Abstract
A series of 3-3-{2-[2-3-methyl-4-phenyl-2,3-dihydro-1,3-thiazol-2-ylidene]hydrazin-1-ylidene-2,3-dihydro-1H-indol-2-one derivatives has been designed and synthesized to study their activity on both HIV-1 (Human Immunodeficiency Virus type 1) RT (Reverse Transcriptase) associated functions. These derivatives are analogs of previously reported series whose biological activity and mode of action have been investigated. In this work we investigated the influence of the introduction of a methyl group in the position 3 of the dihydrothiazole ring and of a chlorine atom in the position 5 of the isatin nucleus. The new synthesized compounds are active towards both DNA polymerase and ribonuclease H in the µM range. The nature of the aromatic group in the position 4 of the thiazole was relevant in determining the biological activity.
Keywords: HIV-1 therapeutic agents, Isatin derivatives, RT dual inhibitors
Introduction
HIV-1 (Human Immunodeficiency Virus type 1) is one of the major causes of death now days. There are different approaches to keep under control this infection. The current approved treatment is based on the highly active antiretroviral therapy (HAART), which associates a combination of antiviral agents, targeting different steps of the virus replication cycle1–3. This multidrug therapeutic regimen leads to the reduction of the amount of circulating virus, in some cases below the current blood testing techniques detectable level, and allows high control of the infection. Moreover, it leads to the reduction of drug resistance occurrence, decrease of mortality and morbidity rates, and an overall improvement of patients quality of life4.
However, there is not a therapeutic regimen capable of completely eradicate the virus from the host and, therefore, due to the chronic nature of HIV infection, a lifelong therapy is required. Hence both adherence to treatment and the management of drug-related toxicities are issues to deal with. In the light of the above the design and synthesis of new and more effective antiviral agents is an attractive open field for medicinal chemists.
In this respect, the identification of a multiple-acting molecule, able to inhibit different steps of the virus replication cycle, appears as a promising approach for the design of new and likely more efficient antiviral agents. Such agent should combine a dual function targeting capability in only one molecule5–8. Albeit HIV-1 Reverse Transcriptase (RT) has been the first and most investigated target for the therapy of HIV-1 infected patients, RT inhibitors (RTIs) are within the most represented components of HAART3. RT plays a key role in the HIV-1 replication cycle and is therefore a strategic target for the design of new therapeutic agents to combat the virus infection9. The molecular aspects of the RT/drug interaction have been reviewed by some of us and indication for the design of dual-acting RT inhibitors outlined10. Moreover the identification, design and synthesis of small molecules, able to simultaneously inhibit the two RT associated enzymatic functions, namely DNA/RNA dependent polymerase (DDDP and RDDP) and ribonuclease H (RNase H) functions, has been reported11–14. Isatin based molecular hybrids have been reported as a valid scaffold for the design of multi-target agents15–19, and in particular for the dual inhibition of RT associated enzymatic functions12,14. Hence, in continuation with our previous studies, we report on the synthesis and the structure-activity relationships of a series of 3-[2-(3-methyl-4-aryl-1,3-thiazol-2-ylidene)hydrazin-1-ylidene]-1H-indol-2-ones whose activity has been evaluated towards both RT associated enzymatic functions DDDP/RDDP and RNase H.
Methods
Materials and apparatus
Starting materials and reagents were obtained from commercial suppliers and were used without purification. Chemical reagents were purchased form Sigma-Aldrich (St. Louis, MO). RNA–DNA labeled sequences were purchased from Metabion international AG.
