Abstract
Background
Alcohol's reinforcement is mediated by dopamine signaling in the ventral striatum, which is modulated by the dopamine transporter (DAT). We hypothesized that methylomic variation in the DAT gene (SLC6A3) affects DAT expression thus contributing to differences in brain reward circuitry in individuals with alcohol dependence (ALC).
Methods
Blood from 45 recently detoxified ALC and 45 healthy control (HC) individuals was used to assess DNA methylation across 5 functional regions of SLC6A3. Participants completed the Monetary Incentive Delay task in a 3-Tesla MRI scanner. Employing regression models, we examined effects of SLC6A3 methylation on Nucleus Accumbens (NAc) BOLD responses during anticipation of high/low reward/loss.
Results
Results showed that decreased methylation of the promoter region of SLC6A3 predicted NAc activation during high loss anticipation (p = 0.028) and low loss anticipation (at trend-level; p = 0.057) in HC but not in individuals with ALC. Specifically, percentage of methylation at two CpG sites, located -1001 and -993 base pairs from the transcription start site, accounted for significant variability in NAc activation in the HC group during high (ps ≤ 0.010) and low (ps ≤ 0.006) loss anticipation. There was no effect on reward anticipation. Furthermore, promoter methylation was positively associated with age, which replicates previous findings.
Conclusions
Our data suggest that methylation in the promoter region of SLC6A3 predicts NAc activation during the anticipation of monetary loss in HCs. However, this effect was not present in the ALC group, suggesting that epigenetic regulation of striatal DAT expression might be disrupted in ALC, which may contribute to previously reported differences in sensitivity to reward and punishment in this population. Alternatively, it is possible that a similar relationship in the ALC group remained undetected possibly due to methodological limitations inherent in fMRI (e.g., poor spatial resolution, low signal to noise ratio) that generally restrict interpretations regarding mechanisms of epigenetic factors involved in group differences in BOLD responses. Future neuroimaging studies are needed to further elucidate the relationship between SLC6A3 methylation and NAc activation in ALC.
Keywords: DAT1/SLC6A3, epigenetics, reward processing, alcohol dependence, methylation
Introduction
Alcohol use disorder is a chronically relapsing disorder affecting 11.3 million Americans aged 26 or older (Center for Behavioral Health Statistics and Quality, 2016). It represents one of the leading preventable causes for morbidity and mortality in the United States (Kochanek et al., 2016). The underlying etiology is complex with environmental and genetic factors playing a role (Tawa et al., 2016). Cognitive functioning including decision-making is often negatively affected by alcohol use disorder. Reward processing, specifically reward/loss anticipation, has been proposed to play a key role in adaptive decision-making because it occurs immediately before a decision is made, thus exerting crucial influence on the evaluation of different options and their respective consequences. Brain regions involved in reward processing include the ventral striatum, in particular the Nucleus Accumbens (NAc; Carter et al., 2009), which has been shown to be activated during the anticipation of rewards and losses in the Monetary Incentive Delay (MID) task (Knutson et al., 2000, Balodis and Potenza, 2015). However, there is substantial variability between individuals and the underlying neurobiological mechanisms remain unknown.
Alcohol's reinforcing effects are mediated in part by dopaminergic mesolimbic reward pathways; more specifically, by increased dopamine release in the NAc (Volkow et al., 2007). Since striatal dopamine signaling is directly influenced by dopamine transporter (DAT) availability (Shen et al., 2004, e.g., Giros et al., 1996, Heinz et al., 1999), it has been hypothesized that genetic variation in the gene encoding the DAT (DAT1/SLC6A3) contributes to differences in reward processing (Lohoff et al., 2010, Dreher et al., 2009, Hahn et al., 2011). While several studies have indeed found associations of SLC6A3 variation with various alcohol use disorder phenotypes and endophenotypes (Ma et al., 2016, Ray et al., 2014, Wernicke et al., 2002, Le Strat et al., 2008, Gorwood et al., 2003), other studies were unable to replicate results (e.g., Du et al., 2011, Kohnke et al., 2005, Xu and Lin, 2011). Genetic variation is one important factor influencing interindividual differences in reward processing; however, it is likely that other mechanisms affecting DAT expression or function may also contribute. Epigenetic mechanisms, such as DNA methylation, histone modification, or chromatin restructuring can dynamically regulate gene expression in response to environmental influences, such as alcohol use. Preclinical evidence indicates that increased DAT gene methylation is associated with reduced DAT expression in the rodent striatum (Kim et al., 2014). Since DNA methylation has generally been shown to be inversely associated with protein expression, these data are in line with previous findings that chronic alcohol use causes a reduction of striatal DAT expression (Heinz et al., 2004, Heinz et al., 2000).
Only a small number of studies to date have investigated how epigenetic regulation of the DAT gene may be associated with alcohol use in humans. Lee et al. (2015) found decreased SLC6A3 promoter methylation in babies of binge-drinking fathers, as well as an association between the level of alcohol consumption in socially drinking parents and decreased SLC6A3 methylation. Nieratschker et al. (2014) and Jasiewicz et al. (2015) found no difference in promoter methylation levels between patients with alcohol dependence (ALC) and healthy controls (HC). However, Nieratschker et al. (2014) demonstrated a significant positive association of SLC6A3 promoter methylation with age in the alcohol-dependent but not in the control group. In contrast, two other studies found a significant elevation of SLC6A3 promoter methylation in the ALC compared to the HC group, which was associated with alcohol craving (Wiers et al., 2015, Hillemacher et al., 2009).
To improve our understanding of underlying neurobiological mechanisms in reward processing in individuals with alcohol use disorders, we investigated how epigenetic variation in SLC6A3 may contribute to individual differences in brain reward circuitry as assessed by neuronal activation in the NAc during the MID task, which has been shown to be sensitive to the effects of ALC (Beck et al., 2009, Wrase et al., 2007, van Holst et al., 2014, Murphy et al., 2017). We hypothesized that methylomic variation in functional regions of SLC6A3 would affect striatal DAT expression, which would in turn contribute to differences in brain reward circuitry between individuals with ALC and HC.
Materials and Methods
Participants
For the present study, a total of 90 (45 ALC and 45 HC) right-handed individuals were recruited to the National Institute on Alcohol Abuse and Alcoholism from the Washington D.C. metropolitan area between July 2014 and September 2016 using newspaper (e.g., Express) and web-based (e.g. Craigslist) advertisements. The mean age of study participants was 39.78 (SD = 11.33) and 46.7% of participants identified as Black/African American, 36.7% identified as White, 6.7% identified as Asian, 4.4% identified as multiracial, and 5.6% chose not to respond. Participant characteristics are displayed in Table 1. All participants provided written informed consent, which complied with the Declaration of Helsinki and was approved by the Institutional Review Board of the NIH/National Institute on Alcohol Abuse and Alcoholism. Participants were administered the Structured Clinical Interview for DSM-IV-TR Axis I Disorders (SCID). Participants were excluded from study participation if they were left-handed, pregnant, reported claustrophobia, or presented with significant neurological or medical diagnoses. Control subjects were excluded from study participation if they met DSM-IV-TR criteria for any current or past alcohol dependence and if they presented with a positive urine drug screen or alcohol breathalyzer on the day of the scan. Among the HC group, seven individuals met DSM-IV-TR criteria for past (but not current) alcohol abuse and none of them were alcohol abstainers. All participants were free from any psychotropic medications on the day of the scan. All participants completed a modified version of the original MID task (Knutson et al., 2000) in which all subjects were successful in approximately 70% of the trials. The subjects also provided DNA samples for methylation assessments.
