SUMMARY
The heavily glycosylated native-like envelope (Env) trimer of HIV-1 is expected to have low immunogenicity, whereas misfolded forms are often highly immunogenic. High-quality correctly folded Envs may therefore be critical for developing a vaccine that induces broadly neutralizing antibodies. Moreover, the high variability of Env may require immunizations with multiple Envs. Here, we report a universal strategy that provides for correctly folded Env trimers of high quality and yield through a repair-and-stabilize approach. In the repair stage, we utilized a consensus strategy that substituted rare strain-specific residues with more prevalent ones. The stabilization stage involved structure-based design and experimental assessment confirmed by crystallographic feedback. Regions important for the refolding of Env were targeted for stabilization. Notably, the α9-helix and an intersubunit β sheet proved to be critical for trimer stability. Our approach provides a means to produce prefusion-closed Env trimers from diverse HIV-1 strains, a substantial advance for vaccine development.
Graphical Abstract
In Brief: Rutten et al. describe a universal repair and stabilize approach that corrects rare mutations and stabilizes refolding regions to obtain high-quality HIV Envs with high yields. The crystal structure shows how the optimization of the trimer interface between α9, α6, and the intersubunit β-sheet stabilizes the membrane-proximal base.
INTRODUCTION
The mature HIV-1 envelope (Env) spike is a trimer of gp120 and gp41 heterodimeric subunits. The gp120 subunit initiates fusion by binding to CD4 receptors and CCR5/CXCR4 coreceptors and transmitting this binding through conformational changes to the transmembrane gp41 subunit. The HIV-1 fusion machinery is sequestered in the prefusion conformation of gp41, which refolds into a postfusion conformation (Melikyan et al., 2000). In the prefusion conformation, Env trimers spontaneously sample distinct conformational states (Munro et al., 2014). Ligands can alter the proportion of molecules in these conformational states, with broadly neutralizing antibodies generally stabilizing a closed state and with CD4 stabilizing an open state (Liu et al., 2008; Munro et al., 2014; Ozorowski et al., 2017; Wang et al., 2016). The relative proportion of these prefusion-conformational states is strain-dependent (Munro et al., 2014), with typical tier 1 (neutralization sensitive) strains adopting a higher proportion of the open state compared to a typical tier 2 (neutralization resistant) virus (Guttman et al., 2014; Liu et al., 2008; Munro et al., 2014; Ozorowski et al., 2017).
Trimer instability has been a substantial barrier to the development of an HIV vaccine that aims to induce broadly neutralizing antibodies (Feng et al., 2016; Guttman et al., 2015; Kwon et al., 2015; Sanders et al., 2013, 2016). Moreover, low expression levels decrease prospects for successful protein subunit development or for the application of a vector based approach. Therefore, stable Env trimers are needed that fold efficiently into a prefusion-closed state and that fully arrest conformational changes to more open states. The introduction of the so-called SOSIP substitutions was an important step to obtain native trimers (Binley et al., 2000; Sanders et al., 2002), but the fraction of prefusion-closed trimer remains less than optimal (Julien et al., 2015) and/or the resultant trimers open in the presence of CD4 (de Taeye et al., 2015). Although elucidation of the trimer structure facilitated substantial progress in recent years to stabilize the conformation of Env based on disfavoring the postfusion conformation and to prevent breathing by introducing disulfide bridges, optimizing packing, utilizing stabilizing substitutions known from other Envs (Binley et al., 2000; Chuang et al., 2017; de Taeye et al., 2015; Dey et al., 2009; Garces et al., 2015; Guenaga et al., 2015, 2017; Julien et al., 2013; Kesavardhana and Varadarajan, 2014; Kong et al., 2016; Kulp et al., 2017; Kwon et al., 2015; Lyumkis et al., 2013; Pancera et al., 2014; Sanders et al., 2002; Steichen et al., 2016; Stewart-Jones et al., 2016; Sullivan et al., 2017; Torrents de la Peña et al., 2017) or by using of a chimeric strategy (Joyce et al., 2017), a general approach to produce prefusion-closed Env trimers of high quality and yield, universally applicable to most HIV-1 strains, would be an important advancement for the field.
As sequential immunizations with heterologous stable high-quality Envs may be needed to induce broadly neutralizing antibodies, we sought a universal approach to deliver high expression of diverse high-quality prefusion-closed trimers. We hypothesized that (1) a consensus or mosaic sequence would provide a superior starting template for universal adaptation, and (2) the stabilization of specific subunit and protomer interfaces that move between closed and open conformations would limit breathing and prevent irreversible refolding. We showed our two-pronged “repair and stabilize” procedure to yield prefusion-closed HIV-1 Env trimers with high expression, stability, and desired antigenicity for Envs—for which SOSIP mutations yielded only unstable, low expressing Envs with poor antigenicity.
RESULTS
Selection of Clade C Env
Because HIV-1 clade C comprises the most common circulating isolates, we evaluated expression and folding of a consensus clade C-based Env stabilized with SOSIP mutations.
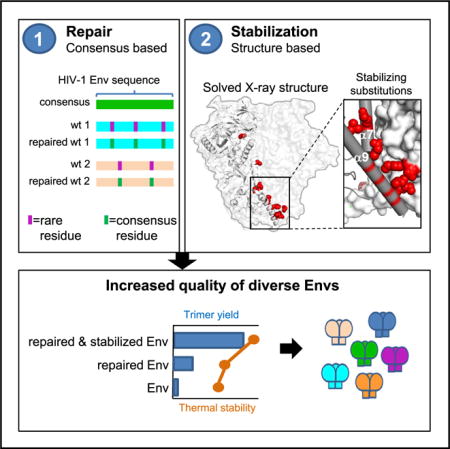
This so-called ConC_SOSIP Env showed higher trimer content compared to the closest matched wild-type (WT) Envs in the database (Figure 1A) suggesting a superior folding for ConC_SOSIP. Similarly, the mosaic clade C Env (DS_sC4_SOSIP), based on a compilation of Env sequences with a broad coverage of T cell epitopes (Fischer et al., 2007), showed higher trimer content compared to its closest wild-type sequence matches (Figure 1B). The mosaic Env did not bind to the trimer-specific apex-binding antibodies PGT145 and PGDM1400 (Sok et al., 2014), whereas ConC_SOSIP showed slightly weaker PGDM1400 but otherwise comparable binding to these monoclonal antibodies (mAbs) relative to that of BG505_SOSIP (data not shown). Furthermore, ConC_SOSIP was almost fully cleaved (Figure 1C) and formed homogeneous trimeric population as assessed by negatively stained electron microscopy (Figure 1D). Therefore, the consensus C-based Env was selected as a template for further stabilization.
Figure 1. HIV-1 Env Selection for Stabilization.
(A and B) SEC profiles of GN lectin-purified HIV Env SOSIPs of (A) clade C consensus (ConC_SOSIP) and closest wild-type Env sequences indicated with GenBank numbers. The trimer peak is indicated with an asterisk, gp140 monomer peak is indicated with a degree symbol, gp120 monomer is indicated with a diamond, and aggregates are indicated with an at symbol. (B) clade C mosaic SOSIP 201C-433C (DS_sC4_SOSIP) and closest wild-type Env sequences indicated with the GenBank numbers. The trimer peak is indicated with an asterisk and gp140 monomer peak with a degree symbol.