All melting points were determined on a Stuart SMP11 melting points apparatus and are uncorrected. Electron ionization mass spectra were obtained by a Fisons QMD 1000 mass spectrometer (70 eV, 200 mA, ion source temperature 200 °C). Samples were directly introduced into the ion source. Found mass values are in agreement with theoretical ones. Melting points, yield of reactions and descriptive data of derivatives EMAC 3039–3064 are reported in Table 1.
Table 1.
Analytical data of derivatives EMAC 3039–3064.
| C–H–N |
|||||||
|---|---|---|---|---|---|---|---|
| Compound | M.w. | Yield % | M.p. °C. | Crystals color | Cryst. solvent | Calc. | Found |
| EMAC 3039 | 449.75 | 71.15 | >250d | Orange | Water/ethanol | C, 58.61; H, 3.55; N, 15.19 | C, 58.57; H, 3.53; N, 15.17 |
| EMAC 3040 | 433.29 | 84.25 | 210–212 | Orange | Water/ethanol | C, 61.35; H, 3.72; N, 15.90 | C, 61.45; H, 3.70; N, 15.87 |
| EMAC 3041 | 494.20 | 91.70 | >250d | Orange | Water/ethanol | C, 52.31; H, 3.17; N, 13.56 | C, 52.33; H, 3.15; N, 13.53 |
| EMAC 3042 | 460.30 | 76.05 | >250d | Orange | Water/ethanol | C, 56.98; H, 3.45; N, 18.46 | C, 56.95; H, 3.41; N, 18.43 |
| EMAC 3043 | 491.40 | 82.40 | >250d | Orange | Water/ethanol | C, 70.22; H, 4.42; N, 13.65 | C, 70.27; H, 4.43; N, 13.62 |
| EMAC 3044 | 440.31 | 81.75 | >250d | Orange | Water/ethanol | C, 63.49; H, 3.65; N, 19.49 | C, 63.51; H, 3.62; N, 19.44 |
| EMAC 3045 | 406.84 | 73.75 | >250d | Orange | Water/ethanol | C, 58.37; H, 3.27; N, 15.13 | C, 58.38; H, 3.23; N, 15.11 |
| EMAC 3046 | 460.30 | 79.30 | >250d | Orange | Water/ethanol | C, 56.98; H, 3.45; N, 18.46 | C, 57.02; H, 3.44; N, 18.44 |
| EMAC 3047 | 484.19 | 84.65 | >250d | Yellow | Water/ethanol | C, 53.61; H, 3.00; N, 13.89 | C, 53.60; H, 2.98; N, 13.87 |
| EMAC 3048 | 429.33 | 59.40 | >250d | Light orange | Water/ethanol | C, 65.50; H, 4.63; N, 16.08 | C, 65.48; H, 4.61; N, 16.06 |
| EMAC 3049 | 445.33 | 83.00 | >250d | Orange | Water/ethanol | C, 62.62; H, 4.43; N, 15.37 | C, 62.61; H, 4.45; N, 15.33 |
| EMAC 3050 | 415.30 | 61.40 | >250d | Ocher yellow | Water/ethanol | C, 64.65; H, 4.22; N, 16.75 | C, 64.63; H, 4.23; N, 16.68 |
| EMAC 3051 | 439.75 | 69.35 | >250d | Yellow | Water/ethanol | C, 53.61; H, 3.00; N, 13.89 | C, 53.58; H, 3.01; N, 13.85 |
| EMAC 3052 | 484.19 | 59.56 | >250d | Orange | Water/ethanol | C, 53.61; H, 3.00; N, 13.89 | C, 53.61; H, 2.99; N, 13.88 |
| EMAC 3053 | 467.74 | 57.70 | >250d | Orange-yellow | Water/ethanol | C, 55.89; H, 3.13; N, 14.48 | C, 55.88; H, 3.14; N, 14.44 |
| EMAC 3054 | 528.65 | 70.00 | >250d | Orange | Water/ethanol | C, 48.29; H, 2.70; N, 12.51 | C, 48.31; H, 2.67; N, 12.48 |
| EMAC 3055 | 494.75 | 66.70 | >250d | Orange | Water/ethanol | C, 52.24; H, 2.92; N, 16.92 | C, 52.20; H, 2.94; N, 16.89 |
| EMAC 3056 | 525.85 | 58.95 | >250d | Orange | Water/ethanol | C, 64.79; H, 3.85; N, 12.59 | C, 64.76; H, 3.84; N, 12.57 |
| EMAC 3057 | 474.76 | 53.70 | >250d | Orange-yellow | Water/ethanol | C, 57.94; H, 3.07; N, 17.78 | C, 58.02; H, 3.05; N, 17.77 |
| EMAC 3058 | 441.28 | 27.20 | >250d | Yellow | Water/ethanol | C, 53.40; H, 2.74; N, 13.84 | C, 53.38; H, 2.76; N, 13.80 |
| EMAC 3059 | 494.65 | 57.60 | >250° | Orange | Water/ethanol | C, 52.24; H, 2.92; N, 16.92 | C, 52.21; H, 2.94; N, 16.88 |
| EMAC 3060 | 518.54 | 29.89 | >250° | Red | Water/ethanol | C, 49.39; H, 2.53; N, 12.80 | C, 49.37; H, 2.50; N, 12.74 |
| EMAC 3061 | 463.68 | 59.10 | >250° | Orange-red | Water/ethanol | C, 59.60; H, 3.95; N, 14.63 | C, 59.58; H, 3.97; N, 14.60 |
| EMAC 3062 | 479.68 | 51.00 | >250° | Red | Water/ethanol | C, 57.21; H, 3.79; N, 14.05 | C, 57.18; H, 3.81; N, 14.00 |
| EMAC 3063 | 449.65 | 55.60 | >250° | Orange | Water/ethanol | C, 58.61; H, 3.55; N, 15.19 | C, 58.56; H, 3.54; N, 15.16 |