Table 1. Demographics and Characteristics.
| Alcohol Dependence | Healthy Controls | P-Value | |
|---|---|---|---|
| Gender, N, (%) | .004 | ||
| Male | 35 (77.8) | 22 (48.9) | |
| Female | 10 (22.2) | 23 (51.1) | |
|
| |||
| Age, mean years (SD) | 43.25 (10.73) | 36.30 (10.94) | .003 |
|
| |||
| Ethnicity, N, (%) | .088 | ||
| Black/African American | 24 (53.3) | 18 (40.0) | |
| European American | 17 (37.8) | 16 (35.6) | |
| Asian | 0 (0.0) | 6 (13.3) | |
| Multiracial | 1 (2.2) | 3 (6.7) | |
| Unknown | 3 (6.7) | 2 (4.4) | |
|
| |||
| Smokers, N, (%) | 25 (55.6) | 0 (0.0) | <.0001 |
|
| |||
| Alcohol Dependence Severity Score, mean (SD) | 19.68 (7.70) (N = 38) | 1.13 (2.25) (N = 31) | <.0001 |
|
| |||
| Average Number of Drinks per Drinking Day, mean (SD) | 13.11 (8.84) | 1.80 (1.78) | <.0001 |
Experimental Paradigm
The experimental task was a modification of the MID task (Knutson et al., 2000), where participants were required to respond by pressing a button in order to win monetary reward or to avoid losing it. Each trial started with a fixation crosshair, followed by a cue shape for 2 seconds, which indicated the trial type (Reward, Loss, or Neutral). A jittered delay of 0.75–1.75 seconds between cue and the target box appeared. Then a feedback was presented after 0.75 to 3 seconds following the target box. This feedback showed the amount lost or won in the trial, as well as the total amount accumulated up to that point.
Five conditions were presented: Neutral (no reward/no loss), low loss, high loss, low reward and high reward. Participants had an average of a 10-min session, viewing 30 trials of each condition. Participants were informed that any reward or loss would be added or subtracted from their final payment. Low reward/loss trials were signaled by a single line in the cue shape. High reward/loss trials were signaled by three lines in the cue shape. Participants played a practice round outside of the scanner to familiarize themselves with the task. The task was projected onto a screen behind the scanner, which was viewable by a mirror attached to the head coil.
MRI Data Acquisition and Preprocessing
Structural (MPRGE) and functional data were acquired using a Siemens 3T Skyra scanner. Whole-brain functional data were collected with echo-planar imaging sequence (36 axial slices, 3.8 × 3.8 × 3.8 mm, 64 × 64 matrix and TR = 2000 ms; TE = 30 ms, Flip Angle = 90). We preprocessed and analyzed the MRI data using AFNI software package (Cox, 1996). Large transients were removed through interpolation (AFNI's 3dDespike). Volumes were then slice-time corrected. All nuisance time series were de-trended. Nuisance variables for each voxel included the six parameter estimates for head motion. Motion higher than 0.3 mm/o was censored out, and if a participant had censored more than 20% of a condition (regressor) of interest, that participant would be excluded from the analysis. No subjects were excluded for head motion. The functional echo-planar images (EPI) were then coregistered to the anatomical scan, resampled to 2-mm isotropic voxels, smoothed with an isometric 4-mm full-width-half-maximum Gaussian kernel, normalized by the mean signal intensity in each voxel to reflect percent-signal change, and transformed into the standardized Talairach and Tournoux (1988) volume for the purposes of group analyses (Talairach and Tournoux, 1988).
BOLD fMRI Data Analysis
Single-subject level analysis on blood-oxygen-level dependent (BOLD) responses was performed using the following experimental conditions: Neutral (no reward/no loss), low loss, high loss, low reward and high reward.
ROI Analyses
The ROI mask of the bilateral NAc (MNI coordinates Left: -12, 8, -8; Right: 12, 8, -8) was generated with the AFNI atlas using the DrawDataset plugins. To follow previously published literature and subtracting all nuisance effects (such as pre-motor activations), except reward/loss response, we then computed average percent signal change (beta weights) of the following comparisons under NAc ROI: high reward > neutral, low reward > neutral, high loss > neutral, low loss > neutral. For simplification purposes, we refer to these as high reward, low reward, high loss, low loss, respectively.
DNA Extraction and Methylation Analysis
Genomic DNA was extracted from participants' blood samples. DNA methylation levels of 5 functional regions across SLC6A3 were assessed using quantitative bisulfite pyrosequencing by EpigenDx following EpigenDx protocol. Briefly, EZ DNA methylation kits (Zymo Research, Inc., CA) were used to treat 500 ng of genomic DNA for each analysis and the manufacturer's protocol was followed for DNA purification eluting samples to 46 μL. One μL of bisulfite treated DNA were combined with .2 μM of each primer for the polymerase chain reaction (PCR). One of the primers was labeled with biotin and purified with HPLC for the purpose of purifying the final PCR product with Sepharose beads. Next, the PCR product was bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences) and the now immobilized PCR products were purified, washed, denatured with a .2 μM NaOH solution. Following the manufacturer's protocol, the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Qiagen) was used for rewashing, .5 μM of sequencing primer was then annealed to the purified single stranded PCR products, and 10 μL of the PCR product were sequenced on the PSQ96 HS System (Pyrosequencing, Qiagen).
QCpG software (Pyrosequencing, Qiagen) was used to assess methylation at each CpG site as an artificial C/T SNP. The percentage of methylated alleles was divided by the total number of alleles (methylated and unmethylated) to determine the level of methylation at each CpG site. Regional methylation was calculated by taking the average of all CpG site methylation levels measured within each region. To ensure quality control, each experiment had non-CpG cytosines serving as internal controls to ensure complete bisulfite conversion of the input DNA. Furthermore, low, medium, and high methylated DNA was included as controls in each run. Finally, EpigenDx performed PCR bias testing by combining unmethylated control DNA with in vitro methylated DNA following different ratios (0%, 5%, 10%, 25%, 50%, 75%, 100%) and conducting bisulfite modification, PCR, and Pyrosequencing analysis.
The SLC6A3 assays analyzed 48 CpG dinucleotides across the gene ranging from -886 to 50969 base pairs from the transcription start site (TSS), based on Ensembl Transcript ID ENST00000270349. Table S1 provides more details on the location of the CpG sites, including their genomic location and position relative to the TSS. Table S2 provides more information on the methylation assays.
Statistical Analysis
Data analyses were conducted in a personal computer-based software package (IBM SPSS Statistics® 20, IBM Corp., Armonk, NY).