(C) Analysis of GN lectin-purified ConC_SOSIP on SDS-PAGE under reducing (+DTT) and non-reducing (−DTT) conditions compared with markers (M).
(D) Negatively stained EM of ConC_SOSIP trimers with 2D-averaged classes (right).
Design Considerations for Stabilizing Substitutions
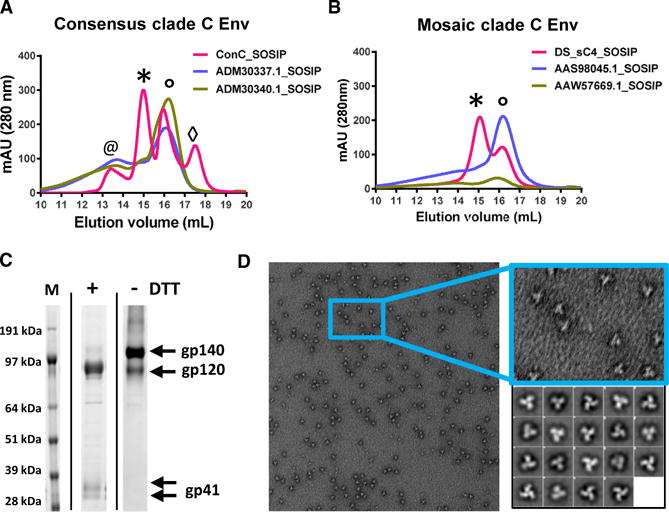
Knowledge of the correct folding of Env and a detailed understanding of the interactions that underlie the refolding mechanism can provide guidance for design of prefusion-closed Env trimers. To stabilize the Env trimer, we took into consideration the overall fusion process in which Env shows reversible breathing to different states. At a critical point in the successive conformational changes, a tipping point is reached in which folding to the postfusion state is irreversible. In refolding region 1 of gp41 (Figure 2A), the loop between prefusion α6–α7 helices refolds into a single helix, which is assembled on the base helix α7 together with α6 and the N-terminal fusion peptide making one long extended trimeric helical coiled coil with the other protomers of the trimer. Subsequently, additional gp41:gp120 interactions are lost and helices α8 and α9 refold and dock against the coiled coil region to complete the six-helix bundle of the post-fusion gp41.
Figure 2. Single Point Mutations that Enhance Yield and Stability of ConC SOSIP Trimers.
(A) Schematic definition of regions of HIV-1 Env that move more than 5 Å between prefusion closed and CD4-bound forms are shown blue for gp120, green for gp41, and black if not defined in either conformation. Residues 31–511 form the mature gp120. Refolding region 1 encompasses residues 512–570. The base helix is residues 571–595. Refolding region 2 encompasses residues 596–664. Previously defined HR1 (residues 540–590) and HR2 (residues 624–664) are also indicated.
(B) Location of structure-based mutation in the prefusion closed Env trimer. Two protomers are shown in surface representation (pink and peach) and the third protomer is shown in ribbon representation, with residues that move more than 5 Å between prefusion-closed and CD4-bound conformations in blue (for gp120) or in green (for gp41) and otherwise gray or black (if not defined in either conformation).
(C) Comparison of expression level plotted against PGT145 binding for single point mutation variants of ConC_SOSIP Env using AlphaLISA in cell culture supernatant 3 days after transfection. Substitutions with highest increase in trimer yield and trimer fraction are labeled. Substitutions at the same positions have identical color and largest dot was selected for further evaluation.
(D) SEC profiles using crude cell culture supernatants 3 days after transfection with the best single point mutation variants compared with backbone ConC_SOSIP (dotted gray line) (colored as in C). The trimer peak is indicated with an asterisk and gp140 monomer peak is indicated with a degree symbol.
See also Table S1.
For a further understanding of the relation of Env instability and viral fusion kinetics, directed evolution for increased (Leaman and Zwick, 2013) or decreased stability allows for the identification of essential contact regions that modulate irreversible refolding; K574R and I535M are associated with increased Env stability in different clades (Dey et al., 2008; Leaman and Zwick, 2013) and Q590E and K601E are predicted to accelerate refolding (Eggink et al., 2009). Here, we observed that mutations Q590E and K601E did indeed decrease Env stability (Figure S1A). Residue Q590 in the base of the α7 helix is directed toward the trimer axis between three α7 chains; if mutated to a Glu, it likely causes charge repulsion between trimer protomers. In most published crystal structures, no side chain density is visible for residue 601, but in PDB: 5FYJ (Stewart-Jones et al., 2016) clear side chain density is visible for R601, which makes ionic interactions with E654 and E657 in α9 of a neighboring protomer. The large reduction in trimer formation upon replacing K601 with Glu is likely due to charge repulsion and concomitant loss of interaction with the α9 helix. Therefore, the interaction of α9 with the remainder of the protein appears to be of importance to the integrity of the entire trimer ectodomain.
Regions of interest for stabilizing the prefusion-closed conformation of the Env ectodomain trimer appeared to be the α9-helix (particularly the interactions made by α9 to α6 and to β27) and the α7-helix (particularly the interactions between α7 and α7 on neighboring protomer and between α7 and gp120). Because interprotomer and gp41-gp120 contacts at the membrane-proximal base of the trimer were expected to be essential for stability, a component of our design efforts focused on stabilizing the α9 trimer interface with the intersubunit sheet composed of gp41 and gp120 . Additional design efforts focused on other regions including (1) the disordered hinge loop (α6 α7 loop) in refolding region 1, (2) other parts of the trimer interface, and (3) between layers 2 and 3 of the inner domain of gp120 (Figure 2B).
Production and Screening of Stabilized Variants
Based on considerations of the fusion process and the prefusion-closed conformation of Env detailed above, a panel of ~150 ConC_SOSIP variants with single amino acid substitutions were designed and synthesized (Table S1). Substitutions were introduced at positions of conserved residues at the trimer and gp120/gp41 interfaces that are (1) buried hydrophilic residues without salt bridges or hydrogen bonds, (2) exposed hydrophobic residues, and (3) involved in charge repulsions. Furthermore, hydrophobic packing was optimized at the trimer and gp120/gp41 interface and at the layer 2-layer 3 interface (Pancera et al., 2010).
The panel of single amino acid substitutions was screened for increased expression and trimer content by AlphaLISA using a panel of type-specific and non-neutralizing Abs and a panel of broadly neutralizing antibodies (bNAbs) including the trimer-specific PGT145 to measure the formation of native-like prefusion-closed trimers (Figure 2C), and the performance was compared to several previously published point mutations (Table S1). 25 AlphaLISA hits were selected for expression in 30 mL scale cell culture and purified with lectin and analyzed with SEC-MALS of which top 11 with highest expression and PGT145 binding are shown in Figure 2D. Strongest increase in PGT145 binding and trimer content based on SEC-MALS was observed for substitutions K658V, L556P, E647F, N651F, K655I, I535N, D589V, I573F, and K588E/Q in gp41 and A204I in gp120.