1H-NMR spectra (Table 2) were registered on a Varian 500 MHz spectrometer (Palo Alto, CA). All samples were measured in DMSO-d6 and CDCl3. Chemical shifts are reported referenced to the solvent in which they were measured. Coupling constants J are expressed in hertz (Hz). Elemental analyzes were obtained on a Perkin–Elmer 240 B microanalyser (Waltham, MA). Analytical data of the synthesized compounds are in agreement within ±0.4% of the theoretical values. TLC chromatography was performed using silica gel plates (Merck F 254, Kenilworth, NJ), spots were visualized by UV light.
Table 2.
1H NMR data of derivatives EMAC 3039–3064.
| Compound | 1H NMR |
|---|---|
| EMAC 3039 | 1H NMR (CDCl3) δ(ppm): 8.29 (d, 1H) (dm; J = 7.48 Hz; 1H); 8.10 (s; 1H); 7.49 (d, 1H) (dm, J = 7.95 Hz; 1H); 7.36 (d, 1H); (dm; J = 8.03 Hz; 1H); 7.24 (s, 1H); 7.04 (t, 1H) (dm; J = 7.60 Hz; 1H); 6.89 (d, 1H) (dm; J = 7.64 Hz; 1H); 6.31 (s, 1H); 3.58 (s, 3H). |
| EMAC 3040 | 1H NMR (DMSO-d6) δ(ppm): 10.48 (s, 1H); 8.22 (d; 1H) (dm, J = 7.39 Hz; 1H); 7.64 (dt, 1H) (dm, J = 5.35, 8.25 Hz; 1H); 7.49 (d, 1H) (dm, J = 7.45 Hz; 1H); 7.38 (td, 1H) (dm; J = 1.63,8.74 Hz; 1H); 7.25 (td, 1H) (dm, J = 1.48, 7.69 Hz; 1H); 6.99 (q, 1H) (dm, J = 7.20 Hz; 1H); 6.84 (m, 1H); 3.50 (s, 3H). |
| EMAC 3041 | 1H NMR (DMSO-d6) δ(ppm): 10.48 (s, 1H); 8.22 (d, 1H) (dm, J = 7.54 Hz; 1H); 7.74 (d, 1H) (dm, J = 8.43 Hz; 1H); 7.54 (d, 1H) (dm; J = 8.12 Hz; 1H); 7.47 (d, 1H) (dm, J = 7.01 Hz; 1H); 7.24 (m, 1H); 6.98(dt, 1H) (dm, J = 7.61, 15.13 Hz; 1H); 6.83(m, 1H); 3.53 (s, 3H). |
| EMAC 3042 | 1H NMR (CDCl3) δ(ppm): 8.38 (d, 1H) (dm, J = 8.20 Hz; 1H); 8.29 (d, 1H) (dm, J = 7.54 Hz; 1H); 7.63 (d, 1H) (dm, J = 8.23 Hz; 1H); 7.50 (s, 1H); 7.26 (m, 1H); 7.06 (t, 1H) (dm; J = 7.36 Hz; 1H); 6.86 (d, 1H) (dm; J = 7.84 Hz; 1H); 6.46 (s, 1H); 3.63 (s, 3H). |
| EMAC 3043 | 1H NMR (DMSO-d6) δ(ppm): 10.56 (s, 1H); 8.24 (d, 1H) (dm, J = 7.52 Hz; 1H); 7.83 (dd, 1H) (dm, J = 4.30,8.21 Hz; 1H); 7.75 (m, 1H); 7.68 (dd, 1H) (dm, J = 5.55,7.98 Hz; 1H); 7.50 (m, 1H); 7.42 (t, 1H) (dm, J = 7.37 Hz; 1H); 7.25 (m, 1H); 7.00 (dt, 1H); (dm, J = 7.51, 12.06 Hz; 1H); 6.89 (t, 1H) (dm, J = 9.92 Hz; 1H); 6.84 (t, 1H) (dm; J = 7.75, 18.98 Hz; 1H); 3.57 (s, 3H). |
| EMAC 3044 | 1H NMR (DMSO-d6) δ(ppm): 10.51 (s, 1H); 8.22 (d, 1H); (dm, J = 7.46 Hz; 1H); 8.01 (dd, 1H) (dm, J = 6.34,8.30 Hz; 1H); 7.81 (dd, 1H) (dm, J = 5.59, 8.10 Hz; 1H); 7.47 (d, 1H); (dm, J = 7.29 Hz; 1H); 7.25(m, 1H); 7.00(m, 1H); 6.85(m, 1H); 3.42 (s, 3H). |
| EMAC 3045 | 1H NMR (CDCl3) δ(ppm): 8.47(s, 1H); 8.29 (d, 1H) (dm; J = 7.51 Hz; 1H); 7.40 (q, 1H) (dm, J = 7.86 Hz; 1H); 7.25 (m, 1H); 7.04 (m, 1H); 6.99 (m, 1H); 6.91(d, 1H); (dm, J = 7.68 Hz; 1H); 6.37(s, 1H); 3.52 (s, 3H). |
| EMAC 3046 | 1H NMR (DMSO-d6) δ(ppm): 1052 (s, 1H); 8.39 (s, 1H); 8.36 (dm, 1H); 8.05 (d, J = 8 Hz; 1H); 7.83 (t, J = 8; 1H); 7.47 (d, J = 7.5 Hz; 1H); 7.23 (t, J = 7.5 Hz; 1H); 7.01 (s, 1H); 6.97 (t, J = 7.5 Hz; 1H); 6.80 (d, J = 8 Hz; 1H); 3.52 (s, 3H). |
| EMAC 3047 | 1H NMR (DMSO-d6) δ(ppm): 10.49 (s, 1H); 8.22 (d, 1H) (dm, J = 7.56 Hz; 1H); 7.91(dd, 1H) (dm, J = 2.06, 6.23 Hz; 1H); 7.81 (dd, 1H) (dm, J = 7.19,8.31 Hz; 1H); 7.60 (ddd, 1H) (dm, J = 2.10, 5.88, 8.30 Hz; 1H); 7.47 (d, 1H) (dm, J = 7.39 Hz; 1H); 7.23(m, 1H); 6.97(m, 1H); 6.80(d, 1H) (dm; J = 7.79 Hz; 1H); 3.51 (s, 3H). |
| EMAC 3048 | 1H NMR (CDCl3) δ(ppm): 8.30 (m, 1H); 8.11 (s, 1H); 7.86(s, 1H); 7.30 (q, 1H); 7.24 (dd, 3H); (dm, J = 1.35, 7.69 Hz; 1H); 7.04 (m, 1H); 6.88(d, 1H); 6.28(s, 1H); 6.37(s, 1H); 3.52 (s, 3H). |
| EMAC 3049 | 1H NMR (CDCl3) δ(ppm): 8.30 (d, 1H); (dm, J = 7.54 Hz; 1H); 7.94 (s, 1H); 7.34(s, 1H) (dm, J = 8.77 Hz; 1H); 7.24 (m, 1H); 7.05 (d, 1H) (dm, J = 7.49 Hz; 1H); 7.01 (m, 1H) (dm, J = 8.75 Hz; 1H); 6.86 (d, 1H) (dm, J = 7.66 Hz; 1H); 6.25(s, 1H); 3.88 (s, 3H); 3.58 (s, 3H). |
| EMAC 3050 | 1H NMR (CDCl3) δ(ppm): 8.30(d, 1H); (dm, J = 7.42 Hz; 1H); 7.98(s, 1H); 7.50 (q, 1H) (dm, J = 3.69 Hz; 1H); 7.42 (dd, 1H) (dm, J = 2.97, 6.43 Hz; 1H); 7.25 (t, 1H); (dm, J = 7.83 Hz; 1H); 7.05 (t, 1H) (dm, J = 7.42 Hz; 1H); 6.88 (d, 1H) (dm, J = 7.64 Hz; 1H); 6.31 (s, 1H); 3.60 (s, 3H). |