One-way Analyses of Covariance (ANCOVA) were performed to examine differences in SLC6A3 cluster methylation, defined as the average percentage of methylated cytosines from all assessed CpG sites in a functional cluster, between the ALC and HC group controlling for age and gender, as previous research has reported an association between age and DNA methylation (Jung and Pfeifer, 2015, Nieratschker et al., 2014), as well as between gender and methylation (El-Maarri et al., 2007). T-tests and chi-square tests were used to examine baseline differences between the two groups on all demographic variables. One-way ANCOVAs were performed to examine differences in NAc BOLD responses between the ALC and HC group while controlling for age and gender.
The primary dependent measure was NAc activation as measured by BOLD fMRI in the NAc. All BOLD fMRI outcomes were normally distributed as examined by Kolmogorov-Smirnov test. For the main analysis, hierarchical linear regression models were employed to examine associations between the percentage of methylation of functional regions of SLC6A3 and NAc BOLD responses during anticipation of high and low rewards and losses in both groups while controlling for age and gender. Next, analyses were run for the single CpG sites of the cluster that was significantly associated with NAc outcomes. Between-group differences in regression slopes for the ALC and HC groups were tested by adding interaction terms (group x SLC6A3 cluster/CpG site) into the regression models. Statistical significance was set at p < .05 (two-tailed).
Results
DAT DNA Methylation between Cases and Controls
ANCOVAs showed no significant differences between the ALC and HC group on cluster methylation when controlling for age and gender (see Table 2).
Table 2.
Summary of ANCOVAs examining differences between cases and controls on cluster methylation and NAc MID task percent signal change (β).
| Alcohol-Dependent Patients (n = 45) | Healthy Controls (n = 45) | F | P-Value | Eta-squared | |
|---|---|---|---|---|---|
| Percent Methylation | |||||
| Cluster A | 9.11 (8.53, 9.69) | 9.42 (8.84, 9.99) | .510 | .477 | .006 |
| Cluster B | 8.38 (7.89, 8.87) | 8.44 (7.95, 8.93) | .026 | .872 | .000 |
| Cluster C | 62.35 (61.25, 63.44) | 61.86 (60.75, 62.97) | .349 | .556 | .004 |
| Cluster D | 89.22 (88.92, 89.53) | 89.17 (88.86, 89.48) | .053 | .819 | .001 |
| Cluster E | 77.96 (77.35, 78.57) | 78.22 (77.61, 78.83) | .338 | .563 | .004 |
|
| |||||
| NAc Activation | |||||
| High Reward | .172 (.038, .307) | -.059 (-.195, .077) | 5.33 | .023 | .059 |
| Low Reward | -.031(-.153, .091) | -.220 (-.343 -.096) | 4.27 | .042 | .048 |
| High Loss | .078 (-.037, .192) | -.160 (-.275, -.044) | 7.74 | .007 | .083 |
| Low Loss | -.026 (-.140, .088) | -.224 (-.338, -.110) | 5.48 | .022 | .061 |
Note. Adjusted means and confidence intervals.
Monetary Incentive Delay Task
All MID task BOLD responses were normally distributed as examined by Kolmogorov-Smirnov tests (all ps > .165). ANCOVAs were conducted to examine differences in MID task BOLD activation in the NAc between cases and controls whilst controlling for age and gender (see Table 2). Analyses showed that there was a significant difference between cases and controls in BOLD responses in the NAc during anticipation of high (F(1,86) = 7.744, p = .007, ηp2 = .083) and low (F(1,86) = 5.477, p = .022, ηp2 = .061) loss. Furthermore, there was a significant difference between cases and controls in BOLD responses in the NAc during anticipation of high (F(1,86) = 5.326, p = .023, ηp2 = .059) and low (F(1,86) = 4.270, p = .042, ηp2 = .048) rewards, with ALC showing higher NAc activations than HC (see Table 2).
MID Task Activation and Effects of SLC6A3 Methylation
Table 3 displays detailed results from hierarchical linear regression analyses examining the association between methylation of functional SLC6A3 clusters and NAc BOLD responses while controlling for effects of age and gender. The main result was that methylation in Cluster A significantly predicted BOLD response during the anticipation of high loss (β = -.33, p = .028) in the HC but not the ALC group (p = .471). This between-group difference was statistically significant (p = .031) with a decrease in methylation of the promoter region of SLC6A3 (Cluster A) predicting greater high loss anticipation-related activity in the NAc in HC but not in ALC individuals (see Fig. 1). Furthermore, NAc activation during anticipation of low loss was predicted by Cluster A at trend-level in the HC (β = -.31, p = .057) but not the ALC group (p = .205). Between-group analyses demonstrated that this difference was significant (p = .015).
Table 3.
Summary of results from hierarchical linear regression models predicting Nucleus Accumbens activation during high and low reward/loss anticipation for each of the 5 clusters.
| Cluster Distance to TSS | Nucleus Accumbens | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High Reward | Low Reward | High Loss | Low Loss | ||||||||||||||
| ALC | HC | ALC | HC | ALC | HC | ALC | HC | ||||||||||
| β | p | β | p | β | p | β | p | β | p | β | p | β | p | β | p | ||
| A | -1101 to -886 | -.05 | .782 | -.10 | .510 | -.08 | .619 | -.13 | .411 | .12 | .471 | -.33 | .028 | .21 | .205 | -.31 | .057 |
| B | -423 to -321 | -.05 | .780 | -.12 | .444 | -.10 | .531 | -.06 | .702 | .01 | .941 | -.26 | .110 | -.04 | .804 | -.30 | .069 |
| C | 29042 to 29089 | .21 | .167 | .23 | .137 | .13 | .399 | .137 | .377 | .09 | .565 | .15 | .345 | .02 | .920 | -.04 | .801 |
| D | 29188 to 29292 | -.11 | .478 | .04 | .817 | -.05 | .769 | .12 | .513 | -.17 | .267 | .17 | .359 | -.12 | .429 | .29 | .126 |
| E | 50868 to 50969 | .14 | .366 | -.18 | .246 | .19 | .221 | -.24 | .123 | -.11 | .476 | -.19 | .226 | .05 | .765 | -.16 | .331 |
Note: ALC = alcohol-dependent group, HC = healthy control group. Boldface indicates significant differences between ALC and HC group as indicated by a significant interaction between group and cluster methylation in linear regression model (alpha set at p < .05).
Fig. 1.
