Abstract
A new Keap1–Nrf2 protein–protein interaction (PPI) inhibitor ZJ01 was identified from our compound library by fluorescence polarization assay, surface plasmon resonance, molecular docking and molecular dynamics simulation. ZJ01 could in vitro trigger Nrf2 nuclear translocation, subsequently resulting in increased mRNA levels of Nrf2 target genes HO-1 and NQO1. Meanwhile, ZJ01 suppressed LPS-induced production of ROS and the mRNA levels of pro-inflammatory cytokines TNF-α, IL-1β and IL-6 in H9c2 cardiac cells. Moreover, in an in vivo mouse model of septic cardiomyopathy induced by intraperitoneal injection of lipopolysaccharide, ZJ01 demonstrated a cytoprotective effect, upregulated Nrf2 protein nuclear accumulation, and remarkably suppressed the abovementioned cytokine levels in cardiomyocytes. The results presented herein provided a novel chemotype for the development of direct Keap1–Nrf2 PPI inhibitors and suggested that compound ZJ01 is a promising drug lead for septic cardiomyopathy treatment.
ZJ01 was identified as a new Keap1–Nrf2 PPI inhibitor and drug lead for septic cardiomyopathy treatment by in vitro and in vivo experiments.
Keywords: Keap1, Nrf2, protein–protein interaction inhibitor, cytoprotective, septic cardiomyopathy
Introduction
Sepsis, a major cause of death worldwide, is a systemic inflammatory syndrome caused by serious infections 1 . According to the statistics, up to 70% of patients with sepsis are affected by septic cardiomyopathy, and the dysfunction of cardiomyocytes contributes to a high mortality rate 2 . Currently, septic cardiomyopathy remains a great challenge for clinical treatment. Lipopolysaccharide (LPS), the major component of the outer membrane of gram-negative bacteria, has proven to be a key stimulator of sepsis 3 . Under septic conditions, LPS can evoke an inflammatory response in innate immune cells, as well as non-immune cells (e.g. cardiomyocytes) expressing LPS-pattern recognition receptors 4 . More importantly, cardiac dysfunction frequently occurs in patients with sepsis and animals injected with LPS 5 , 6 . Increasing evidence indicates that control of the oxidative stress and inflammatory response in cardiomyocytes could alleviate LPS-induced cardiac injury 7–9 .
It has been known that nuclear factor erythroid 2-related factor 2 (Nrf2) can protect cells against oxidative stress and repress inflammation 10 , 11 . In the anti-oxidant defense system, the expression of a number of key genes, such as NAD(P)H: quinone oxidoreductase 1 (NQO1), heme oxygenase-1 (HO-1), superoxide dismutase (SOD) and glutathione S-transferase (GST) 12 , is mainly regulated by three components: Nrf2, Kelch-like ECH-associated protein 1 (Keap1) and anti-oxidant response elements (ARE) 13 . Nrf2 activity is regulated mainly by Keap1 14 . Under unstressed conditions, Nrf2 is negatively regulated by Keap1 via proteasomal degradation to remain at low cellular concentrations. Upon oxidative stress, reactive oxygen species (ROS) and/or electrophilic agents modify the reactive cysteine residues in Keap1, resulting in conformational changes and deactivation of Keap1. Subsequently, Nrf2 is activated after escaping from Keap1-mediated degradation and then translocate into the nucleus where it induces the transcription of the aforementioned downstream genes by binding to ARE 14 . More interestingly, a recent study demonstrated that Nrf2 could inhibit pro-inflammatory cytokine transcription through direct DNA binding 15 . Interestingly, other studies have highlighted the important role of Nrf2 activation in protecting cardiomyocytes from LPS-induced injuries 16 . Thus, to activate Nrf2 via disrupting the Keap1–Nrf2 protein–protein interaction (PPI) represents an attractive strategy to develop drugs for diseases involving oxidative stress and inflammation, such as cardiomyopathy 16 , 17 .
Recently, small molecules (Figure 1) directly targeting the Keap1–Nrf2 PPI have been reported by several research groups 18–22 . However, the structural diversity of current Keap1–Nrf2 PPI inhibitors is still limited. In our project to discover Keap1–Nrf2 PPI inhibitors, the novel small molecule ZJ01 with an iminocoumarin-benzothiazole scaffold (Figure 2) was recently screened out from our in-house compound library based on fluorescence polarization (FP) assay. In the present study, we describe the identification of ZJ01 as a new Keap1–Nrf2 PPI inhibitor and further investigate its in vitro and in vivo cytoprotective effects on LPS-induced cardiomyopathy.
Figure 1.
Recently reported small-molecule Keap1–Nrf2 PPI inhibitors.
Figure 2.
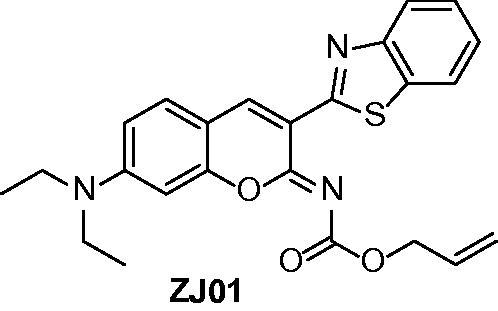
Structure of ZJ01.
Methods
Chemistry
General experimental procedures
Commercially available reagents were used without further purification. Organic solvents were evaporated with reduced pressure using a Buchi rotary evaporator. Reactions were monitored by TCL using Yantai Jiangyou (China) GF254 silica gel plates. Silica gel column chromatography was performed on silica gel (300–400 mesh) from Qingdao Haiyang (China). The NMR spectra were measured on Bruker Avance 600 spectrometer. Chemical shifts were expressed in δ (ppm) and coupling constants (J) in Hz using solvent signals as internal standards (CDCl3, δH 7.26 ppm; δC 77.0 ppm). ESI-MS was recorded on an Agilent 6460 Triple Quard LC/MS, and HRMS spectrum were recorded on an Agilent Q-ToF 6520.
3-(Benzo[d]thiazol-2-yl)-N,N-diethyl-2-imino-2H-chromen-7-amine (3)
To a solution of (1,3-benzothiazol-2-yl)acetonitrile (1) (165 mg, 0.86 mmol) and 4-4-(diethylamino)-2-hydroxybenzaldehyde (2) (150 mg, 0.86 mmol) in MeOH (10 ml) was added a drop of piperazine. The mixture was stirred at 20 °C for 16 h under N2, the precipitate was collected by filtration, washed with methanol, and dried under high vacuum to afford 3 23 (255 mg, 85%). This compound was used in the next step without further purification. 1H NMR (600 MHz, CDCl3) δ 8.25 (1H, br), 8.01 (1H, d, J = 8.1 Hz), 7.90 (1H, d, J = 7.9 Hz), 7.47 (1H, dd, J = 8.1, 7.1 Hz), 7.35 (1H, dd, J = 7.9, 7.1 Hz), 7.29 (1H, d, J = 8.7 Hz), 6.48 (1H, dd, J = 8.7, 2.4 Hz), 6.40 (1H, brs), 3.42 (4H, q, J = 7.1 Hz), 1.22 (6H, t, J = 7.1 Hz). ESIMS m/z: 350.1 [M + H]+.