| EMAC 3051 | 1H NMR (CDCl3) δ(ppm): 8.29 (d, 1H) (dm, J = 7.45 Hz; 1H); 7.75 (s, 1H); 7.57(d, 1H) (dm, J = 10.05 Hz; 1H); 7.41 (d, 1H) (dm, J = 8.48 Hz; 1H); 7.35 (d, 1H) (dm, J = 8.18 Hz; 1H); 7.24 (d, 1H) (dm, J = 6.97 Hz; 1H); 7.04 (d, 1H) (dm, J = 7.55 Hz; 1H); 6.86 (d, 1H) (dm, J = 7.75 Hz; 1H); 6.35 (s, 1H); 3.46 (s, 3H). |
| EMAC 3052 | 1H NMR (CDCl3) δ(ppm): 8.27(d, 1H) (dm, J = 2.16 Hz; 1H); 7.99 (s, 1H); 7.50 (d, 1H) (dm, J = 8.55 Hz; 1H); 7.37 (d, 1H) (dm, J = 8.49 Hz; 1H); 7.22 (dd, 1H) (dm, J = 2.19, 8.30 Hz; 1H); 6.81 (d, 1H) (dm, J = 8.33 Hz; 1H); 6.36 (s, 1H); 3.61 (s, 3H). |
| EMAC 3053 | 1H NMR (CDCl3) δ(ppm): 8.28(s, 1H); 7.63 (d, 1H) (dm, J = 17.38 Hz; 1H); 7.49 (s, 1H); 7.41 (m, 1H); 7.20 (q, 1H) (dm, J = 8.33 Hz; 1H); 6.79 (d, 1H) (dm, J = 8.32 Hz; 1H); 6.35 (s, 1H); 3.58 (s, 3H); NH not detected |
| EMAC 3054 | 1H NMR (CDCl3) δ(ppm): 8.28 (d, 1H) (dm, J = 2.22 Hz; 1H); 7.65 (t; 1H) (dm; J = 9.12 Hz; 1H); 7.30 (dd, 1H) (dm, J = 6.98, 8.73 Hz; 1H); 7.22 (dd, 1H) (dm, J = 2.20, 8.26 Hz; 1H); 6.80 (d, 1H) (dm, J = 8.26 Hz; 1H); 6.37 (s, 1H); 3.61 (s, 3H); NH not detected. |
| EMAC 3055 | 1H NMR (CDCl3) δ(ppm): 8.39 (d, 1H) (d, J = 8.67 Hz; 1H); 8.28 (s, 1H); 7.64 (m, 1H); 7.49 (s, 1H); 7.24 (d, 1H) (dm; J = 12.22 Hz; 1H); 6.79 (m, 1H); 6.49 (d, 1H) (dm; J = 16.75 Hz; 1H); 3.65 (s, 3H). |
| EMAC 3056 | 1H NMR (CDCl3) δ(ppm): 8.30 (d, 1H); (d, J = 2.21 Hz; 1H); 8.18 (s, 1H); 7.73(d, 1H) (dm, J = 8.17 Hz; 1H); 7.65 (d, 1H) (dm, J = 7.55 Hz; 1H); 7.50 (d, 1H) (dm, J = 7.86 Hz; 1H); 7.41 (t, 1H) (dm; J = 7.29Hz; 1H); 7.21 (m; 1H); 6.83 (d, 1H) (dm, J = 8.21 Hz; 1H); 6.76 (d, 1H) (dm, J = 8.22 Hz; 1H); 6.41 (s, 1H); 3.68 (s, 3H). |
| EMAC 3057 | 1H NMR (DMSO) δ(ppm): 10.61 (s, 1H); 8.18 (d. 1H) (dm, J = 2.25 Hz; 1H); 8.02(dd, 1H) (dm, J = 6.72, 8.38 Hz; 1H); 7.82 (dd, 1H) (dm, J = 6.17, 8.23 Hz; 1H); 7.28 (ddd, 1H) (dm, J = 2.24, 8.22, 26.39 Hz; 1H); 7.04 (s, 1H); 6.84 (dd, 1H) (dm, J = 8.26, 29.93 Hz; 1H); 3.54 (s, 3H). |
| EMAC 3058 | 1H NMR (DMSO) δ(ppm): 10.63 (s, 1H); 8.17 (d, 1H) (dm, J = 2.21 Hz; 1H); 7.68(qd, 1H) (dm, J = 6.35, 8.53 Hz; 1H); 7.53 (tdd, 1H), (dm, J = 2.46, 7.91, 10.16 Hz; 1H); 7.31 (ddt, 1H) (dm, J = 2.76, 5.70, 11.48 Hz; 1H); 7.02 (s, 1H); 6.87 (d, 1H) (dm, J = 8.28 Hz; 1H); 6.81(d, 1H) (dm, J = 8.29 Hz; 1H); 3.47 (s, 3H). |
| EMAC 3059 | 1H NMR (CDCl3) δ(ppm): 8.38 (d, 1H); (d, J = 8.38 Hz; 1H); 7.79 (d; 1H) (dm, J = 7.58 Hz; 1H); 7.74 (t, 1H) (dm, J = 8.02 Hz; 1H); 7.67 (s, 1H); 7.24 (dd, 1H) (dm, J = 2.07, 8.08 Hz; 1H); 6.80 (d, 1H) (dm, J = 8.29 Hz; 1H); 6.75 (d, 1H) (dm, J = 8.27 Hz; 1H); 6.49 (s; 1H); 3.65 (s, 1H). |
| EMAC 3060 | 1H NMR (CDCl3) δ(ppm): 8.27 (d, 1H); (d, J = 2.11 Hz; 1H); 7.92 (s, 1H); 7.60 (d, 1H) (dm, J = 8.37 Hz; 1H); 7.55 (s, 1H); 7.27 (m, 1H); 6.81 (d, 1H) (dm, J = 8.29 Hz; 1H); 6.40 (s, 1H); 3.62 (s, 1H). |
| EMAC 3061 | 1H NMR (DMSO) δ(ppm): 10.60 (s, 1H); 8.17 (s, 1H); 7.47 (d, J = 8 Hz; 2H); 7.34 (d, J = 8 Hz; 2H); 7.29 (d, J = 8 Hz; 1H); 6.85 (d, J = 8 Hz; 1H); 3.56 (s, 3H); 3.29 (s, 3H). |
| EMAC 3062 | 1H NMR (DMSO) δ(ppm): 10.60 (s, 1H); 8.18 (d, 1H) (dm, J = 2.15 Hz; 1H); 7.52(d, 1H) (dm, J = 8.54 Hz; 1H); 7.29 (dd, 1H); (dm, J = 2.22, 8.28 Hz; 1H); 7.09 (d, 1H) (dm, J = 8.53 Hz; 1H); 6.86 (d, 1H) (dm, J = 8.29 Hz; 1H); 6.81 (s, 1H); 3.83 (s, 3H); 3.57 (s, 3H). |
| EMAC 3063 | 1H NMR (CDCl3) δ(ppm): 8.29 (d, 1H) (d; J = 2.14 Hz; 1H); 7.69 (s; 1H); 7.51 (m, 1H); 7.43 (m, 1H); 7.21(m, 1H); 6.79 (d, 1H); (dm, J = 8.25 Hz; 1H); 6.37 (s, 1H); 3.63 (s, 1H); NH not detected. |
Biological assay
HIV-1 RT-associated DNA polymerase-independent RNase H activity determination
The HIV RT-associated RNase H activity was measured as described20. Briefly, 20 ng of HIV-1 wt RT was incubated for 1 h at 37 °C in 100 µL reaction volume containing 50 mM Tris HCl pH 7.8, 6 mM MgCl2, 1 mM dithiothreitol (DTT), 80 mM KCl, 250 nM hybrid RNA/DNA (5′-GTTTTCTTTTCCCCCCTGAC-3′-Fluorescein, 5′-CAAAAGAAAAGGGGGGACUG-3′-Dabcyl). Reactions were stopped by addition of EDTA and products were measured with a with a Perkin?Elmer Victor 3 multilabel counter plate reader at excitation–emission wavelength of 490/528 nm (Table 3).