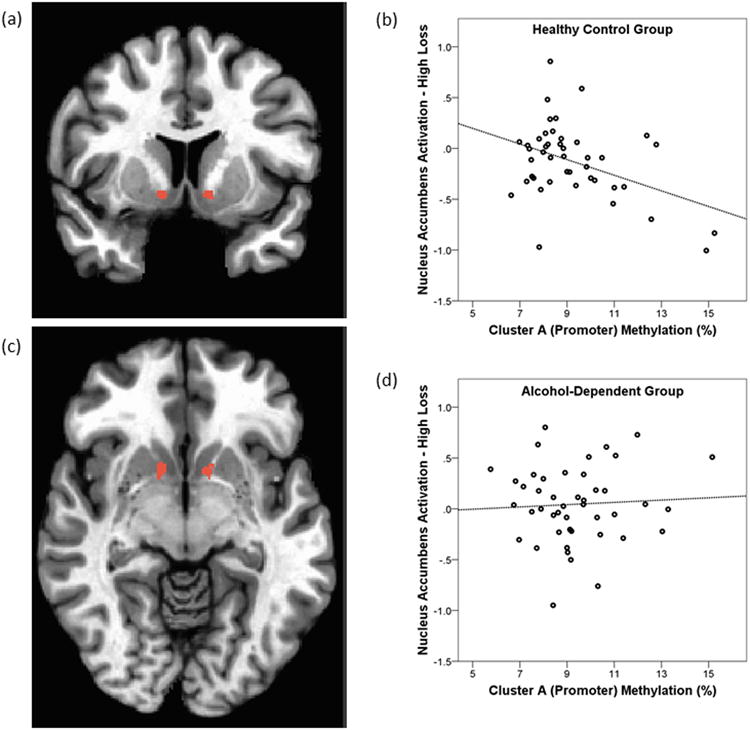
Effects of SLC6A3 promoter methylation on NAc reactivity. Left (a, c): NAc mask defined from Talairach Daemon of AFNI (Peak: x = 12, y = -8, z = -8) used to compute the individual subjects' Beta coefficients for anticipation signals. Right (b, d): Scatterplots of percent methylation of Cluster A (Promoter) and high loss anticipation-related activity in the Nucleus Accumbens in the HC (b) and ALC (d) group.
Exploratory follow-up analyses were run to examine which of the CpG sites included in Cluster A were driving the observed effect on NAc BOLD responsiveness. Table 4 provides detailed results from regression models predicting NAc activation during high and low loss anticipation for each of the identified CpG sites of Cluster A (promoter region). Mainly, these analyses showed that the first two CpG sites in the promoter of SLC6A3, located -1001 (CpG site 242) and -993 (CpG site 241) base pairs from the TSS, predicted high and low loss anticipation-related NAc activation in the HC but not the ALC group. Between-group analyses revealed that this difference was statistically significant for CpG site 242 in both the high (p = .003) and low (p = .001) loss condition. Similarly, the between-group difference was significant for CpG site 241 in the high (p = .038) and low (p = .020) loss condition.
Table 4.
Summary of results from linear regression models predicting Nucleus Accumbens activation during high and low loss anticipation for each of the identified CpG sites in Cluster A.
| Cluster | CpG ID | Distance to TSS | Nucleus Accumbens | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| High Loss | Low Loss | |||||||||
| ALC | HC | ALC | HC | |||||||
| β | p | β | p | β | p | β | p | |||
| A | 242 | -1001 | .26 | .096 | -.37 | .010 | .21 | .176 | -.43 | .004 |
| A | 241 | -993 | -.05 | .770 | -.43 | .003 | .06 | .695 | -.43 | .006 |
| A | 240 | -973 | .02 | .898 | -.12 | .427 | .29 | .064 | -.21 | .202 |
| A | 239 | -964 | .08 | .646 | -.21 | .167 | .18 | .266 | -.18 | .258 |
| A | 238 | -954 | .02 | .892 | -.28 | .072 | .27 | .087 | -.27 | .100 |
| A | 237 | -944 | .02 | .903 | -.18 | .259 | -.01 | .940 | -.17 | .293 |
| A | 236 | -941 | .08 | .626 | .05 | .741 | .29 | .062 | .06 | .713 |
| A | 235 | -934 | .11 | .480 | -.21 | .182 | .21 | .172 | -.19 | .258 |
| A | 234 | -930 | -.09 | .573 | -.34 | .026 | -.03 | .860 | -.28 | .080 |
| A | 233 | -918 | .06 | .746 | -.34 | .029 | -.01 | .960 | -.22 | .180 |
| A | 232 | -915 | -.02 | .908 | -.33 | .032 | .18 | .286 | -.30 | .065 |
| A | 231 | -901 | .04 | .816 | -.28 | .062 | .11 | .481 | -.19 | .223 |
| A | 230 | -899 | .05 | .743 | -.29 | .054 | .02 | .913 | -.23 | .151 |
| A | 229 | -886 | .26 | .114 | -.22 | .154 | -.11 | .498 | -.18 | .265 |
Note: ALC = alcohol-dependent group, HC = healthy control group. Boldface indicates significant differences between ALC and HC group as indicated by a significant interaction between group and CpG methylation in linear regression model (alpha set at p < .05).
Regional SLC6A3 methylation was not associated with BOLD responses in the NAc during anticipation of high (all ps > .137) and low (all ps > .123) reward in the HC and ALC group (see Table 3).
Promoter Methylation and Age
Cluster A (r = .27, p = .009) and Cluster B (r = .31, p = .003) methylation was significantly correlated with age, which is in line with a previous report (Nieratschker et al., 2014). Separate analyses showed that this association was driven by ALC patients (Cluster A: r = .36, p = .016; Cluster B: r = .34, p = .024).
Discussion
The present study investigated how epigenetic variation in the gene coding for the DAT may contribute to individual differences in neural reward circuitry in alcohol-dependent and healthy individuals. Our main finding suggests that decreased methylation of the promoter region of SLC6A3 predicts NAc activation during the anticipation of monetary loss in a group of 45 healthy volunteers. The effects of promoter methylation were significant for both high and low loss anticipation, and were driven by the first two CpG sites we assessed in the promoter of SLC6A3, located -1001 and -993 base pairs from the TSS. In a group of ALC patients, however, these associations were not present, even when effects of age and gender were controlled for. There were no group differences in DAT methylation, but ALC patients had stronger NAc activations than controls during both anticipation of losses and rewards on the MID task (all ps < .05). Our findings hence suggest promoter methylation of SLC6A3 as a biomarker for striatal activation during MID task performance in healthy volunteers, and a disruption of this system in ALC patients. That is, DAT promoter methylation has been associated with DAT expression in a preclinical study (Kim et al., 2014), and DAT availability has been shown to influence dopamine signaling (Shen et al., 2004, Giros et al., 1996, Heinz et al., 1999), which is important for striatal activation during anticipation of rewards and losses (Knutson et al., 2000, Balodis and Potenza, 2015). To the best of our knowledge, the present study is the first to report an association between SLC6A3 methylation and striatal activation during reward processing.
The DAT plays a key role in dopamine-mediated reward processing as it regulates the duration and intensity of dopamine signaling in the striatum. Different genetic variants, most notably the variable number tandem repeat (VNTR) polymorphism in the 3′ region of SLC6A3, have been found to be associated with striatal DAT expression, however, findings have been mixed. Jacobsen et al. (2000) found decreased DAT density in individuals with the 10/10 repeat genotype; other studies found an increase in DAT availability in the 10/10 repeat genotype (Fuke et al., 2001, Heinz et al., 2000), and two other studies found no significant association between genotype and DAT expression (Greenwood and Kelsoe, 2003, Martinez et al., 2001). These discrepant results could be explained by gene-environment interactions. Epigenetic mechanisms dynamically regulate gene expression in response to environmental influences, such as stress or drug and alcohol use. Our study thus expands on previous genetic studies, as epigenetic markers may provide more dynamic insight into gene-environment interaction effects on brain function. In a recent imaging epigenetics study, DNA methylation of a promoter region was shown to alter gene expression and predict brain function as assessed by BOLD fMRI independent of genotype, indicating the potential of exploring epigenetic mechanisms for individual differences in clinically relevant phenotypes (Nikolova et al., 2014).
