Abstract
An extracellular thermostable alkaline serine protease enzyme from Aeribacillus pallidus C10 (GenBank No: KC333049), was purified 4.85 and 17. 32-fold with a yield of 26.9 and 19.56%, respectively, through DE52 anion exchange and Probond affinity chromatography. The molecular mass of the enzyme was determined through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), with approximately 38.35 kDa. The enzyme exhibited optimum activity at pH 9 and at temperature 60 °C. It was determined that the enzyme had remained stable at the range of pH 7.0–10.0, and that it had preserved more than 80% of its activity at a broad temperature range (20–80 °C). The enzyme activity was found to retain more than 70% and 55% in the presence of organic solvents and commercial detergents, respectively. In addition, it was observed that the enzyme activity had increased in the presence of 5% SDS. K M and V max values were calculated as 0.197 mg/mL and 7.29 μmol.mL− 1.min− 1, respectively.
Keywords: Thermotolerant, Aeribacillus pallidus, alkaline serine protease, biochemical characterization, purification
Introduction
The largest class of commercial enzymes on which industrial applications are done comprise proteolytic enzymes. Proteases or peptidases form the hydrolytic enzyme group which hydrolyzes the peptide bonds within the structure of proteins and catalyzes the peptide synthesis in the presence of an organic solvent1 , 2. They are classified as acidic, alkaline and neutral proteases according to the pH ranges they are active at; and as serine, aspartic, cysteine and metalloproteases according to the functional group found in its active area3 , 4. Sixty to sixty-five percent of the global enzyme market, the largest part of which consists of alkaline proteases, is comprised of proteases5. Alkaline proteases are commonly used in a number of fields, such as leather, textile, waste management industries as well as food and pharmaceutical industries, notably the detergent industry2 , 6 , 7. In the presence of oxidizing agents and anionic surfactants, which are the additives used commonly in the modern detergent formulation, alkaline proteases that are both stable and active in the solutions with high pH, as well, are of importance in the detergent industry in particular8. For this reason, in the presence of high specificity, thermostability, alkaline pH and organic solvents, the industrial demand for quite active preparates of proteolytic enyzmes with high stability urges those involved in this field to investigate alternative protease resources.
Scientists are quite intensively interested in alkaline proteases that are produced by microorganisms and are quite important in terms of biotechnology9. The reason for this is that the enyzmes originating from microorganisms have high-catalytic activities, that they do not cause any undesired by-product, that their growth conditions can be easily optimized, that they are quite inexpensive, and that they can be produced in quite large amounts with high purity and at a single production process10 , 11. Apart from the fact that alkaline proteases are produced by a large microorganism group in which fungi and bacteria are also included, a great majority of commercial alkaline proteases in particular are obtained from the bacteria of Bacillus sp. species12 , 13. Thermophilic microorganisms are quite important in terms of industry, since they are capable of adapting to extreme conditions, such as high temperature, pressure, pH and salt concentration14. Aeribacillus pallidus, a thermophilic bacterium, belongs to the class of Bacilli species, and its optimum growth temperature is 55–60 °C15. There are many studies in the literature in which the characterization of A. pallidus, a type species of this new genus, is performed through conventional and molecular methods16–20. Cihan et al.18 identified the Aeribacillus strain and stated that it had no protease activity, whereas in our study, we determined that the strain we used had a high-protease activity.
No enzymatic research regarding this new species has been found in the literature, and for the first time with our study, a research into the purification, characterization and biotechnological applicability of the protease enzyme from A. pallidus C10 strain was performed.
Materials and methods
Identification of thermophilic bacteria
The isolation and identification of the test strain used in this study was reported elsewhere20.
16S rRNA gene sequence analysis
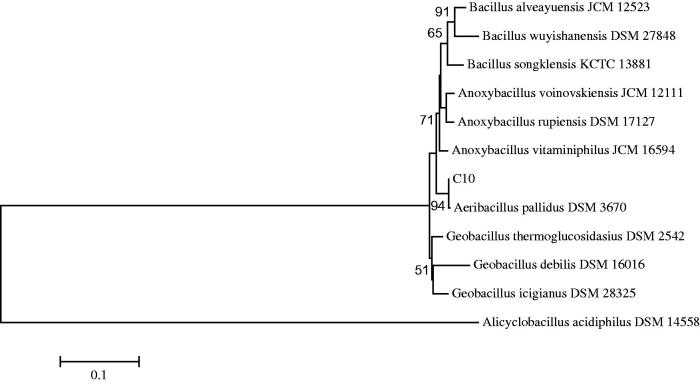
16S rRNA, which is the evolutionary gene of the prokaryotic ribosome, from the genomic DNA of the test isolate, was amplified by PCR with the forward primer UNI16S-L: (5′-ATTCTAGAGTTTGATCATGGCTCA-3′) and the reverse primer UNI16S-R: (5′-ATGGTACCGTGTGACGGGCGG TGTGTA-3′). The amplified fragment was cloned to a vector system (pGEM-T, Promega, the UK); and then, the clone was sequenced (Macrogen, Amsterdam, the Netherlands). The result of 16S rRNA gene sequencing was analyzed using the GenBank and EzTaxon (http://blast.ncbi.nlm.nih.gov/blast.cgi and http://www.eztaxon.org) server. Considering the results of the study, a phylogenetic tree was formed via the Neighbor Joining Method (Figure 1) using the MEGA 4.0 software package (Phoenix, AZ)21.
Figure 1.
Neighbor-joining phylogenetic tree on the basis of 16S rRNA gene sequence data of the thermophilic bacteria. Alicyclobacillus acidiphilus DSM 14558 was used as out-group. Bootstrap values based on 1000 replications are listed as percentages at branching points. Only bootstrap values >50% are shown at nodes. The scale bar represented 1% divergence.
Enzyme and protein assay
The protease activity was determined through the modification of the method defined by Takami et al.22. By adding 2.5 mL from 0.65% of casein solution prepared within 100 mM Tris-HCl buffer (pH 8.5) onto 0.5 mL enzyme solution, the reaction solution was kept at 37 °C for incubation for 10 min. A blank was performed in the same procedure; but this time, 0.5 ml of 100 mM Tris-HCl buffer (pH 8.5) was used instead of the purified enzyme. At the end of the incubation, the reaction was stopped by adding 2.5 mL from 110 mM trichloroacetic acid (TCA). After this mixture was kept at 37 °C in water bath for 30 min, it was centrifuged at 10,000 × g for 10 min. By adding 2.5 mL from 0.5 M Na2CO3 solution and 0.5 mL from 0.5 M Folin-Ciocalteu reagent into 1 mL of the supernatant, the mixture was kept at 37 °C for 30 min after the absorbance was measured at 660 nm. One unit of protease activity was defined as the amount of enzyme required to liberate l μg/min-tyrosine at 37 °C and at pH 8.5. The protein concentration was determined according to the Bradford method, in which bovine serum albumin (BSA) was used as the standard23.
Protease enzyme purification
Ammonium sulfate precipitation and dialysis
The cell isolates of A. pallidus C10 were centrifuged at 10,000 × g for 10 min, and the culture supernatant containing extracellular protease was subjected to ammonium sulfate precipitation. The mixture precipitated through the ammonium sulfate at the range of 0–90% was centrifuged at 5000 × g for 15 min. The precipitate obtained was dissolved through 100 mM Tris-HCl (pH = 8.5) buffer solution at minimum volume and was dialyzed against the same buffer.
