Abstract
Carbonic anhydrases (CAs) III and VII are two cytosolic isoforms of the α-CA family which catalyze the physiological reaction of carbon dioxide hydration to bicarbonate and proton. Despite these two enzymes share a 49% sequence identity and present a very similar three-dimensional structure, they show profound differences when comparing the specific activity for CO2 hydration reaction, with CA VII being much more active than CA III. Recently, CA III and CA VII have been proposed to play a new role as scavenger enzymes in cells where oxidative damage occurs. Here, we will examine functional and structural features of these two isoforms giving insights into their newly proposed protective role against oxidative stress.
Keywords: Carbonic anhydrase III, carbonic anhydrase VII, S-glutathiolation, oxidative stress, reactive cysteines, sulfhydryl groups
Introduction
Carbonic anhydrases (CAs) are ubiquitous metalloenzymes, present throughout most living organisms and encoded by six unrelated gene families: the α-, β-, γ-, δ-, η-, and ζ-CAs. These enzymes contain a Zn(II) ion in the active site that can be replaced by Fe(II) or Co(II) in the gamma family and by Cd(II) in the zeta one1–6. Human CAs (hCAs) belong to the α class which consists of 15 isoforms, among which 12 are enzymatically active (CAs I, II, III, IV, VA, VB, VI, VII, IX, XII, XIII, and XIV) and 3, called CA-related proteins (CARPs VIII, X, and XI), catalytically inactive. Catalytically active hCAs have different cellular localization; in particular, 5 are cytosolic (CAs I–III, VII, and XIII), 4 are membrane-associated (CAs IV, IX, XII, and XIV), 2 are mitochondrial (CAs VA and VB), and 1 is a secretory protein present in milk and saliva (CA VI)1,2,7.
CAs catalyze a very simple reaction: the hydration of carbon dioxide to bicarbonate and proton. These enzymes are expressed in many tissues where they participate in numerous physiological processes such as acid–base balance, respiration, ureagenesis, dioxide and ion transport, bone resorption, gluconeogenesis, body fluid generation, and lipogenesis1,7. Interestingly, apart from these biological activities, CAs are also associated to several pathological processes when abnormal levels and/or activities are registered. For instance, glaucoma and epilepsy are CA-related diseases where CA II and CA VII are involved8,9. Other CAs, such as CA IX and CA XII, have been associated to tumors where they cause extracellular pH lowering, thus helping the progression of malignant cells10,11.
CA III and CA VII are two of the least understood CA isoforms. These two cytosolic enzymes are both localized in tissues that have a high oxygen consumption rate, such as skeletal muscle, liver, and brain; moreover, they share 49% sequence identity and present a very similar three-dimensional structure12,13. Despite these similarities, these isoforms present a very different catalytic efficiency for CO2 hydration reaction, being CA VII one of the more efficient isoforms and CA III the least efficient (CA III activity is 0.35% compared to CA VII activity)14. Recently it has been suggested that these enzymes could participate in defense processes in cells where oxidative damage occurs upon the generation of reactive oxygen species (ROS)15–17. ROS are generated by normal metabolic activity, as well as lifestyle factors such as smoking, exercise, and diet. Their overproduction can be induced by different factors and perturb the normal cell redox balance, shifting cells into a state of oxidative stress. Since in conditions of severe stress, survival of the cells depends on their ability to adjust or resist to stress18–20, cells have developed an antioxidant defense system, involving gluthathione, antioxidant vitamins, sulfhydryl groups, and antioxidant enzymes21. Recently, it has been suggested that CA III and CA VII could be part of this antioxidant defense system. Indeed, the comparative analysis of the amino acid sequence of CA III and CA VII with that of the other cytosolic hCAs clearly showed the presence of a higher number of cysteine residues (Figure 1). Biochemical and structural studies have indicated for some of these cysteines a particularly high reactivity, thus suggesting that both enzymes could have a role in scavenging reactive species through their reactive sulfhydryl groups14,17,22,23. This is a new interesting finding considering that CA VII has been principally studied as an antiepileptic target being involved into the GABAergic transmission24.
Figure 1.
Multiple sequence alignment of cytosolic CAs. All cysteine residues are indicated with an asterisk; the reactive cysteines of isoforms III and VII are highlighted in dark gray, whereas those involved into hCA VII intramolecular disulfide bridge are highlighted in light gray. The sequence of bovine (b), rat (r) and human (h) CA III are reported showing that all cysteine residues are conserved. The alignment has been performed using Clustal Omega server84.
In this review, by examining CA III and CA VII functional and structural features, we will provide insights into their newly proposed protective role against oxidative stress.
CA III biochemical and functional features
The distribution pattern of CA III in the different tissues has been largely investigated by means of both Western blotting and immunohistochemistry experiments25, showing that CA III is abundantly expressed in liver and skeletal muscle. Several other tissues contain detectable quantities of CA III although in a smaller quantity25–33. The great amount of CA III in tissues with a great oxidation potential suggested that this protein could function in vivo as an oxyradical scavenger, protecting cells from oxidative damage. In agreement with this hypothesis, it was demonstrated that in NIH/3T3 cells, transfected with rat CA III cDNA, the intracellular steady-state level of ROS was lower with respect to the parental cells, and addition of exogenous hydrogen peroxide (H2O2) did not induce an increase of ROS contrary to what observed for not transfected cells. Moreover, in proliferation assays CA III overexpressing cells grew faster and were more resistant to cytotoxic concentration of H2O2 with respect to not-transfected cells34.
