Abstract
We have developed a method for turning on and off the expression of transgenes within Drosophila in both time and space. Two different enhancer detector elements carrying an RU486-inducible form of the yeast transcription factor GAL4 were constructed and used to generate enhancer detector lines. These lines were screened for RU486-inducible reporter gene expression in the adult head. We identified lines that exhibit inducible expression in many cell and tissue types, verifying that the elements respond to nearby enhancers. No expression was detected in the absence of the ligand. The P{Switch1} element responded to genomic enhancers less efficiently than P{Switch2} but produced more specific patterns of expression. Two P{Switch} lines were used to ablate fat body tissue in adult females through the induced expression of diphtheria toxin. These females were sterile, which correlates with fat body loss, and they died prematurely.
Forward genetic studies have contributed much to our understanding of the development, physiology, and behavior of Drosophila. To extract their full value, however, such studies need to be complemented with modern transgenic methodology, especially methods that allow for both spatial and temporal control over transgene expression. For the temporal control of transgene expression, surprising success has been obtained by using the heat shock (hsp70) promoter. For instance, we have used the hsp70 promoter to drive the expression of several cDNAs for the phenotypic rescue of learning mutants (1–3). This promoter does not provide for spatial control, however, and is expressed in essentially all cells. Significant control of transgene expression in space has been achieved by using the yeast GAL4 system (4). The GAL4 system uses a defined promoter or proximity to a genomic enhancer (because of the transposon insertion site) to drive expression of the yeast transcription factor, GAL4, in a spatially restricted manner. When combined in the genome with a transgene that has the GAL4 upstream activating sequences (UAS) next to a target gene, the target gene is expressed in tissues that express GAL4. Many research groups have used this technique. For instance, Waddell et al. (5) recently used the GAL4 system to drive the expression of an amnesiac (amn) cDNA in DPM neurons of amn mutants and observed rescue of the learning defect. Nevertheless, the technique provides no control over the timing of GAL4 expression. The FLP/FRT-recombinase system can be added to the GAL4 system to induce the expression of GAL4 as a function of FLP-recombinase expression (6), but this is of limited utility, in part because it can be applied only to dividing and not to postmitotic cells.
One system that offers potential for regulating genes in both time and space is based on a tetracycline-regulated transactivator (7). There are two forms of a tetracycline transactivator (tTA), which is a factor that binds to the tetracycline operator (tetO) in the absence of tetracycline and the reverse transactivator (rtTA), which binds to tetO in the presence of tetracycline. The administration or withdrawal of the tetracycline analog, doxycycline, can thus regulate a transgene flanked by tetO. The tTA system has been reported to function in Drosophila (8). The tTA was placed under the control of an eye-specific enhancer and used to drive the expression of a tetO-lacZ transgene on withdrawal of tetracycline from the food. The expression was first detected 6 h after tetracycline withdrawal, with a day or more required to achieve full expression. The rtTA system has also been engineered for use in Drosophila (9, 10). There are, however, significant disadvantages associated with this methodology. The tTA system requires the continuous presence of doxycycline to repress the target transgene, and chronic administration can have adverse effects (11). For Drosophila, it suffers a strong disadvantage over any two-part GAL4-based system. Each gene that is already available as a UAS construct in Drosophila transgenics for use with the GAL4 system would need to be reconstructed behind tetO and new transformants obtained.
There are two GAL4-based systems that offer promise for temporal and spatial control of transgene expression. One is based on estrogen activation of GAL4 activity (ref. 12; see Discussion). The other is based on a chimeric gene (Gene-Switch) that encodes the GAL4 DNA-binding domain, the human progesterone receptor-ligand-binding domain, and the activation domain from the human protein, p65 (13). In the presence of the antiprogestin, RU486, the chimeric molecule binds to a UAS and provides for ligand-inducible transactivation of downstream target genes. In a test of Gene-Switch in mammalian cells, the chimeric regulator was delivered to cultured cells along with a target gene, human growth hormone gene (UAS-hGH), carried on a adenovirus carrier vector (13). The transfected cells expressed hGH in an RU486-dependent manner. In addition, mice were infected with the adenovirus vector and found to exhibit rapid hGH induction with RU486. Constitutive and long-term expression (weeks) was achieved by implanting RU486 pellets into the animals, showing in addition that the expression system did not desensitize from the continuous presence of ligand or RU-Switch expression. Moreover, repetitive induction of hGH was achieved by RU application followed by withdrawal and reapplication. We have used Gene-Switch for an alternative Drosophila gene expression system. We show that enhancer detector elements constructed with a Gene-Switch cassette provide for experimental control of transgene expression in both time and space. In a companion paper, Osterwalder et al. (14) have used defined promoters to drive Gene-Switch in muscles and neurons.
