Abstract
Purpose
The detection of enzyme activities and evaluation of enzyme inhibitors have been challenging with magnetic resonance imaging (MRI). To address this need, we have developed a diamagnetic, nonmetallic contrast agent and a protocol known as catalyCEST MRI that uses chemical exchange saturation transfer (CEST) to detect enzyme activity as well as enzyme inhibition.
Procedures
We synthesized a diamagnetic MRI contrast agent that has enzyme responsive and enzyme unresponsive CEST signals. We tested the ability of this agent to detect the activity of kallikrein 6 (KLK6) in biochemical solutions, in vitro and in vivo, with and without a KLK6 inhibitor.
Results
The agent detected KLK6 activity in solution and also detected KLK6 inhibition by antithrombin III. KLK6 activity was detected during in vitro studies with HCT116 colon cancer cells, relative to the detection of almost no activity in a KLK6-knockdown HCT116 cell line and HCT116 cells treated with antithrombin III inhibitor. Finally, strong enzyme activity was detected within an in vivo HCT116 tumor model, while lower enzyme activity was detected in a KLK6 knockdown tumor model and in the HCT116 tumor model treated with antithrombin III inhibitor. In all cases, comparisons of the enzyme responsive and enzyme unresponsive CEST signals were critical for the detection of enzyme activity.
Conclusions
This study has established that catalyCEST MRI with an exogenous diaCEST agent can evaluate enzyme activity and inhibition in solution, in vitro and in vivo.
Keywords: CEST MRI, Enzyme activity, Enzyme inhibition, Colon cancer, Molecular imaging
Introduction
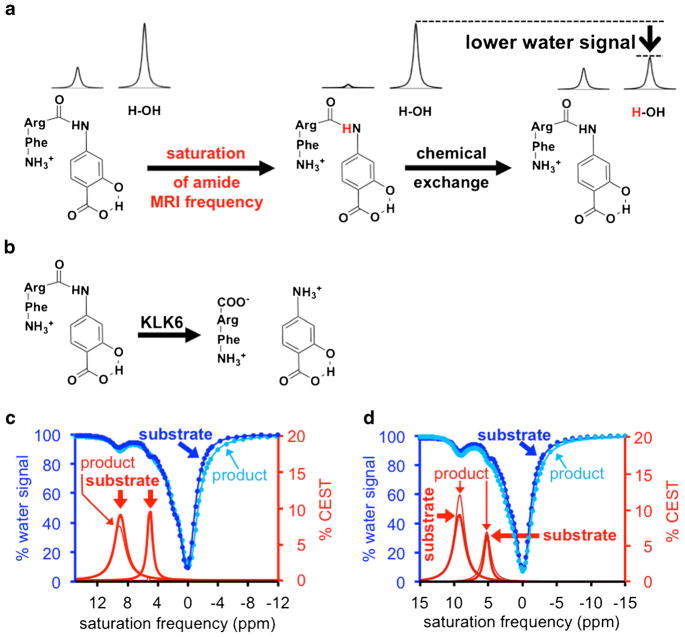
Enzymes are important disease biomarkers and can assess early response to treatment [1]. Many assays can detect enzyme expression during in vitro cell studies and ex vivo tissue studies. However, enzyme activity may more accurately indicate the status of a pathology or drug effect [2]. To address this problem, we have previously developed chemical exchange saturation transfer (CEST) MRI to detect enzyme activity [3]. CEST MRI involves selective saturation of the MR signal of a proton on an agent (Fig. 1a, left) followed by chemical exchange of the agent’s proton to a water molecule (Fig. 1a, right) which then transfers the saturation to the MR signal of water that can be detected using standard MRI acquisition methods [4]. Enzyme catalysis can change the chemical moieties on the agent, which changes the chemical exchange rate of the agent and changes the CEST MRI contrast (Fig. 1b) [5]. Therefore, monitoring the CEST MRI contrast can detect enzyme catalysis using a protocol known as catalyCEST MRI.
Fig. 1.
CatalyCEST MRI of KLK6 activity in solution. a The MR frequency of the amide proton of the agent, FRSA, is saturated (left), and the agent’s proton then exchanges with a proton of a water molecule (right), which transfers the saturation to the water and causes a loss of water MR signal. b Cleavage of the CEST agent by KLK6 converts the aryl amide to an aryl amine, which has a chemical exchange rate that is too fast to generate a CEST signal. c The substrate produced two CEST signals at 5.3 and 9.5 ppm, while the product after KLK6 catalysis produced only one CEST signal at 9.5 ppm. d Inhibition of the KLK6 enzyme activity prevented the disappearance of the CEST signal at 5.3 ppm.
CEST MRI contrast agents that detect the activity of proteases and other enzymes have been developed [6, 7]. However, other conditions can affect CEST contrast, including the concentration of the agent. To address this potential pitfall, we have developed a single CEST agent with both enzyme responsive and unresponsive signals. The ratiometric comparison of these signals can detect enzyme activity in a way that is concentration-independent [8]. We have shown that a single CEST agent can detect enzyme activity in vivo within a mouse tumor model [9].