Ion exchange chromatography
The dialyzate was loaded into DEAE-cellulose (DE52, Whatman) column balanced with 50 mM Tris-HCl buffer (pH 8.5), and bound proteins were eluted with a linear gradient of 50–500 mM NaCl in the same buffer. Fractions of 5 mL were collected and analyzed for enzyme activity and protein concentration. Fractions showing maximum activity were pooled and stored at 4 °C for further analysis.
Affinity chromatography
A second purification method was performed on the protease enzyme through Probond Nickel Chelating Resin (Invitrogen) affinity chromatography. After ammonium sulfate precipitation had been performed, the dialyzed sample was connected to Probond affinity column equalized with a natural-binding buffer (0.5 M NaCl and 50 mM NaH2PO4, pH 8.0). After the removal of the proteins which did not contain histidine amino acid by using a wash buffer (0.5 M NaCl, 20 mM imidazole, and 50 mM NaH2PO4, pH 8.0), the protease enzyme was eluated from the column by using an elution buffer (0.5 M NaCl, 250 mM imidazole, and 50 mM NaH2PO4, pH 8.0). Enzyme and protein assays were performed on the collected fractions.
SDS-polyacrylamide gel electrophoresis and zymography
For the control of the purity and determination of molecular weight of the enzyme, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to the Laemmli method24. The protein bands were determined through the silver-staining process. For the zymogram analysis of alkaline protease enzyme, SDS-PAGE was performed according to a method modified by Garcia-Carreno et al.25. After the electrophoresis, the gel was kept in 2.5% TritonX-100 solution, which was prepared with 100 mM pH 8.5 Tris-HCl, in order to remove the SDS in shaker at 37 °C with 150 rpm for 30 min. Afterwards, the gel was washed up three times with 100 mM Tris-HCl (pH 8.5) buffer, and then TritonX-100 was removed. Finally, the gel was incubated in 0.65% casein solution prepared within 100 mM Tris-HCl (pH 8.5) buffer, in a shaker operating at 37 °C and 150 rpm speed. After incubation, silver-staining process was performed.
The effect of pH on protease enzyme activity and stability
For the purpose of determining the optimum pH of the enzyme, the protease activity was performed by using sodium phosphate (pH 6.0–8.0), Tris-HCl (pH 8.0–9.0) and glycine-NaOH (pH 9.0–11.0) buffer at 0.1 M different pH values containing 0.65% casein. The enzyme was incubated in various buffer solutions (pH 6–11) at room temperature for 2 h for pH stability study, and the residual activity was determined in standard experimental conditions.
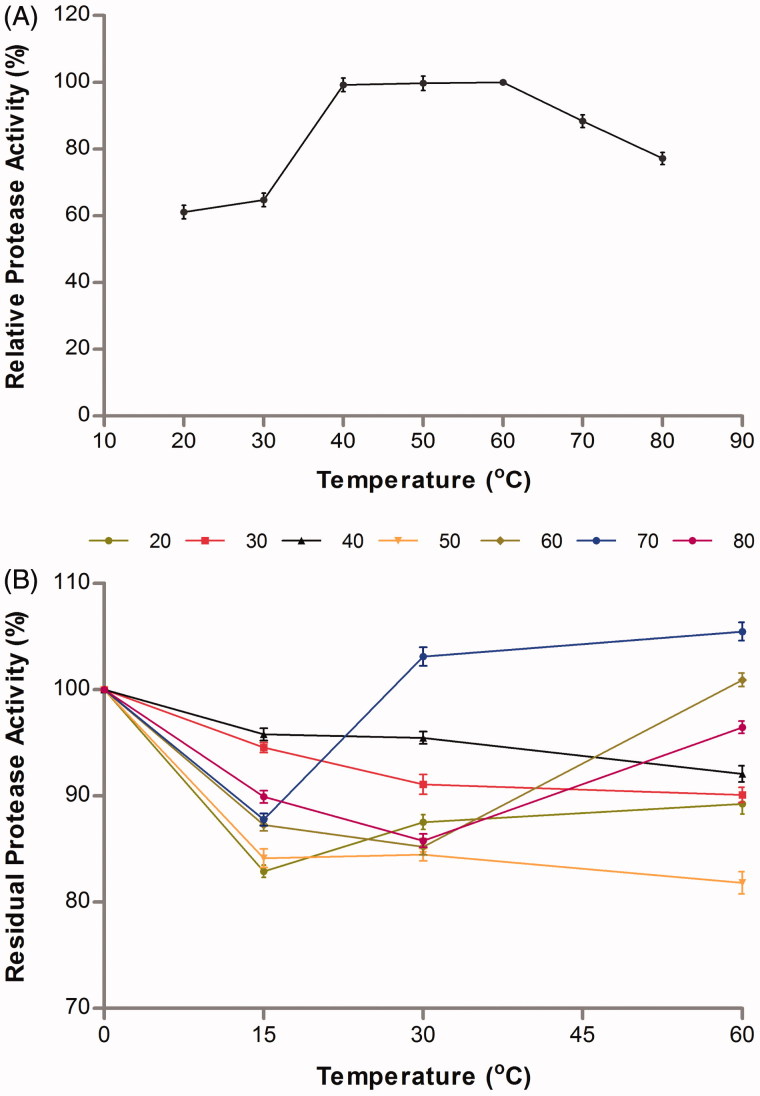
The effect of temperature on protease enzyme activity and stability
The effect of temperature on the enzyme activity was determined by measuring the enzyme activity at pH 8.5 for 10 minutes at different temperatures, from 20 °C up to 80 °C. To determine the temperature stability of the enzyme; the activity was measured by leaving the enzyme solution for incubation at different temperatures (20–80 °C) for 15, 30 and 60-min periods. After the amount of activity prior to the incubation at different temperatures had been accepted as 100, the values obtained in the wake of the incubation were expressed as the residual activity percentage (%).
Enzyme kinetics
In order to determine K M and V max values of the protease enzyme purified from A. pallidus C10, the activities of the casein substrate at the range of 0.02–0.6% at different concentrations were measured. With the obtained values, the Lineweaver–Burk graphic was drawn, and by making use of this graphic, the kinetic parameters, such as K M and V max, were calculated24 , 26.
Substrate specificity
The substrate specificity of C10 protease enzyme was determined by using casein, azocasein, hemoglobin, bovine serum albumin (BSA) and gelatin substrates. 0.65%-solutions pertaining to the substrates were prepared in a 100 mM Tris-HCl (pH 8.5) buffer. Later on, 0.5 mL of enzyme and 2.5 mL of substrate solution were mixed, and the activity was measured. The activity of the enzyme along with the substrate through which it showed the highest activity was accepted as 100%, and the relative enzyme activities were calculated.
The effect of some inhibitors and metal ions on C10 protease activity
The effect of enzyme inhibitors on the protease activity was determined by using phenylmethylsulfonyl fluoride (PMSF) (2 and 5 mM), ethylenediaminetetraacetic acid (EDTA) (2 and 5 mM), β-mercaptoethanol (1 and 5%), and ditio-bis-nitrobenzoic acid (DTNB) (2 and 5 mM). The purified enzyme was incubated in the presence of protease inhibitors at 37 °C for 30 min, and the amount of the activity of the enzyme solution containing no inhibitor was accepted as 100%, and the residual enzyme activity was calculated.
The effect of the metal ions on the protease enzyme activity was decided on by measuring the enzyme activity in the presence of 1, 5 and 10 mM concentrations of various metal ions (Co2+, Mg2+, Fe2+, Mn2+, Ca2+, Ni2+, Zn2+, K+, Ag+). The activity amount of the enzyme solution containing no metal ion was accepted as 100%, and the activity of the samples containing any metal ion was calculated as the residual activity.
