Abstract
The expansive dimension of non-coding genes is by now a well-recognized feature of eukaryotes genomes. Over the past decades, in vitro functional studies and in vivo manipulation of non-coding genes through genetically engineered mouse models (GEMMs) have provided compelling evidence that almost every biological phenomenon is regulated, at some level, by non-coding RNA transcripts or by coding RNAs with non-coding functions. In this opinion article, we will discuss how recent discoveries in the field of non-coding RNAs are contributing to advance our understanding of evolution and organismal complexity and its relevance to human diseases.
Introduction
While previously thought to be solely transcriptional noise, evidence has come to light to illustrate the critical functions that non-coding genes can play in cellular homeostasis (1). In fact, non-coding genes represent the vast majority of the genetic information coded within the DNA, while protein-coding genes encompass only a small portion of the mammalian genome (1,2). Furthermore, comparative genome-wide studies have shown that the complexity and evolution of a species does not necessarily correlate with an increase in protein coding genes, but rather correlates with an increase in non-coding genes (1,2,3). While this alone may not be an indication of functional relevance, it clearly suggests that non-coding RNAs may contribute to differences among species or individuals.
Non-coding genes are generally defined as a class of transcribed but not translated genes (4–7). Through comprehensive analysis of the mammalian transcriptome, the existence of a wide range of non-coding genes from short 19–22 nt RNAs, such as microRNAs (4,5), to long non-coding RNAs, such as lncRNAs or pseudogenes that can span hundreds of base pairs (6,7), has been revealed.
Furthermore, non-coding structural RNAs (8), such as rRNAs, tRNAs and small nucleolar RNAs (snoRNAs) (9), traditionally thought to be involved only in fundamental cellular processes, such as protein translation and ribosome biogenesis, have been shown to also exert additional unexpected roles and are perturbed in diseases such as cancer (10,11). More recently, circular RNAs (circRNAs) are emerging as a new large class of functional non-coding RNAs (12–17), while pseudogenes, copies of coding genes that have lost the ability to code for proteins, are yet other forms of non-coding RNAs with relevant biological functions (18–20) (see Table 1).
Table 1.
Types of non-coding genes
| Name | Features | Function | Refs |
|---|---|---|---|
| microRNAs | 19–22 nt long; pri-miRNAs are transcribed by RNA pol II from single or multi cistronic units. The primary transcript is then processed into a hairpin like structure by Drosha. Maturation of pre-miRNA into mature microRNAs occurs in the cytoplasm and is mediated by DICER1, which cleave the 70 nt long pri-miR into 20–22 nt long RNAs molecules. These short RNAs are then loaded into Argonaute proteins 1–4 (AGO1–4) and the complex then associates with its mRNA target(s). | Inhibition of mRNA translation or stability by base pairing with complementary seed sequence in 3UTR of target mRNAs. microRNAs can mediate almost any biological function (from cell proliferation to cell death, from oncogenic transformation to tumor suppression) depending on their targets. | 4, 5, 10 |
| Long non-coding RNAs | Long non-coding RNAs (lncRNAs) are generally defined as RNA transcripts longer than 200 bp. They share features with protein coding genes including poliA tail, 5’capping, association with rybosomes. They can be transcribed in both orientations (sense and antisense), from intergenic regions (lncRNAs) overlapping with coding genes, from repetitive sequence within telomeres or from introns. Some encode for small peptides, usually no longer than few hundred aminoacid. | Molecularly, lncRNAs mediate a wide range of functions both in-cis and in-trans including chromatin remodelling, genomic imprinting, regulation of transcription regulation of RNA processing, stability and splicing. lncRNAs can also function as molecular decoy for proteins or sponges for other transcripts. | 6, 7, 10 |
| Circular RNAs | Circular RNAs (circRNAs) are generated through non-canonical splicing events involving non- juxtaposed exons, head to tail splicing of the same exon, and intronic RNAs. Non-canonical splicing of exonic circRNAs is favored by the presence of repeated Alu sequences flanking the spliced exons. | Compared to other classes of non-coding genes, there are fewer examples of functional circRNAs. circRNAs have been shown to function as microRNAs sponges and to modulate protein functions. | 12–16 |