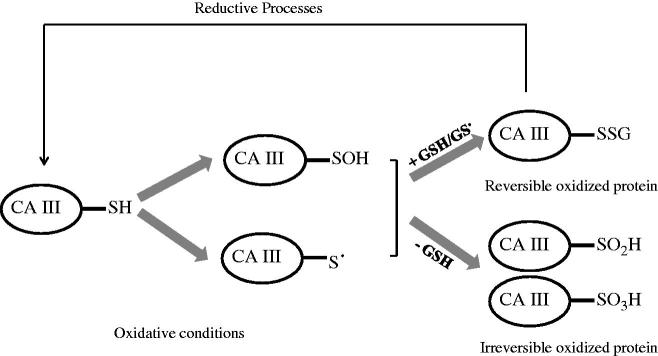
The involvement of CA III in the response to the oxidative stress was also supported by the observation that among the five cysteines present in its sequence (Figure 1), two of them, i.e. Cys183 and Cys188, were shown to form in vivo a disulfide link with glutathione (GSH)35 in a process referred as S-glutathiolation36. Reversible protein S-thiolation is an early cellular response towards oxidative stress due to the partial oxidation of cysteines to sulfenic acid or thiol radicals, which subsequently react with cellular GSH or GS radicals to form disulfide adducts36 (see Scheme 1). The analysis of the 3D structure of S-glutathiolated CA III suggested that the forming bonds between CA III reactive cysteines and GSH moiety were not to be ascribed to a specific recognition of GSH, but rather to the high reactivity of Cys183 and Cys188 and the great abundance of GSH in cell37, which reaches in vivo millimolar concentration36,38,39.
Scheme 1.
Schematic representation of reversible and irreversible oxidation of CA III reactive cysteines.
The behavior of the two reactive sulfhydryl groups of CA III, upon exposure to H2O2, peroxy radicals, or hypochlorous acid (HOCl) in presence or absence of GSH, was investigated in detail by Mallis and coworkers40. These authors found that irreversible oxidation was prevented only when GSH was approximately equimolar to protein thiols, thus allowing the S-gluthathiolation process40. Accordingly, at low GSH concentration, sulfenic acids (cysteine SOH) were irreversibly oxidized to sulfinic or sulfonic acid (cysteine SO2H and SO3H, respectively) which were not reducible by S-disulfide exchange (see Scheme 1). Similar results were obtained in cultured rat hepatocytes when treated with diethyl maleate for GSH depletion and menadione as oxidative agent40. In agreement with these data, protein extracts from liver of aged rats, which presented reduced levels of GSH, contained increased amounts of irreversibly oxidized CA III40,41.
Subsequent investigations on the putative antioxidant role of CA III were performed in 2004 by Zimmerman and coworkers; these authors analyzed the S-glutathiolation and the irreversible oxidation of CA III in skeletal muscle subjected to ischemia or exhaustive exercise. These studies showed that the two reactive sulfhydryl groups of CA III were differentially and progressively oxidized in skeletal muscle when it was exposed to oxidative insult. Under a mild or brief stress, reversible S-glutathiolation of one of CA III reactive cysteines was observed, involving only 20% of skeletal muscle protein, whereas under prolonged or harsh stress both sulfhydryl groups were irreversible oxidized. These results suggested that the high content of CA III in skeletal muscle might serve as a reservoir of reactive sulfhydryl groups able to repair acute and chronic insults. It is worth noticing that less than 10% of S-glutathiolated enzyme was detected in resting skeletal muscle indicative of a regulative physiological function22. The same authors analyzed, by microarray, ca3 knockout and wild-type mice and identified a transcriptional alteration of about 500 genes (out of 12 000), all associated to the GSH mediated anti-oxidative system. However, the ca3 knockout mice exhibited a normal development, fertility, and life span, at least under the standard laboratory conditions used42.
Finally, interesting data were presented on the repression of transcription of CA III gene driven by EVI123. EVI1, a multi-domain protein belonging to cys2hys2 zinc finger family43,44, is involved into cancer progression through several mechanisms, including enhanced cell proliferation, impaired differentiation, and evasion of apoptosis45. It has been shown that high levels of EVI1 expression in Rat1 cells induce an enhanced sensitivity to H2O2-induced apoptosis due to the down-regulation of CA III gene expression. However, the molecular mechanism by which EVI1 acts as CA III repressor has not yet been understood. Authors hypothesized a direct binding of EVI1 to CA III promoter sequence, which was shown to contain cis-regulatory elements necessary for EVI1-mediated transcriptional repression46. Since EVI1 is overexpressed in some human cancers47,48, it would be interesting to assess whether these types of tumors present reduced levels of CA III. If this is the case, a novel strategy could be developed for the treatment of tumors overexpressing EVI1 as they might be vulnerable to therapeutic agents that induce oxidative stress23.
CA VII biochemical and functional features
CA VII is known to be predominantly expressed in the cytosol of rat49 and mice neurons starting around postnatal day 1050. In humans, CA VII is expressed in several normal tissues, including liver, brain, colon, and skeletal muscle51–53, and only few years ago Parkkila’s group found high CA VII expression levels in brain tumor cells, allowing them to suggest CA VII as a tumor marker54. On the other side, Yang and coworkers, by using qPCR and Western blot analysis, recently found that CA VII mRNA and protein levels were down-regulated in colon tumors. Immunohistochemical staining also showed a weak signal in colon tumors with respect to normal colon tissues. Moreover, a correlation between reduced CA VII protein levels and shorter disease-specific survival was also found55. These results confirmed those already obtained by others who, analyzing different samples from colon (normal and tumor tissues) by a gene expression profiling study and a bioinformatics-based analysis, noted that CA VII mRNA was down-regulated in colorectal carcinoma clinical specimens56,57. Altogether, these data provided evidence for the potential utility of CA VII as a prognostic marker for patients with colorectal carcinoma55.