While research on the epigenetic regulation of the human SLC6A3 gene is still limited, a preclinical study has linked differences in DAT gene promoter methylation with decreased striatal DAT expression in the offspring of mice that have been exposed to chronic alcohol treatment comparable to binge drinking in humans (Kim et al., 2014). Consistent with Kim and colleague's (2014) proposal that chronic alcohol exposure may lead to changes in the epigenetic methylation signature of the DAT gene, the present study provides further support for the relevance of SLC6A3 promoter methylation for dopamine signaling in the striatum. Interestingly, while Kim el al. (2014) found an increase in SLC6A3 promoter methylation to be associated with a reduction in striatal DAT expression in rodents, the present study found a decrease in promoter methylation to predict NAc reactivity during loss anticipation in HC. However, Kim et al. (2014) did not specify what part of the striatum they examined. DAT expression likely varies between the dorsal and ventral striatum and previous research has shown differences in DAT expression within the NAc (i.e., core vs. shell; Nirenberg et al., 1997). Given the clinical relevance of dopaminergic neurotransmission in the striatum, this warrants further investigation.
The mechanisms underlying epigenetic regulation are complex and likely vary across genes. Both hyper- and hypomethylation have been shown to lead to changes in gene expression (Nestler, 2014, Lopez-Serra et al., 2006). Therefore, our main findings can be interpreted in different ways. It should be noted that the following interpretations are speculative in nature as spatial resolution in fMRI does not provide differentiation between NAc core and shell, and, furthermore, fMRI BOLD signal merely reflects blood oxygenation in response to neuronal population activity without information on the type of neurotransmitter involved. With this limitation in mind, two different explanations appear to be plausible.
First, our data may indicate that SLC6A3 hypomethylation results in lower DAT transcription and expression in the striatum, which may in turn protract dopamine reuptake and cause increased responsiveness in the NAc. However, a substantial body of evidence indicates that hypermethylation leads to a decrease of gene transcription and expression (Nestler, 2014). Moreover, previous research proposes two ventral striatal pathways that modulate reward-related motivation: one initiates behavioral responses, while the other one inhibits them (Frank, 2005). The direct striatonigral pathway consists of medium spiny neurons (MSNs) expressing low-affinity dopamine D1 receptors that are activated by phasic increases in DA levels, while the striatopallidal MSNs express higher affinity dopamine D2 receptors that are activated by temporary reductions in tonic DA levels (Bromberg-Martin, 2010; Frank, 2005; Volkow & Morales, 2015). Studies have demonstrated that stimulation of the direct pathway mediates reward/approach behavior, while inhibition of the indirect pathway mediates the avoidance of aversive stimuli (Hikida, 2010; Kravitz and Kreitzer, 2012; Frank, 2005). Therefore, an alternative interpretation of our finding may be that the prediction of a future loss may lead to a reduction in dopamine neuron activity. This decrease in activity would lead to a suppression of tonic dopamine that would disinhibit the indirect striatal pathway causing an increase in activity in this pathway. Consistent with this interpretation, our data show that decreased methylation is associated with greater NAc activation during the anticipation of monetary losses in HC, likely because of greater DAT expression in the NAc that more effectively reuptakes dopamine, thus disinhibiting the indirect pathway, which may be reflected in the observed increase in NAc activation. However, this effect was not present in individuals with ALC, which may contribute to impairments in avoidance-based learning often observed in this population. Finally, an alternative explanation may be that an existing relationship between SLC6A3 promoter methylation and NAc reactivity was not detected in the ALC group. This may have been due to technical limitations of BOLD responses in fMRI. As mentioned above, the spatial resolution of BOLD signals with respect to substructures of the striatum is poor and the signal to noise ratio is low, which restricts interpretations regarding mechanisms of epigenetic factors involved in group differences in BOLD responses. Therefore, our findings indicate that future neuroimaging studies are needed to further elucidate the relationship between SLC6A3 methylation and NAc activation in ALC populations.
The observed decorrelation of SLC6A3 and NAc activation in individuals with ALC compared to controls is interesting as it provides further evidence for the notion that epigenetic regulation differs between healthy individuals and those with substance use disorders and may contribute significantly to addiction phenotypes (Nestler, 2014). Our finding could be interpreted as a disruption of the epigenetic regulation of DAT expression in individuals with chronic alcohol use, which may contribute to previously reported differences in sensitivity to reward and punishment in ALC populations (for a review, see Balodis and Potenza, 2015). It is possible that chronic alcohol consumption is the cause for this disruption or, alternatively, that the disruption predisposes individuals to the development of ALC. However, epigenetic regulation of gene expression is complex, such that multiple factors besides alcohol consumption, including smoking, stress, and comorbid psychopathology affect methylation, all of which may contribute to the decorrelation of methylation status and NAc activation in individuals with ALC compared to controls. Given the correlational nature of our data, it is not possible to draw conclusions about the direction of this relationship.
While the present study's finding supports the functional relevance of epigenetic variation in the DAT gene for neuronal phenotypes, another imaging epigenetics study failed to show an association between SLC6A3 methylation and NAc reactivity in ALC during an alcohol cue reactivity task, possibly due to a lack of power (Wiers et al., 2015). Future research is needed to further elucidate the relationship between SLC6A3 methylation and NAc activation in alcohol-dependent and healthy populations.
Another important finding of the present study is that SLC6A3 promoter methylation was positively correlated with age, which replicates findings from a prior study (Nieratschker et al., 2014). These findings are consistent with genome-wide studies that have shown CpG island methylation to increase with age (Hernandez et al., 2011). The positive association with age was driven by the ALC group, which further replicates findings by Nieratschker et al. (2014). While these data are correlational and do not allow for any conclusions about cause and effect, they suggest that age-related alterations in methylation may be enhanced by chronic alcohol use.