| Pseudogenes | Pseudogenes are copies of their corresponding parental genes that have lost their ability to code for proteins. Depending on their biogenesis, there are three distinct classes of pseudogenes: unitary, processed and transcribed. | Pseudogenes mainly regulate their parental gene expression. Examples include: competition for microRNA binding sites, regulation of their parental mRNA stability, crosstalk with the RNAi pathway and modulation of the epigenetic status of parental genes. | 17–20 |
| snoRNAs | Small nucleolar RNAs (snoRNAs) are divided into two classes: box C/D snoRNAs and box H/ACA snoRNAs. They are 70–120 bp long and have complex secondary structures. | snoRSNAs mediate ribosomal RNAs (rRNAs) modifications. Box C/D RNAs mediate 2′-O-methylation, while box H/ACA family of snoRNAs guide pseudouridylation or rRNA. | 8,9 |
| piwiRNAs | PIWI-interacting RNAs or piRNAs are a class of short RNAs (26–31nt long) that are associated with PIWI proteins. They are mainly found in germline cells in both male and females. | They safeguard genomic stability in germline cells. They have been shown to bind to and mediate the silencing of transposable elements post-transcriptionally. Loss of piRNAs or PIWI proteins has the most severe defects in male germline cells. PIWI deficient cells show fertility defects. | 43 |
| tRNAs | tRNAs are the most abundant class of RNAs within the cell. tRNAs are essential constituents of the translational machinery and they function to transport amino acids to the ribosome for the translation of mRNAs into polypeptide chains. Each tRNA is modified at its tail to carry one specific amino acid. The binding between tRNAs and mRNAs involves codon/anticodon pairing. | Recent findings show that tRNAs can also function as regulatory molecules. Distinct classes of tRNAs have been implicated in cancer metastasis. Several mitochondrial diseases including myopathies and encephalopathies, have mutation in mt- tRNAs. Human diseases associated with mutations in enzymes involved in tRNA biogenesis include neurological disease, metabolic syndromes and cancer. | 5, 85–89 |
Thus, as we uncover the increasing diversity and species of non-coding RNAs that exist within the cell, it is becoming clear that the contribution of the non-coding genome in human development and disease pathogenesis is likely significant, and needs to be systematically studied. For example, many microRNAs and lncRNAs have been already shown to be relevant for tissue development (21–26). On the other end, although the analysis of non-coding RNAs in disease it is still in its infancy, mutations to promoter and other genetic elements regulating the expression of non-coding RNAs have already been identified in cancer, suggesting specific targeting of non-coding RNAs in tumorigenesis (27,28). Therefore, there is a critical need to understand the molecular mechanisms through which these diverse classes of non-coding genes exert their functions, and how they are altered in disease.
Non-coding genes in development
Amongst all classes of non-coding genes, microRNAs are perhaps the most well-characterized (Figure 1) of all (4,5). These RNAs function by base pairing with microRNA response elements (MREs) that are preferentially located at the 3’UTR of their target mRNAs, although MREs can be found along the entire mRNA (Figure 1) (4,5). This binding results in either degradation of the transcript or inhibition of protein translation. The role of microRNAs in development is supported by the fact that genetic inactivation of a key enzyme towards their maturation, DICER1, results in early embryonic lethality (21). The conditional deletion of Dicer1 in mice in a lineage specific manner also results in striking defects in almost every tissue that has been examined to date, including skin (22), neuronal development (23), cardiac (24) and muscle tissue (25). While, it has been long established that developmental choices are regulated by signaling pathways such as Notch, Wnt and Hedgehog, fluctuation in the expression of microRNA families that concomitantly targeting one or more components of these pathways represents a powerful means to finely and rapidly regulate expression of these proteins to achieve tissue patterning (29). Despite the ability of microRNAs to exert such functions, Genetically Engineered Mouse Models (GEMMs) have shown that the loss of single microRNAs is often inconsequential for embryonic development and may result in variable phenotypes due to compensatory mechanisms from other classes of genes or due to redundant functions (30).
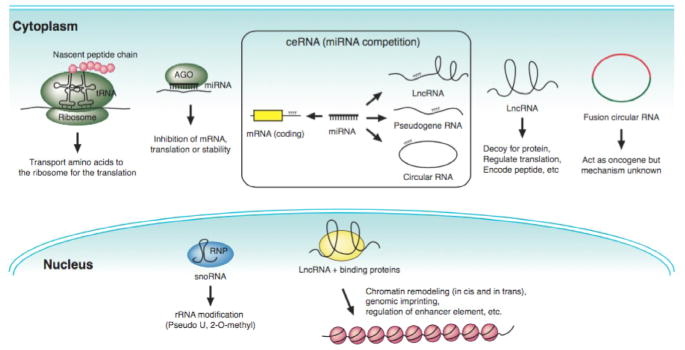
Figure 1. Schematic overview of non-coding genes and their functions.
