h with either 4% BSA or 5% non-fat milk prepared in tris-buffered saline (TBST). Membranes were probed with primary antibodies against GPLD1 (Abcam), PDGFRβ, p-IGF-1Rβ, p-p38 MAPK, and p-NF-κB-p65 from cell signalling technology at 4 °C overnight. The next day, membranes were washed in TBST and probed with horseradish peroxidase-conjugated secondary antibodies goat anti-mouse (Santa Cruz Biotechnology) or goat anti-rabbit (Santa Cruz Biotechnology). Protein bands were visualised using chemiluminescence substrate and ImageQuant LAS 500 (GE Healthcare, Chicago, IL). Following visualisation, protein band density was analysed by Quantity One 1-D Analysis Software (Bio-Rad, Hercules, CA).
Computational methods
Calculated values of logP, n-octanol/water partition coefficients, for synthesised purine and purinomimetics were obtained by ChemAxon algorithm available within MarvinView Ver. 5.2.6. Predictions of biological targets of compound 12b (Supplementary Table S1) were made by web-service PASS (http://www.pharmaexpert.ru/passonline/index.php), which is based on the identification of substructure features typical for active molecules 25 . Available crystal structures of apo- and co-crystallised p38-α kinase with various inhibitors were downloaded from protein data bank 26 , 27 . Representative ligands from the following DFG-in X-ray structure were used: 2GTN and 3GFE, as well as representative ligand from the following DFG-out X-ray structures: 3D83, whereby 2GTN is a Gly flip structure. IC50 inhibition values were collected from BindingDB 28 . The ligand docking studies were carried out using Glide docking protocol 29–32 within Schrodinger suite of software 33 with extra precision (XP). Docking was performed using unconstrained docking. Binding poses were refined and binding energy was estimated using MM-GBSA 34–36 protocol and OPLS3 force-field with flexible residues distance being 5 Å.
Results and discussion
Chemistry
A focussed library of 33 purine and purine isosteres containing an aromatic 1-substituted 1,2,3-triazole moiety (4a, 4c–e, 5c–e, 6e, 7c, 7e, 8a–e, 9c–e, 10a–e, 11a–e, and 12a–e) attached to varied heterocyclic bases: purine, imidazo[4,5-c]pyridine (3-deazapurine), pyrrolo[2,3-d]pyrimidine (7-deazapurine), benzimidazole, and indole were synthesised as shown in Schemes 1–3. N-Alkylation of the corresponding heterocyclic base with propargyl bromide in the presence of NaH, as a base, afforded the N-9- (2a–d) (Schemes 1 and 2) and N-7- (3a and 3b) (Scheme 1) as well as N-1-propargylated (2e–g) (Scheme 3) heterocycles, as key intermediates.
Scheme 1.
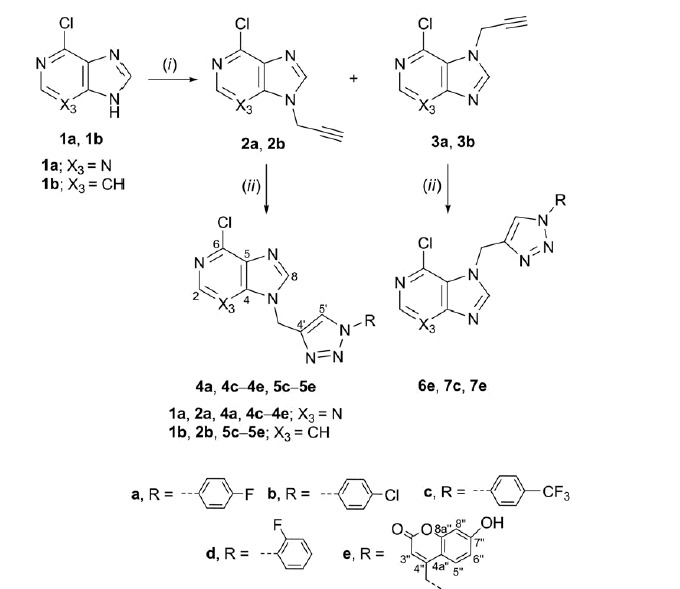
Synthesis of novel purine and 3-deazapurine derivatives with N-1 substituted 1,2,3-triazole. Reagents and conditions: (i) propargyl bromide, NaH, DMF, Ar atmosphere, 60 °C, 24 h; (ii) corresponding azide, Cu, 1 M CuSO4 solution, tert-butanol: H2O = 1: 1, MW 300 W, 80 °C, 45 min.
Scheme 2.
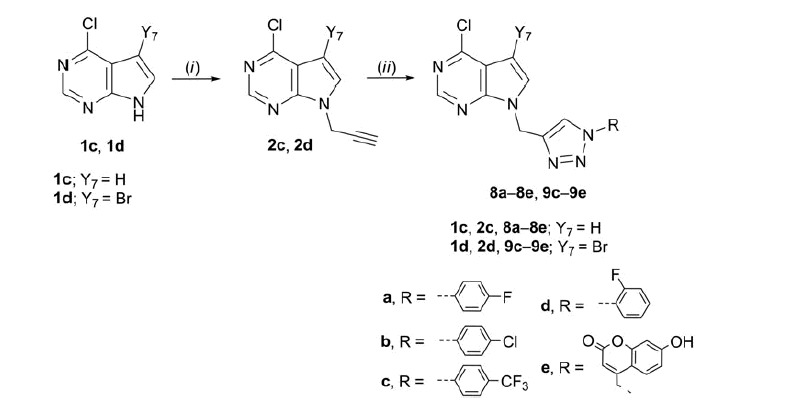
Synthesis of novel 7-deazapurine with N-1 substituted 1,2,3-triazole. Reagents and conditions: (i) propargyl bromide, K2CO3/NaH DMF, Ar atmosphere, 60 °C, 24 h; (ii) corresponding azide, Cu, 1 M CuSO4 solution, tert-butanol: H2O = 1: 1, MW 300 W, 80 °C, 45 min.
Scheme 3.