The practical value of catalyCEST MRI remains unexplored. Therefore, we proposed to employ catalyCEST MRI to detect the activity of kallikrein 6 (KLK6), an important biomarker of colon cancer [10]. KLK6 is a serine protease that is activated in the extracellular tumor microenvironment, and degrades fibro-nectin, laminin, vitronectin, and collagen to promote tumor invasion and metastasis [11]. The detection of KLK6 activity may improve tumor staging in patients with colon cancer. Furthermore, we proposed to use catalyCEST MRI to detect the decrease in KLK6 activity caused by an anti-KLK6 inhibitor, antithrombin III, to investigate whether catalyCEST MRI can provide value for drug studies [12]. Finally, we tested the effect of antithrombin III in biochemical solution, in vitro and in vivo, to evaluate whether a single imaging technology can produce results across each of these three experimental conditions, which may also provide value for drug studies.
Materials and Methods
CatalyCEST MRI Studies in Biochemical Solution
Details regarding chemical syntheses can be found in the Electronic Supplementary Material (ESM; Fig. S1). To study enzyme activity in solution, 2.5 units of KLK6 enzyme (96.15 pmol; 480.7 nM) and 2.5 units of lysyl endopeptidase were added to a 200 μl, 25 mM solution of the contrast agent. The mixture of KLK6 and lysyl endopeptidase was prepared at room temperature 30 min prior to adding the mixture to the solution with the contrast agent so that the lysyl endopeptidase could activate KLK6.
To study enzyme inhibition, 25 μg (436.6 pmol) of antithrombin III and 2.5 units of activated KLK6 enzyme were simultaneously added to 200 μl of agent at 35 mM concentration and pH 7.2. This concentration of inhibitor was 4-fold higher than the concentration of the enzyme, which was less than a 10-fold concentration used in other studies [13].
Samples in a 200-μl conical tube were placed in a customized sample holder at 37.0 ± 0.2 °C. MRI studies were performed with a Biospec MRI scanner operating at 7 T (300 MHz) magnetic field strength with a 72-mm volume transceiver coil (Bruker Biospin, Inc., Billerica, MA). Images were processed and analyzed using ParaVision v5.1 (Bruker Biospin, Inc.) and Matlab v8.4 (Mathworks, Natick, MA). A CEST-FISP MRI acquisition protocol was used with TR = 3.196 ms; TE = 1.598 ms; excitation flip angle = 30°; matrix = 128 × 128; field of view = 8 × 8 cm; in-plane spatial resolution = 625 × 625 μm; 1-mm slice thickness; 1 slice; and 1 average [14]. A 5-s continuous wave saturation pulse was applied with a saturation power of 4 μT. We acquired a series of 85 images with selective saturation from −15 to 15 ppm, in 0.25-ppm increments from 15 to −3 ppm and 1-ppm increments from −4 to −15 ppm, for a total scan time of 7:42 min.
The average signal of the sample and the signal from each pixel were each used to construct a CEST spectrum. Each spectrum was fit with a sum of five Lorentzian line shapes [5]. The center, width, and amplitude of each Lorentzian line were allowed to change to optimize the fit. This method compensated for potential B0 inhomogeneities in the CEST MR images, so that additional MRI studies were not needed for measuring B0 inhomogeneity [15]. The fittings determined the CEST signal amplitudes at 5.3 and 9.5 ppm, which were used to determine the reaction coordinate (Eq. [1]).
| (1) |
CatalyCEST MRI Studies In Vitro
Media were obtained from HCT116 and shKLK6 cell cultures. The HCT116 colon cancer cells had high KLK6 expression (Fig. S2) [10]. HCT116 colon cancer cells with knockdown of KLK6 expression (shKLK6 cells) were generated by stable transfection of SureSilencing shRNA plasmids (SureSilencing shRNAs, QIAGEN, Inc.), targeting the KLK6 gene using short-hairpin RNA (Fig. S2) [16]. Cell growth conditions are listed in the ESM. A volume of 150 μl of the media was added to 200 μl of 25 mM agent at pH 7.3. For in vitro studies of the inhibitor, 25 μg of antithrombin III was added to this mixture, and the concentration of the agent was increased to 50 mM to ensure that the detection was sensitive to the lower KLK6 activity expected from enzyme inhibition. A CEST spectrum was obtained 8 h after adding the media, or media and inhibitor. The reaction coordinate was determined using Eq. [1].
CatalyCEST MRI Studies In Vivo
All in vivo studies were performed with procedures approved by the Institutional Animal Care and Use Committee of the University of Arizona. Studies were performed using four mice with a flank HCT116 tumor and four mice with a flank shKLK6 tumor. Two additional mice bearing a flank HCT116 tumor were injected intravenously with 125 μg of antithrombin III, an anti-KLK6 inhibitor, 2 h before performing the catalyCEST MRI study. Details regarding the tumor models can be found in the ESM.