(Z)-allyl (3-(benzo[d]thiazol-2-yl)-7-(diethylamino)-2H-chromen-2-ylidene)carbamate (ZJ01)
To a solution of 3 (60 mg, 0.17 mmol) in DCM (5 ml) was added Et3N (24 μL, 0.17 mmol). The solution was cooled to 0 °C for 1 h, and then allyl carbonochloridate (10 μL, 0.17 mmol) was added. The mixture was stirred overnight, concentrated, and purified by silica gel column (petroleum ether–ethyl acetate, 4:1) to afford pure ZJ01 as a red powder (18 mg, 25%). mp 148–150 °C. 1H NMR (600 MHz, CDCl3) δ 8.82 (1H, s), 8.02 (1H, brd, J = 8.1 Hz), 7.92 (1H, brd, J = 7.9 Hz), 7.49 (1H, ddd, J = 8.1, 7.2, 1.2 Hz), 7.42 (1H, d, J = 8.8 Hz), 7.35 (1H, ddd, J = 7.9, 7.2, 1.0 Hz), 6.63 (1H, dd, J = 8.8, 2.3 Hz), 6.47 (1H, d, J = 2.3 Hz), 6.10 (1H, ddt, J = 17.2, 10.5, 5.6 Hz), 5.48 (1H, ddt, 17.2, 1.8, 1.4 Hz), 5.31 (1H, ddt, J = 10.5, 1.8, 1.4 Hz), 4.86 (2H, ddd, J = 5.6, 1.4, 1.4 Hz), 3.45 (4H, q, J = 7.1 Hz), 1.24 (6H, t, J = 7.1 Hz). 13C NMR (125 MHz, CDCl3) δ 161.5, 159.6, 155.8, 153.1, 152.4, 151.9, 139.8, 137.0, 132.7, 130.8, 126.1, 124.5, 122.3, 121.6, 118.2, 114.2, 109.9, 108.6, 97.0, 67.0, 45.1, 12.7. ESIMS m/z: 434.1 [M + H]+. HR-ESIMS: [M + H]+ calcd for C24H24N3O3S+ 434.1533, found 434.1546.
(Z)-N-(3-(benzo[d]thiazol-2-yl)-7-(diethylamino)-2H-chromen-2-ylidene)acrylamide (ZJ02)
To a solution of 3 (60 mg, 0.17 mmol) in DCM (5 ml) was added Et3N (24 μL, 0.17 mmol). The solution was cooled to 0 °C for 1 h, and then acryloyl chloride (14 μL, 0.17 mmol) was added. The mixture was stirred overnight, concentrated, and purified by silica gel column (petroleum ether–ethyl acetate, 4:1) to afford pure ZJ01 as a red powder (27 mg, 39%). mp 188–190 °C. 1H NMR (600 MHz, CDCl3) δ 8.80 (1H, s), 8.02 (1H, d, J = 8.1 Hz), 7.91 (1H, d, J = 7.9 Hz), 7.47 (1H, dd, J = 7.4, 7.9 Hz), 7.39 (1H, d, J = 8.8 Hz), 7.35 (1H, dd, J = 7.4, 8.1 Hz), 6.59 (1H, dd, J = 8.8, 2.3 Hz), 6.54–6.46 (2H, m), 6.44 (1H, d, J = 2.0 Hz), 5.97 (1H, dd, J = 2.3, 9.3 Hz), 3.41 (4H, q, J = 7.2 Hz), 1.24 (6H, t, J = 7.2 Hz). 13 C NMR (125 MHz, CDCl3) δ 176.7, 161.7, 155.9, 152.4, 151.9, 150.8, 139.6, 136.9, 134.6, 130.8, 130.3, 126.1, 124.5, 122.3, 121.5, 114.2, 109.8, 108.6, 97.0, 45.1, 12.7. ESIMS m/z: 404.1 [M + H]+. HR-ESIMS: [M + H]+ calcd for C23H22N3O2S+ 404.1427, found 404.1429.
Biology
FP assay
Determination of equilibrium dissociation constants K D2 for the tested compound was performed using the FP assay by fitting the displacement curves as described in our previous study 20 . The FP was measured on Biotek Syngergy 2 microplate reader with 485 nm excitation and 535 nm emission after a 60 min incubation at room temperature. The K D1 value is determined with FITC − βAla − DEETGEF − OH as the fluorescence probe.
Surface plasmon resonance (SPR) analysis
Surface plasmon resonance analysis was performed on a Biacore TM T200 instrument (GE Healthcare, Salem, CT, USA). Keap1 proteins were immobilized on a CM5 chip via EDC/NHS-mediated crosslinking reaction. Tested compound was diluted in PBS at concentrations ranging from 0.25 to 64 µM. The analysis was then performed according to the protocol provided by GE Healthcare. In each analysis, the middle concentration was duplicated at the end of the wash run to confirm the stability of the sensor surface. The parameters of SPR were set as follow: flow rate, 30 µL/min; contact time, 120 s; disassociation time, 120 s. Affinity curve fitting was performed with the Biacore T200 Evaluation Software using a steady-state affinity model (1:1) to calculate the disassociation constant (K d). Detailed SPR measurement data are shown in Table 1.
Table 1.
SPR measurement data of Keap1 with ZJ01.
| Protein | Ligand | Kd (µM) | Rmax (RU) | Offset (RU) | Chi2 (RU2) |
|---|---|---|---|---|---|
| Keap1 | ZJ01 | 48.1 | 58.51 | 0.2069 | 0.236 |
Cell culture and treatment
H9c2 cells, a rat ventricular myoblast cell line, were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The Cells were cultured in Dulbecco’s modified Eagle’s medium with 10% bovine calf serum and then were maintained at 37 °C under humidified conditions with 5% CO2. Cells were seeded in appropriate dishes (30,000 cells/ml). After culture for 24 h, the cells were treated with different concentration of inhibitors and incubated with or without 1 μg/ml of LPS for indicated time.
Mice and treatments
Male C57BL/6 mice (eight weeks old) were purchased from the Experimental Animal Center of Shandong University (Jinan, Shandong, China). All in vivo experiments followed the ARRIVE guidelines 24 . Inhibitors or LPS was dissolved in DMSO: normal saline (1:100). The control group was injected intraperitoneally with equal DMSO and normal saline. C57BL/6 mice were challenged with different concentrations of ZJ01 or S47 overnight for approximately 12 h after being treated intraperitoneally with or without 4 mg/kg of LPS. At the end of treatment, all mice were euthanized by intravenous lateral tail vein injection of keta-mine/xylazine (Sigma-Aldrich, Saint Louis, MO, USA, 150 mg/kg ketamine combined with 10 mg/kg xylazine). The left ventricles were collected for western blotting or real-time PCR assay.
Western blotting assay
Protein Extraction Kit (Beyotime, China) was used to isolate the nuclear and cytosol protein of H9c2 cells and left ventricular cells of C57BL/6 mice according to the protocol. Then the collected protein was stored at −80 °C until use. Equal amounts of protein were applied to 12% SDS-polyacrylamide gel. Proteins in gels were electroblotted onto poly-vinylidene difluoride membranes. After blocking at room temperature for 1 h, the membranes were probed with primary antibodies overnight at 4 °C. After three washes in TBST, membranes were incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature, and proteins were detected by use of an enhanced chemiluminesence detection kit.