Non-coding genes mainly regulate the expression of other transcripts at the post-transcriptional level. Cytoplasmic microRNAs generally bind to the 3’UTR of mRNAs to inhibit protein translation. lncRNAs have the most diverse role. They contribute to gene regulation both in-cis and in-trans through multiple mechanisms, such as chromatin remodeling, protein decoys or molecular sponges. lncRNA can be found in both the nucleus and cytoplasm. snoRNAs are mainly localized to the nucleus where they guide RNPs to specific sites of rRNAs for modifications. Circular transcripts have mainly been described in the cytoplasm where they can act as molecular sponges or bind protein complexes. Fusion circRNAs represent a newly described class of non-coding genes. To date they have only been described in cancer cells. tRNA are another highly abundant class of RNA molecules that have been shown to play a role in cancer and other human diseases.
Similar to microRNAs, piRNAs (or piwiRNAs) represent a distinct class of short RNAs with inhibitory function (31). piRNAs are mainly are expressed in germline cells and maintain genomic stability by inhibiting retrotransposon elements(31). While piRNAs have been show to play a critical role in spermatogenesis, whereby loss of PIWI proteins result in male infertility, expression of piRNAs species outside the germline has not been clearly demonstrated to date and is being actively investigated (32).
The observation that small non-coding RNAs such as microRNAs and piRNAs can play a developmental role suggested that other species of non-coding RNAs could be of important functional relevance during development. As such, long non-coding RNAs (lncRNAs) have recently emerged as major regulators of tissue development in several organisms (33). Systematic studies coupled with biochemical analysis have shown that lncRNAs expressed by embryonic stem cells (ESCs) and associated with specific cell fate choices are highly regulated, both temporally and spatially and are associated with a distinct epigenetic signature despite limited conservation of sequence or expression (34,35). Notably, more than 40% of lncRNAs are expressed in the brain, possibly pointing to a critical role for lncRNAs in brain function (35,36).
Mechanistically, lncRNAs facilitate a wide range of functions, and similar to proteins, it is becoming clear that different functional classes of lncRNA exist, each with distinct roles that are highlighted by the type of proteins and RNAs they interact with. Nuclear lncRNAs are known to act both in-cis and in-trans (Figure 1) (6,7), whereby in-cis acting lncRNAs influence the expression of their nearby genes. The most well-known example of such regulation is represented by XIST, a lncRNA transcribed from the X-chromosome and required for X inactivation and silencing (37). Although the characterization of cell fate choices during development and tissue repair are thought to be mainly orchestrated by master transcription factors that regulate gene expression by binding to specific DNA sequences in the promoter or distal regions (enhancers) of their target genes (38), it is now evident that many transcription factors exist in complexes that incorporate non-coding lncRNA transcripts as outlined further below. However, additional cis-acting divergent lncRNAs can also influence transcription factor activity. Divergent lncRNAs are transcribed in antisense from nearby coding genes and are often found in close proximity to transcription factor binding sites of nearby genes to regulate expression of the same group of genes (39). Additionally, high-throughput genomic studies in differentiated versus undifferentiated cells also identified long non-coding genes specifically transcribed from enhancer regions, namely enhancer associated transcripts (eRNAs) (40,41). eRNAs facilitate the initiation of a developmental program by the recruitment of the transcriptional machinery and participate in nucleosomes re-arrangements or chromatin looping. Notably, hierarchical transcription of two different eRNAs instructs the correct developmental program of muscle cells (42).
The identification of tissue regeneration enhancer elements (TREEs) has also recently been reported (43). These regulatory elements of DNA can transiently induce the expression of genes involved in tissue repair following stress or injury. Whether or not these functions are partially mediated by TREE-associated RNAs, and what the RNAs are specifically, are interesting questions that have yet to be addressed.