The effect of some organic solvents on protease stability
In order to examine the change of the protease enzyme in activity, which had been purified from A. pallidus C10, in the presence of organic solvents; the enzyme was mixed with organic solvents, such as methanol, ethanol, acetone, DMSO, butanol, chloroform, isopropanol, at 15%, 25%, and 50% concentrations, after which it was incubated for 1 and 24 h. The activity amount of the enzyme solution containing no organic solvent was accepted as 100%, and the activity of the samples containing any organic solvent was calculated as the residual activity.
The effect of surfactants, oxidizing agent and commercial detergents on the stability of alkaline protease
The effects of some of the surfactants (SDS, Triton X-100, Tween-20, Tween-80) and the oxidizing agent (H2O2) on the protease activity were determined by subjecting the enzyme along with the surfactants and oxidizing agents to pre-incubation at different concentrations (1 and 5%) at 37 °C for 30 min. The residual activity after incubation was calculated, and the enzyme activity with no additive at all was taken as 100%.
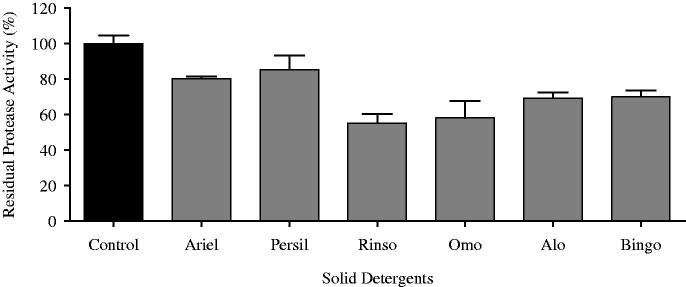
The compliance of the protease purified by using Bingo (Hayat Chemistry, Turkey), Alo (Procter&Gamble, Turkey), Rinso (Unilever, Turkey), Persil (Henkel, Turkey), Ariel (Procter&Gamble, Turkey) and OMO (Unilever, Turkey) with commercial solid laundry detergents was examined. The solutions of the detergents at 1%-concentrations were prepared in tap water. The diluted detergents were heated up at 100 °C for an hour prior to the addition of the enzyme, and the endogenous proteases found within these detergents were inactivated. The enyzme solution along with the detergent solutions prepared was left for incubation at 37 °C for an hour, and the residual activity was calculated. The sample, as a control, in which there was no detergent solution was left for incubation under the same conditions, and the activity of the control was accepted as 100%.
Statistical analysis
All the experiments were repeated three times, and as for the evaluation of the data, The GraphPad Prism-version 5.00 (Graphpad Software, San Diego, CA) statistical program was used. The mean ± standard error (SEM) (n = 3) values seen in all the results were analyzed.
Results and discussion
Characterization of the thermophilic bacteria
Some morphological, physiological and biochemical tests were applied to the test strain (C10), which was isolated from Sirnak–Belkisana hot spring in Turkey. According to the results, the isolate was moving rods which formed aerobic, thermophilic, Gram positive, catalase, oxidase, endospore positive. The optimum pH and temperature for the test strain was detected as 9.0 and 55 °C, respectively. This met the criteria of thermophilic bacteria, which reproduced at temperatures above 50 °C21. The thermophilic bacteria were able to reproduce at a range of salt concentration of 2–6%.
As a result of 16S rRNA sequence analysis, the test isolate was determined to contain about 1427 nucleotides (nt). Later, in consequence of the blast study carried out by using the database in the Genbank and EzTaxon, the thermophilic bacteria were detected to be the strain belonging to the genus of Aeribacillus at a powerful rate of >99% (GenBank No: KC333049).
Purification of A. pallidus C10 protease
The protease enzyme from A. pallidus C10, after the ammonium sulfate precipitation (90% w/v), was purified 4.85 and 17.32-fold at 26.9 and 19.56% efficiency, respectively, through the use of DE52 anion-exchange chromatography and Probond affinity chromatography. The purification profile of the protease enzyme is summarized in Table 1. In the purification process performed through two different chromatographic methods in this study, the purification coefficient was observed to have increased when Probond affinity chromatography was used in comparison to the anion-exchange chromatography. It was determined that when these purification results were compared with those in the literature, similar results27 , 28 were seen and that higher efficiency29 , 30 and fold purification27 , 31 were obtained when compared with some other studies. It is seen in the literature that purification processes for proteases have been performed through several different methods, such as ammonium sulfate precipitation, ultrafiltration, DEAE-Sepharose fast flow/Mono Q-Sepharose ion exchange chromatography, Sephadex gel filtration and Sepharose-bacitracin affinity chromatography, or different combinations of these. Nonetheless, although the Nickel-affinity (Probond) column which binds histidine amino acids to the nickel ion found within its structure is used to purify rather the recombinant proteins, it can also be used for the natural proteins that have an affinity on their own to divalent cations32 , 33. This method was used by starting from the fact that purification could be performed through this affinity column in serine proteases that keep histidine amino acid in their catalytic region34.
Table 1.
Summary of the alkaline protease from A. pallidus C10 purification procedure.
| Purification methods | Purification steps | Total activity(U) | Total protein(mg) | Specific activity(U/mg) | Yield (%) | Purificationfold |
|---|---|---|---|---|---|---|
| DE52 anion exchange chromatography | Culture supernatant | 2148 | 42960 | 0.056 | 100 | 1 |
| (NH4)2SO4 precipitation and dialysis | 283.6 | 1610 | 0.176 | 13.02 | 3.14 | |
| Anion exchange chromatography | 76.3 | 280 | 0.272 | 26.9 | 4.85 | |
| Probond affinity chromatography | Culture supernatant | 2473.8 | 40380 | 0.061 | 100 | 1 |
| (NH4)2SO4 precipitation and dialysis | 332.5 | 4088 | 0.081 | 13.44 | 1.33 | |
| Affinity chromatography | 65.04 | 61.05 | 1.057 | 19.56 | 17.32 |
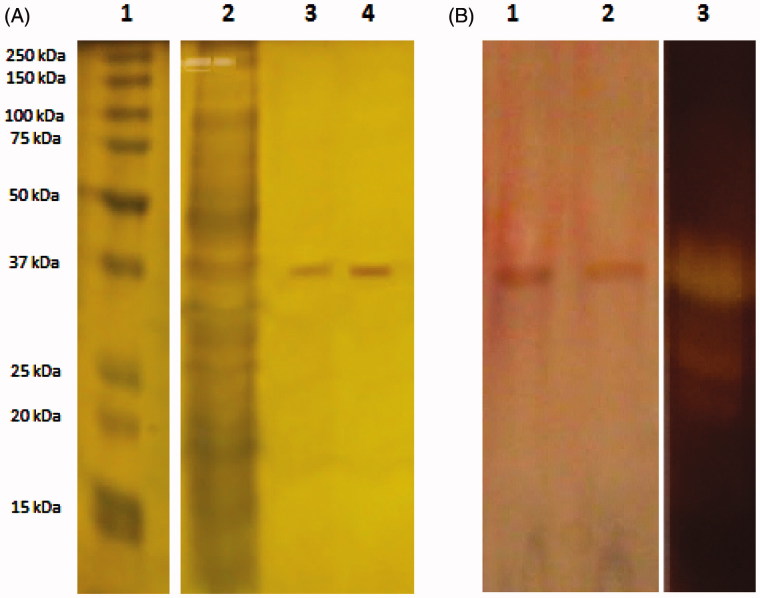
The molecular weight of the purified protease enzyme was estimated to be approximately 38.35 kDa by SDS-PAGE (Figure 2(A,B)). In general, apart form their exceptions; the molecular masses of microbial alkaline proteases vary between 15–36 kDa8. In previously-conducted studies, alkaline proteases with molecular weights similar to A. pallidus C10, such as B. pumilis CBS (34 kDa)35, P. fluorescens (36 kDa)36 , B. pumilus MCAS8 (36 kDa)37 , B. lehensis (39 kDa)38; and alkaline proteases with different molecular weights such as A. nidulans HA-10 (42 kDa)39, B. cereus 1173900 (66 kDa)7 , B. mojavensis A21 (20 kDa)8 , B. subtilis DM-04 (16.9 kDa)27 were reported. The purity and activity of the enzyme was also evaluated using zymogram activity staining. A clear zone of hydrolysis was observed in the gel indicating the presence of proteolytic activity (Figure 2(B)).