To identify the tumor location, images were acquired using a multislice spin echo MRI protocol. The mouse was removed from the magnet, a solution of 200 mM of the contrast agent in 200 μl of Tris buffer was subcutaneously injected into the mouse within 5 mm of the tumor, and the mouse was reinserted into the magnet. A lapse of 10 min between injection of the agent and data acquisition was allowed for the agent to infuse into the tumor. Then, CEST MR images were acquired with a saturation power of 4 μT and saturation time of 3 s [14]. Five 600-ms continuous wave saturation pulses were used at each saturation frequency. For each scan, a series of 51 saturation frequencies were acquired at ppm values of +16, +15 to 3 (0.4-ppm increments), +3.0 to −3.0 (0.5-ppm increments), and −5 to −15 (2-ppm increments). Ten catalyCEST MRI scans were acquired for 35 min. The mouse was then removed from the magnet and cradle and allowed to recover.
For each saturation frequency, the images from the 10 catalyCEST MRI scans were averaged, and a Gaussian spatial filter was applied with a 3 × 3 pixel matrix and a σ value of 1 pixel [15]. The CEST spectrum from each pixel in each region of interest was obtained from the MR images. A function of seven Lorentzian line shapes was fit to each CEST spectrum to measure the signal amplitudes of the CEST effects at 9.5 and 5.3 ppm, two CEST effects from the peptidyl ligand, direct saturation of water, the endogenous effect of amide proton transfer from mobile proteins, and the magnetization transfer effect from less-mobile proteins [5]. The saturation frequency, width, and amplitude of the Lorentzian line shape that fit the direct saturation of water was allowed to change during the fitting procedure. The width and amplitude of the four Lorentzian line shapes that fit the CEST effects of the agent were also allowed to change, but the saturation frequencies were fixed at 9.5, 5.3, 3.0, and 1.8 ppm relative to the saturation frequency of the direct saturation of water. Parametric maps of the CEST signal amplitudes at 9.5 and 5.3 ppm were constructed to evaluate the CEST signals in the tumor and tissues surrounding the tumor. Pixels were retained that had a % CEST signal amplitude above a 95 % probability threshold of being real (details are included in the ESM) [17]. These pixel-wise CEST signals were used to construct parametric maps of the reaction coordinate (Eq[2]). The pixel-wise CEST spectra that had agent detected with significantly sufficient CEST signal were averaged to create a single CEST spectrum that was used to represent the tumor, which was fit with Lorentzian line shapes.
| (2) |
Eq. [2] is identical to Eq. [1] if [% CEST at 5.3 ppm]before = [% CEST at 9.5 ppm]before.
Results
Design and Synthesis of the CEST Agent
Our CEST agent, phenylalanine-arginine-4-aminosalicylic acid (FRSA), was synthesized in five steps with a 20 % yield by combining salicylic acid with a substrate for KLK6 (Fig. S1) [10, 18, 19]. Details of the chemical synthesis can be found in the ESM.
Detection of KLK6 Activity and Inhibition in Solution
FRSA generated CEST signals at MR frequencies of 5.3 ppm from the aryl amide and 9.5 ppm from the salicylic acid, which matched the MR frequencies of similar CEST agents [8, 9, 18, 20]. The agent also generated CEST signals at 1.8 and 3.0 ppm from the peptidyl ligand, which were similar to results from previous studies [8]. These CEST signals from the peptidyl ligand were not used for subsequent analyses due to their proximity to the MR frequency of water (defined as 0 ppm).
To evaluate the detection of KLK6 enzyme activity with catalyCEST MRI, KLK6 activated with lysyl endopeptidase was added to the contrast agent. After incubation for 8 h, the same CEST-FISP MRI protocol was used to obtain CEST spectra after KLK6 enzyme catalysis (Fig. 1c). A shorter incubation time may have been sufficient for this study, yet we incubated for a long time to promote a high level of conversion of a substrate to product. The 7.5 % CEST signal at 5.3 ppm was reduced to a nearly negligible 0.6 % after enzyme reaction, indicating that aryl amide bond was cleaved by the KLK6 enzyme. The CEST signal at 9.5 ppm from the salicylic acid had a minor change from 8.8 to 8.3 % after the enzyme was added, showing that this moiety was largely unresponsive to KLK6 activity and behaved as an internal control. These CEST signal amplitudes were used to determine that the reaction coordinate was 0.92, establishing that catalyCEST MRI can detect KLK6 activity.
We tested the ability of catalyCEST MRI to evaluate the effects of the KLK6 inhibitor, Antithrombin III. CEST spectra was acquired before and after adding the inhibitor using the same CEST-FISP MRI protocol (Fig. 1d). The 6.4 % CEST signal at 5.3 ppm only reduced to 6.2 % after treatment with the inhibited enzyme, while the CEST signal at 9.5 ppm changed from 9.8 % to 10.6 % after the enzyme was added. The reaction coordinate was 0.10, demonstrating that catalyCEST MRI could successfully detect KLK6 inhibition by Antithrombin III.