Table 3.
Activity of compounds EMAC 3039–3063 on HIV-1 RT-associated enzymatic functions RDDP and RNase H.
| Compound | R | RNase H – IC50 (μM) | RDDP – IC50 (μM) | Compound | RNase H – IC50 (μM) | RDDP – IC50 (μM) |
|---|---|---|---|---|---|---|
| EMAC 3039 | 4 Cl | 17.5 ± 1.3 | 24 ± 3.0 | EMAC 3052 | 24 ± 0.8 | 39 ± 3.0 |
| EMAC 3040 | 4 F | 28 ± 1.0 | 30 ± 5.0 | EMAC 3053 | 18.9 ± 1.1 | 31 ± 2.6 |
| EMAC 3041 | 4 Br | 29 ± 1.0 | 100 ± 2.0 | EMAC 3054 | 23 ± 1.0 | 34 ± 2.8 |
| EMAC 3042 | 4 NO2 | 17.2 ± 1.1 | 15.6 ± 1.6 | EMAC 3055 | 21 ± 0.5 | 26 ± 1.5 |
| EMAC 3043 | 4C6H5 | 25 ±1.3 | 35 ± 9.0 | EMAC 3056 | 10.0 ± 0.5 | 9.5 ± 1.5 |
| EMAC 3044 | 4 CN | 21 ± 2.1 | 27 ± 0.7 | EMAC 3057 | 27 ± 3.0 | 28 ± 2.3 |
| EMAC 3045 | 2-4 F | 12.5 ± 0.6 | 30 ± 2.0 | EMAC 3058 | 21 ± 0.9 | 54 ± 5.0 |
| EMAC 3046 | 3 NO2 | 24 ± 0.5 | 69 ± 4.0 | EMAC 3059 | 17.5 ± 2.2 | 60 ± 3.2 |
| EMAC 3047 | 3-4 Cl | 24 ± 0.4 | 50 ± 3.0 | EMAC 3060 | 16.7 ± 1.0 | 98 ± 2.0 |
| EMAC 3048 | 4 CH3 | 15.9 ± 1.2 | 29 ± 1.0 | EMAC 3061 | 19.0 ± 1.0 | 85 ± 7.0 |
| EMAC 3049 | 4 OCH3 | 27 ± 2.0 | 17.9 ± 3.0 | EMAC 3062 | 15.1 ± 5.1 | 28 ± 0.5 |
| EMAC 3050 | H | 19.9 ± 5.4 | 15.0 ± 6.0 | EMAC 3063 | 25 ± 5.0 | 100 ± 10 |
| EMAC 3051 | 2-4 Cl | 15.0 ± 0.3 | 19.6 ± 0.6 | Not synthesized | // | // |
HIV-1 RT-associated RNA dependent DNA polymerase activity determination
The HIV-1 RT-associated RNA-Dependent DP activity was measured as previously described21. Briefly, 20 ng of HIV-1 wt RT was incubated for 30 min at 37 °C in 25 µL volume containing 60 mM Tris–HCl pH 8.1, 8 mM MgCl2, 60 mM KCl, 13 mM DTT, 2.5 µM poly(A)-oligo(dT), 100 µM dTTP. Enzymatic reaction was stopped by addition of EDTA. Reaction products were detected by picogreen addition and measured with a Perkin?Elmer Victor 3 multilabel counter plate reader at excitation–emission wavelength of 502/523 nm.
Molecular modelling studies
Ligand preparation
Theoretical 3D models of the compounds were built by means of Maestro GUI22. Considering the configuration of double bonds described in literature23. The lowest energy conformers were considered for the following studies. These were obtained from the starting conformations through a conformational search by means of MacroModel version 7.2 (Schrödinger LLC, New York, NY)24, considering MMFFs25 as force field and solvent effects by adopting the implicit solvation model Generalized Born/Surface Area (GB/SA) water26. The simulations were performed allowing 1000 steps Monte Carlo analysis with Polak–Ribier Coniugate Gradient (PRCG) method and a convergence criterion of 0.05 kcal/(molÅ).
Protein preparation
The coordinates for RT enzymes were taken from the RCSB Protein Data Bank27 (PDB codes 1vrt28, 2zd129, 1ep430, 3qo931, 1rti28, 1tv632, 3lp233). The proteins were prepared by using the Maestro Protein Preparation Wizard protocol22.
Docking experiments
QMPL default settings were applied34. The docking grids were defined by centering on W229 and Q500. The grid boxes of the same size (46 × 46 × 46 Å) covered overall the whole p66 subunit. Best solutions were subjected to post-docking procedure and analysed.
5000 steps of the Polak–Ribier conjugate gradient (PRCG) minimization method were conducted on the top ranked theoretical complexes using AMBER force field35. The optimization process was performed up to the derivative convergence criterion equal to 0.1 kJ/(mol*Å)−1. The binding free energies were computed applying molecular mechanics and continuum solvation models with the molecular mechanics generalized Born/surface area (MM-GBSA) method36.