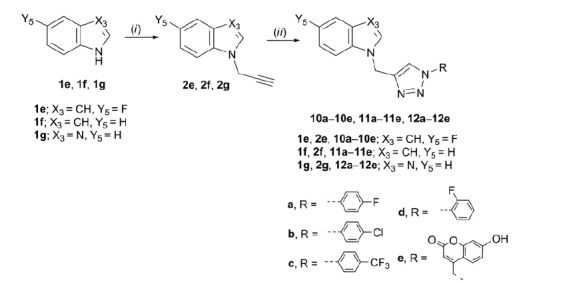
Synthesis of novel benzimidazole and indole derivatives with N-1 substituted 1,2,3-triazole. Reagents and conditions: (i) propargyl bromide, NaH, DMF, Ar atmosphere, 60 °C, 24 h; (ii) corresponding azide, Cu, 1 M CuSO4 solution, tert-butanol: H2O = 1: 1, MW 300 W, 80 °C, 45 min.
Target regioselective 1,4-disubstituted 1,2,3-triazoles (4a, 4c–e, 5c–e, 6e, 7c, 7e, 8a–e, 9c–e, 10a–e, 11a–e, and 12a–e) were subsequently prepared by copper(I)-catalysed Huisgen 1,3-dipolar cyclo-addition of alkynyl derivatives of purine and purine isosteres with diverse halogen-substituted phenyl azides under microwave irradiation using copper(II) sulphate and metallic copper. Inspired by the recent discovery of coumarin–1,2,3-triazole–2-methylbenzimidazole hybrid that exerted strong cytotoxicity against HCC HepG2 cells 37 , the structural diversity of target compounds was further expanded to the synthesis of heterocyclic bases linked through 1,2,3-triazole to 7-hydroxycoumarin 4e–12e. Therefore, click chemistry of 4-azidomethylcoumarin derivative and corresponding alkynes in the presence of Cu(I) afforded compounds 4e–12e.
Generally, from the halogen-substituted phenyl azides, we found that strong electron-withdrawing trifluoromethyl substituent on the phenyl ring improved the yields in the click reaction of the target p-(trifluoromethyl)phenyl-1,2,3-triazole-tagged purine and purine isosteres 4c–12c. Although reactions of N-7 regioisomers of alkynyl purine 3a and 3-deazapurine 3 b were performed with chosen phenyl azides only corresponding coumarin-1,2,3-triazolyl derivatives of N-7-(6-chloropurine) 6e and N-7-(3-deazapurine) 7e, and 4-(trifluoromethyl)phenyl-1,2,3-triazolyl derivative of N-7-(3-deazapurine) 7c were successfully isolated indicating lower reactivity of N-7 regioisomers 3a and 3 b with respect to their N-9 counterparts. The synthesis of aryl-1,2,3-triazolyl N-1-benzimidazole 12a–e was described in our previous reports 21 . In this study, we extended our investigations of these compounds towards evaluation of their cytostatic activities.
NMR structure determination of regioisomeric purine and purine bioisosteres
The chemical structures of novel purine isosteres were confirmed by 1D and 2D NMR techniques. 1H and 13C NMR data are included in Supplementary Material. H2 and H3 protons of purine and 1H-imidazo[4,5-c]pyridine moiety in regioisomers 5c, 5e, 7c, and 7e showed characteristic chemical shifts between at δ H 8.14–8.17 and 7.70–7.79 ppm, respectively. In addition, N7 regioisomers 7c and 7e exhibited more deshielded chemical shifts of H8 (δ H 8.73–8.76 ppm) in comparison to N9 regioisomers 5c and 5e (δ H 8.59–8.63 ppm). N7 and N9 regioisomers were further unambiguously distinguished by 13C NMR chemical shifts and their unique long-range correlation signals observed in 1H–13C HMBC spectra. N7 regioisomers showed 13C chemical shifts of C4 atoms at δ C 150–151 ppm, whereas N9 regioisomers exhibited 13C chemical shifts at δ C 140 ppm. In addition, C5 atoms were observed at δ C 127–128 ppm and 137 ppm for N7 and N9 regioisomers, respectively. N7 regioisomers were confirmed by long-range correlation signals between methylene group (δ H 5.7–6.0 ppm) and C5 atom, whereas methylene group of N9 regioisomers showed correlation signal with C4 atom.
Biological evaluations
Antiproliferative evaluations
Results of antiproliferative evaluations of compounds 4a, 4c–e, 5c–e, 6e, 7c, 7e, 8a–e, 9c–e, 10a–e, 11a–e, and 12a–e on human tumour cell lines including lung adenocarcinoma (A549), ductal pancreatic adenocarcinoma (CFPAC-1), cervical carcinoma (HeLa), and colorectal adenocarcinoma, metastatic (SW620), as well as on normal human lung (WI38) and foreskin (HFF-1) fibroblasts are presented in Table 1.
Table 1.
The growth-inhibition effects in vitro of synthesised compounds on selected tumour and normal cell lines.