Methods
Enhancer Detector Elements.
The pP{Switch1} vector was created by replacing the 2.9-kb GAL4 HindIII fragment from pP{GawB} (4) with the 2.5-kb KpnI/BamHI fragment from the Gene-Switch vector PAPCMV-GL914p65-SV (13) through MluI linkers. LoxP sites were synthesized as overlapping oligonucleotides and ligated into EcoRI and KpnI sites. The pP{Carnegie4} vector was used as the starting P-element for the P{Switch2} transposon. The amino terminus of Gene-Switch was modified by PCR to contain an EcoRI site just 5′ to the ATG translational start site (gaattcgATG) and the modified gene cloned as a EcoRI/XbaI fragment into pP{Carnegie4}. A 4.1-kb SpeI fragment containing the white+mC selectable marker was then cloned from pP{CasPer4} into the XbaI site to generate pP{Switch2}. The fusion at the EcoRI site of the P-transposase gene produces a new protein fusion containing 125 amino acids of the P-transposase.
Stocks and Genetics.
Flies were raised at 23°C on standard cornmeal, sucrose and yeast food, unless noted otherwise. The pP{Switch1} and pP{Switch2} plasmids were individually coinjected into w1118 embryos with the pturbo plasmid as the source for transposase (15). For additional insertions, the X-chromosomal P{Switch1}lines, S162 and S159, were mobilized by crossing with a TMS, Sb,Δ 2–3/TM6B line. Males of the genotype P{Switch1}/Y; TMS, Sb,Δ 2–3/+ were isolated and crossed to w1118 females. Male progeny with a w+ phenotype were selected and mapped. Fly lines designated S1 # carry a P{Switch1} element, and S2 # carry a P{Switch2} element.
RU486 Administration and Fecundity Assays.
A 10 mM stock solution of RU486 (mifepristone; Sigma) was made in 80% ethanol. For most experiments, 5–25 flies that were 1–4 days old were starved overnight in an empty food vial containing one Kimwipe (Structure Probe, West Chester, PA) wetted with 2 ml of water. RU486 was diluted to the final concentration in 2% sucrose; flies were treated as for starvation except that 2 ml of RU486 in 2% sucrose was added. Control flies were fed on 2% sucrose without RU486. Flies were fed at 25°C and 65% relative humidity, unless otherwise noted.
For fecundity assays, treated females were transferred individually to food vials with three wild-type males. For these assays, RU486 and control feeding solutions were supplemented with 0.5% Casamino acids (Difco). Females were placed with males for 48 h and then the vial was cleared of flies. Progeny were counted 2 weeks later.
Histology.
Cryosectioning and staining for β-galactosidase (β-gal) activity was performed as previously described (16). Staining of tissue with other stains followed standardized protocols. For fluorescent immunohistochemistry, flies were fixed in 3% PLP (3% paraformaldehyde/10 mM NaIO4/0.75 M lysine/37 mM phosphate buffer, pH 7.2) for 3 h at 4°C. They were cryoprotected in 25% sucrose overnight before cryosectioning. The rabbit anti-β-gal (Cappel) and rat anti-embryonic lethal abnormal vision (ELAV) (Developemental Studies Hybridoma Bank) primary antibodies were used at a dilution of 1:200 and 1:10, respectively. The Cy3 anti-rat (The Jackson Laboratory), and Alexa288 anti-rabbit (Molecular Probes) secondary antibodies were used at a 1:200 dilution.
Results
Generation of P{Switch} Enhancer Detector Elements.