Combinations of Stabilizing Substitutions
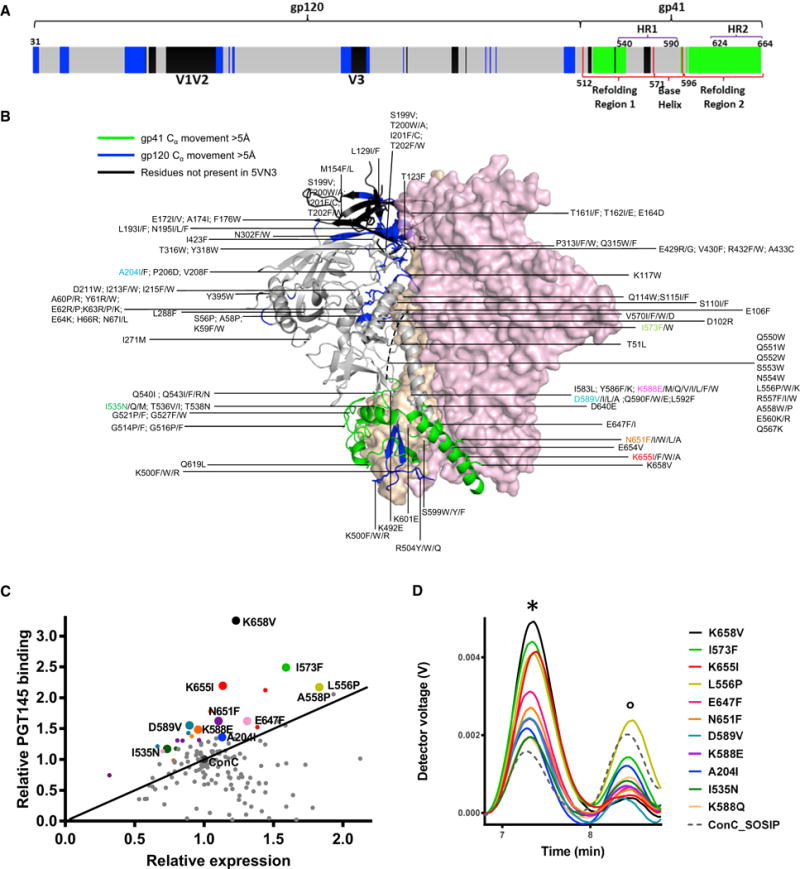
Next, stabilizing substitutions in Env were combined to determine additive effect on stability, antigenicity, and expression. All combinations of substitutions resulted in variants with higher trimer content and higher trimer yield than any of the corresponding single substitutions based on SEC-MALS (Figure 3A). All multiple substitution variants have higher melting temperatures than the ConC_SOSIP backbone and most single cumulative substitutions increase melting temperatures. The addition of both D589V (data not shown) and I573F resulted in melting curves that display two separate melting events, labeled Tm1 and Tm2 (Figures 3A and 3B).
Figure 3. Combined Effect of Cumulative Substitutions.
(A) SEC-MALS analysis (left) and temperature stability as measured by DSC (right) of Env variants with cumulative combination of amino acid substitutions in ConC_SOSIP. The trimer peak is indicated with an asterisk. For constructs with two melting event, a Tm1 (minor) and Tm2 (main) is indicated. Construct number 10 (ConC_base0) has a Glu instead of Gln at position 588.
(B) DSC pattern of ConC_base0.
(C) 17b binding in the presence (solid lines) and absence of CD4 (dotted lines).
(D) Antibody binding levels measured with Octet after 0 and 4 weeks incubation at 37°C of ConC_base0.
(E) SEC-MALS of purified ConC_base0 with SEC (purple) and molar mass trace.
See also Figure S1B.
Because L556P increased the trimer percentage only slightly and because E647F hampered PGT151 binding, a variant was produced with just 7 stabilizing substitutions (termed ConC_base0), which was slightly more stable than the variant with 9 substitutions (Figure 3A, #10). ConC_base0 trimer showed greatly reduced CD4 induction as assessed by binding of the CD4-induced antibody 17b (Figure 3C), remained stable at 37°C for at least 4 weeks based on antigenic evaluation (Figure 3D), and was measured to be ~320 kDa (Figure 3E). The removal of SOS from ConC_base0 hardly affected its trimer yield, suggesting its 7 mutations to stabilize the base region of the Env timer even in the absence of the SOS disulfide (Figure S1B). Back mutation of P559I in the α6–α7 hinge loop did, however, reduce trimer yield, and a proline at position 556 or 558 was able to compensate for the instability in refolding region 1 (Figure S1B).
Crystal Structure of ConC_base0 Variant
To provide a structural basis for the introduced mutations, we determined the crystal structure of the ConC_base0 variant (Figure 4). The structure determination of this variant was facilitated by use of antibodies PGT122 and 35O22, which we modified to enhance crystallization. Data from a single crystal of ConC_base0 in complex with 3H+109L (a PGT122 family member) and a 35O22 variant diffracted anisotropically (to 2.7 Å–3.8 Å resolution), with a nominal resolution of 3.5 Å resolution (Table S2). The resulting electron density allowed for relatively precise positioning of side chains in most regions in the structure.
Figure 4. Crystal Structure of ConC_base0.
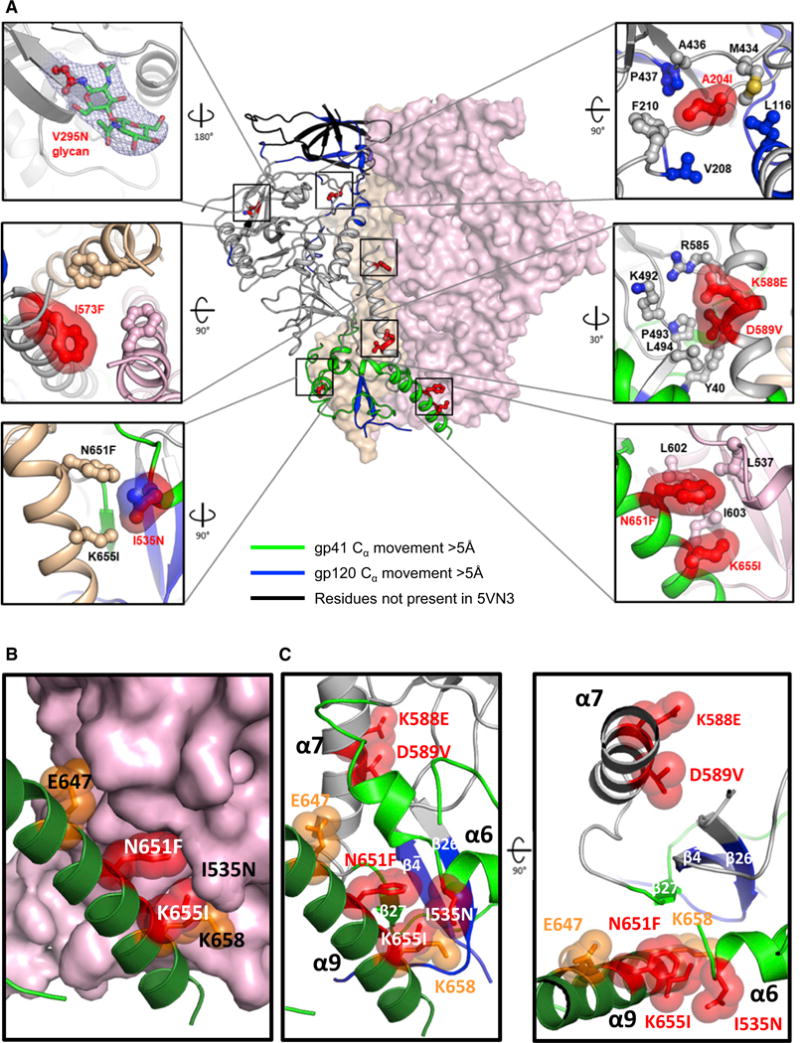
(A) Mutations in ConC_base0 are shown in red ball-and-stick with molecular surface. gp120 and gp41 are shown in ribbon for one protomer with the color scheme shown in Figure 2B. The other two protomers are shown in surface representation, colored peach and pink. Residues on gp120 and gp41 potentially interacting with the stabilizing mutations are shown in ball-and-stick without molecular surface.