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| IC50
a
(µM) |
||||||||
| Compounds | Purine or pseudopurine | R | A549 | CFPAC-1 | HeLa | SW620 | WI38/HFF-1 b | ClogP c |
| 4a | 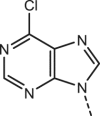 |
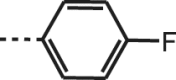 |
53.22 | 33.39 | 34.21 | 38.94 | 5.21 | 2.68 |
| 4c | 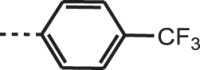 |
15.91 | 7.90 | 12.30 | 14.69 | 0.75 | 3.44 | |
| 4d | 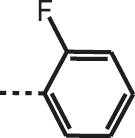 |
54.65 | 31.10 | 33.21 | 41.75 | 23.88 | 2.68 | |
| 4e | 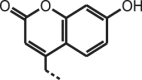 |
88.95 | >100 | 57.96 | 92.50 | 6.58 | 1.68 | |
| 5c | 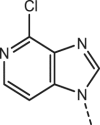 |
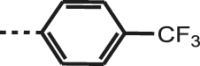 |
9.43 | 7.39 | 22.52 | 11.77 | 5.21 | 3.29 |
| 5d | 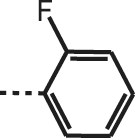 |
73.86 | >100 | >100 | 90.25 | 23.89 | 2.53 | |
| 5e | 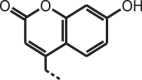 |
>100 | >100 | >100 | >100 | 60.49 | 1.53 | |
| 6e | 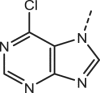 |
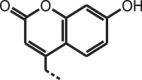 |
>100 | >100 | >100 | >100 | 73.63 | 2.14 |
| 7c | 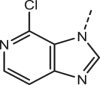 |
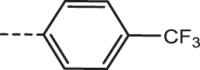 |
>100 | 89.24 | 36.80 | 68.95 | 19.43 | 3.29 |
| 7e |  |
>100 | >100 | 8.77 | >100 | 7.62 | 1.53 | |
| 8a |  |
 |
85.36 | 48.37 | 39.41 | 8.50 | 32.18 | 3.44 |
| 8b |  |
64.52 | 47.69 | 65.46 | 44.64 | 21.02 | 3.84 | |
| 8c | 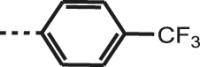 |
46.91 | 46.55 | 38.83 | 37.05 | 4.62 | 4.20 | |
| 8d | 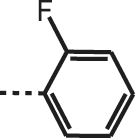 |
69.02 | 35.19 | 83.32 | 65.45 | 22.53 | 3.44 | |
| 8e | 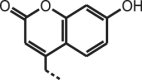 |
69.19 | 36.27 | 33.59 | 73.48 | 39.93 | 2.44 | |
| 9c | 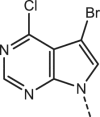 |
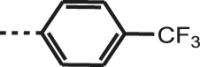 |
66.36 | 62.77 | 32.89 | 55.74 | 57.54 | 5.03 |
| 9d | 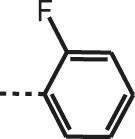 |
86.36 | 65.10 | 20.03 | 65.90 | 48.47 | 4.27 | |
| 9e | 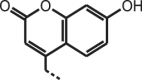 |
45.45 | 47.30 | 24.50 | 45.09 | 10.78 | 3.27 | |
| 10a | 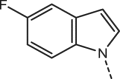 |
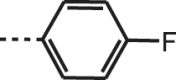 |
>100 | >100 | >100 | >100 | >100 | 3.89 |
| 10b | 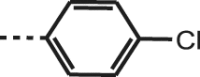 |
40.16 | >100 | 31.01 | 45.84 | 58.07 | 4.29 | |
| 10c | 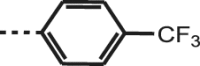 |
71.57 | 64.71 | 53.01 | 54.25 | 24.03 | 4.65 | |
| 10d | 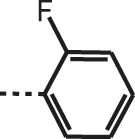 |
>100 | >100 | 82.92 | >100 | >100 | 3.89 | |
| 10e | 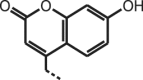 |
51.93 | 77.08 | 47.63 | 57.55 | 45.70 | 2.89 | |
| 11a | 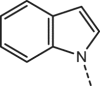 |
 |
44.01 | >100 | >100 | 43.29 | >100 b | 3.95 |
| 11b | 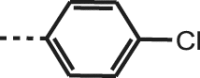 |
>100 | >100 | >100 | >100 | >100 b | 4.41 | |
| 11c | 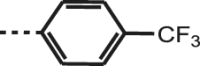 |
>100 | >100 | >100 | >100 | >100 b | 4.68 | |
| 11d | 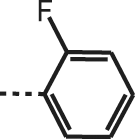 |
51.27 | 85.56 | 71.15 | 44.83 | 40.42 b | 3.95 | |
| 11e | 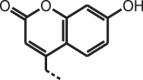 |
75.38 | >100 | 83.01 | 68.49 | 40.00 b | 3.17 | |
| 12a | 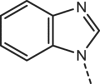 |
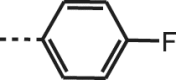 |
76.26 | 86.52 | 71.70 | 99.92 | 91.91 | 3.14 |
| 12b | 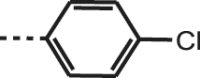 |
0.79 | 52.55 | 42.17 | 91.23 | 38.10 | 3.60 | |
| 12c | 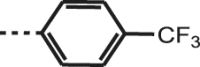 |
14.18 | 56.43 | 30.61 | >100 | >100 | 3.87 | |
| 12d | 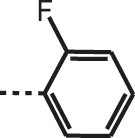 |
36.25 | 34.68 | 21.94 | >100 | 30.63 | 3.14 | |
| 12e |  |
>100 | >100 | >100 | >100 | >100 | 2.35 | |
50% inhibitory concentration or compound concentration required inhibiting tumour cell proliferation by 50%.
Compounds 11a–e were tested in HFF-1 cell line.
Values of n-octanol/water partition coefficients logP were calculated using ChemAxon algorithm (MarvinView Ver. 6.2.2.).