Immunofluorescence analysis
Treated cells were fixed in 4% paraformaldehyde (w/v) for 30 min at room temperature, then incubated with normal goat serum (1:30) for 20 min and Nrf2 antibodies (1:100) overnight at 4 °C. Cells were washed with PBS for three times, then incubated with corresponding secondary antibodies (1:200) for 1 h at 37 °C. Fluorescence was detected by laser scanning confocal microscopy (Leica, Wetzlar, Germany).
DCFH-DA staining for analysis of intracellular ROS activity level
H9c2 Cells (1 × 104 per well) were seeded in black bottomed 96-well culture plate and cultured for 24 h in a CO2 incubator at 37 °C. After treatment, cells were incubated with 10 mM DCFH-DA for 30 min at 37 °C. After washing with PBS for three times, fluorescence intensity was measured with a multi-well microplate reader at an emission wavelength of 528 nm and at an excitation wavelength of 485 nm. All the values were expressed as percentage fluorescence intensity relative to the control.
Real-time PCR
Total RNAs were extracted from treated cells or left ventricle of C57BL/6 mice with TriZol Reagent (Invitrogen Life Technologies, Waltham, MA, USA). RNA (250–500 ng) was reverse-transcribed using the Prime Script RT reagent kit with gDNA Eraser (DRR047, TAKARA) according to the manufacturer’s instructions. The RT-PCR reactions were performed using QuantiTect SYBR Green PCR kit (QIAgen, Dusseldorf, Germany) and LightCycler 2.0 system (Roche Diagnostics, Shanghai, China). Reactions were carried out in a 25 μl volume containing 12.5 μl of 2 × SYBR Green PCMaster Mix. The fold-changes for RNA level were calculated using the MxPro software (Version 4.00, Stratagene, San Diego, CA, USA).
Molecular docking simulation
To obtain the starting structure of Keap1/ZJ01 for simulation, molecular docking was performed with Autodock-4 25 . For the docking calculations we obtained the initial Keap1 complex crystal structures from the Protein Data Bank (www.pdb.org)-(PDB id: 4IQK). Prior to docking, all the water molecules have been removed from Keap1 as none of them plays any role in inhibitor binding. AutoDockTools have been used to prepare the enzyme prior to the docking. Gasteiger 26 partial charges have been assigned to both the inhibitor and enzyme atoms. The docking sampled the ligands in a 126 × 126 × 126 grid with 0.375 Å resolution that was positioned to encompass the Keap1 binding gorge. The quality of the re-docking has been quantified by the root meansquare deviation (RMSD) between the top-scoring docked pose from each docking method and the corresponding X-ray crystal structure. Prior to docking all X-ray ligand structures were compared with PubChem (www.pubchem.com) structures for any missing atoms and then optimized using the program LigPrep from the Schrödinger (Schrödinger Release 2015–2: LigPrep, Schrödinger, LLC, New York, NY, 2015). The optimization is based on molecular mechanics and used the MMFFS force field 27 . The docking results were analysed with the programs AutoDockTools 25 , DOCKRES 28 and VMD 29 . Molecular graphics figures were prepared with the LigPlot + program 30 .
Molecular dynamics simulations
Molecular dynamics simulations were performed on the Keap1/ZJ01 complex obtained from molecular docking. Before simulations, the protonation states of ionisable residues were determined using the H++ program 31 . The complex model was surrounded by a periodic box of transferable intermolecular potential 3P 32 water molecules that extend 12 Å from the protein atoms. Counter-ions were added to neutralize the simulation system. Molecular dynamics simulations were performed using the GROMACS 5.0.4 package 33 with isothermal-isobaric (NPT) ensemble and periodic boundary condition. The CHARMM36-CAMP force field 34 was used for the protein, ions and water molecules. Atom charges of ZJ01 were calculated using the restrained electrostatic potential method 35 encoded in the AMBER suite of programs 36 at the RHF/6-31G* level. Covalent and non-bonded parameters for ZJ01 atoms were assigned by analogy or interpolation from those already present in the AMBER force field.
Energy minimizations were first performed to relieve unfavourable contacts in the system, followed by equilibration steps of 1 ns in total to equilibrate the complex. Subsequently, 50 ns production runs were performed. The temperature of the system was maintained at 300 K using the v-rescale method 37 with a coupling time of 0.1 ps. The pressure was kept at 1 bar using the Parrinello–Rahman 38 with τ p = 1.0 ps and a compressibility of 4.5 × 10−5 bar−1. SETTLE 39 constraints and LINCS 40 constraints were applied to the hydrogen-involved covalent bonds in water molecules and in other molecules, respectively, and the time step was set to 2 fs. Electrostatic interactions were calculated with the particle-mesh Ewald (PME) algorithm 41 with a real-space cut-off of 1.2 nm.
Binding free energy calculation by MMPBSA method
Based on the equilibrated dynamic trajectory, the binding free energy of the complex was calculated using the MM-PBSA method encoded in the AMBER 14.0 program. A total of 2500 snapshots from the trajectory were extracted every 20 ps, and the MM-PBSA calculation was performed on each snapshot using the MMPBSA.py module in AMBER 14.0.
Results
Chemistry
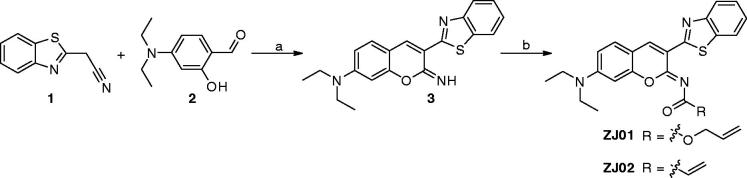
The synthetic route for ZJ01 is outlined in Scheme 1. Briefly, condensation of (1,3-benzothiazol-2-yl)acetonitrile (1) with 4-(diethylamino)-2-hydroxybenzaldehyde (2) readily produced the intermediate 3 23 , which was then converted to the target compounds by reaction with allyl carbonochloridate or acryloyl chloride.
Scheme 1.
Reagents and conditions: (a) a drop of piperazine, MeOH, r.t., 16 h; (b) DCM, Et3N, allyl carbonochloridate, 0 °C, overnight for ZJ01; DCM, Et3N, acryloyl chloride, 0 °C, overnight for ZJ02.