(B) Detail of trimer base showing the functionally critical α-9 helix in dark green. Interaction of stabilizing residues 651F and 655I at the trimer interface (red). Residue E647 and K658 that when substituted to hydrophobic residues are also stabilizing (Figure 2) are indicated in orange.
(C) Both protomers shown as ribbons showing the functionally critical α-9 helix in dark green of one protomer that interacts with α6 and intersubunit β sheet of the other protomer (colored as in A) Clustering of stabilizing residues indicated on both sides of the intersubunit β sheet.
See also Figures S2–S4 and Table S2.
The overall structure of ConC_base0 was highly similar to the clade A BG505 structure (Garces et al., 2015) (0.78 Å between 547 Cα atoms) (Figure S2A) despite 24.9% of its surface residues being different in sequence (Figure S2B) and was also similar to a recently published 3.9 Å structure of stabilized clade C 16055 Env (0.76 Å between 462 Cα atoms) (Guenaga et al., 2017). By inspecting the ConC_base0 crystal structure, we observed clear density for all 7 stabilizing mutations as well as the introduced glycan at V295N (Figure S3). The local interactions verified the stabilizing effect of these mutations. For example, F651 and I655 located in α9 strengthened the trimer interface by interacting with L537, I603, and L602 of a neighboring gp41 (Figure 4B). N535, located in α6 on the other side of the α6/α9 trimer interface, was exposed so a hydrophilic residue was likely more favorable. Although α7 could be considered as the static base helix on top of which the refolding region 1 will mount after the large conformational change to the pre-hairpin structure, the I573F mutation at the N-terminal top of α7, close to the trimeric axis, stabilized the trimer through increased hydrophobic interaction. In addition to the stabilization between three protomers, stabilization between gp120 and gp41 within one protomer was also observed. Residue K588, D589, R585, E490, and K492 formed a cluster of charged residues in the gp120-gp41 interface that was balanced in the metastable structure but could harbor potential repulsions as a result of small local movements. The positive charge at position 588 could be partly compensated by D589, but in most crystal structures, the charged D589 is partly buried in the hydrophobic interface of gp120-gp41 (Lee et al., 2016; Pancera et al., 2014; Stewart-Jones et al., 2016). The D589V mutation on gp41 has hydrophobic interactions with Y40, P493, and L494 on gp120 while the K588E has electrostatic interactions with K46 and K492 on gp120 as well as R585 on gp41. Interestingly, R585H was recently shown to stabilize the BG505 trimer (Kulp et al., 2017; Steichen et al., 2016). From the ConC_base0 crystal structure, we calculated the difference in energy for each of the stabilizing mutants (Figure S4A) as well as the difference in energy for the mutation in the CD4-bound prefusion structure (Figure S4B). In the prefusion closed structure, the most stabilizing mutation appeared to be from the hydrophobic interactions gained by N651F. In addition, A204I, I535N, I573F, and D589V also showed favorable ΔΔG. From the ΔΔG calculations of the same mutations using the CD4-bound prefusion Env structure (PDB: 5VN3), I535N, I573F, and K588E appeared to destabilize the CD4-bound prefusion conformation, which may explain why K588E had a beneficial effect on formation of a prefusion-closed Env trimer (Figure S4C). In general, the energetic analysis was consistent with the observed stabilization for all mutants except K655I.
Overall, the crystal structure of ConC_base0 verified inter-protomer and intra-protomer stabilization to have synergistic stabilization of the combination variant (Figure 4C).
General Application of the Repair-and-Stabilize Approach for HIV-1 Env
To investigate the universality of our approach, the effect of the stabilizing substitutions was assessed with other Env sequences. Because wild-type sequences from viruses isolated from infected patients may have acquired destabilizing mutations that impede correct folding, wild-type Env sequences of several clade C strains and the mosaic sC4 were first repaired by substituting each rare mutation with the corresponding consensus amino acid according to the conceptual framework described in Figure 5A. Among the selected Envs were typical strains known to express and fold properly (DU422), Envs known for their low expression and quality like ZM233M and CN97001(Julien et al., 2015), and the low quality Env ADM30337.1 that was described in Figure 1A. Next, stabilizing substitutions described in Figure 3 were transferred to the repaired sequence (Table S3).
Figure 5. Prefusion Closed HIV Env_SOSIP Trimers Through Sequence Repair and Mutational Stabilization.

(A) Universal concept for repairing HIV-1 Env sequence illustrated for strain C97ZA. Occurrence of highest consensus residue in the total HIV-1 database (top bars) and strain C97ZA residue (bottom bars) sorted from low to high occurrence percentage of C97ZA residue position. C97ZA sequence positions to be substituted to consensus were selected based on the following criteria: positions with a C97ZA residue that occurs <2% in consensus Env sequence (blue bars); positions with a C97ZA residue that occur between 2% and 7.5% in consensus Env and are buried or partly buried (magenta bars); positions that are exposed and hydrophobic in C97ZA and hydrophilic in consensus (yellow bars); and a position that is a PNGS in the consensus (green bar).
(B) AlphaLISA signals in cell culture supernatant for all SOSIP variants normalized to the ConC_SOSIP for broadly neutralizing (left) and non-neutralizing or strain-specific (right) mAbs. For the non-bNAbs the intensities are further normalized for expression levels.
(C) Analytical SEC profile of control Env_SOSIPs (red line) repaired according to the concept described in a (blue line) and additional stabilizing substitutions according to Table S3 (green line). Analysis was performed on crude cell culture supernatant instead of purified protein. The trimer peaks are indicated with asterisks.
(D) Repaired and stabilized (REP & STAB) C97ZA_SOSIP compared with CZA97_SOSIP.4.2-M6.IT-2 (Ringe et al., 2017) according to the approach previously described. The upper panel shows the analytical SEC pattern on crude supernatant and the lower panel shows the AlphaLISA signals normalized to those of ConC_SOSIP.
See also Figures S5 and S6 and Table S3–S5.
Supernatants of cells transiently transfected with wild-type, repaired, and stabilized Env variants were tested for binding to several trimer-specific neutralizing antibodies directed to the apex as well as several non-neutralizing antibodies. The repair substitutions, and especially the stabilizing substitutions, had a dramatic impact on trimer content (Figures 5B and 5C). Moreover, the antigenicity of the trimers was improved as shown by a reduction of binding to non-neutralizing antibodies and melting temperatures increased (Figures 5B and S5; Table S4). Similarly, the single substitutions D589V, N651F, K655I, and I535N increased trimer yield and content in a consensus clade B SOSIP (Figure S6).