Figures
The resulting best complexes were considered for the binding modes graphical analysis with LigandScout (inte:Ligand, Vienna, Austria)37 and Pymol (Schrödinger LLC, New York, NY)38.
Results and discussion
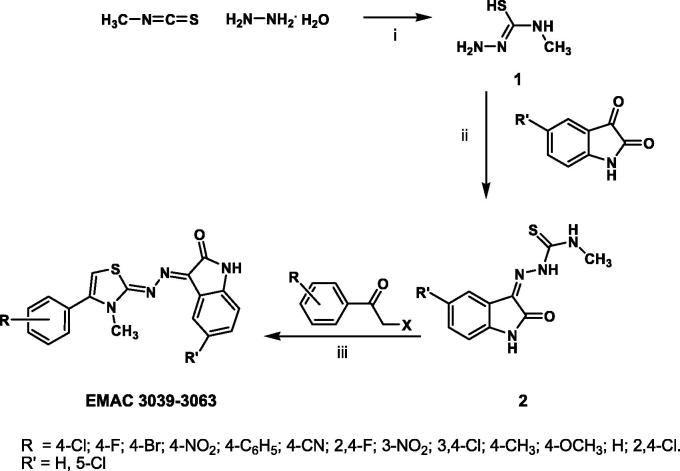
The synthesis of the isatin derivatives EMAC 3039–3063 is illustrated in Figure 1. Firstly, the 1-amino-3-methylisothiourea (1) was obtained by direct reaction between methylisothiocyanate and hydrazine hydrate (ratio 1:1), at rt, using ethanol as solvent. Secondly, the condensation between substituted isatin and compound 1, at reflux condition, using ethanol, gave the desired thiosemicarbazones.
Figure 1.
Synthetic pathway to compounds EMAC 3039–3063; reagents and conditions: (i) methylisothiocyanate, hydrazine hydrate, ethanol, rt; (ii) 1-amino-3-methylisothiourea, substituted isatin, ethanol, reflux; (iii) 2, substituted acetophenones, isopropanol, rt.
Finally, EMAC 3039–3063 derivatives were obtained in good yields by reaction of compound 2 with differently substituted bromo or chloro acetophenones in isopropanol. All the synthesized compounds were submitted to biological assays to evaluate their ability to inhibit both RT-associated enzymatic functions. Results indicates that most of the EMAC isatin derivatives are able to inhibit both RDDP and RNase H functions at µM concentrations, indicating that these derivatives could represent a good starting point for the design of new and more efficient dual HIV-1 RT inhibitors. With the aim of obtaining more information on the structure activity relationships and to achieve insights into the binding mode of compounds EMAC 3039–3063, we performed blind docking studies on the wt-HIV-1 RT heterodimer by applying the QM-polarized ligand docking protocol (QMPLD)34,39 considering two grid box: one centered in W229 (NNRTIBP) and one in Q500 (RNAse domain), in order to include all the p66 subunit39. In particular, we carried out ensemble docking experiments40 using seven different crystal structures to take into account the target flexibility. The same protocol was successfully applied in our previous studies12,13. In fact, the high NNRTIBP plasticity allows different orientations of Y181, Y188, Y183 and primer grip β12–β13 hairpins41. The overall volume of this pocket is 620–720 Å3 and is characterized by a L shape. Its volume is approximately more than twice the one occupied by most of the known NNRTIs. Thus it offers many binding possibilities to small molecules3. This explains the large variety of chemical scaffolds of this class of inhibitors, whose shapes was often described in a creative way: e.g. “butterfly”42, “horseshoe”43 and “dragon”31. These inhibitors are able to lock the enzyme into an inactive conformation.
We focused our attention on the most active compounds of the two series: EMAC 2045 and EMAC 2056.
The best RT complexes obtained by the docking experiments were subjected to a post-docking procedure based on energy minimization and successive binding free energy calculation. The binding free energies were obtained applying molecular mechanics and continuum solvation models using the molecular mechanics generalized Born/surface area (MM-GBSA) method36.
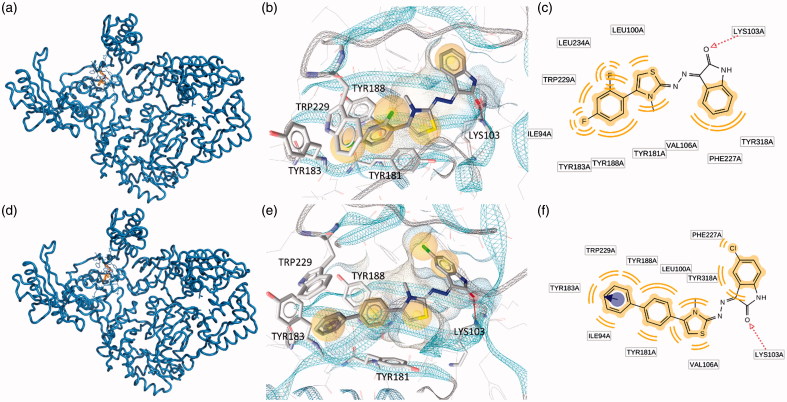
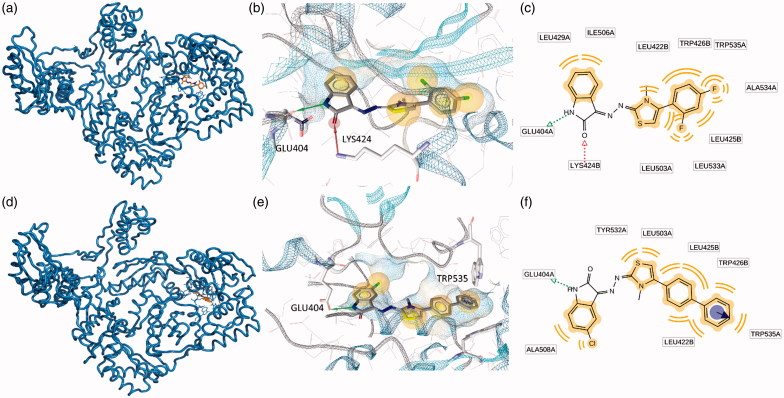
Blind docking calculations indicated the presence of two energetically favored sites that we named as Pocket 1 and Pocket 2. Pocket 1 is located close to the polymerase triad and comprehend the whole L shaped NNRTIBP. Conversely, the second putative binding pocket is located in the RNase H domain, below the RNase H active site. Some new RNase H inhibitors which are able to bind this pocket, have been already described by Felts44 and by our previous studies12–14. Most likely, these compounds are able to prevent the correct anchoring of the nucleic acid in the RNase H domain and, therefore, inhibit the RNase H hydrolytic function. Putative binding modes of EMAC 2045 and EMAC 2056 are depicted in Figures 2 and 3. The two functions inhibition seems explicable by the binding to at least two sites: one (pocket 1) responsible for the polymerase activity, and the second (pocket 2), responsible for the RNase H inhibitory activity. In particular, EMAC 2056 is better accommodated compared to EMAC 2045 into the pocket 1 since is able, with its di-phenyl substituent, to take contact with Tyr188, Tyr181, Tyr183 Trp229 by π − π interactions. Overall the complexes are mainly stabilized by several hydrophobic interactions.