Detection of KLK6 Activity and Inhibition In Vitro
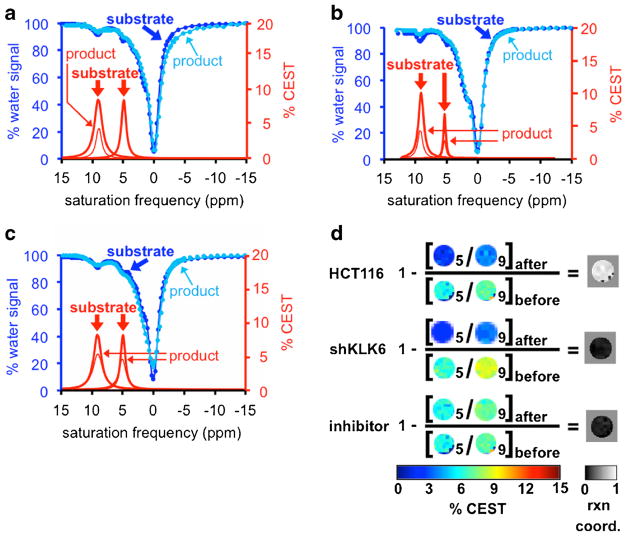
We studied the detection of KLK6 activity in vitro using the HCT116 colon cancer cell line that had high KLK6 expression (Fig. S2) [10]. The CEST signal at 9.5 ppm showed a decrease from 7.8 to 4.0 %. Although sample dilution would be expected to cause the CEST signal to decrease to 4.45 %, the difference between 4.45 and 4.0 % lies within the 1.16 % CEST signal variability that is described below. Therefore, this decrease in CEST signal was largely attributed to the dilution of the agent from 200 to 350 μl (Fig. 2a). The 7.7 % CEST signal at 5.3 ppm before adding media had vanished after adding media, indicating high enzyme activity in this media. The ratio of the two CEST peaks before and after adding media was used to determine a reaction coordinate of 1.0 (Eq. [1]). This study demonstrated that catalyCEST MRI can detect enzyme activity in vitro while accommodating the dilution of the agent.
Fig. 2.
CatalyCEST MRI of KLK6 activity in vitro. a Incubation of the substrate in HCT116 cell media with high KLK6 expression caused the CEST signal at 5.3 ppm to disappear. The decrease in CEST signal at 9.5 ppm was attributed to sample dilution. b Incubation of the substrate in shKLK6 cell media with low KLK6 expression, and c Incubation in HCT116 cell media pre-treated with inhibitor, each did not cause the CEST signal at 5.3 ppm to disappear while the decrease in both CEST signals was attributed to sample dilution. d Parametric maps of the CEST signal amplitudes were used to visualize the KLK6 enzyme reaction coordinate. Images labeled as “5” represent the CEST signal amplitude at 5.3 ppm, and images labeled as “9” represent the CEST signal amplitude at 9.5 ppm.
An identical in vitro study was performed with the shKLK6 cell line with knockdown of KLK6 expression (Fig. 2b) [21]. The CEST signal at 9.5 ppm decreased from 9.2 to 3.9 % after adding media, which was largely caused by the dilution of the sample within experimental variability. The CEST signal at 5.3 ppm decreased from 6.2 to 2.5 % after adding media. Most importantly, the ratio of the two CEST signals was 67.5 % before adding media and 64.5 % after adding media, resulting to a reaction coordinate of 0.04 (Eq. [1]) that indicated a lack of KLK6 activity in the shKLK6 media.
A similar in vitro study was performed with the KLK6 inhibitor, antithrombin III (Fig. 2c). A CEST spectrum was acquired immediately before addition of the media and inhibitor to the agent, which showed a 96.2 % ratio of CEST signals at 5.3 ppm (7.48 %) and 9.5 ppm (7.77 %). A CEST spectrum acquired 8 h after addition of the media showed an 86.9 % ratio of CEST signals from the amide (5.46 %) and salicylic acid (6.28 %). This change in the CEST signal ratio indicated that the reaction coordinate was 0.10, indicating that the inhibitor had successfully reduced KLK6 activity in the media.
The imaging results were processed on a pixel-wise basis and using Eq. [1] to demonstrate that catalyCEST MRI can produce spatial maps of KLK6 activity (Fig. 2d). The large majority of the reaction coordinate map for the HCT116 in vitro study showed a very high reaction coordinate, while the reaction coordinate maps of the in vitro studies with shKLK6 and antithrombin III inhibitor showed a very low reaction coordinate.
Evaluation of Parameters for CatalyCEST MRI Studies In Vivo
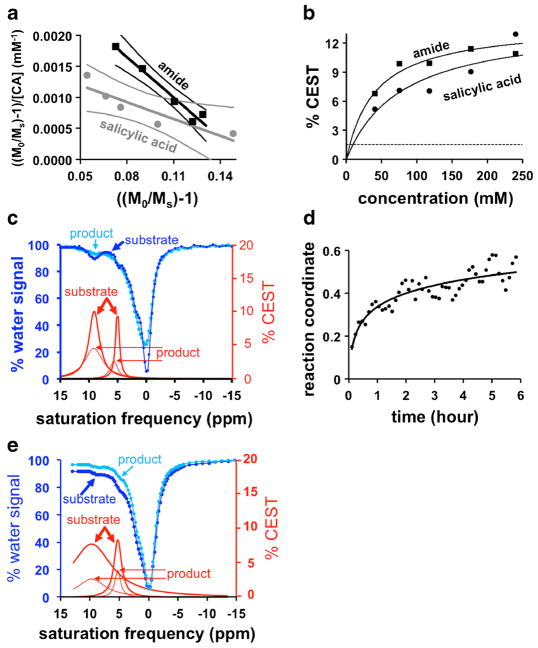
To correlate CEST signal amplitudes with the concentration of the agent, CEST MRI was performed and analyzed with a HW-Conc linear analysis method (Fig. 3a,b) [22]. This correlation was non-linear, although the correlation could be approximated to be linear for CEST signals ≤10 %. The nonlinear correlation at very high concentration can cause the reaction coordinate to be underestimated, because the ratio of CEST signals will experience little change while the high amide concentration decreases due to enzyme catalysis. In addition, a 1.16 % CEST signal has a 95 % probability of being real based on the signal-to-noise of our in vivo images (details are included in the ESM) [17]. The salicylic acid moiety can generate 1.16 % CEST signal at 7.0-mM concentration, while the aryl amide can generate this level of CEST signal with 3.8 mM of contrast agent. Based on these results, we sought a delivery method that could accumulate at least 7.0 mM of agent in tumor tissue for adequate detection of both CEST signals.