It can be noted that among purine-1,2,3-triazole hybrids (4a–e, 6e), purine analogue 4c with p-(trifluoromethyl)phenyl-substituted 1,2,3-triazole exhibited the highest antitumor activity, particularly in CFPAC-1 cells (IC50 = 7.90 µM). Besides rather non-selective inhibitory effects against all tested tumour cell lines, this compound was also toxic to normal fibroblasts WI38. Similarly, among the 1,2,3-triazole-tagged 3-deazapurines (5c–e, 7c, 7e), 1-p-(trifluoromethyl)phenyl-1,2,3-triazole in 5c contributed to strong cytostatic potency on CFPAC-1 (IC50 = 7.39 µM) along with the inhibitory effect on normal fibroblasts WI38. N-7 regioisomer 7c showed marked reduction in activity (IC50 = 89.24 µM) compared to its N-9 counterpart. In contrast, N-7 regioisomer of 3-deazapurine 7e with coumarin attached to 1,2,3-triazole displayed strong antiproliferative activity (IC50 = 8.77 µM) on HeLa cells, whereas its N-9 analogue 5e was devoid of any antitumor effects. From 7-deazapurine series (8a–e), compound 8a bearing p-fluorophenyl-substituted 1,2,3-triazole showed the highest inhibitory effect on SW620 (IC50 = 8.50 µM). Introduction of bromine at C-7 of 7-deazapurine in 9c–e did not have a considerable effect on cytostatic activity with respect to the corresponding congeners 8c–e. Of the 5-fluoroindole (10a–e) and indole (11a–e) 1,2,3-triazole conjugates, some compounds exhibited only marginal activities. Among 1,2,3-triazole-tagged benzimidazoles (12a–e), benzimidazole 12 b with p-chlorophenyl-substituted 1,2,3-triazole displayed significant and selective antiproliferative effect (IC50 = 0.79 µM) on A549 cells. Correlation between lipophilicity and antiproliferative effect showed that compounds 4c, 5c, 8a, and 12 b with marked cytostatic effects (IC50 < 10 µM) had C logP values in the range of 3.3–3.6. The only exception was 3-deazapurine–7-hydroxycoumarin hybrid 7e that exhibited lower Clog P value of 1.5.
Apoptosis detection
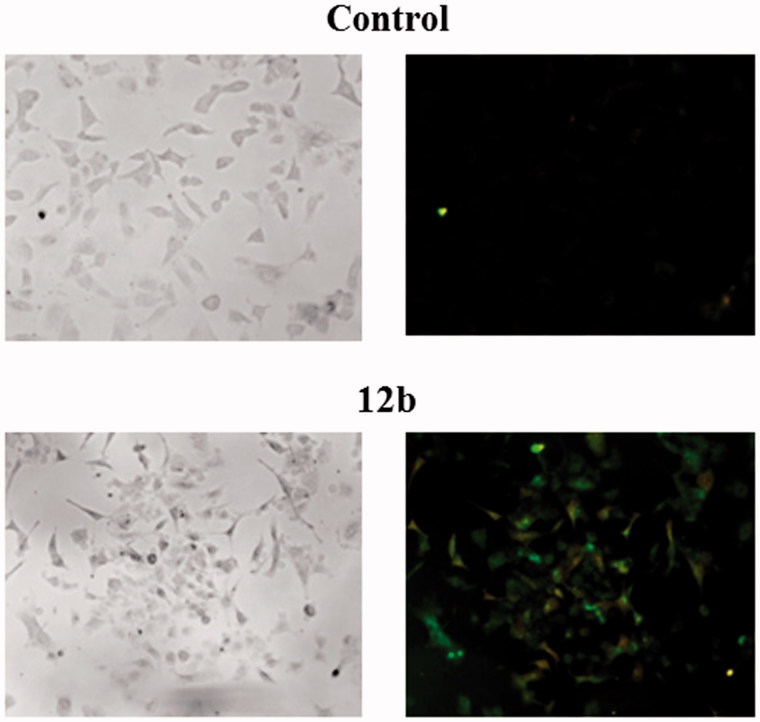
Further biological evaluations of compound 12b which was identified as a candidate were performed in order to investigate whether its antiproliferative effect in non-small cell lung cancer (A549) could be associated with induction of apoptosis. Therefore, annexin V assay was performed as previously described 38 and obtained data are presented in Figure 2 and Table 2.
Figure 2.
Detection of apoptosis induced by compound 12b in non-small cell lung cancer cell line A549 using Annexin V-FITC assay. Cells were visualised by fluorescence microscope at 40× magnification before and after treatment with the concentration of 2 × IC50 for 48 h. PI staining was used as a nuclear marker. Shown here are bright-field images (left micrographs) and late apoptotic/primary necrotic cells (right micrographs).
Table 2.
Annexin V assay for apoptosis detection.
| A549 | Control (%) | 12b (%) |
|---|---|---|
| Secondary necrotic cells | 0 | 2.39 |
| Early apoptotic cells | 0.52 | 31.14 |
| Viable cells | 99.48 | 46.71 |
| Late apoptotic/Primary necrotic cells | 0 | 19.76 |
The percentages of viable cells (PI−/Ann V−), early apoptotic cells (PI−/Ann V+), late apoptotic/primary necrotic cells (PI+/Ann V+) and secondary necrotic cells (PI+) after 48 h treatment with compound 12b at 2 × IC50 value are shown.
Treatment with compound 12b led to a significant reduction in the viable cell population by 55.72% concomitant with a marked increase in both, early apoptotic and late apoptotic/necrotic cell population by 30.62 and 19.76%, respectively. These findings clearly showed that compound 12b induced apoptosis in A549 cells.
Western blot analysis
In addition, the Prediction of Activity Spectra for Substances (PASS) 25 analysis was performed to reveal the probable biological activities and protein targets of 12b. The PASS analysis predicted glycosylphosphatidylinositol specific phospholipase D1 (GPLD1) and p38 mitogen-activated protein kinase (p38 MAPK) as potential targets of 12b (Supplementary Table S1), which was further confirmed in vitro by Western blot method.
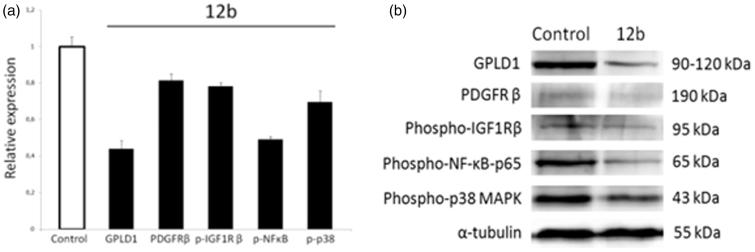
Glycosylphosphatidylinositol-specific phospholipase D1 (GPLD1) is a secreted mammalian enzyme that specifically cleaves the inositol phosphate linkage in proteins anchored by phosphatidylinositol glycans, thereby releasing the attached protein from the plasma membrane. Previous in vitro study has suggested that GPLD1 expression could be associated with tumour progression and malignancy 39 , and that some GPLD1 inhibitors exert growth-inhibitory activity in cancer cells at concentrations similar to those required to inhibit the enzyme 40 . Our results revealed that compound 12b dramatically reduced the expression levels of GPLD1 protein in human non-small cell lung cancer A549, which confirmed the PASS prediction of GPLD1 protein as a potential target of 12b (Figure 3).