Biological evaluation
Hits identification and confirmation
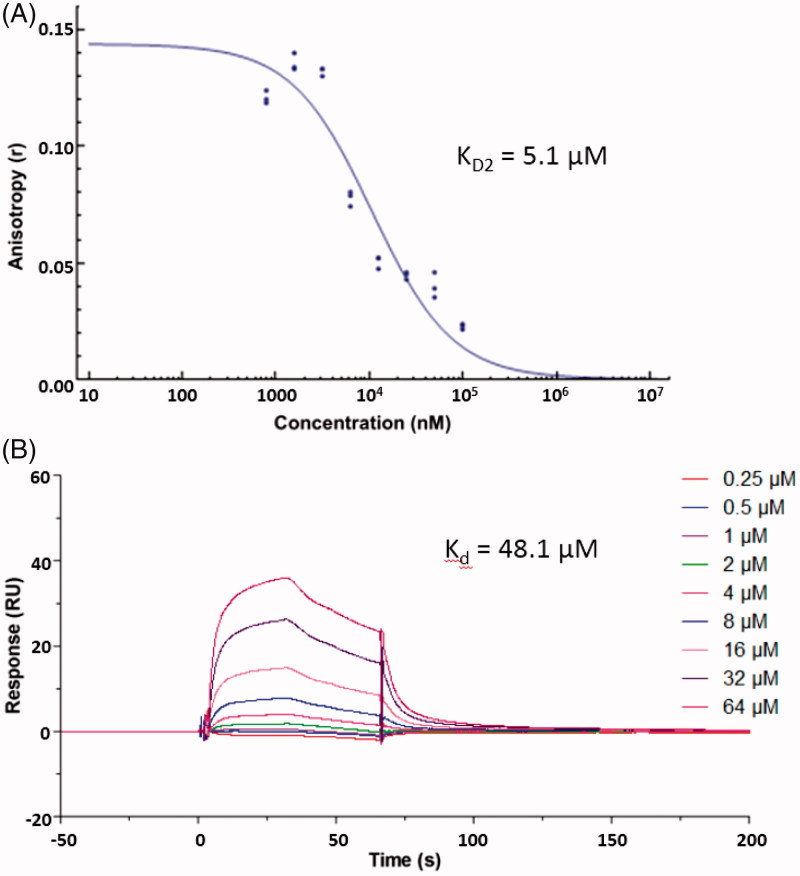
To identify Keap1–Nrf2 PPI inhibitors, our in-house library of 569 compounds was screened for their activity (K D2, K D1 is the equilibrium dissociation constant of the fluorescent probe) in FP assay 20 at the concentration of 100 μM, with the previously identified Keap1–Nrf2 PPI inhibitor S47 20 as a positive control. The assay results revealed synthetic compound ZJ01 to have the best inhibitory activity, demonstrating 81.25% inhibition of Keap1-Nrf2 PPI interaction at 100 μM. However, an analogue (ZJ02, Scheme 1) with the same core structure displayed only 20% inhibition in the initial testing, indicating that the side chain play a vital role in their activity. Compound ZJ01 was then retested to determine its K D2, which was found to be 5.1 μM (Figure 3(A)), slightly better than that of S47 (K D2 ≈ 7.6 μM, close to the reported 2.9 μM) 20 . Later, a surface plasmon resonance (SPR) competition assay 42 was employed to confirm the interaction between ZJ01 and the Keap1 Kelch domain. As shown in Figure 3(B), ZJ01 displayed an extracellular binding affinity with Keap1 with an equilibrium dissociation constant (K d) of 48.1 μM, somewhat weaker than that of S47 (K d ≈ 39.4 μM). The equilibrium binding curve fits are shown in Figures S14 and S15 (ESI†). These results indicated that ZJ01 can directly bind to Keap1 and disrupt the Keap1–Nrf2 interaction.
Figure 3.
(A) Dose–response curve of ZJ01 in FP assay. (B) SPR sensorgrams of ZJ01.
Molecular modelling study
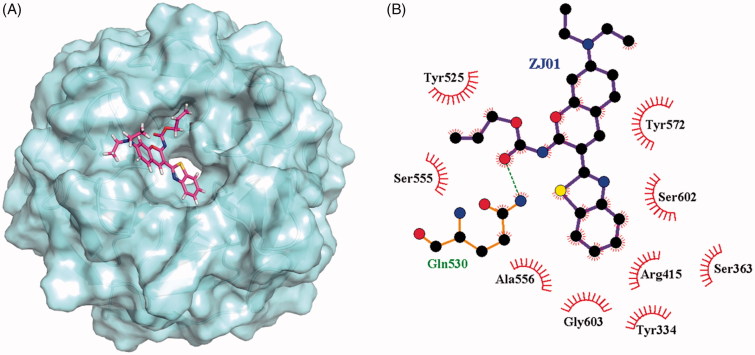
To investigate the possible binding mode of ZJ01 to the Keap1 Kelch domain, molecular docking was conducted using Autodock-4 25 . The binding mode of ZJ01 to Keap1 is shown in Figure 4. A 50 ns molecular dynamics (MD) study, discussed in the following section, did not indicate any instability of the docked Keap1/ZJ01 complex. Similar docking was also performed for Keap1/ZJ02. Compared with ZJ01, the result (Figure S16, ESI†) indicated that ZJ02 had a totally different spatial orientation at the Keap1–Nrf2 interaction surface, which possibly resulted from the protonated nitrogen atom of amide in ZJ02, which remained unprotonated in ZJ01. This was speculated to be the reason that these two compounds exhibited different activities in the FP assay.
Figure 4.
(A) Putative binding mode of ZJ01 to Keap1. (B) Schematic diagram showing interactions between ZJ01 and Keap1. Residues involved in hydrogen bonds and hydrophobic interactions are shown as sticks and starbursts, respectively. Molecular graphics figures were prepared with the LigPlot + program.
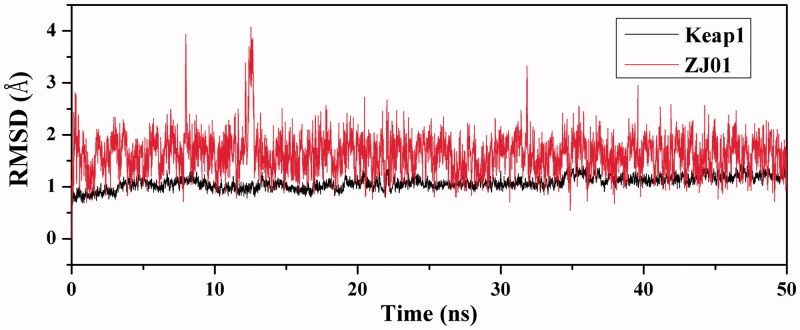
MD simulation
Steady RMSDs values indicate that the MD trajectory is reliable
To explore the dynamic traits of the Keap1/ZJ01 complex, a 50 ns MD simulation was performed on the complex model obtained by molecular docking. To examine the structural stability of the complex during MD simulations, the time evolution of weighted root-mean-square deviations (RMSDs) for backbone atoms of the Keap1 protein and heavy atoms of ZJ01 from their initial positions (t = 0) was calculated. As illustrated in Figure 5, RMSD values of the protein backbone were found to be between 0.5 and 1.2 Å during simulations. Steady RMSD values for the heavy atoms of the protein and ZJ01 indicated well-equilibrated states of the system. The steady RMSD values showed the reliability of the MD trajectories and suggested the suitability for post analysis.
Figure 5.
Time dependencies of RMSDs for the backbone atoms of the Keap1–ZJ01 complex.