Fig. 3.
CatalyCEST MRI conditions. a The relationship between concentration and CEST signal was fit with a linear HW-Conc analysis method (29). The R2 values were 0.97 and 0.82 for the lines for the amide and salicylic acid, respectively. b This linear fitting was converted to show the relationship between CEST signal versus concentration. The dashed line represents the minimum CEST signal during in vivo studies that can be reliably measured with 95 % probability. c The CEST spectra showed the ratio of the CEST signals before and 6 h after incubation of the agent in plasma, which showed enzyme activity and slow degradation of the agent. d The reaction coordinate increased over 6 h, which matched a monoexponential function. e The incubation of the agent in plasma pre-treated with inhibitor showed no enzyme activity.
To evaluate the stability of the contrast agent in plasma, we added 20 mM of agent to blood plasma from mice bearing a HCT116 tumor, and monitored the CEST signal amplitudes for 6 h (Fig. 3c). These two CEST signals were approximately the same at 8.50 and 9.21 % at the start of the study, and had decreased to 2.94 and 4.08 % by the end of the study. The temporal change in the ratio of the CEST signals was used to determine that the reaction coordinate matched an exponential function, which indicated that the reaction was first order as anticipated (Fig. 3d). The results at the end of the study showed a reaction coordinate of 0.5, demonstrating that the plasma had enzyme activity that could cleave the peptidyl ligand of the agent. In addition, the decrease of the CEST signal at 9.5 ppm suggested that the plasma caused some general degradation of the agent during the 6 h of incubation. Based on this result, we performed subcutaneous injections during our in vivo studies, rather than performing intravenous injections that may cause cleavage of our agent in the blood during delivery to the tumor. An identical study was performed with the addition of antithrombin III inhibitor (Fig. 3e). These results showed no evidence for cleavage of the agent, indicating successful inhibition of relevant enzyme activity in the plasma.
Conditions that may affect CEST signals were evaluated to determine their effect on the catalyCEST MRI detection of enzyme activity (details are provided in the ESM). The reaction coordinate was found to be independent of saturation time (Fig. S3a–c), which matched results with similar ratiometric CEST MRI methods [23]. A low saturation power can underestimate the reaction coordinate because incomplete saturation of each labile proton on an agent causes their CEST signal amplitudes to become more equivalent, so we performed in vivo studies at a relatively high 4-μT saturation power (Fig. S3d–f) [24]. The reaction coordinate showed only a minor dependence on temperature of 3.2 % °C−1 (Fig. S4). The reaction coordinate was also underestimated with decreasing pH because a lower pH causes the chemical exchange rates to decrease and become more equivalent (Fig. S5).
Detection of KLK6 Activity and Inhibition In Vivo
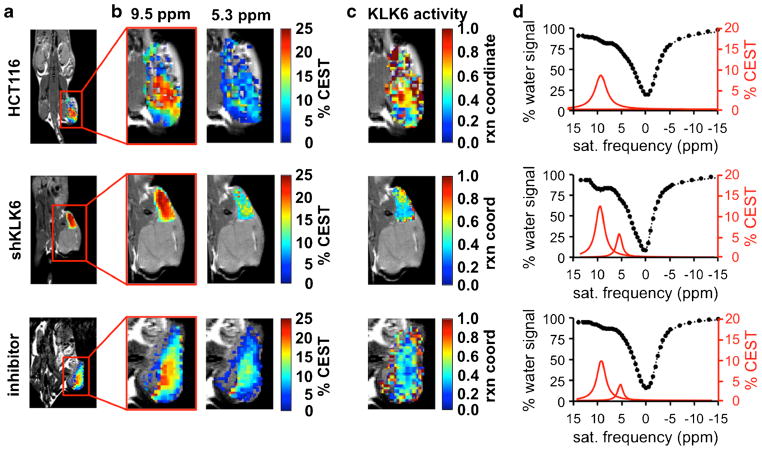
CatalyCEST MRI studies were performed in vivo with the HCT116 tumor model, shKLK6 model, and the HCT116 model treated with inhibitor (Figs. 4, S6). The results with the HCT116 model showed an average pixel-wise 5.5 % CEST signal at 9.5 ppm in the tumor, demonstrating sufficient delivery of the agent into the tumor for catalyCEST MRI detection. The HCT116 model treated with antithrombin III inhibitor showed an average 4.6 % CEST signal at 9.5 ppm, which also demonstrated good delivery of the agent into the tumor. The similarity of these results suggested that the inhibitor did not affect agent uptake. For comparison, the shKLK6 model showed an average 13.2 % CEST signal at 9.5 ppm in the tumor, demonstrating excellent delivery of the agent to the tumor of this model. The rationale for the greater uptake of agent in the shKLK6 model relative to the HCT116 model is unknown. The CEST amplitude at 9.5 ppm among all 10 mice studied ranged from 2.3 to 14.3 % with a standard deviation of 4.8 % CEST amplitude. Despite this range of uptake among the tumors studied, the reaction coordinate could be determined in a concentration-independent manner due to the ratiometric approach using an enzyme responsive and an unresponsive CEST signal from the same agent. In each case, CEST signals from tissues surrounding the tumor were negligible, indicating washout of the agent from non-tumor regions before and/or during acquisition to a concentration level that was too low for reliable CEST signal detection (Fig. S7).