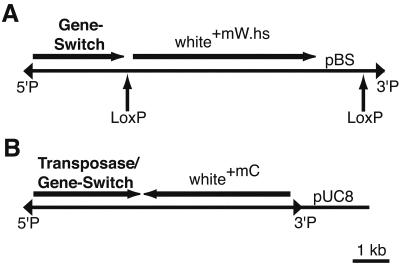
We constructed two enhancer detector elements that use the Gene-Switch fusion protein (Fig. 1; ref. 17). For the first, P{Switch1}, we replaced the GAL4 cassette of the enhancer detector vector, p{GawB} with a Gene-Switch cassette. In this vector, the Gene-Switch cassette was fused to the 5′-untranslated region of the P-transposase gene, so that translation is initiated from the first AUG of Gene-Switch, located 3′ to the P-transposase promoter and transcriptional start site. LoxP sites were inserted to flank the white+mW.hs selectable marker and pBluescript, to allow for their removal by CRE recombinase in transgenic flies if desired. For the second, P{Switch2}, we generated a protein fusion of Gene-Switch to the amino terminus of the P-element transposase within the P{Car4} transposon. The P{Switch2} element contains the P-transposase promoter, complete 5′-untranslated region of the transposase gene and its transcriptional start site, nuclear localization sequence, and first intron. This fusion is analogous to the P-transposase/β-gal fusions that are present in the enhancer detector elements, P{lArB} and P{Z}. The P{Switch2} transposon contains the white+mC as a selectable marker (Fig. 1B). A more detailed description of these vectors is given in Figs. 6 and 7, which are published as supporting information on the PNAS web site, www.pnas.org.
Figure 1.
Structure of the P{Switch} enhancer detector elements. (A) P{Switch1}. The Gene-Switch cassette was inserted downstream of the P-transposase promoter and transcriptional start site. The translational start site is within the Gene-Switch cassette. The positions of the LoxP sites are shown below the transposon and are marked by arrows. The 5′ and 3′ P-element inverted repeats are illustrated as triangles. (B) P{Switch2}. The Gene-Switch cassette was cloned in frame at an EcoRI site within the P-transposase gene.
A total of 56 independent transformants were obtained by injection of the P{Switch1} vector into embryos. Some of the insertions were mobilized to obtain 268 additional transpositions. The frequency of P{Switch1} interchromosomal transposition was 0.8%. A total of 58 independent P{Switch2} lines were generated by embryo injection.
P{Switch} Expression Patterns and RU486 Induction.
A total of 276 (Table 1) of the P{Switch} lines were crossed to P{UAS-LacZ.B} bearing animals. Progeny from these crosses were fed for 1 day on 500 μM RU486 in 2% sucrose or on 2% sucrose without the antiprogestin. In the initial screen of 218 P{Switch1} lines, we detected 25 lines that expressed β-gal within the head (Table 1). The reporter expression depended on RU486 administration in every line tested (Fig. 2). A higher percentage of the P{Switch2} lines expressed β-gal within the head (Table 1), potentially reflecting a higher sensitivity of this vector to genomic enhancers and/or a higher expression level of Gene-Switch. Diverse expression patterns were observed for both P{Switch} elements (Table 2; Fig. 3), although none were found with ubiquitous expression of β-gal in the head. The majority of P{Switch1}lines were relatively specific for expression in only one head structure or tissue type (30 distinct patterns in 25 lines), whereas the P{Switch2} lines frequently had broader expression (47 patterns in 22 lines; Table 2).
Table 1.
P{Switch} lines showing β-gal expression in the adult head
| Lines screened | Positive lines | Expression frequency | |
|---|---|---|---|
| P{Switch1} | 218 | 25 | 12% |
| P{Switch2} | 58 | 22 | 38% |
| Totals | 276 | 47 | 17% |
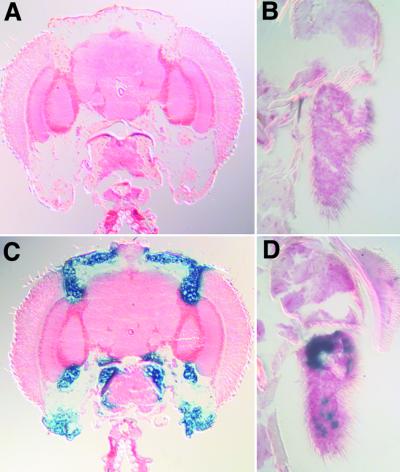
Figure 2.
P{Switch} elements provide inducible gene expression. Frontal head sections of S132/P{UAS-lacZ.B}, uninduced (A) and induced (C). Robust fat body expression is observed after induction. The third antennal segment of S1155/+; P{UAS-lacZ.B}, uninduced (B) and induced (D).
Table 2.