Figure 3.
Western blot analysis of predicted protein targets of compound 12b. Representative Western blots are shown detecting the cellular levels of selected proteins before and after treatment of A549 cells with indicated compound at 2 × IC50 value for 48 h. Approximate molecular weights (kDa) are indicated. Relative protein expressions, as determined by densitometric analysis of protein bands and normalised to the alpha-tubulin loading control. Two independent experiments were performed with similar results. Data are presented as mean values ± SD. Statistically significant (p < .05) differences in the expression levels were marked by an asterisk.
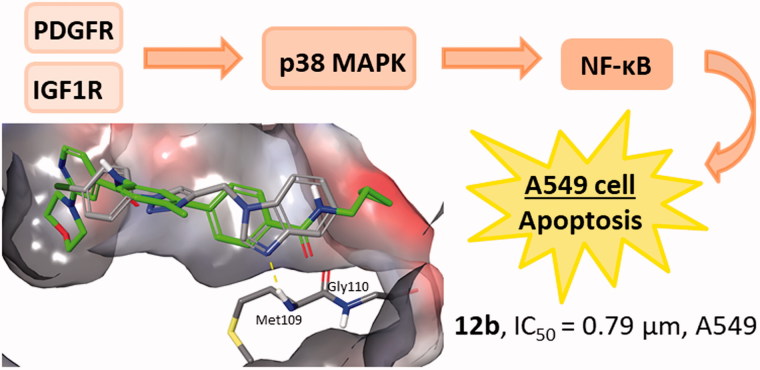
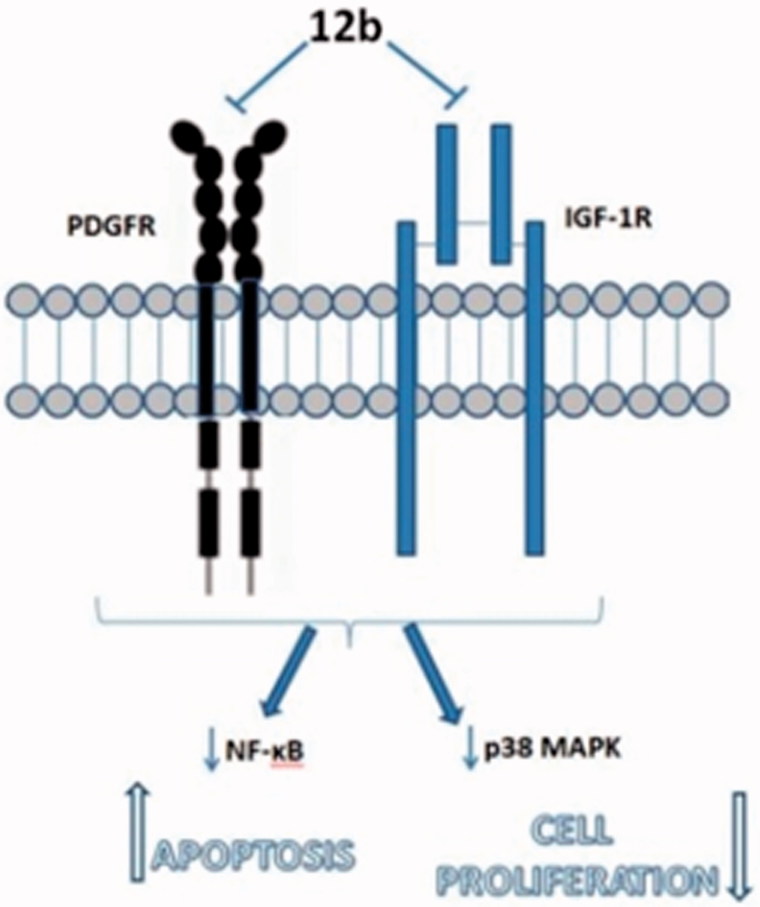
Importantly, this finding indicates that compound 12b targets cell surface and suggests that 12b could potentially modulate plasma membrane signalling to initiate a cascade of events inside the cell leading to growth inhibition and induction of apoptosis. To further substantiate this hypothesis, we examined the expression levels of two plasma membrane receptors known to promote non-small cell lung cancer cell proliferation and survival 41 , 42 , namely, PDGFR and IGF-1 R. We found that compound 12b downregulated the expression of PDGFR and p-IGF-1Rβ, which supports the involvement of plasma membrane signalling in the response of A549 cells to compound 12b. Impairment in the signalling mediated by these two receptors was further substantiated by a significant reduction in the p-NF-κB-p65 subunit expression level indicative of suppression of the activity of anti-apoptotic transcription factor NF-κB, a downstream target of both PDGFR and IGF-1 R signaling 43 , 44 , which could account for observed induction of apoptosis revealed by Annexin V assay. In addition, we detected a marked decline in the expression level of p-p38 MAP kinase indicative of inhibition of its activity in cells treated with 12b. This kinase transduces signals generated by growth factors to drive cell proliferation, and its activity was previously found to be increased in human non-small cell lung cancer 45 . Thus, anti-proliferative effects of 12b revealed by the MTT assay could be attributed to the inhibition of p38 MAPK activity, which was also in the agreement with in silico prediction of p38 MAPK as a putative target of 12b. Having in mind that stimulation of PDGF receptor as well as IGF-1R both could lead to activation of p38 MAP kinase 46 , 47 , we may presume that inhibition of p38 kinase activity resulted from abrogation of PDGFR and IGF-1 R signalling (Figure 4).
Figure 4.
Observed antiproliferative and antiapoptotic effects of 12b in non-small cell lung cancer cells A549 are associated with inhibition of specific plasma membrane receptors resulting in the blockade of downstream signalling propagated by p38 MAPK and NF-κb.