Fig. 4.
CatalyCEST MRI of KLK6 activity in vivo. a Anatomical images showed the locations of flank tumors in the HCT116 model, the shKLK6 model, and the HCT116 model treated with inhibitor. b The CEST signal amplitudes of the tumors after saturation at 9.5 or 5.3 ppm were overlaid on anatomical images. c These CEST signal amplitudes and Eq. [2] were used to determine parametric maps of the reaction coordinate of KLK6 activity. d CEST spectra (black) and Lorentzian line shapes (red) show the average CEST signal amplitudes within the tumors. For comparison, Fig. S6 shows similar results from all mice tested in this study. Fig. S7 shows the parametric maps of the CEST signals from the tumor and tissues surrounding the tumor.
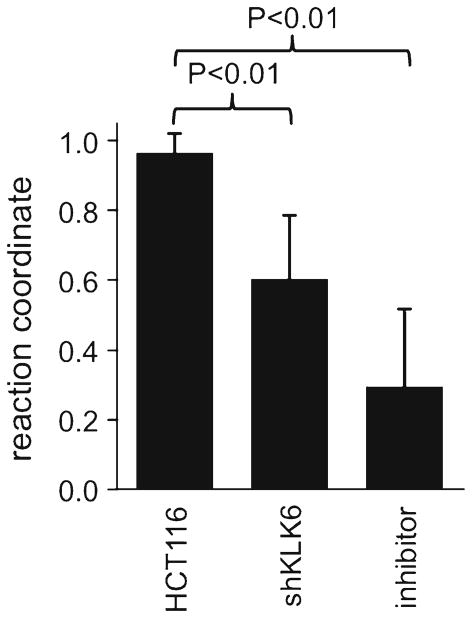
The HCT116 tumor model showed an average pixel-wise CEST signal of (0.07 ± 0.06)% and (5.6 ± 1.9)% at 5.3 and 9.5 ppm, respectively. This low CEST signal at 5.3 ppm resulted in an average reaction coordinate of 0.98 ± 0.03 that indicated strong KLK6 activity (Fig. 5). These results were also reflected in the activity map that showed the distribution of the reaction coordinate in the HCT116 model. For the shKLK6 tumor model, the average amplitude of the CEST signal at 5.3 and 9.5 ppm were (5.1 ± 2.8)% and (13.2 ± 3.3)%, which resulted in an average reaction coordinate of 0.62 ± 0.19. Therefore, catalyCEST MRI was able to detect a lower enzyme activity in this model with knockdown KLK6 expression, although some enzyme activity that cleaved the agent was still present. Furthermore, the standard deviation of 0.19 indicated a range of enzyme activities in this tumor model, relative to the HCT116 model that showed more consistent results. Finally, the HCT116 tumor model treated with antithrombin III inhibitor had CEST signals of (2.5 ± 0.7) and (4.6 ± 2.2)% at 5.3 and 9.5 ppm, engendering an average reaction coordinate of 0.33 ± 0.22. The average KLK6 activity detected in the HCT116 model was significantly higher (P < 0.01) than the average activity detected in the shKLK6 model or the model treated with inhibitor. This result showed that catalyCEST MRI can detect the effects of enzyme inhibition within the in vivo tumor model.
Fig. 5.
In vivo KLK6 enzyme activities assessed with catalyCEST MRI. The average reaction coordinate from the HCT116 model was statistically different compared to the average reaction coordinates from the shKLK6 model and HCT116 model treated with inhibitor (P < 0.01). Error bars represent the standard deviation of the reaction coordinate values.