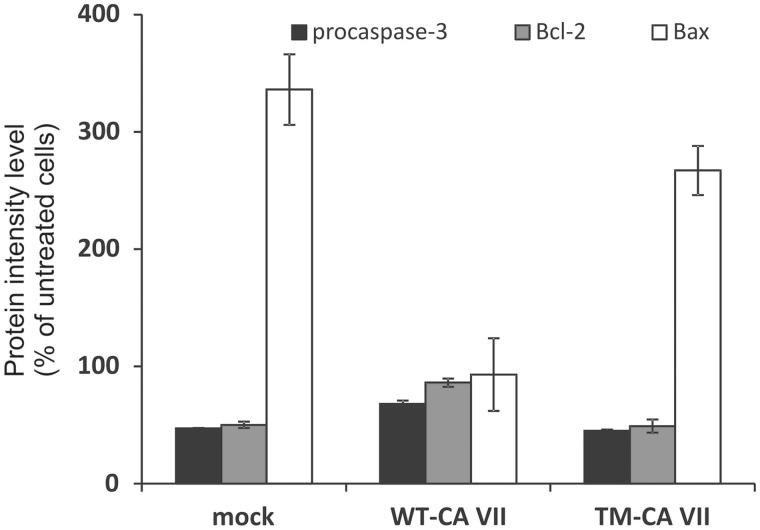
A recent study from our group unveiled for this enzyme another potential role as an oxygen radical scavenger for protecting cells from oxidative damage17. Indeed, we found that among the 4 cysteine residues present in the amino acid sequence of the enzyme (Figure 1), those in position 183 and 217 were particularly reactive, being quantitatively S-glutathiolated during the in vitro purification14. The glutathiolated protein, as well as a variant in which the two reactive cysteine residues were substituted by serines (C183S/C217S), showed an enzymatic activity similar to that reported for the wild-type protein (Kcat/KM of 6.5 × 107 M−1s−1 for the variant, 8.0 × 107 M−1s−1 for the glutathiolated protein and 7.2 × 107 M−1s−1 for the wild-type enzyme). The same results were obtained for the inhibition constants using sulfonamide inhibitor acetazolamide (KI of 2.8, 3.0, and 2.7 nM for wild-type, C183S/C217S and the glutathiolated protein, respectively). Altogether these results were indicative of two sulfhydryl groups very reactive and not involved in the enzyme catalytic mechanism, thus suggesting, in analogy with what observed for CA III, that CA VII could function in vivo as an oxygen radical scavenger through its reactive cysteines, protecting cells from oxidative damage. To verify this hypothesis, the CA VII ability to protect human cells from oxidative stress was investigated17. In particular, HeLa cells, that do not express endogenous CA VII, were transiently transfected with a plasmid encoding the wild-type protein and then stressed by using sodium arsenite (SA). It was observed that CA VII was able to protect cells from death, as the induction of apoptosis was lower in cells expressing CA VII with respect to mock-transfected cells17. In particular, in mock-transfected cells the percentage of apoptotic cells increased from 15% to 49% after SA treatment, whereas cells expressing CA VII showed a lower percentage of apoptotic cells after induction of oxidative stress (from 18% to 39%). The activation of apoptosis mediated by mitochondria was then analyzed by Western blotting, showing that CA VII was able to protect HeLa cells from oxidative stress (Figure 2). In fact, levels of both Bcl-2 (Figure 2, gray bars) and Bax (Figure 2, white bars), which are, respectively, anti- and pro-apoptotic markers involved in mitochondria-mediated apoptosis, were almost unchanged in SA-treated cells expressing WT-CA VII with respect to untreated cells (86% and 93%, respectively, bars in the middle of Figure 2). By contrast, mock-transfected cells showed a significant alteration in Bcl-2 and Bax levels after induction of oxidative stress (50% and 336%, respectively, left side of Figure 2), as well as in the decrease of procaspase-3 levels, one of the final effectors of apoptosis (47% and 68%, for mock-transfected and CA VII-expressing cells, black bars in Figure 2). It is worth to note that, after treatment with SA, a stronger CA VII signal was observed probably due to a protective attempt of the cell to contrast oxidative stress injury17.
Figure 2.
Quantitative analysis of apoptotic marker levels in transfected HeLa cells after treatment with SA for 16 h. In each histogram, the protein intensity level was normalized to endogenous actin and expressed as percentage with respect to untreated cells. Black histograms refer to procaspase-3 levels, gray histograms to Bcl-2 and white histograms to Bax levels.
The importance of cysteine residues was validated by performing the same set of experiments in cells transfected with a CA VII mutant (TM-CA VII), in which all the cysteines present in the amino acid sequence (Cys54, Cys178, Cys183, and Cys217) were replaced by serine residues. Noteworthy, it was observed that cells expressing the TM-CA VII were susceptible to oxidative stress, as the increase in Bax levels as well the decrease in Bcl-2 and procaspase-3 levels were similar to those reported for mock-transfected cells (267%, 49%, and 45%, respectively, right side of Figure 2). These findings indicated lack of protection from oxidative stress in the presence of the mutated protein. Since this mutated enzyme possessed the same catalytic activity as the native protein (see above), it was possible to conclude that the protective role of CA VII against oxidative stress was not related to the enzyme catalytic activity, but rather to the presence of very reactive cysteine residues, which could act as oxygen radical scavenger.
A great amount of data support the idea that in vivo oxidative stress and the accompanying ROS are genotoxic and contribute to the development of several human cancers including colorectal carcinoma58–61. Since the latter presents a reduced CA VII expression55, it is tempting to speculate that the absence of the antioxidant action of CA VII could be responsible of a higher sensibility to the oxidative stress and consequently of the disease progression. Thus, the reported protective role of CA VII might suggest a novel tumor-suppressing function for this enzyme.