Structural analysis of possible interactions of compound 12b with p38 MAPK
Based on the Western blot analysis revealing strong reduction in the expression level of p38 MAP kinase induced by compound 12b (Figure 3), p38 MAPK was selected for further in silico molecular binding study. Results are supported by structural similarity to purinomimetic inhibitor, ralimetinib (LY2228820) 4 , as a selective inhibitor of p38 MAPK that has been evaluated in clinical trials in patients with advanced cancer.
The possible interactions with the p38 MAPK active site have been investigated in several steps to elucidate the most probable binding mode as described in the ‘Materials and methods’ section. The ligand docking studies were carried out using the p38 complexed with purine based inhibitor (pdb: 2GTN) and pyrazolopyridinone inhibitor (pdb: 3GFE) for DFG-in binding mode and p38 in complex with a biphenyl amide inhibitor (pdb: 3D83) to explore DFG-out binding mode. In the 2GTN structure Gyl flip was observed which might be relevant for the binding of 12b. Binding poses of 12b collected from different docking experiments were further refined using Embrace protocol and binding energies were estimated using MM-GBSA protocol as described in the ‘Materials and methods’ section.
Complexes of 12b with 2GTN and 3D83 are similarly stable, while the interactions with 3GFE structure were somewhat weaker. Best predicted binding pose of compound 12b in comparison with 2GTN ligand is shown in Figure 5.
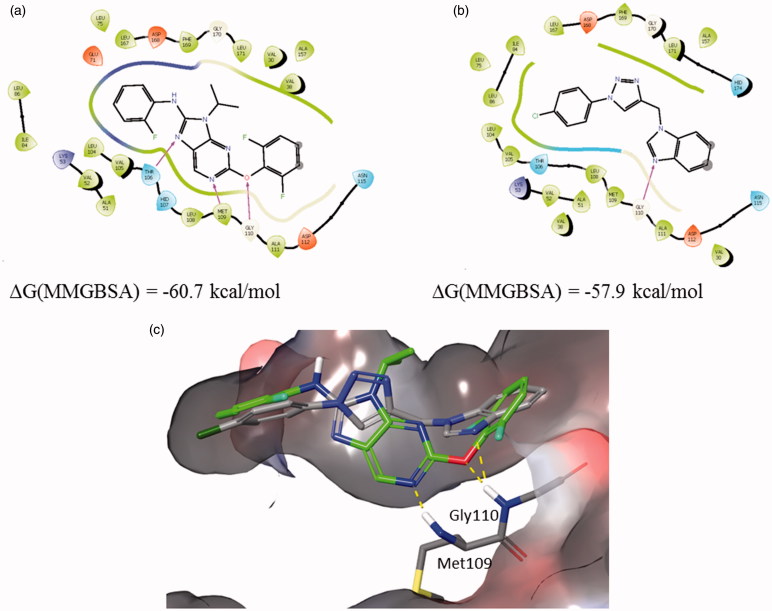
Figure 5.
Binding interactions of (a) 2GTN X-ray structure with purine like ligand, (b) 12b docked into 2GTN protein structure, and (c) structures from a and b superimposed within the active site pocket (green-2GTN ligand).
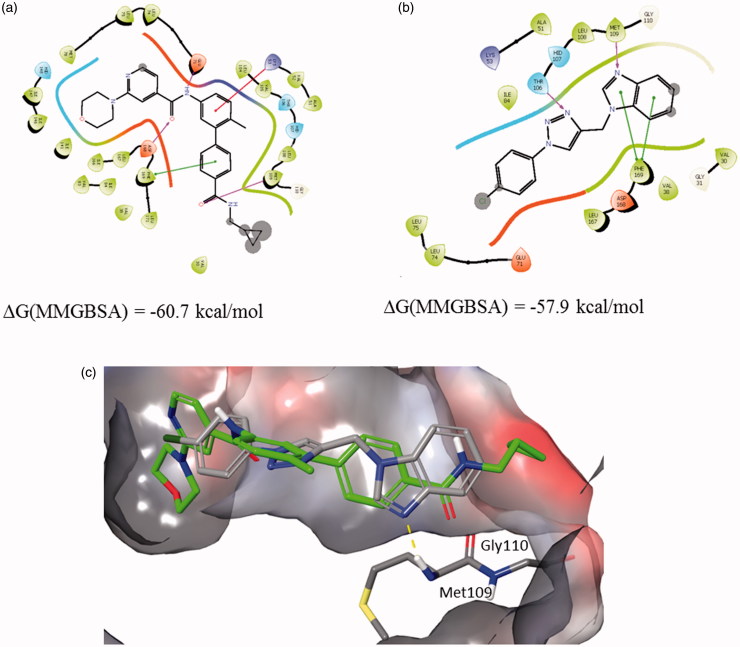
The benzimidazole moiety forms an H-bond with the backbone of Gly 110 in the hinge region, while the p-Cl-phenyl moiety linked to the triazole is placed in the hydrophobic environment. Best predicted binding pose of compound 12b in comparison with 3D83 ligand is shown in Figure 6. The benzimidazole moiety forms an H-bond with Met109 in the hinge region and π–π stacking with the Phe169, while the triazole moiety forms an H-bond with Thr106. Using established correlation of calculated ΔG energies and experimental binding activities 18 , 48 activity range of 12b is predicted to be below 100 nM.
Figure 6.
Binding interactions of (a) 3D83 X-ray structure, (b) 12b docked into 3D83 protein structure, and (c) structures from a and b superimposed within the active site pocket (green-3D83 ligand).