Discussion
Our studies in biochemical solution and in vitro showed almost no enzyme activity after inhibition with antithrombin III. However, our in vivo studies still detected some remaining enzyme activity after administration of antithrombin III, which may be due to incomplete inhibition of KLK6 in vivo. Similarly, some remaining enzyme activity was detected in the shKLK6 tumor model with knockdown KLK6 expression, which may be attributed to the well-known incomplete elimination of KLK6 expression using short-hairpin RNA technology. In both cases, the remaining enzyme activity may also be due to the activity of other extracellular enzymes in the tumor tissue that could cleave the CEST agent and were not inhibited by antithrombin III. For example, we have previously detected the activity of cathepsin B using the same FRSA agent [8]. Therefore, catalyCEST MRI may have reduced specificity for detecting an intended enzyme in vivo. Yet, this potential problem may also be considered a strength of this diagnostic technique, because evaluating the sum of a specific activity caused by multiple enzymes in vivo is a better indicator of an inhibitor’s ability to neutralize a biological process rather than simply neutralizing a single type of molecule.
The ability to interrogate enzyme activity in solution, in vitro and in vivo with a single imaging technology can mitigate concerns that different results are not a manifestation of a different methodology. However, minor differences exist between these studies. For example, the endogenous T1 relaxation time constant of the water MR signal is typically shorter in vivo than in biochemical solution and in vitro, and a shorter T1 relaxation time can reduce the CEST signal amplitude [25]. CEST signals are also dependent on temperature as shown in this study, and temperature can be more easily controlled in biochemical solutions and in vitro media relative to in vivo tissues. The CEST signal amplitudes are directly dependent on concentration of the agent, and the concentration of the agent is difficult to determine in vivo relative to well-determined preparations of biochemical samples and in vitro media. Fortunately, past studies have shown that two CEST signals can have equal dependencies on T1 relaxation and concentration, so that the ratio of two CEST signals and the reaction coordinate are independent of these characteristics [15]. Similarly, our study has shown that temperature has only a minor effect on the ratio of CEST signals that determine the reaction coordinate of enzyme activity. These evaluations show the merits of including an enzyme unresponsive CEST signal in the study, which improves the comparison of results between biochemical solutions, in vitro conditions and in vivo studies.
As shown in our studies, some experimental conditions can affect the accuracy of the measured reaction coordinate. A very high concentration of agent that produces >10 % CEST can cause an underestimation of the reaction coordinate using Eqs. (1) and (2). Most of the CEST signals measured in our studies were below this signal level. Yet, future studies that measure high CEST signals should incorporate a non-linear CEST-concentration calibration in the analysis, as shown in Fig. 3a. A lower saturation power during CEST MRI studies can cause the reaction coordinate to be underestimated, which may be addressed by performing additional MRI scans that map the variability in saturation power applied in vivo, known as a B1 map [26]. Also, a lower pH causes the reaction coordinate to be underestimated, which should be addressed during in vivo studies of tumors with high metabolic acidosis [27]. For example, we estimated that the shKLK6 model would need to be ≥0.9 pH units lower than the HCT116 model to account for our in vivo catalyCEST MRI results described below, which is unrealistic for these similar tumor cell lines. This issue may be evaluated by mapping the extracellular pH in tumors using acidoCEST MRI [28].
Our studies with KLK6 have expanded the portfolio of enzyme activities that are detected with catalyCEST MRI. Our CEST agent has a modular design, whereby the peptidyl ligand can be modified to become a substrate for other enzymes [10, 11] or the salicylic acid moiety can be modified to be a substrate [29]. Thus, exploiting this modular design to develop additional CEST agents for the detection of enzyme activity is a useful paradigm for molecular imaging.
Supplementary Material
Acknowledgments
The authors thank Ms. Jasmine Acfalle for laboratory assistance. These studies were supported by NIH grants R01CA169774, R01CA157595, and P30CA23074.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11307-017-1092-8) contains supplementary material, which is available to authorized users.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Schomburg I, Chang A, Placzek S, et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA. Nucleic Acids Res. 2013;41:D764–D772. doi: 10.1093/nar/gks1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glanemann C, Loos A, Gorret N, et al. Disparity between changes in mRNA abundance and enzyme activity in Corynebacterium glutamicum: implications for DNA microarray analysis. Appl Microbiol Biotechnol. 2003;61:61–68. doi: 10.1007/s00253-002-1191-5. [DOI] [PubMed] [Google Scholar]
- 3.Yoo B, Pagel MD. A PARACESTMRI contrast agent to detect enzyme activity. J Am Chem Soc. 2006;128:14032–14033. doi: 10.1021/ja063874f. [DOI] [PubMed] [Google Scholar]