Expression patterns of P{Switch} lines
| Pattern | P{Switch1} lines | P{Switch2} lines | Total |
|---|---|---|---|
| Central brain | 4 | 7 | 11 |
| Neurolemma | 7 | 6 | 13 |
| Antenna | 5 | 12 | 17 |
| Maxillary palps | 3 | 9 | 12 |
| Ocellar ganglion | 2 | 3 | 5 |
| Fat body | 7 | 8 | 15 |
| Bristles | 2 | 2 | 4 |
| Totals | 30 | 47 | 77 |
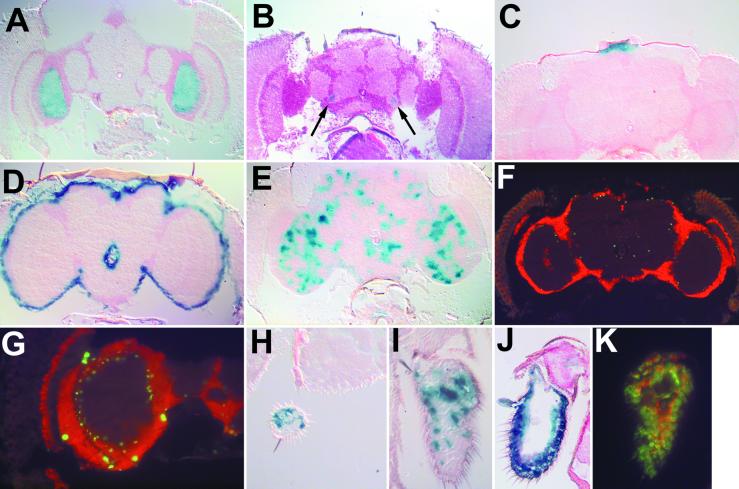
Figure 3.
P{Switch} enhancer detectors induce reporter expression in a variety of tissues and cell types. (A) Line S26 shows expression specifically in the medulla. (B) S21 shows expression near the antennal nerve (arrow). (C) S144 drives expression in the ocellar ganglion. (D) S113 expression is found within the neurolemma. (E) S16 is expressed in a diffuse pattern throughout the central brain and optic lobes. (F, G, and K) Gene-Switch in S16, S1145, and S233 is expressed in nonneuronal cell types. Shown are 20 μM cyrosections in which all neurons are labeled by anti-embryonic lethal abnormal vision (ELAV) in red. Nuclear localized β-gal, corresponding to P{Switch} expression, is labeled in green. (F) S16 frontal section. The β-gal positive nuclei lie just inside the cellular cortex. (G) S1145 optic lobes. Several large nuclei are labeled on the perimeter of the optic lobes with smaller nuclei just inside the layer of neuronal cell bodies. (K) S233 third antennal segment. The β-gal-positive nuclei are distinct from neurons in the antenna. S186 is expressed within both the maxillary palps (H) and the third antennal segment (I). (J) S233 exhibits broad expression in the third antennal segment as detected by β-gal histochemistry
Two lines, S1145 and S16, showed interesting but diffuse staining patterns within the central brain as detected with a cytoplasmic β-gal reporter, which diffuses into the processes of expressing neurons (see S16, Fig. 3E). S1145, but not S16, also exhibited expression within the neurolemma. To explore these patterns in more detail, we induced the expression of a nuclearly localized β-gal in these lines and identified expressing nuclei through immunofluorescence (Fig. 3 F, G, and K). In this experiment, the nuclei of neurons were identified with an antibody against the pan-neural protein embryonic lethal abnormal vision (ELAV). For both S16 and S1145, we observed nonneuronal cells just within the neural cell body layer (Fig. 3 F and G); in a separate experiment, these cells were colabeled with anti-repo, a glial-specific marker (not shown). In S1145, there are also large nuclei on the surface of the brain that were colabeled with anti-repo antibodies; these nuclei are probably within neurolemma (Fig. 3G). Additionally, we found nonneuronal staining within the third antennal segment of S233 (Fig. 3K). Thus, these P{Switch} transposons are capable of detecting a number of cell- and tissue-specific enhancers, and RU486 is capable of penetrating these cells and activating the Gene-Switch protein.
Kinetics of Gene-Switch Induction.