Conclusion
In summary, target halogenated purine and pseudopurines with regioselective 1,4-disubstituted 1,2,3-triazoles (4a, 4c–e, 5c–e, 6e, 7c, 7e, 8a–e, 9c–e, 10a–e, 11a–e, and 12a–e) were prepared by environmentally friendly click reactions under microwave irradiation using Cu(I) catalyst. Results of antiproliferative evaluations showed that p-(trifluoromethyl)-substituted 1,2,3-triazole in N-9 alkylated purine 4c and 3-deazapurine 5c was critical for strong albeit unselective activity in CFPAC-1 cells, whereas 1-(p-fluorophenyl)-1,2,3-triazole derivative of 7-deazapurine 8a showed cytostatic effect on SW620 cells. Among purinomimetics with coumarin-substituted 1,2,3-triazoles, only N-7 regioisomer of 3-deazapurine 7e exhibited strong cytostatic activity on HeLa cells. Notably, 1-(4-chlorophenyl)-1,2,3-triazole-tagged benzimidazole 12b showed the most pronounced and selective inhibitory effect on A549 cells. Further in silico and Western blot analyses revealed that 12b targets molecular processes at the extracellular side and inside the plasma membrane regulated by GPLD1 and growth factor receptors PDGFR and IGF-1R leading to the inhibition of cell proliferation and induction of apoptosis mediated by p38 MAP kinase and NF-κB, respectively. Based on these findings, we propose benzimidazole scaffold with aryl-substituted 1,2,3-triazole ring as new and promising chemical entity with potent cytostatic activity against A549 cells. Further structural optimisation of this compound may lead to reduction of its toxicity in normal cells without compromising its antitumor effects to develop a novel agent for treatment of non-small lung cancer cells.
Supplementary Material
Funding Statement
We greatly appreciate the financial support of the Croatian Science Foundation [grant number IP–2013–11–5596], University of Rijeka research grants [13.11.1.1.11. and 13.11.2.1.12.], and the access to equipment owned by the University of Rijeka within the project RISK ‘Development of University of Rijeka campus laboratory research infrastructure’, financed by the European Regional Development Fund (ERDF).
References
- 1. Binétruy B, Heasley L, Bost F, et al. Concise review: regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells 2007;25:1090–5. [DOI] [PubMed] [Google Scholar]
- 2. Huang C, Jacobson K, Schaller MD.. MAP kinases and cell migration. J Cell Sci 2004;117:4619–28. [DOI] [PubMed] [Google Scholar]
- 3. Dhillon AS, Hagan S, Rath O, Kolch W.. MAP kinase signalling pathways in cancer. Oncogene 2007;26:3279–90. [DOI] [PubMed] [Google Scholar]
- 4. Patnaik A, Haluska P, Tolcher AW, et al. First-in-human phase I study of the oral p38 MAPK inhibitor, Ralimetinib (LY2228820 Dimesylate), in patients with advanced cancer. Clin Cancer Res 2016;22:1095–102. [DOI] [PubMed] [Google Scholar]
- 5. Bendell JC, Javle M, Bekaii-Saab TS, et al. A phase 1 dose-escalation and expansion study of binimetinib (MEK162), a potent and selective oral MEK1/2 inhibitor. Br J Cancer 2017;116:575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol 2016;14:249–56. [DOI] [PubMed] [Google Scholar]
- 7. Watanabe K, Otsu S, Hirashima Y, et al. A phase I study of binimetinib (MEK162) in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol 2016;77:1157–64. [DOI] [PubMed] [Google Scholar]
- 8. LoRusso PM, Infante JR, Kim KB, et al. A phase I dose-escalation study of selumetinib in combination with docetaxel or dacarbazine in patients with advanced solid tumors. BMC Cancer 2017;17:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tai WM, Yong WP, Lim C, et al. A phase Ib study of selumetinib (AZD6244, ARRY-142886) in combination with sorafenib in advanced hepatocellular carcinoma (HCC). Ann Oncol 2016;27:2210–15. [DOI] [PubMed] [Google Scholar]
- 10. Bachegowda L, Morrone K, Winski SL, et al. Pexmetinib: a novel dual inhibitor of Tie2 and p38 MAPK with efficacy in preclinical models of myelodysplastic syndromes and acute myeloid leukemia. Cancer Res 2016;76:4841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garcia-Manero G, Khoury HJ, Jabbour E, et al. A phase I study of oral ARRY-614, a p38 MAPK/Tie2 dual inhibitor, in patients with low or intermediate-1 risk myelodysplastic syndromes. Clin Cancer Res 2015;21:985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Auffinger P, Hays FA, Westhof E, Ho PS.. Halogen bonds in biological molecules. Proc Natl Acad Sci USA 2004;101:16789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu Y, Shi T, Wang Y, et al. Halogen bonding – a novel interaction for rational drug design? J Med Chem 2009;52:2854–62. [DOI] [PubMed] [Google Scholar]
- 14. Wilcken R, Zimmermann MO, Lange A, et al. Principles and applications of halogen bonding in medicinal chemistry and chemical biology. J Med Chem 2013;56:1363–88. [DOI] [PubMed] [Google Scholar]
- 15. Bradham C, McClay DR.. p38 MAPK in development and cancer. Cell Cycle 2006;5:824–8. [DOI] [PubMed] [Google Scholar]
- 16. Wagner EF, Nebreda AR.. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009;9:537–49. [DOI] [PubMed] [Google Scholar]
- 17. Iyoda K, Sasaki Y, Horimoto M, et al. Involvement of the p38 mitogen-activated protein kinase cascade in hepatocellular carcinoma. Cancer 2003;97:3017–26. [DOI] [PubMed] [Google Scholar]
- 18. Bistrović A, Krstulović L, Harej A, et al. Design, synthesis and biological evaluation of novel benzimidazole amidines as potent multi-target inhibitors for the treatment of non-small cell lung cancer. Eur J Med Chem 2017. pii: S0223-5234(17)30863-2. doi: 10.1016/j.ejmech.2017.10.061. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19. Yan R, Sander K, Galante E, et al. A one-pot three-component radiochemical reaction for rapid assembly of 125I-labeled molecular probes. J Am Chem Soc 2013;135:703–9. [DOI] [PubMed] [Google Scholar]