- 4.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hingorani DV, Randtke EA, Pagel MD. A catalyCEST MRI contrast agent that detects the enyme-catalyzed creation of a covalent bond. J Am Chem Soc. 2013;135:6396–6398. doi: 10.1021/ja400254e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoo B, Pagel MD. An overview of responsive MRI contrast agents for molecular imaging. Front Biosci. 2008;13:1733–1752. doi: 10.2741/2796. [DOI] [PubMed] [Google Scholar]
- 7.Hingorani DV, Bernstein AS, Pagel MD. A review of responsive MRI contrast agents: 2005–2014. Contrast Media Mol Imaging. 2015;10:245–265. doi: 10.1002/cmmi.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hingorani DV, Montano LA, Randtke EA, et al. A single diamagnetic catalyCEST MRI contrast agent that detects cathepsin B enzyme activity by using a ratio of two CEST signals. Contrast Media Mol Imaging. 2016;11:130–138. doi: 10.1002/cmmi.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinharay S, Randtke EA, Jones KM, et al. Noninvasive detection of enzyme activity in tumor models of human ovarian cancer using catalyCEST MRI. Magn Reson Med. 2017;77:2005–2014. doi: 10.1002/mrm.26278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henkhaus RS, Gerner EW, Ignatenko NA. Kallikrein 6 is a mediator of K-RAS-dependent migration of colon carcinoma cells. Biol Chem. 2008;389:757–764. doi: 10.1515/BC.2008.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayani J, Diamandis EP. The physiology and pathobiology of human callikrein-related peptidase 6 (KLK6) Clin Chem Lab Med. 2012;50:211–233. doi: 10.1515/CCLM.2011.750. [DOI] [PubMed] [Google Scholar]
- 12.Prassas I, Eissa A, Poda G, Diamandis EP. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov. 2015;14:183–202. doi: 10.1038/nrd4534. [DOI] [PubMed] [Google Scholar]
- 13.Travis J, Salvesen GS. Human plasma proteinase inhibitors. Annu Rev Biochem. 1983;52:655–709. doi: 10.1146/annurev.bi.52.070183.003255. [DOI] [PubMed] [Google Scholar]
- 14.Shah T, Lu L, Dell KM, et al. CEST-FISP: a novel technique for rapid chemical exchange saturation transfer MRI at 7 T. Magn Reson Med. 2011;65:432–437. doi: 10.1002/mrm.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen LQ, Randtke EA, Jones KM, et al. Evaluations of tumor acidosis within in vivo tumor models using parametric maps generated with acidoCEST MRI. Mol Imaging Biol. 2015;17:488–496. doi: 10.1007/s11307-014-0816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sells E, Pandey R, Chen HW, Skovan BA, Cui H, Ignatenko NA. Specific microRNA-mRNA regulatory network of colon cancer invasion mediated by tissue kallikrein-related peptidase 6. Neoplasia. 2017;19:396–411. doi: 10.1016/j.neo.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu G, Ali MM, Yoo B, et al. PARACEST MRI with improved temporal resolution. Magn Reson Med. 2009;61:399–408. doi: 10.1002/mrm.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang X, Song X, Li Y, et al. Salicylic acid and analogues as diaCEST MRI contrast agents with highly shifted exchangeable proton frequencies. Angew Chem Int Ed. 2013;52:8116–8119. doi: 10.1002/anie.201302764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magklara A, Mellati AA, Wasney GA, et al. Characterization of the enzymatic activity of human kallikrein 6: Autoactivation, substrate specificity, and regulation by inhibitors. Biochem Biophys Res Commun. 2003;307:948–955. doi: 10.1016/s0006-291x(03)01271-3. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, Yadav NN, Song X, et al. Tuning phenols with intra-molecular bond shifted HYdrogens (IM-SHY) as diaCEST MRI contrast agents. Chem Eur J. 2014;20:15824–15832. doi: 10.1002/chem.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basu Roy UK, Henkhaus RS, Loupakis F, et al. Caveolin-1 is a novel regulator of K-RAS-dependent migration in colon carcinogenesis. Int J Cancer. 2013;133:43–57. doi: 10.1002/ijc.28001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali MM, Liu G, Shah T, et al. Using two chemical exchange saturation transfer magnetic resonance imaging contrast agents for molecular imaging studies. Acc Chem Res. 2009;42:915–924. doi: 10.1021/ar8002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Randtke EA, Chen LQ, Pagel MD. The reciprocal linear QUEST analysis method facilitates the measurements of chemical exchange rates with CEST MRI. Contrast Media Mol Imaging. 2014;9:252–258. doi: 10.1002/cmmi.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randtke EA, Chen LQ, Corrales LR, Pagel MD. The Hanes-Woolf linear QUESP method improves the measurements of fast chemical exchange rates with CEST MRI. Magn Reson Med. 2014;71:1603–1612. doi: 10.1002/mrm.24792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woessner DE, Zhang S, Merritt ME, Sherry AD. Numerical solution of the Bloch equations provides insights into the optimum design of PARACEST agents for MRI. Magn Reson Med. 2005;53:790–799. doi: 10.1002/mrm.20408. [DOI] [PubMed] [Google Scholar]
- 26.Duan Q, van Gelderen P, Duyn J. Improved Bloch-Siegert based B1 mapping by reducing off-resonance shift. NMR Biomed. 2013;26:1070–1078. doi: 10.1002/nbm.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen LQ, Pagel MD. Evaluating pH in the extracellular tumor microenvironment using CEST MRI and other imaging methods. Adv Radiol. 2015;2015:206405. doi: 10.1155/2015/206405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LQ, Randtke EA, Jones KM, et al. Evaluations of extracellular pH within in vivo tumors using acidoCEST MRI. Magn Reson Med. 2014;72:1408–1417. doi: 10.1002/mrm.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Cuervo G, Sinharay S, Pagel MD. A catalyCEST MRI contrast agent that can simultaneously detect the two enzyme activities. Chembiochem. 2016;17:383–387. doi: 10.1002/cbic.201500586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.