Figure 2.
SDS-PAGE of the purified protease from A. pallidus C10. (A) Lane 1: Standard protein molecular mass markers, lane 2: (NH4)2SO4 precipitated proteins, lane 3–4: Purified protease from DE52 anion exchange chromatography, (B) Lane 1–2: Purified enzyme from Probond affinity chromatography, lane 3: Zymography of the purified enzyme.
The effect of pH on alkaline protease activity and stability
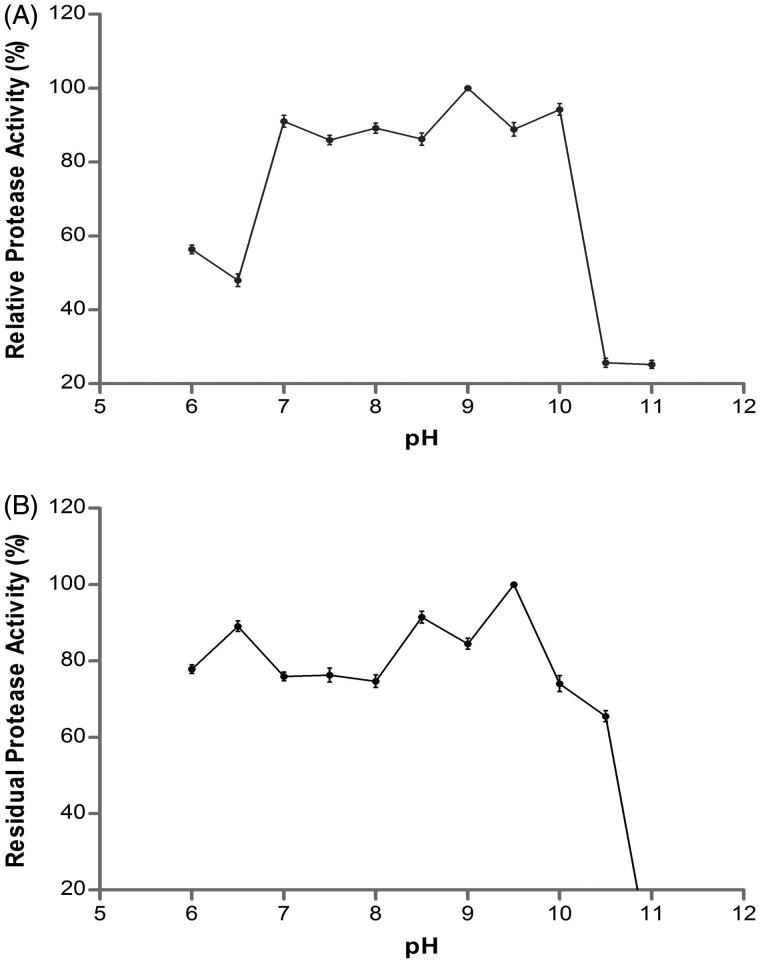
The optimum pH of the protease enzyme purified from A. pallidus C10 with high activity at the range of pH 7–10 was determined as 9.0 (Figure 3(A)). The fact that the enzyme preserves more than 85% of its activity at the range of alkaline pH (7.0–10.0) indicates to the fact that the enzyme in question is an alkaline protease. These findings are in accordance with the proteases, the optimum pH of which was at the range of 7.0–10.0 in the previously reported studies37 , 40 , 41.
Figure 3.
Effect of pH on activity (A) and stability (B) of the purified protease from A. pallidus C10.
When the pH stability profile for the protease enzyme purified from A. pallidus C10 was examined; it was determined that the enzyme had preserved its activity by more than 70% at the range of pH 6.0–10.5 after the 2-h-incubation at room temperature. The pH stability results showed that the protease was 100% stable at pH 9.5 and that it lost its activity at the rate of 11, 9, 16 and 30% at pH 6.5, 8.5, 9.0, and 10.0, respectively (Figure 3(B)). Some alkaline proteases with similar pH stability ranges have also been reported, such as B. Pumilus CBS35, B. amyloliquefaciens SP142, Bacillus strain HUTBS7143, and B. licheniformis P00344. In detergent formulation, when the significance of proteases at high-pH values that are particularly stable at 8.5–10.0 pH range is taken into consideration12 , 45 , 46, the fact that A. pallidus C10 protease enzyme has an activity and pH stability under high-alkaline conditions indicates to its potential applicability for the detergent industry.
The effect of temperature on alkaline protease activity and stability
The effect of temperature on the protease enzyme purified from A. pallidus C10 was examined at different temperatures, and the optimum temperature was determined to be 60 °C. It was ascertained that C10 protease enzyme was active at a broad temperature range, from 40 to 80 °C, and that it had 99, 99, 88 and 77% activity, respectively, at the temperatures of 40, 50, 70 and 80 °C (Figure 4(A)). These results suggest that this enzyme is a thermophilic protease. Some proteases, the optimum temperature of which was 60 °C, have also been reported, which are B. licheniformis ATCC 2142447, B. mojavensis A218, Bacillus sp. SM201446, and A.veronii PG0148. Nevertheless, the alkaline proteases with lower optimum temperature value than the purified protease were reported in the previously-conducted studies, as well27 , 39 , 44.
Figure 4.
Effect of temperature on activity (A) and stability (B) of the purified protease from A. pallidus C10.
The thermal stability of the protease enzyme purified from A. pallidus C10 was determined by performing an activity measurement at 20–80 °C temperature range at 15, 30, and 60 min. When the temperature stability profile was examined, it was determined that the enzyme had not lost more than 80% of its activity at all temperatures (Figure 4(B)). Most of the cold-active alkaline proteases fail to meet industrial requirements due to the fact that they are not stable at the temperatures above 20 °C and due to their low-product yields in large-scale productions49 , 50. On the other hand, most of the thermostable proteases lose their stability at low temperatures in spite of the fact that they show catalytic efficiency and stability at higher temperatures8 , 51. Therefore, the alkaline proteases stable at a quite broad temperature range, which are capable of remaining stable under hot, normal and cold conditions, have quite an important place in terms of industry. It is seen that A. pallidus C10 protease purified in this study is a powerful candidate for this sector as it is capable of preserving more than 80% of its activity at a broad temperature range, from 20 to 80 °C. Gupta and Khare52, in a study they conducted on P. aeruginosa PseA, stated that the activity that remained from the initial activity at the end of 30 minutes and at 45, 50, 55 °C proved to be 100%, 91%, and 80%. Despite this, the protease enzyme purified from A. pallidus C10 had preserved 92% of its initial activity at the end of 1 h at and at 40 °C, while it preserved 81% of its initial activity at the end of 1 h and at 50 °C. On the other hand, Charles et al.39 determined that the protease enzyme obtained from A. nidulans HA-10 had showed the lowest activity at 70 °C as the result of 1 h-incubation and that it had, later, become inactive at 80 °C. Heidari et al.53, with their study, reported that the enzyme they had purified had lost its activity when the temperature was above 60 °C. Divakar et al.48, however, reported that the enzyme they had isolated from A. veronii PG01 had preserved 53% of its activity at 70 °C at the end of 1 h. Jayakumar et al.37, in their study, reported that the enzymes had preserved their activities by 50% at 30–70 °C temperature range at the end of 30 minutes. On the other hand, it was determined that the protease enzyme purified from A. pallidus C10 had showed 105% activity at 70 °C and 96% activity at 80 °C at the end of one hour. The results obtained in this study suggest that the protease enzyme purified from A. pallidus C10 exhibited a high level of stability against temperature for quite a long time, and when compared with the other studies found in the literature, its tolerance to temperature is seen to be quite high.