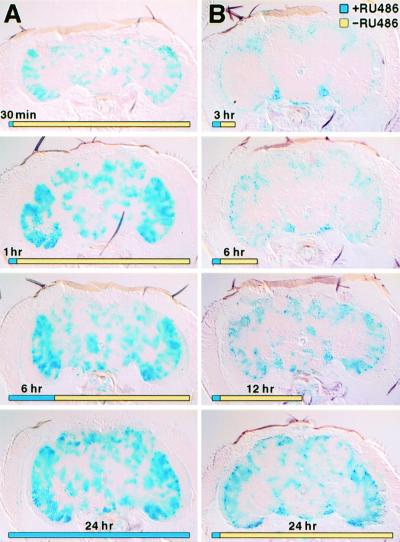
We studied the kinetics of Gene-Switch induction in several enhancer detector lines with β-gal expression in the central brain by histochemical detection of β-gal activity. Flies of the genotype S16/P{UAS-lacZ.B} and S152/P{UAS-lacZ.B} were fed for 24 h with either 2% sucrose alone or 2% sucrose containing RU486 in concentrations from 90 μM to 2 mM. After the feeding, flies were immediately sectioned and stained for β-gal. The concentration of 500 μM RU486 induced maximal β-gal activity. Next, we examined the time requirement of RU486 administration by feeding S16/P{UAS-lacZ.B} flies for varying periods of time and then transferring the flies to cornmeal/sucrose food without RU486. These flies were sectioned 24 h after initial feeding (Fig. 4A). Flies fed RU486 for 1 h displayed as robust β-gal activity as flies fed for a full 24 h, suggesting that RU486 uptake reached saturating levels within the first hour. In our next experiment, we fed S16/P{UAS-lacZ.B} flies RU486 for 1 h and then transferred these flies to cornmeal/sucrose food without RU486. After 3, 6, 12, or 24 h, we sectioned the flies and stained for β-gal activity. After 3 or 6 h, β-gal activity was detectable and localized principally as punctate clusters in the cellular cortex (Fig. 4B). By 12 h, the activity was found in the neuropil and was widespread after 24 h (Fig. 4B). Similar results were found for the S152 line. As an alternative to feeding, we also completely immersed adult flies in a sucrose solution with RU486. Surprisingly, flies immersed completely for 1 h still recover when removed to a food vial. However, the induction of Gene-Switch by this mechanism, which presumably involves diffusion through the cuticle and perhaps uptake through the trachea, occurred no more rapidly than with feeding.
Figure 4.
The induction of β-gal in P{Switch1} line S16. In A, S16/P{UAS-lacZ.B} flies were fed RU486 for the indicated times (blue portion of timeline) and then transferred to food without RU486 (yellow portion of timeline). All heads were cut at 24 h. In B, all flies were fed for 1 h and then transferred to food without RU486. Heads were sectioned at the time indicated.
In contrast to the rapid induction of the reporter, the decay of reporter activity after RU486 withdrawal was slow. Flies were administered RU486 for 24 h and then transferred to a food source without RU486. Histochemical assays for β-gal activity in the head at several times after treatment revealed that the flies still showed significant activity even after 6 days of withdrawal. It is not presently known whether this is because of the stability of the reporter, or whether there remains a persistent activation of the Gene-Switch after withdrawal.
These results indicate that the fly rapidly assimilates RU486 and activates gene expression within as little as 3 h. The uptake of RU486 is not the rate-limiting step for induction, because 1- and 24-h feeding periods activate reporter to similar levels (compare 1- and 24-h timepoints, Fig. 4A). Rather, the rate-limiting step must be the activation of the Gene-Switch molecule or the maturation of β-gal.
Induced Lethality and Ablation of Fat Bodies by P{Switch} Expressed Diphtheria Toxin.
The S1106 and S230 lines both drive reporter expression within adult fat bodies. In S230, expression is also detected in the oenocytes and hindgut. Both of these lines also show expression in the midgut, although the substantial amount of endogenous β-gal activity in the tissue makes the level of induction difficult to quantify. We used these lines to express the diphtheria toxin A chain from the P{UAS-DTI} transposon (Table 3; Fig. 5). These transheterozygotes at 25°C showed no change in viability in the absence of RU486 relative to control siblings. Also at 25°C, S230/P{UAS-DTI} flies, given chronic administration of RU486, began dying at 4 days, and nearly all flies were dead by the fifth day. The S1106/+; P{UAS-DTI}/+ induced flies began dying at 5 days of treatment and were all dead by the sixth day (Table 3). At 18°C, S1106/+; P{UAS-DTI}/+ flies treated with RU486 survived to 7 days but were female sterile (Table 4). The enhanced survival at 18°C may be because of reduced Gene-Switch activity at low temperature. Thus, temperature may also provide for finer control over gene activation with P{Switch} transposons.
Table 3.