H-bond and hydrophobic interaction analyses revealed a well-defined substrate pocket
To probe the molecular interactions between Keap1 and ZJ01, the H-bond and hydrophobic interactions of the complex were analysed based on MD trajectories. As shown in Table 2, there is only one conserved H-bond between Keap1 and ZJ01 in the complex. Additionally, the non-polar contacts between active site residues and ZJ01 cannot be ignored due to the fact that Keap1 has a relatively hydrophobic pocket. Thus, the occupancy rates of hydrophobic interactions between active site residues and ZJ01 were calculated based on MD trajectories. The result demonstrated that residues Tyr334, Arg415, Tyr525, Ala556, Tyr572, and Ser602 have more intense hydrophobic interactions with ZJ01 (Table 3). Taken together, these results indicated extensive polar and non-polar interactions between Keap1 and ZJ01.
Table 2.
Hydrogen bond existing in the complex and its occupancy during MD simulation.
| H-bond donor | H-bond acceptor | Occupancy rate (%) a |
|---|---|---|
| Gln530:NE2 | ZJ01:O2 | 99 |
Only H-bond occupancies >50% are shown.
Table 3.
Residues involved in hydrophobic interactions with ZJ01 during MD simulation and their corresponding occupancies rates.
| Residue | Occupancy rate (%) a |
|---|---|
| Tyr 334 | 94 |
| Arg 415 | 81 |
| Tyr 525 | 96 |
| Ala 556 | 89 |
| Tyr 572 | 99 |
| Ser 602 | 51 |
Only hydrophobic interaction occupancies >50% are shown.
Binding free energy analysis
To further explore the molecular interactions between Keap1 and ZJ01, the binding free energy values of the Keap1/ZJ01 complex were calculated using the MM-PBSA method encoded in the AMBER 14 program (in Table 4) 43 . The binding free energy (ΔG) value of –26.10 kcal/mol suggested a stable binding between ZJ01 and Keap1.
Table 4.
Binding free energy values of the Keap1/ZJ01 complex.
All calculated values are given in kcal/mol.
ΔG gas represents the binding free energy in vacuum.
ΔG solv represents the solvation free energy change calculated by the MM-PBSA method.
ΔG = ΔG gas + ΔG solv.
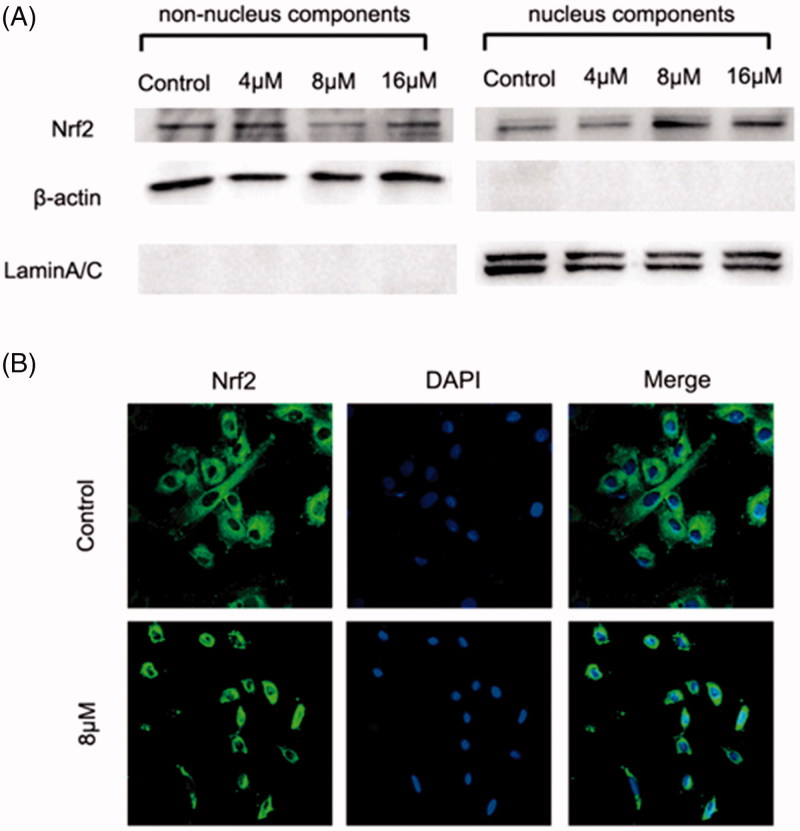
Nrf2–ARE pathway activation in H9c2 cardiac cells
The above results demonstrated that ZJ01 was an effective direct Keap1–Nrf2 PPI inhibitor. Previous studies have confirmed that Keap1–Nrf2 PPI inhibitors can make Nrf2 detach from Keap1 and subsequently translocate to the nucleus to activate the anti-oxidant defense system 20 , 21 . Therefore, the effect of ZJ01 on the distribution of Nrf2 in H9c2 cardiac cells was investigated. Western blotting analysis indicated that ZJ01 increased the nuclear Nrf2 protein levels and decreased the non-nuclear Nrf2 protein levels at 8 μM (Figure 6(A)). Meanwhile, the immunofluorescence assay demonstrated that Nrf2 translocated into the nucleus after the cells were treated with ZJ01 (Figure 6(B)) or S47 (Figure S17).
Figure 6.
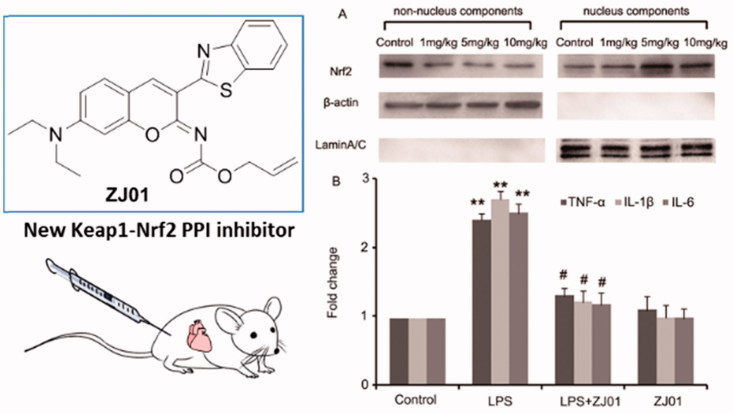
ZJ01 induced nuclear translocation of Nrf2 in H9c2 cells. H9c2 cells were treated with different concentrations of ZJ01 for 6 h. (A) Western blotting analysis of nuclear and non-nuclear Nrf2 protein levels. (B) Immunofluoresence staining analysis of Nrf2 localization. Nuclei were counterstained with PI.