Kinetic parameters of alkaline protease
By using different concentrations of casein, K M and V max values for the alkaline protease purified from A. pallidus C10 were determined as 0.197 mg/mL and 7.29 μmol.mL− 1.min− 1, respectively. When compared with the alkaline proteases reported in the literature, such as B. lehensis (K M of 0.35 mg.mL− 1)38, B. circulans M34 (KM of 0.96 mg.mL− 1)54, P. aeruginosa PseA (K M of 2.69 mg.mL− 1)52, B. circulans (KM of 0.597 mg.mL− 1)26, and haloalkalophilic Bacillus sp. (K M of 2 mg.mL− 1)55, A. pallidus C10 alkaline protease was determined to have a higher affinity for the casein substrate. When the V max/K M value is quite high, it means that this enzyme has a powerful catalytic activity to hydrolyze the substrate54. The fact that A. pallidus C10 protease has also a high affinity and catalytic efficiency is an important characteristic for various biotechnological applications.
Substrate specificity of alkaline protease
As the result of the activity measurements performed by using 0.65% (w/v) BSA, casein, azocasein, gelatin and hemoglobin for the purpose of determining the specificity of the alkaline protease enzyme purified from A. pallidus C10 to the natural substrates, the enzyme was determined to have shown the highest activity in the presence of casein (100%). The specificity of the enzyme in the face of the other substrates was determined as: gelatin (25%), azocasein (15%), BSA (5%) and hemoglobin (0%). Similarly, the alkaline proteases like B. circulans MTCC 794232, B. pumilus MCAS837, A. veronii PG0148, and B. laterosporus-AK141 were also reported to have shown the highest activity against casein.
The effect of inhibitors and metal ions on alkaline protease activity
The effect of different inhibitors on the protease enzyme obtained from A. pallidus C10 has been shown in Table 2. The fact that it is totally inhibited in the presence of 5 mM PMSF suggests that the purified enzyme belongs to serine protease family. In the presence of DTNB, a weak inhibition was determined. Separately, the increase in the enzyme activity in the presence of β-mercaptoethanol suggests that the enzyme is thiol dependent56. The fact that the protease enzyme shows high stability in the presence of EDTA is of great significance in terms of being used in the detergent industry, since the chelating agents like EDTA are among the most commonly found components in detergent formulations, which are used for the purpose of softening water and assisting in removing stains8 , 32. The stability of the purified C10 protease enzyme against EDTA indicates that it has a great advantage in terms of being the desired situation for detergent formulation and in terms of the use of the enzyme in the detergent industry due this characteristic.
Table 2.
The effect of inhibitors on A. pallidus C10 alkaline protease activity.
| Inhibitor | Concentration(mM) | Residual activity (%) | p Values |
|---|---|---|---|
| Control | 0 | 100 | – |
| PMSF | 1 | 69.54 ± 0.29 | <0.0001 a |
| 5 | 0 | – | |
| EDTA | 1 | 79.92 ± 0.08 | <0.0001 a |
| 5 | 100.4 ± 0.42 | 0.1277ns | |
| DTNB | 1 | 92.43 ± 0.26 | <0.0001 a |
| 5 | 77.21 ± 0.05 | <0.0001 a | |
| β-mercaptoethanol | 1 | 119 ± 0.57 | <0.0001 a |
| 5 | 605.7 ± 0.33 | <0.0001 a |
p > 0.05 (not significant, ns).
p < 0.0001.
When the effect of the metal ions on the protease enzyme purified from A. pallidus C10 was examined; it was seen that the enzyme had enhanced its activity in the presence of 1, 5, 10 mM of Ca2+ ve Mg2+ metal ions, whereas in the presence of Co2+ and Mn2+, almost more than 80% of the enzyme activity was seen to have remained. However, at 10 mM concentrations in which 1 and 5 mM Ag+ and K+ metal ions had activated the enzyme, the activity of the enzyme was determined to have decreased by 10%. 1 mM Fe2+ and Ni2+ ions of the enzyme were determined to have been activated by 5 and 10 mM Zn2+ (Table 3). Similarly, it was reported in the literature that the protease enzyme had been activated in the presence of Ca2+ and Mg2+ ions8 , 26 , 29 , 32 , 48.
Table 3.
The effect of metal ions on A. pallidus C10 alkaline protease activity.
| Metal ions | Concentration(mM) | Residual activity (%) | p Values |
|---|---|---|---|
| Control | 0 | 100 | – |
| Fe2+ | 1 | 109 ± 2.50 | 0.0032 b |
| 5 | 95.41 ± 5.68 | 0.2692ns | |
| 10 | 101.2 ± 1.50 | 0.1647ns | |
| Mg2+ | 1 | 120.8 ± 0.38 | <0.0001 d |
| 5 | 113.1 ± 1.83 | 0.0003 c | |
| 10 | 113.6 ± 0.52 | <0.0001 d | |
| Ca2+ | 1 | 115.3 ± 2.17 | 0.0003 c |
| 5 | 120.6 ± 0.24 | <0.0001 d | |
| 10 | 108.8 ± 0.69 | <0.0001 d | |
| Zn2+ | 1 | 83.11 ± 7.28 | 0.0172 a |
| 5 | 114.8 ± 1.45 | <0.0001 d | |
| 10 | 111.6 ± 2.67 | 0.0016 b | |
| Co2+ | 1 | 96.61 ± 0.97 | 0.0093 b |
| 5 | 87.45 ± 1.67 | 0.0003 c | |
| 10 | 83.08 ± 7.77 | 0.0212 a | |
| Ni2+ | 1 | 107.1 ± 0.77 | 0.0002 c |
| 5 | 94.30 ± 1.79 | 0.0079 b | |
| 10 | 91.54 ± 1.64 | 0.0013 b | |
| Mn2+ | 1 | 83.08 ± 7.77 | 0.0212 a |
| 5 | 84.92 ± 0.69 | <0.0001 d | |
| 10 | 80.58 ± 4.39 | 0.0017 b | |
| Ag+ | 1 | 108.2 ± 0.57 | <0.0001 d |
| 5 | 124.3 ± 2.70 | 0.0001 c | |
| 10 | 90.78 ± 8.61 | 0.1501ns | |
| K+ | 1 | 149.3 ± 5.23 | <0.0001 d |
| 5 | 129.9 ± 4.61 | 0.0004 c | |
| 10 | 91.60 ± 2.29 | 0.0041 b |
p > 0.05 (not significant, ns).
p < 0.05.
p < 0.01.
p < 0.001.
p < 0.0001.