Induced expression of diptheria toxin within adult fat bodies produces lethality
| Genotype | % lethality
|
|
|---|---|---|
| −RU486 (n) | +RU486 (n) | |
| Wild type | 7.1 (70) | 5.7 (70) |
| S1106/+ | 8.0 (50) | 4.0 (50) |
| S1106/+;UAS-DTI/+ | 9.6 (52) | 100.0 (50) |
| S230/+ | 7.5 (40) | 5.0 (50) |
| S230/UAS-DTI | 10.0 (50) | 100.0 (50) |
| UAS-DTI/+ | 6.0 (50) | 6.0 (50) |
Lethality was measured after chronic administration for 7 days at 25°C. n, number assayed.
Figure 5.
Induction of diphtheria toxin (DTI) within adult fat bodies produces tissue ablation. Sagittal cryosections of P{Switch} abdomens are shown. Arrowheads point to areas that contain the dispersed fat body tissue. A–D are stained for β-gal activity. E–H are stained with hematoxylin and eosin. I and J are fixed and stained with osmium tetroxide. (A) S1106/P{UAS-lacZ.B}, uninduced (B) P{UAS-lacZ.B}/+; S230/+ uninduced. (C) S1106/P{UAS-lacZ.B} induced. (D) P{UAS-lacZ.B}/+; S230/+ induced. (E) S1106/P{UAS-DTI} uninduced. (F) P{UAS-DTI}/+; S230/+ uninduced. (G) S1106/P{UAS-DTI } induced. (H) P{UAS-DTI}/+; S230/+ induced. (I) S1106/P{UAS-DTI} uninduced. (J) S1106/P{UAS-DTI} induced.
Table 4.
Expression of diphtheria toxin within fat bodies at 18°C results in female sterility
| Genotype | % lethality
|
Fecundity*
|
||
|---|---|---|---|---|
| −RU486 | +RU486 | −RU486 | +RU486 | |
| Wild type | 8.0 (50) | 6.0 (50) | 14.5 ± 1.1 | 13.7 ± 1.4 |
| S1106/+ | 9.5 (21) | 10.0 (20) | 12.9 ± 1.5 | 12.3 ± 1.3 |
| UAS-DTI/+ | 8.0 (50) | 6.0 (50) | 12.4 ± 1.0 | 12.8 ± 0.9 |
| S1106/+;UAS-DTI/+ | 6.0 (50) | 8.0 (50) | 13.3 ± 0.9 | 0 |
Defined as the average number of progeny produced for 2 days after treatment.
We stained abdominal sections of female S230/P{UAS-DTI} and S1106/+; P{UAS-DTI}/+ flies after 3 days of RU486 treatment at 25°C. In 11 of 12 diphtheria toxin-expressing flies, the fat bodies were completely ablated, whereas they remained present in all 16 of the control flies. These data indicate that both P{Switch1} and P{Switch2} elements can drive the expression of diphtheria toxin to levels capable of producing lethality or destroying a complete organ system.
Discussion
A common limitation in many genetic studies of physiology, development, or behavior is the inability to simultaneously control gene expression in a spatial and temporal way. In this paper, we describe a gene expression system for Drosophila that allows for control in both dimensions. Temporal control was achieved through the use of Gene-Switch, a fusion protein containing the GAL4 DNA-binding domain fused to the ligand-binding domain of the human progesterone receptor and the p65 transcriptional activation domain (13). The binding of antiprogestins such as RU486 creates a conformation change in Gene-Switch resulting in nuclear translocation and a transcriptional activation from a GAL4 promoter (18, 19). The activation is rapid. A maximal rate of accumulation of β-gal was achieved by feeding adults RU486 for as little as 1 h, and reporter activity was observed just 3 h after the start of feeding. Ceiling levels of activity required between 24 and 48 h of incubation.
Spatial control of Gene-Switch expression was provided through enhancer detection. The quasirandom integration of P-factors into the genome and the ability of the P-factor promoter to be regulated by nearby enhancers and other regulatory elements is a well-characterized and powerful tool in Drosophila genetics (20–22). Many different expression patterns were detected among the several hundred lines that we have generated so far, and the cloning of several of these have revealed the integration of the element into distinct genomic sites. Large-scale enhancer detector screens similar to those performed with more conventional enhancer detector elements (4, 21–23) should allow for the isolation of GAL4 drivers with virtually any spatial pattern of expression along with temporal control. Alternatively, well-defined promoters or enhancers could be used to drive the expression of Gene-Switch (ref. 14; and G.R., unpublished work). These reagents, along with the hundreds of UAS transgenes that already exist among Drosophila researchers, should allow for a powerful new way to manipulate gene expression.