Figure 2.
Putative binding modes of EMAC2045 and EMAC2056 and critical residues individuated for their binding in the pocket 1: (a, d) EMAC2045-HIV-1 RT complex and EMAC2056-HIV-1 RT complex; (b and e) close-up into the EMAC2045 and EMAC2056 binding site; (c and f) 2D depiction of EMAC2045 and its respective interactions with RT residues. pale yellow sphere indicates hydrophobic interactions with lipophilic residues. Red arrow indicates a hydrogen bond (HB) acceptor interaction, while the violet sphere represents the aromatic π − π stacking interaction.
Figure 3.
Putative binding modes of EMAC2045 and EMAC2056 and critical residues individuated for their binding in the pocket 2: (a, d) EMAC2045-HIV-1 RT complex and EMAC2056-HIV-1 RT complex; (b and e) close-up into the EMAC2045 and EMAC2056 binding site; (c and f) 2D depiction of EMAC2045 and its respective interactions with RT residues. pale yellow sphere indicates hydrophobic interactions with lipophilic residues. Red arrow indicates a hydrogen bond (HB) acceptor interaction, green HB donor, while the violet sphere represents the aromatic π − π stacking interaction.
In pocket 2 the compounds are sandwiched between the two subunits p66 and p51. In fact, the complexes are stabilized by interactions with both A (p66) and B (p51) chains (Figure 3).
Conclusions
In this research we have designed and synthesized a library of isatin based derivatives EMAC 3039–3063 to evaluate their activity towards both RT-associated enzymatic functions RDDP and RNase H. Our data indicate that the isatin derivatives are generally able to inhibit both RDDP and RNase H functions al µM concentrations and that these molecules are a good starting point for the design of new dual HIV-1 RT inhibitors. Nevertheless, considering the high potential of the isatin scaffold we will pursue in our efforts to identify new isatin based anti-viral agents.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- 1.Zhan P, Liu X, Li Z, et al. . Design strategies of novel NNRTIs to overcome drug resistance. Curr Med Chem 2009;16:3903–17. [DOI] [PubMed] [Google Scholar]
- 2.Morphy R, Rankovic Z.. Designed multiple ligands. An emerging drug discovery paradigm. J Med Chem 2005;48:6523–43. [DOI] [PubMed] [Google Scholar]
- 3.Mehellou Y, De Clercq E.. Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J Med Chem 2010;53:521–38. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch MS.Initiating therapy: when to start, what to use. J Infect Dis 2008;197:S252–60. [DOI] [PubMed] [Google Scholar]
- 5.Bornot A, Bauer U, Brown A, et al. . Systematic exploration of dual-acting modulators from a combined medicinal chemistry and biology perspective. J Med Chem 2013;56:1197–210. [DOI] [PubMed] [Google Scholar]
- 6.Rosini M.Polypharmacology: the rise of multitarget drugs over combination therapies. Future Med Chem 2014;6:485–7. [DOI] [PubMed] [Google Scholar]
- 7.Besnard J, Hopkins AL, editors. De novo design of ligands against multitarget profiles. Weinheim, Berlin, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2014. [Google Scholar]
- 8.Anighoro A, Bajorath J, Rastelli G.. Polypharmacology: challenges and opportunities in drug discovery. J Med Chem 2014;57:7874–87. [DOI] [PubMed] [Google Scholar]
- 9.Sluis-Cremer N, Wainberg MA, Schinazi RF.. Resistance to reverse transcriptase inhibitors used in the treatment and prevention of HIV-1 infection. Future Microbiol 2015;10:1773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distinto S, Maccioni E, Meleddu R, et al. . Molecular aspects of the RT/drug interactions. Perspective of dual inhibitors. Curr Pharm Des 2013;19:1850–9. [DOI] [PubMed] [Google Scholar]
- 11.Distinto S, Esposito F, Kirchmair J, et al. . Identification of HIV-1 reverse transcriptase dual inhibitors by a combined shape-, 2D-fingerprint- and pharmacophore-based virtual screening approach. Eur J Med Chem 2012;50:216–29. [DOI] [PubMed] [Google Scholar]
- 12.Corona A, Meleddu R, Esposito F, et al. . Ribonuclease H/DNA polymerase HIV-1 reverse transcriptase dual inhibitor: mechanistic studies on the allosteric mode of action of isatin-based compound RMNC6. PLoS One 2016;11:e0147225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meleddu R, Cannas V, Distinto S, et al. . Design, synthesis, and biological evaluation of 1,3-diarylpropenones as dual inhibitors of HIV-1 reverse transcriptase. Chem Med Chem 2014;9:1869–79. [DOI] [PubMed] [Google Scholar]
- 14.Meleddu R, Distinto S, Corona A, et al. . (3Z)-3-(2-[4-(aryl)-1,3-thiazol-2-yl]hydrazin-1-ylidene)-2,3-dihydro-1H-indol-2-one derivatives as dual inhibitors of HIV-1 reverse transcriptase. Eur J Med Chem 2015;93:452–60. [DOI] [PubMed] [Google Scholar]
- 15.Agamennone M, Belov DS, Laghezza A, et al. . Fragment-based discovery of 5-arylisatin-based inhibitors of matrix metalloproteinases 2 and 13. Chem Med Chem. 2016;11:1892–8. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Ning Y, Zhao W, et al. . Synthesis, neuro-protection and anti-cancer activities of simple isatin Mannich and Schiff bases. Lett Drug Des Discov 2016;13:395–400. [Google Scholar]