- 20. Chittepu P, Sirivolu VR, Seela F.. Nucleosides and oligonucleotides containing 1,2,3-triazole residues with nucleobase tethers: synthesis via the azide-alkyne ‘click’ reaction. Bioorg Med Chem 2008;16:8427–39. [DOI] [PubMed] [Google Scholar]
- 21. Maračić S, Kraljević TG, Paljetak HČ, et al. 1,2,3-Triazole pharmacophore-based benzofused nitrogen/sulfur heterocycles with potential anti-Moraxella catarrhalis activity. Bioorg Med Chem 2015;23:7448–63. [DOI] [PubMed] [Google Scholar]
- 22. Dürüst Y, Sağırlı A, Kariuki BM, Knight DW.. [1,3]-Dipolar cycloaddition of N-aryl sydnones to benzothiophene 1,1-dioxide, 1-cyclopropylprop-2-yn-1-ol and 1-(prop-2-ynyl)-1H-indole. Tetrahedron 2014;70:6012–19. [Google Scholar]
- 23. Duan YC, Ma YC, Zhang E, et al. Design and synthesis of novel 1,2,3-triazole-dithiocarbamate hybrids as potential anticancer agents. Eur J Med Chem 2013;62:11–19. [DOI] [PubMed] [Google Scholar]
- 24. Gazivoda T, Raić-Malić S, Krištafor V, et al. Synthesis, cytostatic and anti-HIV evaluations of the new unsaturated acyclic C-5 pyrimidine nucleoside analogues. Bioorg Med Chem 2008;16:5624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Filimonov DA, Poroikov VV.. Probabilistic approaches in activity prediction In: Varnek A, Tropsha A, eds. Chemoinformatics approaches to virtual screening. Cambridge, UK: RSC Publishing, 2008:182–216. [Google Scholar]
- 26. RSC PDB Available from: http://www.rcsb.org/pdb/, 2017. [last accessed 31 Mar 2017]. [Google Scholar]
- 27. Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen X, Lin Y, Gilson MK.. The binding database: overview and user's guide. Biopolymers 2001;61:127–41. [DOI] [PubMed] [Google Scholar]
- 29. Torch, version 10.4.1. , Cresset, Litlington, Cambridgeshire, UK. Available from: http://www.cresset-group.com/torch/ [last accessed 1 Sep 2017]. [Google Scholar]
- 30. Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein − ligand complexes. J Med Chem 2006;49:6177–96. [DOI] [PubMed] [Google Scholar]
- 31. Halgren TA, Murphy RB, Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 2004;47:1750–9. [DOI] [PubMed] [Google Scholar]
- 32. Friesner RA, Banks JL, Murphy RB, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 2004;47:1739–49. [DOI] [PubMed] [Google Scholar]
- 33. Schrödinger Release 2017-1: MacroModel, Schrödinger, New York, NY: LLC, 2017. [Google Scholar]
- 34. Li J, Abel R, Zhu K, et al. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins Struct Funct Bioinformatics 2011;79:2794–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jorgensen WL, Maxwell DS, Tirado-Rives J.. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 1996;118:11225–36. [Google Scholar]
- 36. Shivakumar D, Williams J, Wu Y, et al. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 2010;6:1509–19. [DOI] [PubMed] [Google Scholar]
- 37. Kraljević TG, Harej A, Sedić M, et al. Synthesis, in vitro anticancer and antibacterial activities and in silico studies of new 4-substituted 1,2,3-triazole–coumarin hybrids. Eur J Med Chem 2016;124:794–808. [DOI] [PubMed] [Google Scholar]
- 38. Gregorić T, Sedić M, Grbčić P, et al. Novel pyrimidine-2,4-dione–1,2,3-triazole and furo[2,3-d]pyrimidine-2-one–1,2,3-triazole hybrids as potential anti-cancer agents: synthesis, computational and X-ray analysis and biological evaluation. Eur J Med Chem 2017;125:1247–67. [DOI] [PubMed] [Google Scholar]
- 39. He XT, Hannocks MJ, Hampson I, Brunner G.. GPI-specific phospholipase D mRNA expression in tumor cells of different malignancy. Clin Exp Metastasis 2002;19:291–9. [DOI] [PubMed] [Google Scholar]
- 40. Brunner G, Zalkow L, Burgess E, et al. Inhibition of glycosylphosphatidylinositol (GPI) phospholipase D by suramin-like compounds. Anticancer Res 1996;16:2513–16. [PubMed] [Google Scholar]
- 41. Wang P, Song L, Ge H, et al. Crenolanib, a PDGFR inhibitor, suppresses lung cancer cell proliferation and inhibits tumor growth in vivo. Oncotargets Ther 2014;7:1761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim JS, Kim ES, Liu D, et al. Activation of insulin-like growth factor 1 receptor in patients with non-small cell lung cancer. Oncotarget 2015;6:16746–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Romashkova JA, Makarov SS. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature 1999;401:86–90. [DOI] [PubMed] [Google Scholar]
- 44. Zhao D, Bakirtzi K, Zhan Y, et al. Insulin-like growth factor-1 receptor transactivation modulates the inflammatory and proliferative responses of neurotensin in human colonic epithelial cells. J Biol Chem 2011;286:6092–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greenberg AK, Basu S, Hu J, et al. Selective p38 activation in human non-small cell lung cancer. Am J Respir Cell Mol Biol 2002;26:558–64. [DOI] [PubMed] [Google Scholar]
- 46. Matsumoto T, Yokote K, Tamura K, et al. Platelet-derived growth factor activates p38 mitogen-activated protein kinase through a Ras-dependent pathway that is important for actin reorganization and cell migration. J Biol Chem 1999;274:13954–60. [DOI] [PubMed] [Google Scholar]
- 47. Cosaceanu D, Budiu RA, Carapancea M, et al. Ionizing radiation activates IGF-1R triggering a cytoprotective signaling by interfering with Ku-DNA binding and by modulating Ku86 expression via a p38 kinase-dependent mechanism. Oncogene 2007;26:2423–34. [DOI] [PubMed] [Google Scholar]
- 48. Greenidge PA, Kramer C, Mozziconacci JC, Wolf RM.. MM/GBSA binding energy prediction on the PDBbind data set: successes, failures, and directions for further improvement. J Chem Inf Model 2012;53:201–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.