X-ray structural studies on CA III and CA VII
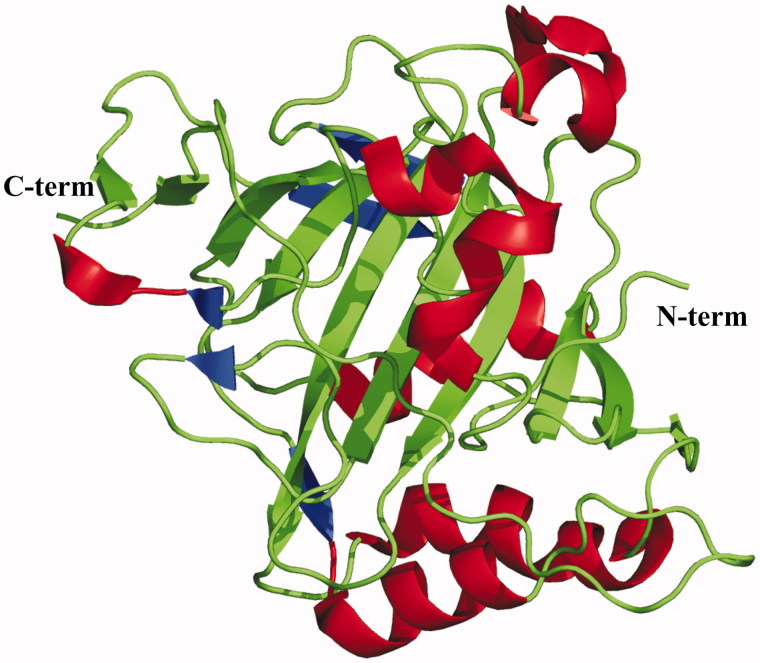
Several crystallographic structural studies have been reported on CA III12,37,62,63, the first one being on the bovine isoform which was solved in 199312, showing that the enzyme three-dimensional structure presents the typical α-CA fold13,64–72, characterized by a central 10-stranded β-sheet surrounded by several helices and additional β-strands (Figure 3). Structural data on the human isoform have been reported later, together with different site-specific variants aimed at identifying which residues were responsible for the low catalytic efficiency of CA III62,63.
Figure 3.
Ribbon representation of the overall fold of CA III.
In particular, McKenna’s group reported a kinetic and structural characterization of three hCA III mutants where an active site residue was substituted by a histidine one, namely mutants K64H, R67H, and K64H-R67N63. It is well known that the rate-limiting step of CO2 hydration reaction, catalyzed by α-CAs, is the proton transfer reaction from the zinc-bound water molecule to the external medium, to regenerate the zinc-bound hydroxide which is the reactive species1. This reaction is assisted by the proton shuttle His64 in hCA II73, the most catalytically active member of α-CA class2. Interestingly, the same position is occupied by a lysine in hCA III enzyme, thus suggesting that restoring a histidine in such position could greatly improve the CA III catalytic efficiency. Surprisingly, the kinetic data of K64H variant revealed only a small increase in the rate constant for proton transfer from proton donors to the zinc-bound hydroxide (kB values were 3.0 and 20 ms−1 for native hCA III and K64H variant, respectively)63. Moreover His64 in K64H variant had a capacity for proton transfer that was only 2.5% that of His64 in hCA II. Comparison of the crystal structures of K64H hCA III mutant and hCA II suggested that the different ability in proton transfer could be related, at least in part, to the limited conformational mobility of His64 observed in the mutant compared to hCA II, where His64 adopts two different conformations (referred to as inward and outward)73. In agreement with this hypothesis, it was previously reported that the high conformational mobility of His64 in hCA II would be an important feature for its ability in proton transfer74.
A higher reaction rate for proton transfer was achieved with R67H and K64H-R67N mutants (kB value of 81 and 100 ms−1 for R67H and K64H-R67N, respectively), indicating that other residues of the active site cavity can be involved into catalytic mechanism. Additional site-specific mutagenesis studies also showed the influence of amino acid replacement at position 198, suggesting that the hydrophobicity of Phe198 can affect the pKa of the zinc-bound water lowering CA III catalytic efficiency75.
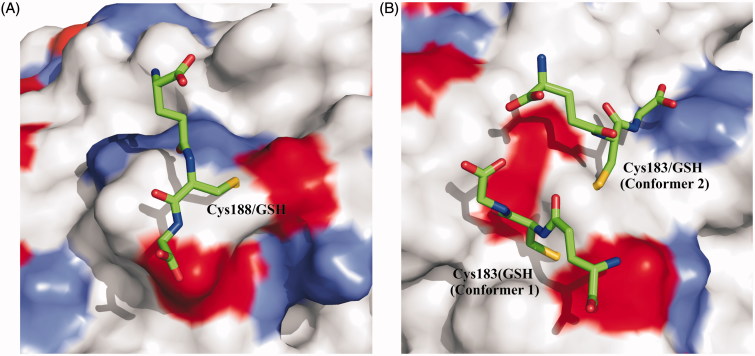
The X-ray structure of CA III extracted from rat liver was also solved providing very interesting data for understanding the molecular determinants responsible for the above described high reactivity of Cys183 and Cys18837. These two residues are located on the molecular surface of the protein and were found S-glutathiolated in the crystal structure. The analysis of the structure revealed that the presence of disulfide linkages between cysteine residues and GSH molecule did not alter the overall structure of the protein, nor the conformation of residues located near to Cys183 and Cys188. This observation was in agreement with the experimental data that S-glutathiolation has no effect on the catalytic activity of the enzyme, as the GSH moieties are distant from enzyme active site37. Moreover, the analysis of electron density in correspondence of Cys-GSH adducts indicated conformational flexibility of the glutathionyl moieties, with the disulfide bridge involving Cys183 adopting two different conformations (conformation 1 and 2), and that involving Cys188 with only one orientation observed.
Previously reported data indicated Cys188 as the most reactive cysteine76; this was explained by the examination of S-glutathiolated CA III structure. In fact, Cys188 was located in an environment characterized by a lower negative charge that could justify its greater propensity to react with uncharged or charged species (Figure 4)35,76,77. On the contrary, Cys183 was located in a depression of the surface showing a greater negative charge making this residue less reactive. Further clarifications on the Cys188 high reactivity were obtained from site-specific mutagenesis studies78. Oxidation of a thiol group involves its ionization to the thiolate. As the pKa of free cysteine is approximately 8.5, it was expected that, at physiological pH, cysteine was almost completely in its protonated form. However, within a protein 3D structure, the cysteine pKa can be altered and, in particular, lowered by the interaction with basic amino acids. Several charged residues were identified from CA III crystal structure that could affect the pKa of Cys188 (i.e. Lys213, Arg189, Asp190, and Glu214, numbering referring to that used for the crystal structure of CA III from rat liver). In order to elucidate their role, site-specific mutants were produced and characterized showing that Lys213 was the major responsible for the lowering of the pKa , whereas Arg189 seemed not to affect it. Notably, the acidic Asp190 and Glu214 reduced the reactivity of Cys188, thus suggesting that their conformational rearrangement could serve to modulate the CA III propensity to glutathiolation78.