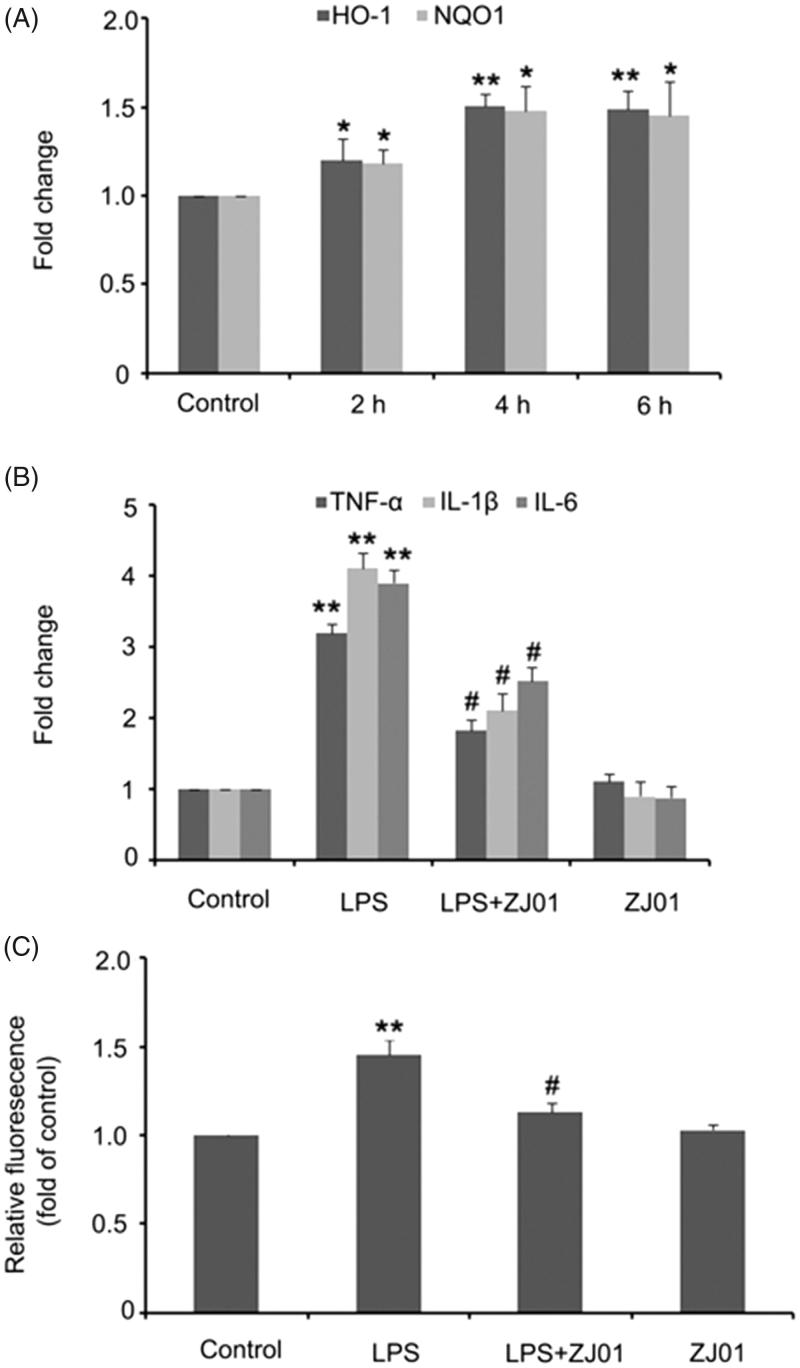
The nuclear Nrf2 could bind to the ARE of target genes, which then resulted in transcriptional induction and chemoprotection effects. Thus, the mRNA levels of Nrf2 target anti-oxidant genes, such as HO-1 and NQO1, were further examined in H9C2 cells after exposure to ZJ01. It was found that ZJ01 could significantly increase the HO-1 and NQO1 mRNA levels (Figure 7(A)). Altogether, these results revealed that ZJ01 activated the Nrf2–ARE pathway in H9c2 cardiac cells and might be a protective agent in H9c2 cardiac cells.
Figure 7.
ZJ01 increased the expression of anti-oxidant genes and alleviated LPS-induced production of pro-inflammatory cytokines and ROS in H9c2 cells. (A) RT-PCR analysis of anti-oxidant genes HO-1 and HQO1 after treatment of H9c2 cells with 8 μM ZJ01 for 6h. (B,C) H9c2 cells were stimulated with 1 μg/ml LPS and treated with or without 8 μM ZJ01 for 6 h. (B) The expression levels of pro-inflammatory cytokines TNF-α, IL-1β and IL-6 were determined by RT-PCR. (C) The intracellular ROS levels were examined by DCHF. *p < .05, **p < .01 vs. control, #p < .05 vs. LPS group. n = 3.
Anti-inflammatory response and anti-oxidative stress ability assay in H9c2 cardiac cells
Oxidative stress and inflammatory responses are responsible for the deterioration of sepsis-induced cardiomyopathy. LPS has been used to induce an inflammatory response and oxidative stress in H9c2 cells, which are typically used for in vitro investigations 4 , 44 . Inflammatory cytokines, including tumour necrosis factor (TNF)-α, interleukin (IL)-1α, IL-1β, and IL-6, promote the development of sepsis-induced cardiomyopathy 45 . Previously published studies indicated that LPS could increase the production of IL-1β, IL-6, and TNF-α in H9c2 cardiomyocytes and primary cardiomyocytes 4 , 46 , and additional evidence postulated that LPS-induced ROS could lead to the activation of intracellular signalling pathways and transcription factors, and subsequently induce the production of inflammatory mediators, including TNF-α, IL-1β and IL-6 7 , 47–49 . Therefore, inhibiting oxidative stress and blocking inflammatory signalling may produce beneficial effects in the dysfunctional heart. The effects of ZJ01 on the production of TNF-α, IL-1β, IL-6 and ROS were thus examined in LPS-stimulated H9c2 cells as well as the in vitro model.
Consistent with previously published data from other groups, our results showed that 1 μg/ml LPS induced a significant increase in mRNA levels of TNF-α, IL-1β and IL-6 in H9c2 cardiac cells. The 8 μM volume of ZJ01 or S47, which could effectively activate Nrf2, was applied to test its inhibitory effect on the production of TNF-α, IL-1β and IL-6 expression after exposure to LPS. It was observed that ZJ01 and S47 markedly alleviated LPS-induced production of these pro-inflammatory cytokines at the mRNA level (Figure 7(B); Figure S18A). Intracellular ROS levels were measured by dichloro-dihydro-fluorescein diacetate (DCFH-DA). The results showed that the ROS levels were significantly upregulated after treatment with 1 μg/ml LPS, and this increase could be blocked by 8 μM ZJ01 (Figure 7(C)). However, 8 μM S47 could not inhibit the elevated ROS levels induced by LPS (Figure S18B). The above results revealed that ZJ01 and S47 had the same anti-inflammatory effect, while ZJ01 had better anti-oxidant effects on LPS-treated myocardial cells. As ZJ01 is an effective Keap1–Nrf2 PPI inhibitor, we speculated ZJ01 might inhibit LPS-induced cardiomyopathy through disrupting Keap1–Nrf2 PPI which could induce Nrf2 nuclear accumulation and thus blocking oxidative and inflammatory signalling.