The yields of repaired and stabilized C97ZA_SOSIP was more than 10 mg/L after transient transfection in Expi293F culture, which is higher compared with the optimized C97ZA Env previously described (Ringe et al., 2017). Because cell type, signal peptide, and the residue at position 152 are different for both proteins, we compared the expression levels of both optimized proteins with identical signal peptide based on ConC (Table S3) and Lys at position 152 (Table S5). Our repaired and stabilized C97ZA_SOSIP yielded approximately four times more native-like closed trimers (Figure 5D).
DISCUSSION
HIV-1 Env is highly variable in sequence and conformation, which are two major hurdles that may need to be addressed to obtain an efficacious vaccine that induces bNAbs against HIV. Human antibodies preferentially recognize immunogenic non-glycosylated Env regions, such as those exposed on open conformations; even minor imperfections in Env folding may hamper the induction of the desired glycan shield-penetrating antibodies. A prerequisite for an effective HIV vaccine that focuses on induction of broadly neutralizing antibodies may thus be high expression of multiple closed trimers with high-quality folding and stability.
In this study, improved quality and stability of the native trimer were pursued by repairing folding through a consensus approach and stabilizing trimer in the prefusion-closed conformation through a structure-based screening approach that optimized regions critical to the folding process. In specific, we hypothesized that a consensus sequence might possess characteristics of founder virus Envs, which have higher fitness and more optimal folding than chronic virus Envs (Carlson et al., 2014; Tully et al., 2016). This study showed that a consensus clade C sequence and repair of selected accumulated mutations of wild-type to consensus, yields superior expression, trimer formation, and antigenicity compared with the closest wild-type Env sequences. Because the prefusion spike is a highly dynamic and metastable fusion machine, the repaired sequence was further stabilized by modifications that arrest conformational changes needed for the fusion process. We therefore systematically searched for conserved weak interactions between the protomer interface and subunit interface based on the known X-ray and electron microscopy (EM) structures (Lee et al., 2016; Pancera et al., 2014; Stewart-Jones et al., 2016; Wang et al., 2016).
Although several regions were identified in this study that stabilized the closed prefusion conformation of the trimer, several of the most successful stabilizing substitutions were located at the membrane-proximal base of the trimer, in the lower part of the fusion subunit gp41 where all the essential contacts between gp120 termini and gp41 refolding region 1 and 2 are located. Several stabilizing substitutions (i.e., E647F, N651F, K655I, and D658V) in α9 improved the interaction at the trimer interface with α6 and β27, locking refolding region 1 and 2 and preventing the dissociation of α9 with α6 and β27 (Figures 4B and 4C). Stabilization of the closed conformation was supported by the observation that CD4 induction leading to 17b binding was reduced in the stabilized ConC_base0 (Figure 3C). β27 is part of refolding region 2 and is the only structural element that is part of a secondary structure that also involves gp120. This intersubunit gp120/gp41 β sheet connects gp41 with the N and C terminus of gp120 and stabilizes the prefusion conformation of refolding region 2, preventing it from unfolding. This intersubunit β sheet has a pivotal role in Env stability, and we found stabilizing substitutions on both sides of the β sheet that keep β27 in place: the aforementioned stabilizing substitutions in α9 on one side and stabilizing substitutions in the highly charged region on the other side. Substitutions 588E and 589V improved the interaction of gp41 with the gp120 termini, stabilizing Y40 ensuring the intersubunit sheet to not be disrupted (Figures 4A and 4C). The stabilizing substitutions we identified in the base are in line with the structure of the CD4-bound prefusion state of Env (Ozorowski et al., 2017; Wang et al., 2016), which shows that both the apex and the base are able to breathe and that, when the gp120 termini and the α6-β27 loop are pulled upward, the interaction between α9 and β27 and interactions in the charged network are lost. Therefore, substitutions may prevent breathing of the base that may also reduce breathing of the apex.
The strong effect on increased prefusion trimer yields by substitutions at the base indicate an important step in the refolding process to be regulated at the trimer base. We envision extreme motions of gp120 to provide freedom for the loop between α6 and α7 to hinge: when β27 contacts are lost, the gp120 N and C termini would be pulled from the gp41 clamp, and refolding regions 1 and 2 would be liberated. Because the hinge loop preceding the base helix is a common element of refolding region 1 in all type I fusion proteins, it had previously been suggested that stabilizing the hinge loop by introduction of prolines may be a general solution to stabilize this metastable element (Krarup et al., 2015), which also has been successfully applied to other type I fusion proteins (Hastie et al., 2017; Pallesen et al., 2017). The current study thus draws attention to a conserved structural element essential for the stability of refolding region 2, which comprises the intersubunit β sheet composed of β strands of the head and the fusion domain. Detachment or “splaying” of the head of the type I fusion proteins is expected ultimately to disrupt the intersubunit sheet and to destabilize contacts with refolding region 2 (Figure 6). Therefore, stabilizing the intersubunit sheet may be a general strategy for conformationally fixing the prefusion state of other metastable type I fusion proteins, a potentially important step in their use as vaccine immunogens.
Figure 6. Intersubunit Sheets of Head Domain and Fusion Domain in Type I Fusion Proteins.
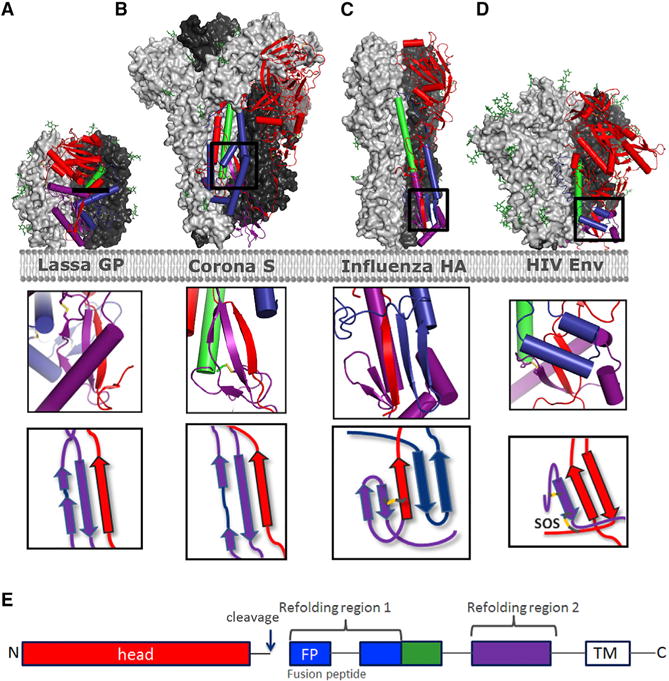
(A–D) Prefusion trimer structures are shown with 2 protomers as light and dark gray surface representations and one protomer in ribbon with helices represented as cylinders for Lassa GP (A) (Hastie et al., 2017), MERS S (B) (Pallesen et al., 2017), Influenza HA (C), and HIV Env (6CK9) (D). Color coding of structural elements in the fusion protein is based on refolding region 1 (blue), base helix (green), refolding region 2 (purple), and head domain (red) according to (E), with inserts depicting the intersubunit sheet composed of part of the head region (red) and refolding region 2 or both refolding regions. Detachment or splaying of the head is expected to destabilize the hybrid sheet and to effect refolding.
(E) Color scheme.