To date, research on the role of epigenetic regulation of the DAT in ALCs has produced mixed results; however, it should be noted that study designs varied widely from an imaging epigenetics approach (Wiers et al., 2015) to case-control comparisons in clinical studies without imaging components (Jasiewicz et al., 2015, Nieratschker et al., 2014, Hillemacher et al., 2009). Two studies found methylation in the promoter region of SLC6A3 to be significantly higher in alcohol-dependent compared to healthy individuals (Hillemacher et al., 2009, Wiers et al., 2015). In contrast, the present study found no significant difference in SLC6A3 methylation levels between groups which is in accordance with two prior studies (Nieratschker et al., 2014, Jasiewicz et al., 2015). This may be explained by differences in methodology (i.e., study design, methylation quantification, data analytic strategies) or a lack of power. Future studies with larger sample sizes are needed for clarification. Furthermore, Wiers et al. (2015) found a positive association between SLC6A3 promoter methylation and craving while Hillemacher et al. (2009) found a negative association. While these discrepant results may be explained by differences in methodology and outcome measures, an alternative explanation might be the timing of the assessments. Hillemacher et al. (2009) obtained blood samples and craving assessments immediately upon admission to an inpatient detoxification treatment, while Wiers et al. (2015) studied recently detoxified individuals with abstinence averaging 57 and 39 days in their two alcohol-dependent groups. These studies' findings are in line with evidence of fluctuations in DAT availability during alcohol use, withdrawal, and abstinence (e.g., Laine et al., 1999) and underline the importance of careful characterization of study samples and assessment timelines, as well as the need for longitudinal investigations of gene methylation in alcohol-dependent populations.
There are some limitations that should be noted for the interpretation of the present study's findings. Cases and controls were not matched exactly for age and gender. Despite significant efforts to target and recruit young adults with alcohol dependence to conduct meaningful case-control comparisons, the average age of the ALC group remained higher than that of the control group. Therefore, all main analyses corrected for possible confounding effects of age, as well as gender. Additionally, while all participants were free from psychoactive medications on the day of the scan, medication history of participants was unknown and should be considered as a possible confound for the outcome. Since the instructions of the MID task stated that task performance would influence participants' final payment, it is possible that differences in motivation due to differences in socioeconomic status may have affected the outcome. However, comparisons between cases and controls showed that income distributions were equal (χ2(8, 90) = 10.36, p = .241). Therefore, it is unlikely that socioeconomic status confounded our results. While there were no smokers in the control group, approximately 56% of the ALC group were smokers, which is common in patients addicted to alcohol (Li et al, 2007). Nevertheless, the group differences in smoking may have influenced our results. That is, similarly to alcohol, smoking a cigarette increases dopamine release in the midbrain (Brody et al, 2004), and chronic smoking leads to disruptions in the dopaminergic system including lower striatal dopamine D2 receptor availability (e.g., Albrecht et al, 2013; Wiers et al, 2017) and neural responses to the MID task (e.g., Fedota et al, 2015; Nestor et al, 2016). Whether smoking also influences SLC6A3 methylation, however, remains unknown. Our analyses showed no statistically significant differences between smokers and non-smokers on any of the methylation or imaging outcomes; neither within the ALC group nor when the HC non-smokers were included. Future studies are necessary to disentangle the effects of smoking on SLC6A3 methylation and its association with neural responses to the MID task in alcoholism. Further, since DAT gene methylation status in rats was shown to be influenced by binge drinking of parents (Kim et al., 2014), the question remains whether our results were influenced by family history of alcoholism. In our sample, family density score (i.e., the proportion of first and second degree relatives identified as having received treatment for a drinking problem or as having experienced several negative consequences of their drinking), as assessed by the Family Tree Questionnaire (Mann et al., 1985), was significantly greater in ALC (M = .143, SD = .164) than in HC (M = .033, SD = .067), p < .001. However, family density score did not correlate with methylation in Cluster A or in the 3 relevant CpG sites; neither in the whole sample nor in the separate groups (all ps > .05). Future studies may explore associations between DAT methylation and parental drinking patterns; ideally with a 2 (ALC diagnosis) × 2 (binge drinking parent) factorial design. Given the exploratory nature of the analyses due to the fact that the literature on imaging epigenetics studies on the DAT is limited (only one study published so far) (Wiers et al., 2015), alpha levels were not adjusted for multiple comparisons. Furthermore, since it is not possible to measure methylation directly in the brain of living participants, DNA extracted from peripheral blood was used for methylation assessments. While there is growing evidence for the relevance of peripheral methylation in providing information about brain function in clinical populations (Wiers et al., 2015, Nikolova et al., 2014, Frodl et al., 2015), future research comparing peripheral methylation with striatal DAT mRNA to clarify whether peripheral methylation mimics methylation in the brain is recommended. Future investigations may employ longitudinal designs to further elucidate the temporal relationship between SLC6A3 methylation, striatal DAT expression, and striatal fMRI BOLD response differences between individuals with and without alcohol use disorder. Finally, it has been proposed that sample sizes > 62 should afford sufficient statistical power to detect the effect of genetic variation on neuroimaging outcomes (Munafò et al., 2008, Mier et al., 2010) and a prior imaging epigenetic study of the SLC6A3 gene has revealed associations at a sample size of 55 (Wiers et al., 2015), therefore, it may be concluded that the present study was adequately powered.
In summary, the present study provides evidence that peripheral SLC6A3 promoter methylation predicts NAc activation during the anticipation of high and low loss in healthy control but not alcohol-dependent individuals. The epigenetic regulating effect observed in the healthy control group may hence be disrupted after chronic alcohol use. These findings add to a growing body of literature indicating that epigenetic variation may be one of the mechanisms by which chronic alcohol exposure modulates striatal neurocircuitry and thus reward processing. Finally, these findings support the relevance of epigenetic variance in the periphery as biomarkers of neuronal phenotypes.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (NIH) intramural funding ZIA-AA000242 (Section on Clinical Genomics and Experimental Therapeutics; to FWL), Division of Intramural Clinical and Biological Research of the National Institute on Alcohol Abuse and Alcoholism (NIAAA).