The effect of some organic solvents on alkaline protease stability
The effects of various organic solvents at different concentrations (15%, 25% and 50%, v/v) on the purified A. pallidus C10 protease stability are summarized in Table 4. In the wake of 1 h-incubation; it was determined that the enzyme had preserved more than 85% of its activity at 15% concentration in ethanol, acetone, DMSO, butanol, chloroform and isopropanol solutions, whereas ethanol and methanol solutions with 25% concentration had enhanced the enzyme activity, and in other organic solutions; however, it did not lose its activity at the rate of almost 70–80%. Nonetheless, it was ascertained that the enzyme retained more than 80% of its activity at 50% concentration in the presence of DMSO and methanol, and at the rate of almost 50% in butanol, chloroform and isopropanol solutions. After 24 h; it was seen that the enzyme had lost its activity at the rate of 10–26% at 15% concentrations in the presence of methanol, ethanol, isopropanol and chloroform, and again, it lost its activity at the rate of 4–50% at 25% concentrations. Enzyme activity was found to be induced by acetone (15%), DMSO (25%), and butanol (25%) by 103%, 102% and 106%, respectively. At 50% concentrations; however, the enzyme was determined to have failed to preserve its stability in the organic solvents mentioned. The organic solvent stability of A. pallidus C10 protease is higher than the other alkaline proteases mentioned, which are Bacillus sp. NPST-AK151, Saccharopolyspora sp. A957, and B. pumilus MCAS837. In the presence of organic solvents, the ability of the natural proteases to remain stable without performing any modification for the stabilization of the enzyme is of great importance for various applications58. The use of proteases for peptide and ester synthesis non-aqueous conditions in the organic media has gained a great deal of importance within the last decade59. Since it exhibits good stability in the presence of organic solvents, the protease enzyme obtained from A. pallidus C10 can be said to have a great advantage in terms of its applicability in these industrial applications.
Table 4.
The effect of organic solvents on A. pallidus C10 alkaline protease activity.
| Organic solvents | Concentration (%) | Residual activity (%) (1h) | p Values | Residual activity (%) (24h) | p Values |
|---|---|---|---|---|---|
| Control | 0 | 100 | – | 100 | – |
| Methanol | 15 | 68.42 ± 1.83 | <0.0001 d | 83.38 ± 1.77 | 0.0001 c |
| 25 | 114 ± 2.79 | 0.0009 c | 50.27 ± 1.39 | <0.0001 d | |
| 50 | 80.24 ± 1.74 | <0.0001 d | 0 | – | |
| Ethanol | 15 | 93.59 ± 1.12 | 0.0011 b | 90.28 ± 1.98 | 0.0014 b |
| 25 | 107.8 ± 1.52 | 0.0009 c | 75.47 ± 2.95 | 0.0002 c | |
| 50 | 103.7 ± 2.07 | 0.0303 a | 0 | – | |
| Acetone | 15 | 86.73 ± 1.29 | <0.0001 d | 102.9 ± 1.92 | 0.0472 a |
| 25 | 88.50 ± 2.02 | 0.0008 c | 72.67 ± 1.45 | <0.0001 d | |
| 50 | 72.74 ± 1.89 | <0.0001 d | 0 | – | |
| DMSO | 15 | 84.73 ± 0.93 | <0.0001 d | 85.86 ± 3.65 | 0.0030 b |
| 25 | 78.10 ± 1.75 | <0.0001 d | 102.2 ± 1.53 | 0.0522ns | |
| 50 | 95.33 ± 2.19 | 0.0299 a | 0 | – | |
| Butanol | 15 | 93.74 ± 2.09 | 0.0091 b | 58.88 ± 2.20 | <0.0001 d |
| 25 | 73.66 ± 1.85 | <0.0001 d | 106.5 ± 3.21 | 0.0217 a | |
| 50 | 52.01 ± 2.44 | <0.0001 d | 0 | – | |
| Chloroform | 15 | 95.94 ± 2.58 | 0.0724ns | 74.56 ± 0.13 | <0.0001 d |
| 25 | 85.02 ± 1.73 | 0.0002 c | 96.41 ± 0.87 | 0.0056 b | |
| 50 | 59.59 ± 1.44 | <0.0001 d | 0 | – | |
| Isopropanol | 15 | 90.21 ± 1.91 | 0.0012 b | 90.13 ± 1.73 | 0.0008 c |
| 25 | 84.46 ± 1.00 | <0.0001 d | 62.91 ± 2.42 | <0.0001 d | |
| 50 | 50.50 ± 1.038 | <0.0001 d | 0 | – |
p > 0.05 (not significant, ns).
p < 0.05.
p < 0.01.
p < 0.001.
p < 0.0001.
The effects of surfactants, oxidizing agent and commercial detergents on alkaline protease stability
It was determined that the protease enzyme purified from A. pallidus C10 could not preserve its stability in the presence of all the compounds studied on apart from SDS. While the enzyme showed weak stability in the presence of the oxidizing agent, H2O2, it was determined to have preserved its activity by 79% at 1% SDS concentration, and it enhanced its activity by 16% in the presence of 5% SDS (Table 5). While the number of studies in the literature in which stable enzymes are found in the presence of SDS are few37 , 38 , 46, the fact that the protease enzyme purified in this study has high stability in the presence of SDS is important.
Table 5.
The effect of various surfactants and the oxidant agent on A. pallidus C10 alkaline protease activity.
| Surfactants/oxidizingagent | Concentration(%) (v/v) | Residual activity (%) | p Values |
|---|---|---|---|
| Control | 0 | 100 | – |
| SDS | 1 (w/v) | 79.33 ± 2.65 | 0.0002 a |
| SDS | 5 (w/v) | 116.7 ± 1.76 | <0.0001 b |
| Triton X-100 | 1 | 41.33 ± 1.85 | <0.0001 b |
| Triton X-100 | 5 | 63.0 ± 1.52 | <0.0001 b |
| Tween-20 | 1 | 20.0 ± 1.15 | <0.0001 b |
| Tween-20 | 5 | 37.67 ± 1.45 | <0.0001 b |
| Tween-80 | 1 | 31.0 ± 2.08 | <0.0001 b |
| Tween-80 | 5 | 29.0 ± 2.08 | <0.0001 b |
| H2O2 | 1 | 9.66 ± 0.88 | <0.0001 b |
| H2O2 | 5 | 11.67 ± 0.88 | <0.0001 b |
p < 0.001.
p < 0.0001.
As seen in Figure 5, it was ascertained that the enzyme had not lost its activity by 80.25% and 85.28%, respectively, in the presence of Ariel and Persil; whereas, in the presence of other detergents, it was seen to have preserved more than 55% of its initial activity. Rai and Mukherjee27 reported that B. subtilis DM-04 protease enzyme had showed 60% activity in the presence of Ariel at 37 °C during 1 h incubation. Joshi and Satyanarayana38 also determined that B. lehensis protease enzyme had preserved its activity by 80% in 0.5% Ariel solution and in 1% Ariel solution.
Figure 5.
Stability of the alkaline protease from A. pallidus C10 in the presence of various commercial detergents.