In summary, we describe a “repair-and-stabilize” approach— that could be generally applicable to all HIV-1 clades—to obtain high levels of stable prefusion-closed HIV Env trimers. Our approach utilized optimization of prefusion-closed folding based on the Env fitness expected for a consensus sequence and stabilization based on the arrest of refolding events at the base of the trimer critical to the fusion process.
EXPERIMENTAL PROCEDURES
Design of HIV Envelope Antigen Sequences
C4 is a mosaic sequence we previously created during research toward synthetic Env antigens suitable for expression in adenoviral vectors (Langedijk, 2017). For the consensus C (based on clade C) an alignment was downloaded (HIV Sequence Alignments. 3,434 sequences in total 7DEC14) from the Los Alamos database (https://www.hiv.lanl.gov/content/index). From this alignment, we selected C-clade ENVs (1,252 sequences), and these were used in the consensus maker in the Los Alamos HIV database website. Because the consensus contains a few question marks, the consensus sequence was used to Blast the closest wild-type homologs, which was then also used to substitute the amino acids that have a question mark in the consensus sequence. The resulting sequence was designated ConC (Langedijk, 2017). Furthermore, two wild-type sequences with the highest homology to C4 and two wild-type sequences with the highest homology to ConC were identified with BLAST. The two sequences with the highest homology to C4 are the sequences with GenBank: AAS98045.1 and AAW57669.1. They are 93% and 84% identical to C4, respectively. The two sequences with the highest homology to ConC are the sequences with GenBank: ADM30337.1 and ADM30340.1, both having 90% sequence identity to ConC. The sequences were further modified by addition of the so-called SOSIP substitutions, optimization of the furin site by replacing the furin site (positions 508–511) by 6 Arg residues (R6), a C-terminal truncation of gp160 at residue 664, and a V295N mutation to introduce a PNGS that is present in more than 50% of all HIV-1 stains, resulting in soluble gp140 proteins with the SOSIP.R6.664.V295N mutations (Table S5 for the sequences). The DS_sC4_SOSIP was, however, truncated after 655 and contained the 201C-433C substitution.
Repair of Wild-Type Clade C Sequences
To search for non-optimal mutations in wild-type sequences an alignment of all HIV-1 Env sequences in the UniProt database and the Los Alamos database HIV database (~90,000 sequences) was made, and the amino acid distribution was calculated for each amino acids. In general, a number of relatively rarely occurring amino acids in Env sequences were substituted into more prevalent amino acids (Table S3). Furthermore, two additional substitutions, Y353F and Q171K at the apex of C97ZA_SOSIP, were introduced to possibly improve the binding of apex targeting antibodies, and extra glycan sites were introduced by the substitution of D411N, K236T, and V295N because these PNGS were conserved >50%.
Expression of HIV Env Constructs
The constructs were synthesized and codon-optimized at GenScript (Piscat-away, NJ 08854) and cloned into pCDNA2004 or generated by standard methods involving site-directed mutagenesis and PCR. HEK-Expi293 cells were transiently transfected with 90% of pCDNA2004 plasmid with the Env insert and 10% of Furin-pCDNA2004, according to the manufacturer’s instructions and cultured for 5 days at 37°C and 10% CO2. Culture supernatants were spun for 10 min at 1,250 × g. For expressions in 96-well format, the cells were cultured for 3 days at 37°C and 10% CO2. 4 μL of Opti-MEM was mixed with 4 μL 100 ng/μL DNA, and 8 μL Expi293F mix (54 μL/mL Opti-MEM) as added and incubated for 20 min. Subsequently, 200 μL/well Expi293F cells were added at 2.5 × 10E6 cells/mL. We use special culture plates and incubation system for this. The culture supernatant was harvested and spun for 10 min at 200 × g to remove cells and cellular debris. The spun supernatant was subsequently sterile-filtered using a 0.22 μm vacuum filter and stored at 4°C until use. For crystallization, the protein was produced in HEK293 GnTI−/−cells via transient transfection with 10% of Furin-pCDNA2004.
Purification of HIV gp140 Trimer Using Lectin and SEC
The recombinant HIV Envs were purified by a 2-step purification protocol applying a Galantus nivalis-lectin column (Vectorlabs, AL-1243, lot Z0325) for the initial purification and subsequently a Superdex200 Increase column (GE) for the polishing step to remove residual contaminants. For the lectin step, the culture supernatant was diluted with 40 mM Tris, 500 mM NaCl pH7.5, and passed over a 4 mL CV Tricorn 10–50 lectin agarose column at 300 cm/hr. Subsequently, the column was washed with 4 column volumes (CV) of 40 mM Tris, 500 mM NaCl pH7.5, and eluted with 4 CV of 40 mM Tris, 500 mM NaCl, 1 M mannopyronoside pH 7.5 with an upflow of 120 cm/hr. The eluate was concentrated using a spin concentrator (50 K, Amicon Ultra, Millipore) and the protein was further purified using a Superdex200 Increase 10/300 column using 20 mM citrate, 75 mM NaCl, 5% sucrose, and 0.03% Tween 80 pH 6.0 as running buffer. The second peak (Figure 1) contained the HIV gp140 trimer. The fractions containing this peak were pooled, and the protein concentration was determined using OD280 and stored a 4°C until use. Purified protein samples were analyzed on 4%–12% (w/v) Bis-Tris NuPAGE gels, 1× MOPS (Life Technologies) under reducing or non-reducing conditions. All procedures were performed according to manufacturer’s instructions. The gels were scanned on an Odyssey instrument (Li-Cor) and images were analyzed using Image Studio 3.0 (Li-Cor).
Size Exclusion Chromatography and Multi-angle Light Scattering Analysis
Purity and mass was further confirmed by size exclusion chromatography (SEC) and multi-angle light scattering (MALS) using a high-performance liquid chromatography system (Agilent Technologies) and miniDAWN TREOS (Wyatt) instrument coupled to a Optilab T-rEX Refractive Index Detector (Wyatt). In total, 40 μg of purified protein was applied to a TSK-Gel G3000SWxl column (Tosoh Bioscience) equilibrated in running buffer (150 mM sodium phosphate, 50 mM sodium chloride, pH 7.0) at 1 mL/min. In other experiments, analysis was performed on supernatants instead of purified Env. The data were analyzed by the Astra 6 software package, and molecular weight calculations were derived from the refractive index signal.
AlphaLISA
AlphaLISA (Perkin-Elmer) is a bead-based proximity assay in which singlet oxygen molecules, generated by high energy irradiation of Donor beads, transfers to Acceptor beads, which are within a distance of ~200 nm. It is a sensitive high throughput screening assay that does not require washing steps. A cascading series of chemical reactions results in a chemiluminescent signal (Eglen et al., 2008). For the AlphaLISA assay, the constructs were equipped with a Flag-His tag (AAALPETGGGSDYKDDDDKP(GGGGS)7H6). The HIV constructs were expressed in HEK-Expi293F cells, which were cultured for 3 days in 96-well plates (200 μL/well). Crude supernatants were diluted 120 times in AlphaLISA buffer (PBS + 0.05% Tween-20 + 0.5 mg/mL BSA). For 17b-based assays, supernatants were diluted 12 times. Subsequently, 10 μL of these dilutions were transferred to a half-area 96-well plate and mixed with 40 μL acceptor beads, mixed with donor beads and mAb. The beads were mixed well before use. After 2 hr of incubation at room temperature (RT), non-shaking, the signal was measured with Neo (BioTek) The donor beads were conjugated to ProtA (catalog #AS102M, Perkin Elmer), which could bind to the mAb. The acceptor beads were conjugated to an anti-His antibody (cat-alog #AL112R, Perkin Elmer) to detect the His-tag of the protein. For the quantification of the protein level, a combination of Nickel-conjugated donor beads (catalog #AS101M, Perkin Elmer) together with acceptor beads carrying anti-Flag antibody (catalog #AL112R, Perkin Elmer) were used. For 17b in combination with sCD4-His, a combination of ProtA donor beads and anti-Flag acceptor beads were used. The average signal of mock transfections (no Env) was subtracted from the AlphaLISA counts measured for the different Env proteins. As a reference, the ConC_SOSIP Env plasmid was used. For normalization, the data were divided by the ConC_SOSIP signal. Trimer percentages (referred to as “trimer content” throughout the paper) were obtained by dividing these normalized signals through the normalized signal obtained for the quantification.