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- Albrecht DS, Kareken DA, Yoder KK. Effects of smoking on D2/D3 striatal receptor availability in alcoholics and social drinkers. Brain Imaging Behav. 2013;7:326–334. doi: 10.1007/s11682-013-9233-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balodis IM, Potenza MN. Anticipatory reward processing in addicted populations: a focus on the monetary incentive delay task. Biol Psychiatry. 2015;77:434–444. doi: 10.1016/j.biopsych.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A, Schlagenhauf F, Wüstenberg T, Hein J, Kienast T, Kahnt T, Schmack K, Hägele C, Knutson B, Heinz A. Ventral striatal activation during reward anticipation correlates with impulsivity in alcoholics. Biol Psychiatry. 2009;66:734–742. doi: 10.1016/j.biopsych.2009.04.035. [DOI] [PubMed] [Google Scholar]
- Brody AL, Olmstead RE, London ED, Farahi J, Meyer JH, Grossman P, Lee GS, Huang J, Hahn EL, Mandelkern MA. Smoking-induced ventral striatum dopamine release. Am J Psychiatry. 2004;161:1211–8. doi: 10.1176/appi.ajp.161.7.1211. [DOI] [PubMed] [Google Scholar]
- Bromberg-Martin ES, Matsumoto M, Hikosaka O. Dopamine in motivational control: rewarding, aversive, and alerting. Neuron. 2010;68:815–34. doi: 10.1016/j.neuron.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RM, Macinnes JJ, Huettel SA, Adcock RA. Activation in the VTA and nucleus accumbens increases in anticipation of both gains and losses. Front Behav Neurosci. 2009 doi: 10.3389/neuro.08.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Behavioral Health Statistics and Quality. Key Substance Use and Mental Health Indicators in the United States: Results from the 2015 National Survey on Drug Use and Health. 2016 (HHS Publication No SMA 16-4984, NSDUH Series H-51) Retrieved from http://www.samhsa.gov/data/
- Cox RW. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Dreher JC, Kohn P, Kolachana B, Weinberger DR, Berman KF. Variation in dopamine genes influences responsivity of the human reward system. Proc Natl Acad Sci USA. 2009;106:617–622. doi: 10.1073/pnas.0805517106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Nie Y, Li Y, Wan YJ. The association between the SLC6A3 VNTR 9-repeat allele and alcoholism-a meta-analysis. Alcohol Clin Exp Res. 2011;35:1625–34. doi: 10.1111/j.1530-0277.2011.01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–14. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- Fedota JR, Sutherland MT, Salmeron BJ, Ross TJ, Hong LE, Stein EA. Reward Anticipation Is Differentially Modulated by Varenicline and Nicotine in Smokers. Neuropsychopharmacology. 2015;40:2038–46. doi: 10.1038/npp.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ. Dynamic dopamine modulation in the basal ganglia: a neurocomputational account of cognitive deficits in medicated and nonmedicated Parkinsonism. J Cogn Neurosci. 2005;17:51–72. doi: 10.1162/0898929052880093. [DOI] [PubMed] [Google Scholar]
- Frodl T, Carballedo A, Vaisheva F, Meaney J. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J Psychiatry Neurosci. 2015;40:296. doi: 10.1503/jpn.140180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke S, Suo S, Takahashi N, Koike H, Sasagawa N, Ishiura S. The VNTR polymorphism of the human dopamine transporter (DAT1) gene affects gene expression. Pharmacogenomics J. 2001;1:152–156. doi: 10.1038/sj.tpj.6500026. [DOI] [PubMed] [Google Scholar]
- Giros B, Jaber M, Jones SR, Wightman RM, Caron MG. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature. 1996;379:606–12. doi: 10.1038/379606a0. [DOI] [PubMed] [Google Scholar]
- Gorwood P, Limosin F, Batel P, Hamon M, Ades J, Boni C. The A9 allele of the dopamine transporter gene is associated with delirium tremens and alcohol-withdrawal seizure. Biol Psychiatry. 2003;53:85–92. doi: 10.1016/s0006-3223(02)01440-3. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–20. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Hahn T, Heinzel S, Dresler T, Plichta MM, Renner TJ, Markulin F, Jakob PM, Lesch KP, Fallgatter AJ. Association between reward-related activation in the ventral striatum and trait reward sensitivity is moderated by dopamine transporter genotype. Hum Brain Mapp. 2011;32:1557–65. doi: 10.1002/hbm.21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz A, Goldman D, Gallinat J, Schumann G, Puls I. Pharmacogenetic insights to monoaminergic dysfunction in alcohol dependence. Psychopharmacology (Berl) 2004;174:561–570. doi: 10.1007/s00213-004-1903-x. [DOI] [PubMed] [Google Scholar]
- Heinz A, Goldman D, Jones DW, Palmour R, Hommer D, Gorey JG, Lee KS, Linnoila M, Weinberger DR. Genotype influences in vivo dopamine transporter availability in human striatum. Neuropsychopharmacology. 2000;22:133–139. doi: 10.1016/S0893-133X(99)00099-8. [DOI] [PubMed] [Google Scholar]
- Heinz A, Saunders RC, Kolachana BS, Jones DW, Gorey JG, Bachevalier J, Weinberger DR. Striatal dopamine receptors and transporters in monkeys with neonatal temporal limbic damage. Synapse. 1999;32:71–79. doi: 10.1002/(SICI)1098-2396(199905)32:2<71::AID-SYN1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, Van Der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–72. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikida T, Kimura K, Wada N, Funabiki K, Nakanishi S. Distinct roles of synaptic transmission in direct and indirect striatal pathways to reward and aversive behavior. Neuron. 2010;66:896–907. doi: 10.1016/j.neuron.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J Psychiatr Res. 2009;43:388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Jacobsen LK, Staley JK, Zoghbi SS, Seibyl JP, Kosten TR, Innis RB, Gelernter J. Prediction of dopamine transporter binding availability by genotype: a preliminary report. Am J Psychiatry. 2000;157:1700–1703. doi: 10.1176/appi.ajp.157.10.1700. [DOI] [PubMed] [Google Scholar]
- Jasiewicz A, Rubis B, Samochowiec J, Malecka I, Suchanecka A, Jablonski M, Grzywacz A. DAT1 methylation changes in alcohol-dependent individuals vs. controls. J Psychiatr Res. 2015;64:130–3. doi: 10.1016/j.jpsychires.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, Choi CS, Park JH, Joo SH, Kim SY, Ko HM, Kim KC, Jeon SJ, Park SH, Han SH. Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. J Neurosci Res. 2014;92:658–670. doi: 10.1002/jnr.23275. [DOI] [PubMed] [Google Scholar]
- Knutson B, Westdorp A, Kaiser E, Hommer D. FMRI visualization of brain activity during a monetary incentive delay task. Neuroimage. 2000;12:20–27. doi: 10.1006/nimg.2000.0593. [DOI] [PubMed] [Google Scholar]
- Kochanek KD, Murphy SL, Xu J, Tejada-Vera B. Deaths: final data for 2014. National vital statistics reports: from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2016;65:1. [PubMed] [Google Scholar]
- Kohnke MD, Batra A, Kolb W, Kohnke AM, Lutz U, Schick S, Gaertner I. Association of the dopamine transporter gene with alcoholism. Alcohol Alcohol. 2005;40:339–42. doi: 10.1093/alcalc/agh179. [DOI] [PubMed] [Google Scholar]
- Kravitz AV, Kreitzer AC. Striatal mechanisms underlying movement, reinforcement, and punishment. Physiology. 2012;27:167–177. doi: 10.1152/physiol.00004.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine T, Ahonen A, Torniainen P, Heikkilä J, Pyhtinen J, Räsänen P, Niemelä O, Hillbom M. Dopamine transporters increase in human brain after alcohol withdrawal. Mol Psychiatry. 1999;4:189–191. doi: 10.1038/sj.mp.4000514. [DOI] [PubMed] [Google Scholar]