Conclusion
Alkaline proteases are among the most important hydrolytic enzymes, which have commercial value and which have several fields of practice in various industrial sectors. In this study, the alkaline protease enzyme produced by a new strain called A. pallidus C10 was purified and characterized by using two different chromatographic methods. Although there are studies found in the literature with respect to A. pallidus species and the identification of the isolates belonging to this species, our study is the first one in which the alkaline protease enzyme was purified and characterized. It was determined that the enzyme had become highly active and stable under alkaline conditions (pH 7.0–10.0) and that it had preserved its activity to a large extent at a broad temperature range (20–80 °C). It was ascertained to be more stable particularly at high temperatures like 60 °C, 70 °C and 80 °C when compared with similar studies conducted before. Thus, it is seen that the purified protease enzyme, with this aspect, can be the candidate for reason of preference for the commercial applications that require long-term stability at high temperatures. Separately, the enzyme was observed to have enhanced or preserved its activity in the presence of various metal ions, EDTA, SDS and organic solvents. When all these data are considered together, they indicate to the applicability potential of the thermostable A. pallidus C10 alkaline protease as a great detergent additive in various industrial fields and biotechnological applications, notably the detergent industry.
Disclosure statement
The authors declared that there is no conflict of interests.
Funding
This work was funded by grants from the Scientific Research Project of Ataturk University of Turkey [project number: 2013/297].
References
- 1. Ibrahim AS, Al-Salamah AA, El-Badawi YB, et al. . Detergent-, solvent- and salt-compatible thermoactive alkaline serine protease from halotolerant alkaliphilic Bacillus sp. NPST-AK15: purification and characterization. Extremophiles 2015;19:961–71. [DOI] [PubMed] [Google Scholar]
- 2. Asokan S, Jayanthi C.. Alkaline protease production by Bacillus licheniformis and Bacillus coagulans . J Cell Tissue Res 2010;10:2119–23. [Google Scholar]
- 3. Mahajan RT, Badgujar SM.. Biological aspects of proteolytic enzymes: a review. J Pharm Res 2010;3:2048–68. [Google Scholar]
- 4. Rao MB, Tanksale AM, Ghatge MS, Deshpande VV.. Molecular and biotechnological aspects of microbial proteases. Microbiol Mol Biol Rev 1998; 62:597–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Genckal H, Tari C.. Alkaline protease production from alkalophilic Bacillus sp. isolated from natural habitats. Enzyme Microb Technol 2006;39:703–10. [Google Scholar]
- 6. Lazim H, Houda M, Nedra S, et al. . Production and optimization of thermophilic alkaline protease in solid-state fermentation by Streptomyces sp. CN902. J Ind Microbiol Biotechnol 2009;36:531–7. [DOI] [PubMed] [Google Scholar]
- 7. Ravindran B, Ganesh KA, Aruna BPS, Sekaran G.. Solid-state fermentation for the production of alkaline protease by Bacillus cereus 1173900 using proteinaceous tannery solid waste. Curr Sci 2011;100:726–30. [Google Scholar]
- 8. Haddar A, Bougatef A, Agrebi R, et al. . A novel surfactant-stable alkaline serine-protease from a newly isolated Bacillus mojavensis A21. Purification and characterization. Process Biochem 2009;44:29–35. [Google Scholar]
- 9. Xin Y, Sun Z, Chen Q, et al. . Purification and characterization of a novel extracellular thermostable alkaline protease from Streptomyces sp. M30. J Microbiol Biotechnol 2015;25:1944–53. [DOI] [PubMed] [Google Scholar]
- 10. Chatterjee S. Production and estimation of alkaline protease by immobilized Bacillus licheniformis isolated from poultry farm soil of 24 Parganas and its reusability. J Adv Pharm Technol Res 2015;6:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahto RB, Bose KJ.. Production of alkaline protease from Bacillus subtilis by different entrapment techniques. J Biochem Tech 2012;4:498–501. [Google Scholar]
- 12. Maurer KH. Detergent proteases. Curr Opin Biotechnol 2004;15:330–4. [DOI] [PubMed] [Google Scholar]
- 13. Sanatan PT, Lomate PR, Giri AP, Hivrale VK.. Characterization of a chemostable serine alkaline protease from Periplaneta americana. BMC Biochem 2013;14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taslimi P, Nadaroglu H, Adıguzel A, et al. . Removal of some textile dyes from agueous solution by using catalase-peroxidase from Aeribacillus pallidus (p26). J Pure Appl Microbiol 2013;7:2629–40. [Google Scholar]
- 15. Muhammad SA, Ahmed S.. Production and characterization of a new antibacterial peptide obtained from Aeribacillus pallidus SAT4. Biotechnol Rep 2015;8:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yasawong M, Areekit S, Pakpitchareon A, et al. . Characterization of thermophilic halotolerant Aeribacillus pallidus TD1 from Tao Dam Hot Spring, Thailand. Int J Mol Sci 2011;12:5294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Radchenkova N, Spasen V, Ivan P, et al. . Production and properties of two novel exopolysaccharides synthesized by a thermophilic bacterium Aeribacillus pallidus 418. Appl Biochem Biotechnol 2013;171:31–43. [DOI] [PubMed] [Google Scholar]
- 18. Cihan AC, Ozcan B, Tekin N, Cokmus C.. Phylogenetic diversity of isolates belonging to genera Geobacillus and Aeribacillus isolated from different geothermal regions of Turkey. World J Microbiol Biotechnol 2011;27:2683–96. [Google Scholar]
- 19. Mnif S, Sayadi S, Chamkha M.. Biodegradative potential and characterization of a novelaromatic-degrading bacterium isolated from a geothermal oil field under saline and thermophilic conditions. Int Biodeter Biodegrad 2013;86:258–64. [Google Scholar]
- 20. Yanmış D, Adıgüzel A.. Molecular typing of thermophilic Bacili isolated from different hot springs of Turkey. Res J Biotechnol 2014;9:10. [Google Scholar]
- 21. Baltaci MO, Genc B, Arslan S, et al. . Isolation and characterization of the thermophilic bacteria from geothermal areas in Turkey and preliminary research on biotechnological enzyme potentials. Geomicrobiol J 2016. [Epub ahead of print]. doi:10.1080/01490 451.2015.1137662. [Google Scholar]
- 22. Takami H, Akiba T, Horikosh K.. Production of extremely thermo stable alkaline protease from Bacillus sp. AH-101. Appl Microbiol Biotechnol 1989;30:120–4. [DOI] [PubMed] [Google Scholar]
- 23. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–51. [DOI] [PubMed] [Google Scholar]
- 24. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–5. [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Carreno FL, Dimes LF, Haard NF. Substrate-gel electrophoresis for composition and molecular weight of proteinases or proteinaceous proteinase inhibitors. Anal Biochem 1993;214:65–9. [DOI] [PubMed] [Google Scholar]
- 26. Rao S, Sathish T, Ravichandra P, Prakasham R.. Characterization of thermo- and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem 2009;44:262–8. [Google Scholar]
- 27. Rai S, Mukherjee AK.. Ecological significance and some biotechnological application of an organic solvent stable alkaline serine protease from Bacillus subtilis strain DM-04. Bioresour Technol 2009;100:2642–5. [DOI] [PubMed] [Google Scholar]