Bio-Layer Interferometry (Octet)
Antibodies were immobilized on anti-hIgG (AHC) sensors (FortéBio catalog #18-5060) at a concentration of 1 μg/mL in 10× kinetics buffer (FortéBio catalog #18-1092) in 96-half well black flat bottom polypylene microplates (Forté-Bio catalog #3694). The experiment was performed on an Octet RED384 instrument (Pall-FortéBio) at 30°C, shaking speed 1,000 rpm. Activation was 300 s, immobilization of antibodies 600 s, washing 300 s, and binding the Envs 1,200 s, followed by a dissociation of 1,200 s, all shaking at 1,000 rpm. The data analysis was performed using the FortéBio Data Analysis 8.1 software (FortéBio).
Negative Stain Electron Microscopy
HIV-1 Env DS_ConC_SOSIP trimer that contained a disulfide 201C- 433C (DS) was diluted to between 0.0005–0.05 mg/mL in Tris-buffered saline (TBS), pH 7.4, and adhered onto a carbon-coated 200 Cu mesh grid (EMS CF200-Cu) that had been glow discharged in air 2 × 10−1 mbar in air, 25 mA, 30 s, just before use. Subsequently, a 3 μL drop was applied for 1 min followed by blotting with filter paper (Whatman no. 1 or 4). After drying for 1 min, grids were then stained with 3 μL of 2.3% uranyl acetate (UAc) in filtered milliQ (filtered with Millipore filter 0.22 μm) for 60 s, followed by blotting with filter paper (Whatman no. 1 or 4). Data were collected using an FEI Tecnai F20 electron microscope operating at 120 keV, with a magnification of 25,000× that resulted in a pixel size of 4.68 Å at the specimen plane. Images were acquired with a Gatan BM ultrascan.
Differential Scanning Calorimetry
Melting temperatures for HIV Env were determined using MicroCal capillary differential scanning calorimetry (DSC) system. 400 μL of a 0.5 mg/mL protein sample were used per measurement. The measurement was performed with a start temperature of 20°C and a final temperature of 110°C. The scan rate was 100°C/hr, and the feedback mode was low (= signal amplification). The data were analyzed using the Origin J software (MicroCal VP analysis tool).
Purification of HIV gp140 Trimer Using CAP456-VRC26 Affinity Chromatography and SEC
For crystallization, the ConC_base0 supernatant expressed in Expi 293 cells supplemented with kifunensin was passed over the CAP256-VRC26.09 IgG coupled to NHS-activated Sepharose 4 fast Flow (catalog #17-0906-01), washed with 20 mM Tris, 150 mM NaCl, pH 7.4, and eluted with the elution buffer containing 20 mM Tris, 3 M MgCl2, pH 7.2. The eluate was concentrated using a spin concentrator (50 K, Amicon Ultra, Millipore), and the protein was further purified using a Superdex200 Increase 10/300 column using 50 mM Tris, 150 mM NaCl, pH 7.4 as running buffer.
HIV-1 Env Trimer-Fab Complex Preparation and Crystallization
Purified trimers were mixed with a variant of 3H+109L (Garces et al., 2015) Fab and a variant of 35O22 (Pancera et al., 2014) scFv in a 1:3:3 molar ratio (gp140 protomer:Fab:scFv) and incubated overnight at room temperature. The 3H+109L variant contained two methionine substitutions in the light chain and the 35O22 variant three contained threonine and two serine mutations in the heavy chain. These alterations were designed to enhance the crystallization lattice (Y.-T.L., unpublished data). Complexes were further purified by SEC and concentrated to 10–15 mg/mL for crystallization. Crystals of the ConC_base0 in complex with 3H+109L Fab and 35O22 scFv were grown in 75 mM HEPES, pH 7.5, 2% PEG 8,000 and 10% PEG 400 with a protein-to-reservoir ratio of 0.5 μL to 0.5 μL.
X-Ray Data Collection, Structure Solution, and Model Building
Crystals were harvested and incubated in crystallization condition supplemented with 20% ethylene glycol as cryo-protectant for 10 s before freezing in liquid nitrogen. Diffraction data were collected at SER-CAT beamline of the Advanced Photo Source at the Argonne National Laboratory. HKL-2000 was used to process the diffraction data, followed by anisotropy data truncation by the UCLA anisotropy server (https://services.mbi.ucla.edu/anisoscale/). A structure of BG505.SOSIP in complex with PGT122 and 35O22 (4TVP.pdb) was modified by using Sculptor to change the BG505 sequence in the model to the corresponding ConC_base0 sequence and PGT122 Fab to 3H+109L Fab. Molecular replacement was carried out by using Phaser, utilizing the modified 4TVP.pdb as the search model. Refinement and structure evaluation was carried out in Phenix.
Cα Deviation between Prefusion-Closed and CD4-Bound Conformation
The Cα deviation between prefusion-closed trimer (PDB: 5FYL or the ConC_base0 structure) and CD4-bound prefusion trimer (PDB: 5VN3) were calculated by aligning gp120 using outer-domain residues (residue numbers 252–482) and aligning gp41 using α7 helix (residue numbers 572–595).
Mutational Free Energy Change Calculations
Mutational free energy change (ΔΔG) were calculated using FoldX (Schymkowitz et al., 2005) based on the ConC_base0 structure and a Env trimer structure of strain B41 in the CD4-bound prefusion conformation (PDB: 5VN3), respectively. The input trimer structure was first repaired using FoldX’s RepairPDB command in order to optimize the structure by fixing steric clashes and lowering the global energy. Mutagenesis was performed using FoldX’s BuildModel command. Number of runs was set to five to insure convergence based on optimal rotamer positions. The average ΔΔG over the five runs were reported for each mutation.
Supplementary Material
Highlights.
High-quality HIV-1 Env proteins by universal repair and stabilize strategy
Crystal structure of stabilized clade C trimer explaining stabilization strategy
Identification of ten novel stabilizing substitutions, of which seven are at the base
α9-helix and intersubunit β sheet at the Env base is critical for trimer stability
Acknowledgments
We thank D. Peng for assistance with expression of antibody variants used in crystallization and members of Structural Biology and Structural Bioinformatics Core Sections, Vaccine Research Center for discussions and comments on the manuscript. We thank J.E. Robinson for providing antibody 14e, D.R. Burton and M. Feinberg for template versions of antibody PGT122, and M. Connors for antibody 35O22 that were modified for use in structural analysis. Funding was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases National Institutes of Health (ZIA AI005024-14), by the IAVI Neutralizing Antibody Consortium, and by Janssen Pharmaceuticals. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science (W-31-109-Eng-38).