Figure 4.
Schematic representation of (A) Cys188-GSH adducts and (B) Cys183-GSH. The two Cys183 conformers are indicated as 1 and 2. Surface representation of CA III is also reported showing the positive charged residues (Lys and Arg) colored light gray, while negative ones (Asp and Glu) colored black.
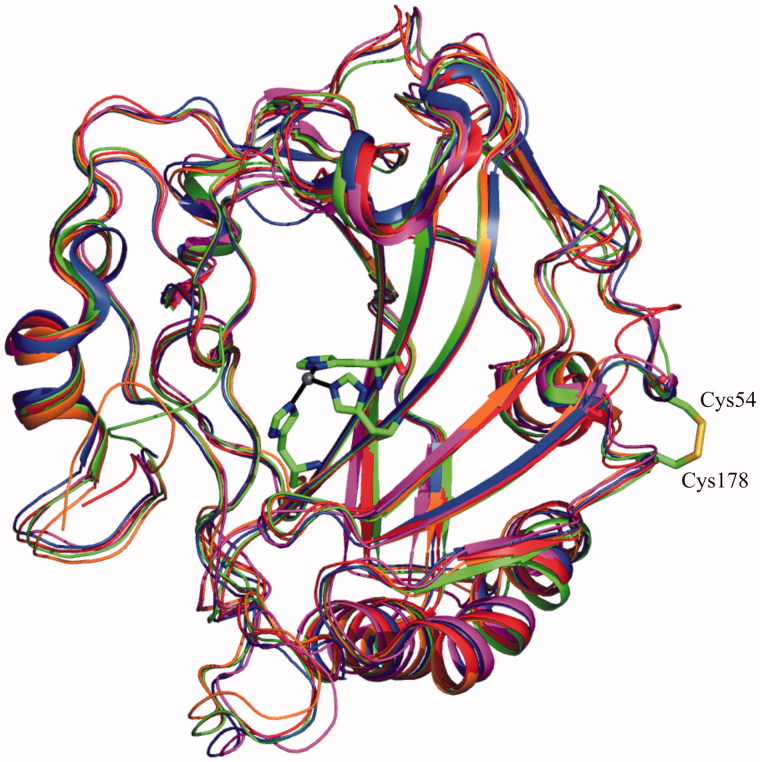
The crystal structure of hCA VII, solved in 2010 by our group13, showed structural features similar to those previously reported for other cytosolic CAs (Figure 5)12,64,65,71. Indeed, the protein was a monomer characterized by a central 10-stranded anti-parallel β-sheet surrounded by additional β-strands and three α- and four 310 helices. The hCA VII active site was located in a conical cavity about 15 Å wide and 15 Å deep, which extended from the surface of the protein to the center of the molecule. The catalytic zinc ion was located at the bottom of this cavity, coordinated by three histidine residues and the deprotonated nitrogen atom of sulfonamide moiety of acetazolamide molecule, which co-crystallized with the enzyme. The active site cavity was divided in two very different portions delimited by hydrophobic or hydrophilic amino acids. In particular, Val121, Leu198, Ala135, Leu141, Val143, Val207, and Phe131 delineated the hydrophobic region, while Asn62, His64, Gln67, Lys91, and Gln92 identified the hydrophilic one. This peculiar active site arrangement, already observed for other members of the α-CA family, was directly correlated to the catalytic mechanism of the enzyme. In particular, the hydrophobic region was supposed to be involved in the sequestration of the CO2 substrate and its opportune orientation for nucleophilic attack by the zinc-bound hydroxide79, whereas the hydrophilic region was supposed to be involved in the formation of a well ordered hydrogen-bonded solvent network, which assists the proton transfer reaction74,80,81.
Figure 5.
Structural superposition of all cytosolic ?-CAs. The zinc ion coordination and intramolecular disulfide bridge of hCA VII are also depicted.
An intramolecular disulfide bond was observed in hCA VII structure between Cys54 and Cys178 (Figure 5). However, the observation that these two cysteines are not conserved within the α-CA family82 and that disulfide bonds are extremely rare in cytosolic proteins83, suggested that this disulfide bond could be a result of the oxidizing conditions that arise during protein handling.
No structural data were available on the two cysteines (Cys183 and Cys217) involved into S-glutathiolation in vitro14, as a variant form, containing the substitution of these cysteines with serines, was used for the structural studies. However, the analysis of the structure of this variant showed that these residues were located on the protein surface and were completely accessible to the solvent. Molecular modeling studies, corroborated by site-specific mutagenesis, should be performed in order to clarify the structural determinants of the high chemical reactivity of these two sulfhydryl groups.
Conclusion
To respond to oxidative stress cells have developed antioxidant defense systems, including gluthathione, antioxidant vitamins, sulfhydryl groups, and antioxidant enzymes. CA III and CA VII have been proposed to be involved in these antioxidant defense systems, acting, through their reactive cysteines, as scavenger enzymes of reactive species in cells where oxidative damage occurs. Biochemical and structural studies of CA III allowed identification of the molecular determinants responsible for the high reactivity of cysteine residues involved in the proposed scavenger function and confirmed the critical role of the charged residues to modulate the reactivity of such residues.