In vivo studies
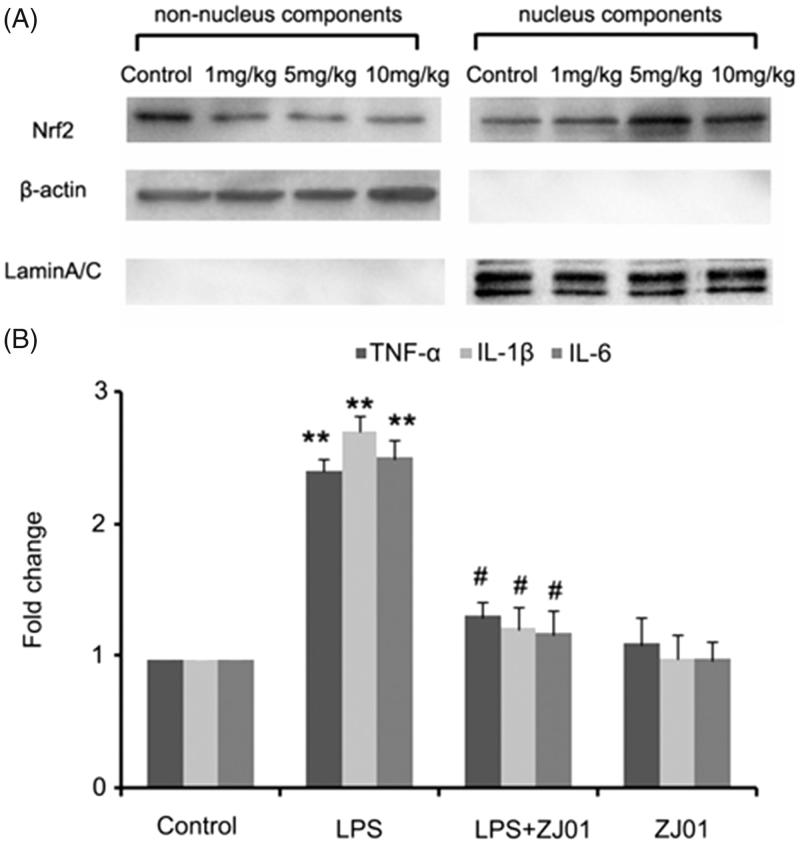
As left ventricular inflammation is an important feature of cardiac injury induced by LPS 16 , to examine whether ZJ01 and S47 were able to confer a cardiac protective effect in vivo, C57BL/6 mice were challenged with different concentrations of ZJ01 or S47 overnight for approximately 12 h after being treated intraperitoneally with or without 4 mg/kg of LPS, a dose commonly used for experimental models of septic cardiomyopathy in animals 16 . Then, the left ventricles were collected for further studies. Consistent with the in vitro experimental results, western blotting assay demonstrated that ZJ01 increased the nuclear Nrf2 protein levels and decreased the non-nuclear Nrf2 protein levels at 5 mg/kg or 10 mg/kg concentration suggesting ZJ01 could induce Nrf2 nuclear accumulation in left ventricular cells (Figure 8(A)). Real-time PCR results showed that inhibit LPS-induced inflammatory cytokines including TNF-α, IL-1β and IL-6 at the mRNA level in in left ventricular cells in vivo (Figure 8(B)). Compared to ZJ01, S47 could induce the Nrf2 nuclear accumulation at a 10 mg/kg concentration (Figure S19), but did not exhibit obvious effects on Nrf2 nuclear accumulation at the lower 5 mg/kg concentration. This result suggested that ZJ01 might be more effective than S47 in activating Nrf2 of cardiomyocytes in vivo.
Figure 8.
ZJ01 induced the Nrf2 nuclear accumulation and inhibited LPS-induced inflammatory cytokines production of left ventricular cells in vivo. (A) C57BL/6 mice were treated intraperitoneally with different concentrations of ZJ01 for 12 h. The nuclear and non-nuclear Nrf2 protein levels of left ventricular cells were determined by Western blotting technology. (B) C57BL/6 mice were treated intraperitoneally with 4 mg/kg of LPS and then with 5 mg/kg ZJ01 for 12 h. RT-PCR analysis of the expression of pro-inflammatory cytokines IL-1β, IL-6, and TNF-α. **p < .01 vs. control, #p < .05 vs. LPS group. n = 3.
Conclusions
In this study, a new Keap1–Nrf2 PPI inhibitor ZJ01 with an iminocoumarin-benzothiazole core was identified via FP and SPR assays. The amide side chain of ZJ01 was found to have an important role in its activity. ZJ01 could inhibit LPS-induced production of pro-inflammatory cytokines (TNF-α, IL-1β and IL-6) and ROS through activating the Nrf2–ARE pathway in H9c2 cells. In vivo, ZJ01 also induced Nrf2 nuclear accumulation and inhibited LPS-induced inflammatory cytokine production in cardiomyocytes. The biological data, together with computational results, indicated that ZJ01 could serve as a new template for generating direct Keap1–Nrf2 PPI inhibitors and possessed great potential in septic cardiomyopathy therapy.
Supplementary Material
Funding Statement
This research work was financially supported by the National Natural Science Foundation of China [Nos. 21672082, 31671214], Shandong Key Development Project [No. 2016GSF201209], the Young Taishan Scholars Program [No. tsqn20161037], Shandong Talents Team Cultivation Plan of University Preponderant Discipline [No. 10027], Fund of Shandong Province Natural Science Foundation [ZR2017BH038], Scientific Research fund of University of Jinan [Nos. XKY1609, 160100202], the Shanghai “ChenGuang” Project [No. 16CG42] and the Shanghai Municipal Commission of Health and Family Planning [No. 2017YQ052], and by the Brazilian Government Agencies FAP/DF [No. 0193.001020/2015] and CNPq [No. 447.628/2014-3].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1. Rackov G, Shokri R, De Mon MÁ, et al. The role of IFN-β during the course of sepsis progression and its therapeutic potential. Front Immunol 2017;8:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martin L, Peters C, Heinbockel L, et al. The synthetic antimicrobial peptide 19-2.5 attenuates mitochondrial dysfunction in cardiomyocytes stimulated with human sepsis serum. Innate Immun 2016;22:612–9. [DOI] [PubMed] [Google Scholar]
- 3. Lakshmikanth CL, Jacob SP, Chaithra VH, et al. Sepsis: in search of cure. Inflamm Res 2016;65:587–602. [DOI] [PubMed] [Google Scholar]
- 4. Yang P, Han Y, Gui L, et al. Gastrodin attenuation of the inflammatory response in H9c2 cardiomyocytes involves inhibition of NF-κB and MAPKs activation via the phosphatidylinositol 3-kinase signaling. Biochem Pharmacol 2013;85:1124–33. [DOI] [PubMed] [Google Scholar]
- 5. Yücel G, Zhao Z, El-Battrawy I, et al. Lipopolysaccharides induced inflammatory responses and electrophysiological dysfunctions in human-induced pluripotent stem cell derived cardiomyocytes. Sci Rep 2017;7:2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Natanson C, Danner RL, Elin RJ, et al. Role of endotoxemia in cardiovascular dysfunction and mortality. Escherichia coli and Staphylococcus aureus challenges in a canine model of human septic shock. J Clin Invest 1989;83:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turdi S, Han X, Huff AF, et al. Cardiac-specific overexpression of catalase attenuates lipopolysaccharide-induced myocardial contractile dysfunction: role of autophagy. Free Radic Biol Med 2012;53:1327–38. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8. Yao X, Carlson D, Sun Y, et al. Mitochondrial ROS induces cardiac inflammation via a pathway through mtDNA damage in a pneumonia-related sepsis model. PLoS One 2015;10:e0139416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suliman HB, Welty-Wolf KE, Carraway M, et al. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc Res 2004;64:279–88. [DOI] [PubMed] [Google Scholar]
- 10. Ahmed SM, Luo L, Namani A, et al. Nrf2 signaling pathway: pivotal roles in inflammation. Biochim Biophys Acta 2017;1863:585–97. [DOI] [PubMed] [Google Scholar]
- 11. Barančík M, Grešová L, Barteková M, Dovinová I.. Nrf2 as a key player of redox regulation in cardiovascular diseases. Physiol Res 2016;65:S1–S10. [DOI] [PubMed] [Google Scholar]