Footnotes
DATA AND SOFTWARE AVAILABILITY
The accession number for the coordinates and structure factors for the crystal structure of ConC-SOSIP_base Env trimer in complex with variants of antibodies PGT122 and 35O22 optimized for crystallization reported in this paper is PDB: 6CK9.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.03.061.
AUTHOR CONTRIBUTIONS
L.R., Y.-T.L., D.T., D.v.M., H.S., P.D.K., and J.P.M.L. designed research and analyzed the data. L.R., Y.-T.L., S.B., N.M.S., D.T., A.K., I.J.M.B., G.-Y.C., and K.F. performed research/experiments. L.R., D.T., and J.P.M.L. performed the structure-based design of stabilizing substitution. Y.-T.L. and P.D.K. determined the Env trimer crystal structure, with K.F and G.Y.C. providing energetic calculations. L.R., Y.-T.L., D.T., P.D.K., and J.P.M.L. wrote the paper, with all authors providing comments or revisions.
DECLARATION OF INTERESTS
These studies were funded by Janssen Vaccines and Prevention. L.R., D.T., D.v.M., S.B., N.M.S., A.K., I.J.M.B., H.S., and J.P.M.L. are employees at Janssen. L.R., D.T., N.M.S., A.K., and J.P.M.L. are inventors on an international patent application describing trimer stabilizing HIV envelope protein mutations. The remaining authors declare no competing interests.
References
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, Deymier MJ, Ende ZS, Klatt NR, DeZiel CE, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345:1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J Virol. 2017;91:e02268–16. doi: 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, et al. Immunogenicity of stabilized HIV-1 envelope trimers with reduced exposure of non-neutralizing epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey AK, David KB, Ray N, Ketas TJ, Klasse PJ, Doms RW, Moore JP. N-terminal substitutions in HIV-1 gp41 reduce the expression of non-trimeric envelope glycoproteins on the virus. Virology. 2008;372:187–200. doi: 10.1016/j.virol.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, et al. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Langedijk JP, Bonvin AM, Deng Y, Lu M, Berkhout B, Sanders RW. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J Biol Chem. 2009;284:26941–26950. doi: 10.1074/jbc.M109.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eglen RM, Reisine T, Roby P, Rouleau N, Illy C, Bossé R, Bielefeld M. The Use of AlphaScreen Technology in HTS: Current Status. Curr Chem Genomics. 2008;1:2–10. doi: 10.2174/1875397300801010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Tran K, Bale S, Kumar S, Guenaga J, Wilson R, de Val N, Arendt H, DeStefano J, Ward AB, Wyatt RT. Thermostability of well-ordered HIV spikes correlates with the elicitation of autologous tier 2 neutralizing antibodies. PLoS Pathog. 2016;12:e1005767. doi: 10.1371/journal.ppat.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Perkins S, Theiler J, Bhattacharya T, Yusim K, Funkhouser R, Kuiken C, Haynes B, Letvin NL, Walker BD, et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med. 2007;13:100–106. doi: 10.1038/nm1461. [DOI] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. Structure-guided redesign increases the propensity of HIV Env to generate highly stable soluble trimers. J Virol. 2015;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, Carrette B, Ward AB, Wilson IA, Wyatt RT. Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity. 2017;46:792–803. doi: 10.1016/j.immuni.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garcia NK, Cupo A, Matsui T, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. CD4-induced activation in a soluble HIV-1 Env trimer. Structure. 2014;22:974–984. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Cupo A, Julien JP, Sanders RW, Wilson IA, Moore JP, Lee KK. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie KM, Zandonatti MA, Kleinfelter LM, Heinrich ML, Rowland MM, Chandran K, Branco LM, Robinson JE, Garry RF, Saphire EO. Structural basis for antibody-mediated neutralization of Lassa virus. Science. 2017;356:923–928. doi: 10.1126/science.aam7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Georgiev IS, Yang Y, Druz A, Geng H, Chuang GY, Kwon YD, Pancera M, Rawi R, Sastry M, et al. Soluble prefusion-closed DS-SOSIP. 664-Env trimers of diverse HIV-1 strains. Cell Rep. 2017;21:2992–3002. doi: 10.1016/j.celrep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Ozorowski G, Hua Y, Torrents de la Peña A, de Taeye SW, Nieusma T, Cupo A, Yasmeen A, Golabek M, et al. Design and structure of two HIV-1 clade C SOSIP.664 trimers that increase the arsenal of native-like Env immunogens. Proc Natl Acad Sci USA. 2015;112:11947–11952. doi: 10.1073/pnas.1507793112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavardhana S, Varadarajan R. Stabilizing the native trimer of HIV-1 Env by destabilizing the heterodimeric interface of the gp41 postfusion six-helix bundle. J Virol. 2014;88:9590–9604. doi: 10.1128/JVI.00494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, et al. Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat Commun. 2016;7:12040. doi: 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JP. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun. 2017;8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22:522–531. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langedijk JPM. Human Immunodeficiency Virus Antigens, Vectors, Compositions, and Methods of Use Thereof. 20170165355. US patent. 2017 filed December 15, 2016, and published June 15, 2017.
- Leaman DP, Zwick MB. Increased functional stability and homogeneity of viral envelope spikes through directed evolution. PLoS Pathog. 2013;9:e1003184. doi: 10.1371/journal.ppat.1003184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, Ward AB. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science. 2016;351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikyan GB, Markosyan RM, Hemmati H, Delmedico MK, Lambert DM, Cohen FS. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature. 2017;547:360–363. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci USA. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Ozorowski G, Yasmeen A, Cupo A, Cruz Portillo VM, Pugach P, Golabek M, Rantalainen K, Holden LG, Cottrell CA, et al. Improving the expression and purification of soluble, recombinant native-like HIV-1 envelope glycoprotein trimers by targeted sequence changes. J Virol. 2017;91:e00264–17. doi: 10.1128/JVI.00264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Wilson IA, Moore JP. HIV’s Achilles’ Heel. Sci Am. 2016;315:50–55. doi: 10.1038/scientificamerican1216-50. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci USA. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity. 2016;45:483–496. doi: 10.1016/j.immuni.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JT, Sulli C, Nilo A, Yasmeen A, Ozorowski G, Sanders RW, Ward AB, Klasse PJ, Moore JP, Doranz BJ. High-throughput protein engineering improves the antigenicity and stability of soluble HIV-1 envelope glycoprotein SOSIP trimers. J Virol. 2017;91 doi: 10.1128/JVI.00862-17. https://doi.org/10.1128/JVI.00862-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrents de la Peña A, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, et al. Improving the immunogenicity of native-like HIV-1 envelope trimers by hyperstabilization. Cell Rep. 2017;20:1805–1817. doi: 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully DC, Ogilvie CB, Batorsky RE, Bean DJ, Power KA, Ghebremichael M, Bedard HE, Gladden AD, Seese AM, Amero MA, et al. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog. 2016;12:e1005619. doi: 10.1371/journal.ppat.1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA. 2016;113:E7151–E7158. doi: 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