On the other hand, fewer data are available for CA VII, which presents many similarities with CA III, starting from its propensity to undergo S-glutathiolation in vitro, ending with its ability to protect cells from oxidative insults. Interestingly, available data clearly indicate that the protective role of CA VII is not related to the enzyme catalytic activity, since a CA VII variant, in which the cysteine residues were replaced by serines, showed the same catalytic activity of the wild-type protein, but was not able to protect cells from oxidative stress injury. Further studies are needed to investigate the influence of the residues surrounding the reactive cysteines in order to highlight the molecular mechanisms regulating this phenomenon.
Disclosure statement
The authors report no declarations of interest.
References
- 1.Alterio V, Di Fiore A, D’Ambrosio K, et al. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem Rev 2012;112:4421–68. [DOI] [PubMed] [Google Scholar]
- 2.Supuran CT.Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81. [DOI] [PubMed] [Google Scholar]
- 3.Dathan NA, Alterio V, Troiano E, et al. Biochemical characterization of the chloroplastic beta-carbonic anhydrase from Flaveria bidentis (L.) “Kuntze”. J Enz Inhib Med Chem 2014;29:500–4. [DOI] [PubMed] [Google Scholar]
- 4.Ferry JG.The gamma class of carbonic anhydrases. Biochim Biophys Acta 2010;1804:374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alterio V, Langella E, De Simone G, Monti SM.. Cadmium-containing carbonic anhydrase CDCA1 in marine diatom Thalassiosira weissflogii. Marine Drugs 2015;13:1688–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Simone G, Di Fiore A, Capasso C, Supuran CT.. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9. [DOI] [PubMed] [Google Scholar]
- 7.Supran CT, De Simone G, eds. Carbonic anhydrases as biocatalysts: from theory to medical and industrial applications. Amsterdam: Elsevier; 2015. [Google Scholar]
- 8.Scozzafava A, Supuran CT.. Glaucoma and the applications of carbonic anhydrase inhibitors. Sub-Cell Biochem 2014;75:349–59. [DOI] [PubMed] [Google Scholar]
- 9.Thiry A, Dogne JM, Supuran CT, Masereel B.. Carbonic anhydrase inhibitors as anticonvulsant agents. Curr Top Med Chem 2007;7:855–64. [DOI] [PubMed] [Google Scholar]
- 10.Pastorek J, Pastorekova S.. Hypoxia-induced carbonic anhydrase IX as a target for cancer therapy: from biology to clinical use. Semin Cancer Biol 2015;31:52–64. [DOI] [PubMed] [Google Scholar]
- 11.Guler OO, De Simone G, Supuran CT.. Drug design studies of the novel antitumor targets carbonic anhydrase IX and XII. Curr Med Chem 2010;17:1516–26. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson AE, Liljas A.. Refined structure of bovine carbonic anhydrase III at 2.0 A resolution. Proteins 1993;16:29–42. [DOI] [PubMed] [Google Scholar]
- 13.Di Fiore A, Truppo E, Supuran CT, et al. Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide . Bioorg Med Chem Lett 2010;20:5023–6. [DOI] [PubMed] [Google Scholar]
- 14.Truppo E, Supuran CT, Sandomenico A, et al. Carbonic anhydrase VII is S-glutathionylated without loss of catalytic activity and affinity for sulfonamide inhibitors. Bioorg Med Chem Lett 2012;22:1560–4. [DOI] [PubMed] [Google Scholar]
- 15.Cabiscol E, Levine RL.. Carbonic anhydrase III. Oxidative modification in vivo and loss of phosphatase activity during aging. J Biol Chem 1995;270:14742–7. [DOI] [PubMed] [Google Scholar]
- 16.Monti SM, Supuran CT, De Simone G, Di Fiore A.. Carbonic anhydrase VII In: Supran CT, De Simone G, eds. Carbonic anhydrases as biocatalysts: from theory to medical and industrial applications. Amsterdam: Elsevier; 2015:151–68. [Google Scholar]
- 17.Del Giudice R, Monti DM, Truppo E, et al. Human carbonic anhydrase VII protects cells from oxidative damage. Biol Chem 2013;394:1343–8. [DOI] [PubMed] [Google Scholar]
- 18.Finkel T, Holbrook NJ.. Oxidants, oxidative stress and the biology of ageing. Nature 2000;408:239–47. [DOI] [PubMed] [Google Scholar]
- 19.Yoshikawa T, Naito Y.. What is oxidative stress? Jpn Med Assoc J 2002;45:271–6. [Google Scholar]
- 20.Uttara B, Singh AV, Zamboni P, Mahajan RT.. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol 2009;7:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machlin LJ, Bendich A.. Free radical tissue damage: protective role of antioxidant nutrients. FASEB J 1987;1:441–5. [PubMed] [Google Scholar]
- 22.Zimmerman UJ, Wang P, Zhang X, et al. Anti-oxidative response of carbonic anhydrase III in skeletal muscle. IUBMB Life 2004;56:343–7. [DOI] [PubMed] [Google Scholar]
- 23.Roy P, Reavey E, Rayne M, et al. Enhanced sensitivity to hydrogen peroxide-induced apoptosis in Evi1 transformed Rat1 fibroblasts due to repression of carbonic anhydrase III. FEBS J 2010;277:441–52. [DOI] [PubMed] [Google Scholar]
- 24.Rivera C, Voipio J, Kaila K. Two developmental switches in GABAergic signalling: the K+-Cl − cotransporter KCC2 and carbonic anhydrase CAVII. J Physiol (Lond) 2005;562:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harju AK, Bootorabi F, Kuuslahti M, et al. Carbonic anhydrase III: a neglected isozyme is stepping into the limelight. J Enz Inhib Med Chem 2013;28:231–9. [DOI] [PubMed] [Google Scholar]
- 26.Wade R, Gunning P, Eddy R, et al. Nucleotide sequence, tissue-specific expression, and chromosome location of human carbonic anhydrase III: the human CAIII gene is located on the same chromosome as the closely linked CAI and CAII genes. Proc Natl Acad Sci USA 1986;83:9571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spicer SS, Ge ZH, Tashian RE, et al. Comparative distribution of carbonic anhydrase isozymes III and II in rodent tissues. Am J Anat 1990;187:55–64. [DOI] [PubMed] [Google Scholar]