- 17.Tavari M, Malan SF, Joubert J.. Design, synthesis, biological evaluation and docking studies of sulfonyl isatin derivatives as monoamine oxidase and caspase-3 inhibitors. Med Chem Commun 2016;7:1628–39. [Google Scholar]
- 18.Rane RA, Karunanidhi S, Jain K, et al. . A recent perspective on discovery and development of diverse therapeutic agents inspired from isatin alkaloids. Curr Top Med Chem (Sharjah, United Arab Emirates) 2016;16:1262–89. [DOI] [PubMed] [Google Scholar]
- 19.Ozgun DO, Yamali C, Gul HI, et al. . Inhibitory effects of isatin Mannich bases on carbonic anhydrases, acetylcholinesterase, and butyrylcholinesterase. J Enzyme Inhib Med Chem. 2016;31:1498–1501. [DOI] [PubMed] [Google Scholar]
- 20.Corona A, Di Leva FS, Thierry S, et al. . Identification of highly conserved residues involved in inhibition of Hiv-1 Rnase H function by diketo acid derivatives. Antimicrob Agents Chemother 2014;58:6101–10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suchaud V, Bailly F, Lion C, et al. . Development of a series of 3-hydroxyquinolin-2(1H)-ones as selective inhibitors of HIV-1 reverse transcriptase associated RNase H activity. Bioorg Med Chem Lett 2012;22:3988–92. [DOI] [PubMed] [Google Scholar]
- 22.Schrödinger LLC Maestro GUI. New York, NY: Schrödinger LLC; 2015. [Google Scholar]
- 23.Pervez H, Ahmad M, Hadda TB, et al. . Synthesis and fluorine-mediated interactions in methanol-encapsulated solid state self-assembly of an isatin-thiazoline hybrid. J Mol Struct 2015;1098:124–9. [Google Scholar]
- 24.Mohamadi F, Richards NGJ, Guida WC, et al. . Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem 1990;11:440–67. [Google Scholar]
- 25.Halgren T.Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J Comput Chem 1996;17:520–52. [Google Scholar]
- 26.Still WC, Tempczyk A, Hawley RC, Hendrickson T.. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc 1990;112:6127–9. [Google Scholar]
- 27.Berman HM, Westbrook J, Feng Z, et al. . The protein data bank. Nucleic Acids Res 2000;28:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren JS, Esnouf R, Garman E, et al. . High-resolution structures of HIV-1 rt from 4 RT-inhibitor complexes. Nat Struct Biol 1995;2:293–302. [DOI] [PubMed] [Google Scholar]
- 29.Das K, Bauman JD, Clark AD, et al. . High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A 2008;105:1466–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren JS, Nichols C, Bird LE, et al. . Binding of the second generation non-nucleoside inhibitor S-1153 to HIV-1 reverse transcriptase involves extensive main chain hydrogen bonding. J Biol Chem 2000;275:14316–20. [DOI] [PubMed] [Google Scholar]
- 31.Das K, Bauman JD, Rim AS, et al. . Crystal structure of tert-butyldimethylsilyl-spiroaminooxathioledioxide-thymine (TSAO-T) in complex with HIV-1 reverse transcriptase (RT) redefines the elastic limits of the non-nucleoside inhibitor-binding pocket. J Med Chem 2011;54:2727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pata JD, Stirtan WG, Goldstein SW, Steitz TA.. Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other nonnucleoside inhibitors. Proc Natl Acad Sci USA 2004;101:10548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himmel DM, Maegley KA, Pauly TA, et al. . Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure (London, England: 1993) 2009;17:1625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrödinger LLC QMPolarized protocol New York, NY: Schrodinger Suite; 2012. [Google Scholar]
- 35.McDonald DQ, Still WC.. AMBER torsional parameters for the peptide backbone. Tetrahedron Lett 1992;33:7743–6. [Google Scholar]
- 36.Kollman PA, Massova I, Reyes C, et al. . Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 2000;33:889–97. [DOI] [PubMed] [Google Scholar]
- 37.InteLigand Software GmbH LigandScout 4.0. Enzersdorf Maria, Austria: Maria Enzersdorf; 2015. [Google Scholar]
- 38.PyMOL Version 1.5.0.4 ed molecular graphics system. New York, NY: Schrödinger, LLC. [Google Scholar]
- 39.Cho AE, Guallar V, Berne BJ, Friesner R.. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem 2005;26:915–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang S-Y, Zou X.. Ensemble docking of multiple protein structures: considering protein structural variations in molecular docking. Proteins 2007;66:399–421. [DOI] [PubMed] [Google Scholar]
- 41.Paris KA, Haq O, Felts AK, et al. . Conformational landscape of the human immunodeficiency virus type 1 reverse transcriptase non-nucleoside inhibitor binding pocket: lessons for inhibitor design from a cluster analysis of many crystal structures. J Med Chem 2009;52:6413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding JP, Das K, Moereels H, et al. . Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors. Nat Struct Biol 1995;2:407–15. [DOI] [PubMed] [Google Scholar]
- 43.Das K, Lewi PJ, Hughes SH, Arnold E.. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of non-nucleoside HIV-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol 2005;88:209–31. [DOI] [PubMed] [Google Scholar]
- 44.Felts AK, Labarge K, Bauman JD, et al. . Identification of alternative binding sites for inhibitors of HIV-1 ribonuclease H through comparative analysis of virtual enrichment studies. J Chem Inform Model 2011;51:1986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]