- 12. Zhuang CL, Wu ZL, Xing CG, Miao ZY.. Small molecules inhibiting Keap1–Nrf2 protein–protein interactions: a novel approach to activate Nrf2 function. MedChemCommun 2017;8:286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999;13:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu MC, Ji JA, Jiang YL, et al. An inhibitor of the Keap1–Nrf2 protein–protein interaction protects NCM460 colonic cells and alleviates experimental colitis. Sci Rep 2016;6:26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobayashi EH, Suzuki T, Funayama R, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun 2016;7:11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hao E, Lang F, Chen Y, et al. Resveratrol alleviates endotoxin-induced myocardial toxicity via the Nrf2 transcription factor. PLoS One 2013;8:e69452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magesh S, Chen Y, Hu L.. Small molecule modulators of Keap1–Nrf2–ARE pathway as potential preventive and therapeutic agents. Med Res Rev 2012;32:687–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abed DA, Goldstein M, Albanyan H, et al. Discovery of direct inhibitors of Keap1–Nrf2 protein–protein interaction as potential therapeutic and preventive agents. Acta Pharm Sin B 2015;5:285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marcotte D, Zeng W, Hus JC, et al. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg Med Chem 2013;21:4011–9. [DOI] [PubMed] [Google Scholar]
- 20. Zhuang C, Narayanapillai S, Zhang W, et al. Rapid identification of Keap1–Nrf2 small-molecule inhibitors through structure-based virtual screening and hit-based substructure search. J Med Chem 2014;57:1121–6. [DOI] [PubMed] [Google Scholar]
- 21. Jiang ZY, Lu MC, Xu LL, et al. Discovery of potent Keap1–Nrf2 protein–protein interaction inhibitor based on molecular binding determinants analysis. J Med Chem 2014;57:2736–45. [DOI] [PubMed] [Google Scholar]
- 22. Davies TG, Wixted WE, Coyle JE, et al. Monoacidic inhibitors of the Kelch-like ECH-associated protein 1: nuclear factor erythroid 2-related factor 2 (KEAP1:NRF2) protein–protein interaction with high cell potency identified by fragment-based discovery. J Med Chem 2016;59:3991–4006. [DOI] [PubMed] [Google Scholar]
- 23. Komatsu K, Urano Y, Kojima H, Nagano T.. Development of an iminocoumarin-based zinc sensor suitable for ratiometric fluorescence imaging of neuronal zinc. J Am Chem Soc 2007;129:13447–54. [DOI] [PubMed] [Google Scholar]
- 24. Kilkenny C, Browne WJ, Cuthill IC, et al. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010;8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem 1998;19:1639–62. [Google Scholar]
- 26. Jorgensen WL, Maxwell DS, Tirado-Rives J.. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 1996;118:11225–36. [Google Scholar]
- 27. Gasteiger J, Marsili M.. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 1980;36:3219–28. [Google Scholar]
- 28. Mezei M, Zhou MM.. Dockres: a computer program that analyzes the output of virtual screening of small molecules. Source Code Biol Med 2010;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Humphrey W, Dalke A, Schulten K.. VMD: visual molecular dynamics. J Mol Graph 1996;14:33–8. 27-8. [DOI] [PubMed] [Google Scholar]
- 30. Laskowski RA, Swindells MB.. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51:2778–86. [DOI] [PubMed] [Google Scholar]
- 31. Gordon JC, Myers JB, Folta T, et al. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 2005;33:W368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jorgensen WL, Chandrasekhar J, Madura ID, et al. Comparison of simple potential functions for simulating liquid water. J Chem Phys 1983;79:926–35. [Google Scholar]
- 33. Pronk S, Páll S, Schulz R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013;29:845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klauda JB, Venable RM, Freites JA, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 2010;114:7830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayly CI, Cieplak P, Cornell WD, Kollman PA.. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 1993;97:10269–80. [Google Scholar]
- 36. Case DA, Cheatham TE 3rd, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem 2005;26:1668–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bussi G, Donadio D, Parrinello M.. Canonical sampling through velocity rescaling. J Chem Phys 2007;126:014101. [DOI] [PubMed] [Google Scholar]
- 38. Parrinello M, Rahman A.. Polymorphic transitions in single crystals: a new molecular dynamic method. J Appl Phys 1981;52:7182–90. [Google Scholar]
- 39. Miyamoto S, Kollman PA.. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 1992;13:952–62. [Google Scholar]
- 40. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM.. LINCS: a linear constraint solver for molecular simulations. J Comput Chem 1997;18:1463–72. [Google Scholar]
- 41. Essmann U, Perera L, Berkowitz ML, et al. A smooth particle mesh ewald method. J Chem Phys 1995;103:8577–93. [Google Scholar]
- 42. Zhang J, Liu H, Zhu K, et al. Antiinfective therapy with a small molecule inhibitor of Staphylococcus aureus sortase. Proc Natl Acad Sci U S A 2014;111:13517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Case DA, Babin V, Berryman JT, et al. 2014, AMBER 14. San Francisco: University of California. [Google Scholar]
- 44. Jing R, Zhou Z, Kuang F, et al. microRNA-99a reduces lipopolysaccharide-induced oxidative injury by activating notch pathway in H9c2 cells. Int Heart J 2017;58:422–7. [DOI] [PubMed] [Google Scholar]
- 45. Marchant DJ, Boyd JH, Lin DC, et al. Inflammation in myocardial diseases. Circ Res 2012;110:126–44. [DOI] [PubMed] [Google Scholar]
- 46. Planavila A, Rodríguez-Calvo R, Jové M, et al. Peroxisome proliferator-activated receptor beta/delta activation inhibits hypertrophy in neonatal rat cardiomyocytes. Cardiovasc Res 2005;65:832–41. [DOI] [PubMed] [Google Scholar]
- 47. Planavila A, Redondo-Angulo I, Ribas F, et al. Fibroblast growth factor 21 protects the heart from oxidative stress. Cardiovasc Res 2015;106:19–31. [DOI] [PubMed] [Google Scholar]
- 48. Pan LL, Liu XH, Gong QH, Zhu YZ.. S-propargyl-cysteine (SPRC) attenuated lipopolysaccharide-induced inflammatory response in H9c2 cells involved in a hydrogen sulfide-dependent mechanism. Amino Acids 2011;41:205–15. [DOI] [PubMed] [Google Scholar]
- 49. Zhou H, Yuan Y, Liu Y, et al. Icariin protects H9c2 cardiomyocytes from lipopolysaccharide–induced injury via inhibition of the reactive oxygen species–dependent c–Jun N–terminal kinases/nuclear factor-κB pathway. Mol Med Rep 2015;11:4327–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.