- 28.Kelly CD, Carter ND, de Boer P, et al. Detection of CAIII mRNA in rat skeletal muscle and liver by in situ hybridization. J Histochem Cytochem 1991;39:1243–7. [DOI] [PubMed] [Google Scholar]
- 29.Shiels A, Jeffery S, Wilson C, Carter N.. Radioimmunoassay of carbonic anhydrase III in rat tissues. Biochem J 1984;218:281–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaananen HK, Paloniemi M, Vuori J.. Purification and localization of human carbonic anhydrase. III. Typing of skeletal muscle fibers in paraffin embedded sections. Histochemistry 1985;83:231–5. [DOI] [PubMed] [Google Scholar]
- 31.Zheng A, Rahkila P, Vuori J, et al. Quantification of carbonic anhydrase III and myoglobin in different fiber types of human psoas muscle. Histochemistry 1992;97:77–81. [DOI] [PubMed] [Google Scholar]
- 32.Jeffery S, Edwards Y, Carter N.. Distribution of CAIII in fetal and adult human tissue. Biochem Genet 1980;18:843–9. [DOI] [PubMed] [Google Scholar]
- 33.Kato K, Mokuno K.. Distribution of immunoreactive carbonic anhydrase III in various human tissues determined by a sensitive enzyme immunoassay method. Clin Chim Acta 1984;141:169–77. [DOI] [PubMed] [Google Scholar]
- 34.Raisanen SR, Lehenkari P, Tasanen M, et al. Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J 1999;13:513–22. [DOI] [PubMed] [Google Scholar]
- 35.Chai YC, Jung CH, Lii CK, et al. Identification of an abundant S-thiolated rat liver protein as carbonic anhydrase III; characterization of S-thiolation and dethiolation reactions. Arch Biochem Biophys 1991;284:270–8. [DOI] [PubMed] [Google Scholar]
- 36.Filomeni G, Rotilio G, Ciriolo MR.. Disulfide relays and phosphorylative cascades: partners in redox-mediated signaling pathways. Cell Death Different 2005;12:1555–63. [DOI] [PubMed] [Google Scholar]
- 37.Mallis RJ, Poland BW, Chatterjee TK, et al. Crystal structure of S-glutathiolated carbonic anhydrase III. FEBS Lett 2000;482:237–41. [DOI] [PubMed] [Google Scholar]
- 38.Mari M, Morales A, Colell A, et al. Mitochondrial glutathione, a key survival antioxidant. Antioxid Redox Signal 2009;11:2685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mari M, Colell A, Morales A, et al. Redox control of liver function in health and disease. Antioxid Redox Signal 2010;12:1295–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallis RJ, Hamann MJ, Zhao W, et al. Irreversible thiol oxidation in carbonic anhydrase III: protection by S-glutathiolation and detection in aging rats. Biol Chem 2002;383:649–62. [DOI] [PubMed] [Google Scholar]
- 41.Thomas JA, Mallis RJ.. Aging and oxidation of reactive protein sulfhydryls. Exp Gerontol 2001;36:1519–26. [DOI] [PubMed] [Google Scholar]
- 42.Kim G, Lee TH, Wetzel P, et al. Carbonic anhydrase III is not required in the mouse for normal growth, development, and life span. Mol Cell Biol 2004;24:9942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morishita K, Parker DS, Mucenski ML, et al. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell 1988;54:831–40. [DOI] [PubMed] [Google Scholar]
- 44.Bartholomew C, Kilbey A, Clark AM, Walker M.. The Evi-1 proto-oncogene encodes a transcriptional repressor activity associated with transformation. Oncogene 1997;14:569–77. [DOI] [PubMed] [Google Scholar]
- 45.Wieser R.The oncogene and developmental regulator EVI1: expression, biochemical properties, and biological functions. Gene 2007;396:346–57. [DOI] [PubMed] [Google Scholar]
- 46.Tweedie S, Morrison K, Charlton J, Edwards YH.. CAIII a marker for early myogenesis: analysis of expression in cultured myogenic cells. Som Cell Mol Genet 1991;17:215–28. [DOI] [PubMed] [Google Scholar]
- 47.Nucifora G, Laricchia-Robbio L, Senyuk V.. EVI1 and hematopoietic disorders: history and perspectives. Gene 2006;368:1–11. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Chen L, Ko TC, et al. Evi1 is a survival factor which conveys resistance to both TGFbeta- and taxol-mediated cell death via PI3K/AKT. Oncogene 2006;25:3565–75. [DOI] [PubMed] [Google Scholar]
- 49.Kaila K, Lamsa K, Smirnov S, et al. Long-lasting GABA-mediated depolarization evoked by high-frequency stimulation in pyramidal neurons of rat hippocampal slice is attributable to a network-driven, bicarbonate-dependent K + transient. J Neurosci 1997;17:7662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruusuvuori E, Huebner AK, Kirilkin I, et al. Neuronal carbonic anhydrase VII provides GABAergic excitatory drive to exacerbate febrile seizures. EMBO J 2013;32:2275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bootorabi F, Janis J, Smith E, et al. Analysis of a shortened form of human carbonic anhydrase VII expressed in vitro compared to the full-length enzyme. Biochimie 2010;92:1072–80. [DOI] [PubMed] [Google Scholar]
- 52.Shah GN, Hewett-Emmett D, Grubb JH, et al. Mitochondrial carbonic anhydrase CA VB: differences in tissue distribution and pattern of evolution from those of CA VA suggest distinct physiological roles. Proc Natl Acad Sci USA 2000;97:1677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkkila S, Kivela AJ, Kaunisto K, et al. The plasma membrane carbonic anhydrase in murine hepatocytes identified as isozyme XIV. BMC Gastroenterol 2002;2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bootorabi F, Haapasalo J, Smith E, et al. Carbonic anhydrase VII – a potential prognostic marker in gliomas. Health 2011;3:6–12. [Google Scholar]