Two of the potential problems that might have limited the effectiveness of this system—an inability to induce expression in many tissue and cell types and a slow time course for induction—can therefore be eliminated. Another potential problem with any inducible expression system is the background level of expression in the absence of the inducer. We have found, however, virtually no expression of reporter (in the head) in the several hundred lines examined so far. Many of the lines have exhibited an extremely faint trace of β-gal expression in the hypodermis of the proboscis, but this is the only common pattern that has been observed repeatedly in the absence of inducer. In contrast, this has been a significant problem with other enhancer detector elements. For example, we observed antennal expression in the vast majority of lines that we produced with the enhancer detector element, P{lArB} (23). In addition, our evaluation of 20 lines of an enhancer detector element made with an estrogen receptor-GAL4 fusion (12) revealed a common and inducible expression pattern in the neurolemma (not shown). These unwanted expression patterns, of course, limit the utility of the system. They are presumably caused by the novel juxtaposition of sequences within the enhancer detector element to produce enhancer activity that is relatively independent of insertion site.
Another limitation of such inducible gene expression systems is unwanted nonspecific effects of the inducer (ligand) itself. We have failed to see significant alterations in the physiology or behavior of animals treated at the doses used here, although we have not explored the effects of administration during development. No increased lethality was observed in control flies treated with RU486 (Tables 3 and 4). Moreover, preliminary tests for several different behaviors, including phototaxis, geotaxis, locomotion, and escape responses, have failed to show any effect. Thus, the only effect of RU486 on flies so far appears to be the induction of Gene-Switch.
In a test of biological function, we ablated the complete fat body organ system through the expression of diphtheria toxin A chain within this tissue. Although several functions have been attributed to this organ, its requirement for Drosophila viability and fecundity has never been directly addressed. Flies missing fat body died within a few days. Because the fat body is responsible for the production of antibiotic peptides, the loss of this system may result in a severe immune deficiency (24). When the induction of diphtheria toxin was performed at 18°C, the female flies survived for a longer period but were sterile. The longer survival at 18°C may reflect a reduced activity of Gene-Switch or a decreased metabolic demand at the lower temperature. The sterility associated with these flies may be because of the absence of yolk protein biosynthesis within this tissue. In Drosophila, there are three yolk protein genes (25, 26). These proteins provide the developing embryo with a source of amino acids. The yolk protein genes are transcribed solely within the female fat bodies and follicle cells; yolk protein 3 is almost entirely synthesized within the fat body (27, 28). Yolk proteins produced in the fat body are translocated into the hemolymph and taken up by the developing oocyte by receptor-mediated endocytosis (26, 28, 29). Mutations in any of the three yolk proteins reduce female fertility, and there is an additive effect such that each gene is required for full fecundity (30, 31). It is also possible that expression of diphtheria toxin within the midgut generates a nutritional deficit. Yolk protein biosynthesis in both fat body and oocytes is induced only after a protein-rich diet, and most amino acids are absorbed through the midgut (32, 33). Thus, after removing the fat bodies as a source of yolk proteins, and the possible reduction in amino acid uptake by these flies, the ovaries appear incapable of producing fertile oocytes. In the absence of RU486, the fat body remained intact in females carrying both the P{Switch} lines and the UAS-DTI transgene. These females displayed normal fertility and fecundity in the absence of RU486, suggesting that fat body function remains unimpaired in the absence of RU486.
Supplementary Material
Acknowledgments
We thank T. Osterwalder, H. Keshishian and colleagues for sharing their results before publication. National Institutes of Health Grants NS19904 and GM63929 from the National Institutes of Health supported this work. Additional support came from the R. P. Doherty-Welch Chair in Science. Fly stocks were kindly provided by the Bloomington Stock Center and L. Stevens (Albert Einstein College of Medicine, Bronx, NY). Jin He provided expert technical assistance.