- 28. Fakhfakh N, Kanoun S, Manni L, Nasri M.. Production and biochemical and moleculer characterization of a keratinolytic serine protease from chicken feather-degrading Bacillus licheniformis RPk. Can J Microbiol 2009;55:427–36. [DOI] [PubMed] [Google Scholar]
- 29. Annamalai N, Rajeswari MV, Thavasi R, et al. . Optimization, purification and characterization of novel thermostable, haloalkaline, solvent stable protease from B. halodurans CAS6 using marine shellfish wastes: a potential additive for detergent and antioxidant synthesis. Bioprocess Biosyst Eng 2013;36:873–83. [DOI] [PubMed] [Google Scholar]
- 30. Nam MS, Whang KS, Choi SH, et al. . Purification, characterization, and propertiesof an alkaline protease produced by Serratia marcescens S3-R1 inhabiting Korean ginseng rhizosphere. J Sci Food Agric 2013;93:3876–82. [DOI] [PubMed] [Google Scholar]
- 31. Miyaji T, Otta Y, Nakagawa T, et al. . Purification and molecular characterization of subtilisin-like alkaline protease BPP-A from Bacillus pumilus strain MS-1. Lett Appl Microbiol 2006;42:242–7. [DOI] [PubMed] [Google Scholar]
- 32. Patil U, Mokashe N, Chaudhari A.. Detergent-compatible, organic solvent-tolerant alkaline protease from Bacillus circulans MTCC 7942: purification and characterization. Prep Biochem Biotechnol 2016;46:56–64. [DOI] [PubMed] [Google Scholar]
- 33. Jónsdóttir G, Bjarnason JB, Gudmundsdóttir A.. Recombinant cold-adapted trypsin I from Atlantic cod-expression, purification, and identification. Protein Expr Purif 2004;33:110–22. [DOI] [PubMed] [Google Scholar]
- 34. Polgar L. The catalytic triad of serine peptidases. Cell Mol Life Sci 2005;62:2161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jaouadi B, Ellouz-Chaabouni S, Rhimi M, Bejar S.. Biochemical and molecular characterization of a detergent-stable serine alkaline protease from Bacillus pumilus CBS with high catalytic efficiency. Biochimie 2008;90:1291–305. [DOI] [PubMed] [Google Scholar]
- 36. Kandasamy N, Punitha V, Amsamani S, et al. . Eco-benign enzymatic dehairing of goatskins utilizing a protease froma Pseudomonas fluorescens species isolated from fish visceral waste. J Clean Prod 2012;25:27–33. [Google Scholar]
- 37. Jayakumar R, Shanmugam J, Balumuri A, Sundaram S.. Characterization of thermostable serine alkaline protease from an alkaliphilic strain Bacillus pumilus MCAS8 and Its Applications. Appl Biochem Biotechnol 2012;168:1849–66. [DOI] [PubMed] [Google Scholar]
- 38. Joshi S, Satyanarayana T.. Characteristics and applications of a recombinant alkaline serine protease from a novel bacterium Bacillus lehensis . Bioresour Technol 2013;131:76–85. [DOI] [PubMed] [Google Scholar]
- 39. Charles P, Devanathan V, Periasamy A, et al. . Purification, characterization and crystallization of an extracellular alkaline protease from Aspergillus nidulans HA-10. J Basic Microbiol 2008;48:347–52. [DOI] [PubMed] [Google Scholar]
- 40. Shivanad P, Jayaraman G.. Production of extracellular protease from halotolerant bacterium, Bacillus aquimaris strain VITP4 isolated from Kumta coast. Process Biochem 2009;44:1088–94. [Google Scholar]
- 41. Arulmani M, Aparanjini K, Vasanthi K, et al. . Purification and partial characterization of serine protease from thermostable alkalophilic Bacillus laterosporus-AK1. World J Microbiol Biotechnol 2007;23:475–81. [Google Scholar]
- 42. Guleria S, Walia A, Chauhan A, Shirkot CK.. Purification and characterization of detergent stable alkaline protease from Bacillus amyloliquefaciens SP1 isolated from apple rhizosphere. J Basic Microbiol 2016;56:138–52. [DOI] [PubMed] [Google Scholar]
- 43. Akel H, Al-Quadan F, Yousef TK.. Characterization of a purified thermostable protease from hyperthermophilic Bacillus strain HUTBS71. Eur J Sci Res 2009;31:280–8. [Google Scholar]
- 44. Sarker PK, Talukdar SA, Deb P, et al. . Optimization and partial characterization of culture conditions for the production of alkaline protease from Bacillus licheniformis P003. Springerplus 2013;2:506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sana B, Ghosh D, Saha M, Mukherjee J.. Purification and characterization of a salt, solvent, detergent and bleach tolerant protease from a new gamma-Proteobacterium isolated from the marine environment of Sunderbans. Process Biochem 2006;41:208–15. [Google Scholar]
- 46. Jain D, Pancha I, Mishra SK, et al. . Purification and characterization of haloalkaline thermoactive, solvent stable and SDS-induced protease from Bacillus sp.: a potential additive for laundry detergents. Bioresour Technol 2012;115:228–36. [DOI] [PubMed] [Google Scholar]
- 47. Bezawada J, Yan S, John RP, et al. . Recovery of Bacillus licheniformis alkaline protease from supernatant of fermented wastewater sludge using ultrafiltration and its characterization. Biotechnol Res Int 2011;2011:238549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Divakar K, Priya JDA, Gautam P.. Purification and characterization of thermostable organic solvent-stable protease from Aeromonas veronii PG01. J Mol Catal B: Enzym 2010;66:311–18. [Google Scholar]
- 49. Gerday C, Aittaleb M, Bentahir M, et al. . Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol 2000;18:103–7. [DOI] [PubMed] [Google Scholar]
- 50. Saek K, Ozaki K, Kobayashi T, Ito S.. Detergent alkaline proteases: enzymatic properties, genes, and crystal structures. J Biosci Bioeng 2007;103:501–8. [DOI] [PubMed] [Google Scholar]
- 51. Deng A, Wu J, Zhang Y, et al. . Purification and characterization of a surfactant-stable high-alkaline protease from Bacillus sp. B001. Bioresour Technol 2010;101:7111–17. [DOI] [PubMed] [Google Scholar]
- 52. Gupta A, Roy I, Khare SK, Gupta MN. Purification and characterization of a solvent stable protease from Pseudomonas aeruginosa PseA. A J Chromatogr A 2005;1069:155–61. [DOI] [PubMed] [Google Scholar]
- 53. Heidari HRK, Amoozegar MA, Hajighasemi M, et al. . Production, optimization and purification of a novel extracellular protease from the moderately halophilic bacterium Halobacillus karajensis . J Ind Microbiol Biotechnol 2008;36:21–7. [DOI] [PubMed] [Google Scholar]
- 54. Sari E, Loğoğlu E, Öktemer A.. Purification and characterization of organic solvent stable serine alkaline protease from newly isolated Bacillus circulans M34. Biomed Chromatogr 2015;29:1356–63. [DOI] [PubMed] [Google Scholar]
- 55. Gupta A, Roy I, Patel RK, et al. . Onestep purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp. J Chromatogr A 2005;20:103–8. [DOI] [PubMed] [Google Scholar]
- 56. Beg QK, Gupta R.. Purification and characterization of an oxidant stable, thiol dependent serine alkaline protease from Bacillus mojavensis . Enzyme Microb Technol 2003;32:294–304. [Google Scholar]
- 57. Raut G, Sachin V, Ren B, et al. . Chandrakant K, Purification and characterization of organic solvent and detergent stable proteaseisolated from marine Saccharopolyspora sp. A9: application of protease forwound healing. Biochem Eng J 2012;69:196–203. [Google Scholar]
- 58. Doukyu N, Ogino H.. Organic solvent tolerant enzymes. Biochem Eng J 2010;48:270–82. [Google Scholar]
- 59. Jellouli K, Ghorbel-Bellaaj O, Ayed H, et al. . Alkaline-protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergents additive and for shrimp waste deproteinization. Process Biochem 2011;46:1248–56. [Google Scholar]