- 55.Yang GZ, Hu L, Cai J, et al. Prognostic value of carbonic anhydrase VII expression in colorectal carcinoma. BMC Cancer 2015;15:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birkenkamp-Demtroder K, Olesen SH, Sorensen FB, et al. Differential gene expression in colon cancer of the caecum versus the sigmoid and rectosigmoid. Gut 2005;54:374–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu CM, Yao CT, Chang YT, et al. Gene expression profiling of colorectal tumors and normal mucosa by microarrays meta-analysis using prediction analysis of microarray, artificial neural network, classification, and regression trees. Dis Mark 2014;2014:634123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sosa V, Moline T, Somoza R, et al. Oxidative stress and cancer: an overview. Ageing Res Rev 2013;12:376–90. [DOI] [PubMed] [Google Scholar]
- 59.Fang J, Seki T, Maeda H.. Therapeutic strategies by modulating oxygen stress in cancer and inflammation. Adv Drug Deliv Rev 2009;61:290–302. [DOI] [PubMed] [Google Scholar]
- 60.Perse M.Oxidative stress in the pathogenesis of colorectal cancer: cause or consequence? Biomed Res Int 2013;2013:725710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stone WL, Krishnan K, Campbell SE, Palau VE.. The role of antioxidants and pro-oxidants in colon cancer. World J Gastrointest Oncol 2014;6:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duda DM, Tu C, Fisher SZ, et al. Human carbonic anhydrase III: structural and kinetic study of catalysis and proton transfer. Biochemistry 2005;44:10046–53. [DOI] [PubMed] [Google Scholar]
- 63.Elder I, Fisher Z, Laipis PJ, et al. Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III. Proteins 2007;68:337–43. [DOI] [PubMed] [Google Scholar]
- 64.Kannan KK, Ramanadham M, Jones TA.. Structure, refinement, and function of carbonic anhydrase isozymes: refinement of human carbonic anhydrase I. Ann N Y Acad Sci 1984;429:49–60. [DOI] [PubMed] [Google Scholar]
- 65.Eriksson AE, Jones TA, Liljas A.. Refined structure of human carbonic anhydrase II at 2.0 A resolution. Proteins 1988;4:274–82. [DOI] [PubMed] [Google Scholar]
- 66.Stams T, Nair SK, Okuyama T, et al. Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-A resolution. Proc Natl Acad Sci USA 1996;93:13589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boriack-Sjodin PA, Heck RW, Laipis PJ, et al. Structure determination of murine mitochondrial carbonic anhydrase V at 2.45-A resolution: implications for catalytic proton transfer and inhibitor design. Proc Natl Acad Sci USA 1995;92:10949–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pilka ES, Kochan G, Oppermann U, Yue WW.. Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development. Biochem Biophys Res Commun 2012;419:485–9. [DOI] [PubMed] [Google Scholar]
- 69.Alterio V, Hilvo M, Di Fiore A, et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc Natl Acad Sci USA 2009;106:16233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whittington DA, Waheed A, Ulmasov B, et al. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc Natl Acad Sci USA 2001;98:9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Fiore A, Monti SM, Hilvo M, et al. Crystal structure of human carbonic anhydrase XIII and its complex with the inhibitor acetazolamide. Proteins 2009;74:164–75. [DOI] [PubMed] [Google Scholar]
- 72.Whittington DA, Grubb JH, Waheed A, et al. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes. J Biol Chem 2004;279:7223–8. [DOI] [PubMed] [Google Scholar]
- 73.Mikulski RL, Silverman DN.. Proton transfer in catalysis and the role of proton shuttles in carbonic anhydrase. Biochim Biophys Acta 2010;1804:422–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Silverman DN, McKenna R.. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc Chem Res 2007;40:669–75. [DOI] [PubMed] [Google Scholar]
- 75.LoGrasso PV, Tu C, Chen X, et al. Influence of amino acid replacement at position 198 on catalytic properties of zinc-bound water in human carbonic anhydrase III. Biochemistry 1993;32:5786–91. [DOI] [PubMed] [Google Scholar]
- 76.Cabiscol E, Levine RL.. The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc Natl Acad Sci USA 1996;93:4170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji Y, Akerboom TP, Sies H, Thomas JA.. Gel electrofocusing method for studying protein S-nitrosylation. Meth Enzymol 1999;301:145–51. [DOI] [PubMed] [Google Scholar]
- 78.Kim G, Levine RL.. Molecular determinants of S-glutathionylation of carbonic anhydrase 3 . Antioxid Redox Signal 2005;7:849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Domsic JF, Avvaru BS, Kim CU, et al. Entrapment of carbon dioxide in the active site of carbonic anhydrase II. J Biol Chem 2008;283:30766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fisher SZ, Maupin CM, Budayova-Spano M, et al. Atomic crystal and molecular dynamics simulation structures of human carbonic anhydrase II: insights into the proton transfer mechanism. Biochemistry 2007;46:2930–7. [DOI] [PubMed] [Google Scholar]
- 81.Roy A, Taraphder S.. Identification of proton-transfer pathways in human carbonic anhydrase II. J Phys Chem B 2007;111:10563–76. [DOI] [PubMed] [Google Scholar]
- 82.Alterio V, Di Fiore A, D’Ambrosio K, et al. X-ray crystallography of carbonic anhydrase inhibitors and its importance in drug design In: Supuran CT, Winum J-Y, eds. Design of zinc-enzyme inhibitors: functional, structural, and disease applications. Hoboken (NJ): Wiley; 2009:73–138. [Google Scholar]
- 83.Kadokura H, Katzen F, Beckwith J.. Protein disulfide bond formation in prokaryotes. Ann Rev Biochem 2003;72:111–35. [DOI] [PubMed] [Google Scholar]
- 84.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]