- Le Strat Y, Ramoz N, Pickering P, Burger V, Boni C, Aubin HJ, Ades J, Batel P, Gorwood P. The 3′ part of the dopamine transporter gene DAT1/SLC6A3 is associated with withdrawal seizures in patients with alcohol dependence. Alcohol Clin Exp Res. 2008;32:27–35. doi: 10.1111/j.1530-0277.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- Lee BY, Park SY, Ryu HM, Shin CY, Ko KN, Han JY, Koren G, Cho YH. Changes in the methylation status of DAT, SERT, and MeCP2 gene promoters in the blood cell in families exposed to alcohol during the periconceptional period. Alcohol Clin Exp Res. 2015;39:239–50. doi: 10.1111/acer.12635. [DOI] [PubMed] [Google Scholar]
- Li TK, Volkow ND, Baler RD, Egli M. The biological bases of nicotine and alcohol co-addiction. Biol Psychiatry. 2007;61:1–3. doi: 10.1016/j.biopsych.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Bloch PJ, Hodge R, Nall AH, Ferraro TN, Kampman KM, Dackis CA, O'brien CP, Pettinati HM, Oslin DW. Association analysis between polymorphisms in the dopamine D2 receptor (DRD2) and dopamine transporter (DAT1) genes with cocaine dependence. Neurosci Lett. 2010;473:87–91. doi: 10.1016/j.neulet.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Ballestar E, Fraga MF, Alaminos M, Setien F, Esteller M. A profile of methyl-CpG binding domain protein occupancy of hypermethylated promoter CpG islands of tumor suppressor genes in human cancer. Cancer Res. 2006;66:8342–6. doi: 10.1158/0008-5472.CAN-06-1932. [DOI] [PubMed] [Google Scholar]
- Ma Y, Fan R, Li MD. Meta-Analysis Reveals Significant Association of the 3′-UTR VNTR in SLC6A3 with Alcohol Dependence. Alcohol Clin Exp Res. 2016;40:1443–53. doi: 10.1111/acer.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RE, Sobell LC, Sobell MB, Pavan D. Reliability of a family tree questionnaire for assessing family history of alcohol problems. Drug Alcohol Depend. 1985;15:61–7. doi: 10.1016/0376-8716(85)90030-4. [DOI] [PubMed] [Google Scholar]
- Martinez D, Gelernter J, Abi-Dargham A, Van Dyck CH, Kegeles L, Innis RB, Laruelle M. The variable number of tandem repeats polymorphism of the dopamine transporter gene is not associated with significant change in dopamine transporter phenotype in humans. Neuropsychopharmacology. 2001;24:553–560. doi: 10.1016/S0893-133X(00)00216-5. [DOI] [PubMed] [Google Scholar]
- Mier D, Kirsch P, Meyer-Lindenberg A. Neural substrates of pleiotropic action of genetic variation in COMT: a meta-analysis. Mol Psychiatry. 2010;15:918–927. doi: 10.1038/mp.2009.36. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Brown SM, Hariri AR. Serotonin transporter (5-HTTLPR) genotype and amygdala activation: a meta-analysis. Biol Psychiatry. 2008;63:852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A, Nestor LJ, Mcgonigle J, Paterson L, Boyapati V, Ersche KD, Flechais R, Kuchibatla S, Metastasio A, Orban C, Passetti F, Reed L, Smith D, Suckling J, Taylor E, Robbins TW, Lingford-Hughes A, Nutt DJ, Deakin JF, Elliott R. Acute D3 Antagonist GSK598809 Selectively Enhances Neural Response During Monetary Reward Anticipation in Drug and Alcohol Dependence. Neuropsychopharmacology. 2017;42:1049–1057. doi: 10.1038/npp.2016.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76:259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor LJ, Mccabe E, Jones J, Clancy L, Garavan H. Smokers and ex-smokers have shared differences in the neural substrates for potential monetary gains and losses. Addict Biol. 2016 doi: 10.1111/adb.12484. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Grosshans M, Frank J, Strohmaier J, Von Der Goltz C, El-Maarri O, Witt SH, Cichon S, Nothen MM, Kiefer F, Rietschel M. Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addict Biol. 2014;19:305–11. doi: 10.1111/j.1369-1600.2012.00459.x. [DOI] [PubMed] [Google Scholar]
- Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, Williamson DE, Hariri AR. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci. 2014;17:1153–1155. doi: 10.1038/nn.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MJ, Chan J, Pohorille A, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. The dopamine transporter: comparative ultrastructure of dopaminergic axons in limbic and motor compartments of the nucleus accumbens. J Neurosci. 1997;17:6899–907. doi: 10.1523/JNEUROSCI.17-18-06899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray LA, Bujarski S, Squeglia LM, Ashenhurst JR, Anton RF. Interactive effects of OPRM1 and DAT1 genetic variation on subjective responses to alcohol. Alcohol Alcohol. 2014;49:261–70. doi: 10.1093/alcalc/agt183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HW, Hagino Y, Kobayashi H, Shinohara-Tanaka K, Ikeda K, Yamamoto H, Yamamoto T, Lesch KP, Murphy DL, Hall FS, Uhl GR, Sora I. Regional differences in extracellular dopamine and serotonin assessed by in vivo microdialysis in mice lacking dopamine and/or serotonin transporters. Neuropsychopharmacology. 2004;29:1790–9. doi: 10.1038/sj.npp.1300476. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain : 3-dimensional proportional system : an approach to cerebral imaging. Stuttgart ; New York, G Thieme ; New York: Thieme Medical Publishers; 1988. [Google Scholar]
- Tawa EA, Hall SD, Lohoff FW. Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol. 2016;51:507–14. doi: 10.1093/alcalc/agw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holst RJ, Clark L, Veltman DJ, Van Den Brink W, Goudriaan AE. Enhanced striatal responses during expectancy coding in alcohol dependence. Drug Alcohol Depend. 2014;142:204–8. doi: 10.1016/j.drugalcdep.2014.06.019. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Morales M. The Brain on Drugs: From Reward to Addiction. Cell. 2015;162:712–25. doi: 10.1016/j.cell.2015.07.046. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Swanson JM, Telang F. Dopamine in drug abuse and addiction: results of imaging studies and treatment implications. Arch Neurol. 2007;64:1575–1579. doi: 10.1001/archneur.64.11.1575. [DOI] [PubMed] [Google Scholar]
- Wernicke C, Smolka M, Gallinat J, Winterer G, Schmidt LG, Rommelspacher H. Evidence for the importance of the human dopamine transporter gene for withdrawal symptomatology of alcoholics in a German population. Neurosci Lett. 2002;333:45–8. doi: 10.1016/s0304-3940(02)00985-0. [DOI] [PubMed] [Google Scholar]
- Wiers CE, Cabrera EA, Tomasi D, Wong CT, Demiral SB, Kim SW, Wang GJ, Volkow ND. Striatal Dopamine D2/D3 Receptor Availability Varies Across Smoking Status. Neuropsychopharmacology. 2017 doi: 10.1038/npp.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiers C, Shumay E, Volkow N, Frieling H, Kotsiari A, Lindenmeyer J, Walter H, Bermpohl F. Effects of depressive symptoms and peripheral DAT methylation on neural reactivity to alcohol cues in alcoholism. Transl Psychiatry. 2015;5:e648. doi: 10.1038/tp.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrase J, Schlagenhauf F, Kienast T, Wüstenberg T, Bermpohl F, Kahnt T, Beck A, Ströhle A, Juckel G, Knutson B. Dysfunction of reward processing correlates with alcohol craving in detoxified alcoholics. Neuroimage. 2007;35:787–794. doi: 10.1016/j.neuroimage.2006.11.043. [DOI] [PubMed] [Google Scholar]
- Xu M, Lin Z. Genetic influences of dopamine transport gene on alcohol dependence: a pooled analysis of 13 studies with 2483 cases and 1753 controls. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1255–60. doi: 10.1016/j.pnpbp.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