Abbreviations
- UAS
upstream activating sequence
- tTA
tetracycline transactivator
- tetO
tetracycline operator
- rtTA
reverse transactivator
- β-gal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dauwalder B, Davis R L. J Neurosci. 1995;15:3490–3499. doi: 10.1523/JNEUROSCI.15-05-03490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grotewiel M S, Beck C D, Wu K H, Zhu X R, Davis R L. Nature (London) 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y, Endo K, Wu K, Rodan A R, Heberlein H, Davis R L. Cell. 2001;105:757–768. doi: 10.1016/s0092-8674(01)00386-5. [DOI] [PubMed] [Google Scholar]
- 4.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 5.Waddell S, Armstrong D J, Kitamoto T, Kaiser K, Quinn W G. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 6.Struhl G, Basler K. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
- 7.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bello B, Resendez-Perez D, Gehring W J. Development (Cambridge, UK) 1998;125:2193–2202. doi: 10.1242/dev.125.12.2193. [DOI] [PubMed] [Google Scholar]
- 9.Bieschke E T, Wheeler J C, Tower J. Mol Gen Genet. 1998;258:571–579. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- 10.Stebbins M J, Urlinger S, Byrne G, Bello B, Hillen W, Yin J C P. Proc Natl Acad Sci USA. 2001;98:10775–10780. doi: 10.1073/pnas.121186498. . (First Published August 21, 2001; 10.1073/pnas.121186498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansuy I M, Winder D G, Moallem T M, Osman M, Mayford M, Hawkins R D, Kandel E R. Neuron. 1998;21:257–265. doi: 10.1016/s0896-6273(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 12.Han D D, Stein D, Stevens L M. Development (Cambridge, UK) 2000;127:573–583. doi: 10.1242/dev.127.3.573. [DOI] [PubMed] [Google Scholar]
- 13.Burcin M M, Schiedner G, Kochanek S, Tsai S Y, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:355–360. doi: 10.1073/pnas.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterwalder T, Yoon K S, White B H, Keshishian H. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomlinson A, Kimmel B E, Rubin G M. Cell. 1988;55:771–784. doi: 10.1016/0092-8674(88)90133-x. [DOI] [PubMed] [Google Scholar]
- 16.Han P L, Levin L R, Reed R R, Davis R L. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Xu J, Pierson T, O'Malley B W, Tsai S Y. Gene Ther. 1997;4:432–441. doi: 10.1038/sj.gt.3300402. [DOI] [PubMed] [Google Scholar]
- 18.Tsai M J, O'Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, O'Malley B W, Jr, Tsai S Y, O'Malley B W. Proc Natl Acad Sci USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Kane C J, Gehring W J. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellen H J, O'Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 22.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 23.Han P L, Meller V, Davis R L. J Neurobiol. 1996;31:88–102. doi: 10.1002/(SICI)1097-4695(199609)31:1<88::AID-NEU8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman J A, Reichhart J-M, Hetru C. Curr Opin Immunol. 1996;8:8–13. doi: 10.1016/s0952-7915(96)80098-7. [DOI] [PubMed] [Google Scholar]
- 25.Bownes M. FEBS Lett. 1979;100:95–98. doi: 10.1016/0014-5793(79)81138-2. [DOI] [PubMed] [Google Scholar]
- 26.Bownes M, Hames B D. J Embryol Exp Morphol. 1978;47:111–120. [PubMed] [Google Scholar]
- 27.Barnett T, Pachl C, Gergen J P, Wensink P C. Cell. 1980;21:729–738. doi: 10.1016/0092-8674(80)90436-5. [DOI] [PubMed] [Google Scholar]
- 28.Williams J L, Bownes M. Eur J Biochem. 1986;161:95–101. doi: 10.1111/j.1432-1033.1986.tb10128.x. [DOI] [PubMed] [Google Scholar]
- 29.Mahowald A P. In: Developmental Systems: Insects. Counce S J, Waddington C H, editors. Vol. 1. New York: Academic; 1972. pp. 1–47. [Google Scholar]
- 30.Postlethwait J H, Kaschnitz R. FEBS Lett. 1978;95:247–251. doi: 10.1016/0014-5793(78)81004-7. [DOI] [PubMed] [Google Scholar]
- 31.Bownes M, Lineruth K, Mauchline D. Mol Gen Genet. 1991;228:324–327. doi: 10.1007/BF00282485. [DOI] [PubMed] [Google Scholar]
- 32.Sondergaard L, Mauchline D, Egetoft P, White N, Wulff P, Bownes M. Mol Gen Genet. 1995;248:25–32. doi: 10.1007/BF02456610. [DOI] [PubMed] [Google Scholar]
- 33.Roman G, Meller V, Wu K H, Davis R L. Am J Physiol. 1998;275:C857–C869. doi: 10.1152/ajpcell.1998.275.3.C857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.