Several lncRNAs have been shown to be an integral component of the polycomb repressor complex-2 (PRC2) and direct its activity (33). A classic example of this is represented by the lncRNA HOTTAIR which recruits PRC2 to the HoxD locus, thus affecting its epigenetic status by favoring H3K27 trymetylation. Similarly it has been shown that the lncRNA Braveheart is important for the specification of the cardiac lineage from the nascent mesoderm, contributing to the activation of multiple genes involved in cardiac differentiation by bringing components of the PRC2 complex in proximity with its promoters (44). Thus, the utilization of non-coding RNAs within transcription complexes may represent an important manner by which to prioritize and determine expression of transcription factor target genes, in a cell type and tissue specific manner.
While one of the defining characteristics of lncRNAs is that they lack open reading frames (ORFs) of greater than 100 amino acids, some reports also describe small functional peptides that originate from small ORFs within annotated lncRNAs. For example, Dwarf and Myoregulin are two small peptides encoded by annotated lncRNAs expressed in skeletal muscle and that are involved in the regulation of muscle contraction/relaxation through modulating SERCA activity (45,46). Such studies demonstrate that careful analysis of lncRNA are required in order to understand whether this is a feature common to many lncRNAs or rather if it represents an exception to the rule, and whether such RNAs contain both coding and non-coding functions.
Finally, it is important to point out that, similar to coding mRNAs, lncRNAs contain MRE and can compete for binding to miRNAs. This is also relevant as we will discuss later (see competing endogenous RNA (ceRNA) below) in the context of modulating developmental functions.
More recently, the presence and generation of circRNAs, an abundant non coding species of RNA derived from both coding and non-coding linear RNA transcripts through back-splicing events, highlights the diversity of regulatory non-coding RNAs within the cell. These transcripts appear to correlate with the acquisition of distinct developmental stages suggesting a functional role of certain circRNAs in tissue development (12–16). While little is known about the extent to which this class of transcripts is functional, two independent reports have described the isolation and characterization of circRNAs highly enriched in the human and the mouse brain (12,13). In these studies cirS-7 (also known as CDR-1) has been shown to harbor more than 70 MREs for miR-7. Over-expression of cirS-7 resulted in the up-regulation of miR-7 target genes and, vice versa, its down-regulation lead to inhibition of miR-7 targets. A similar mechanism has also been reported for SRY circRNA (see ceRNA below) (12,13). While the initial discovery of circRNAs as microRNAs sponges or ceRNAs may have suggested that circRNAs act through a common mechanism of gene regulation, subsequent reports have also shown that circRNAs may have additional (i.e. ceRNA independent) functions, such as regulation of protein complex stability (17), and a great number of circRNA transcripts do not harbor MREs in their sequence (47). As we move forward in the study of this new class of RNAs, a more systematic study of circRNAs in various tissues will help to uncover additional layers of circRNA dependent regulation of gene expression.
Non-coding genes in cancer
Given that much of the molecular machinery that governs early development relates to ability of stem cells to self-renew and the regulation of stem cell fate, it is not surprising that developmental processes are often deregulated in cancer. Hence, understanding the role of non-coding RNA in developmental can inform the pathogenesis of human cancer. Despite the extensive data highlighting important functional roles of non-coding RNAs in regulating fundamental cellular programs, there is an immense lack of efforts focused on understanding how non-coding RNAs are targeted and altered in cancer. For example, tremendous efforts have focused on cataloging genetic alterations and aberrations across the cancer genome, with international consortium that is The Cancer Genome Atlas (TCGA) analyzing hundreds of cancer genomes across an extensive panel of tumor types. While this represents a critical step in understanding the genetics of cancer and informing its treatment (48,49), the majority of these sequencing efforts have focused on identifying specific alterations in protein coding genes. This effort has enabled us establish key driver mutation(s) in the majority of tumors. However, a significant number of tumors do not harbor known driver mutations in protein coding genes. These cancers could indeed harbor mutations or genomic alterations in the non-coding genomic space, when systematically interrogated.
High throughput sequencing of more than 10 different cancer types, including breast, prostate, liver, pancreas and lung carcinoma, has shown that cancer mutations can often occur in regulatory regions governing gene expression. These hotspots for cancer include DNA elements such as promoters, enhancers, or splicing sites amongst others, and can directly impact the expression status of non-coding RNA elements, while in addition the mutation of non-coding genes themselves maybe relevant for cancer initiation and progression (50,51). However, to date, the non-coding RNA dimension is still very poorly annotated in human cancer, and systematic sequencing efforts are needed to fully establish if non coding RNA mutations represent driver mutations in this disease. Indeed, a comprehensive study of lncRNAs across more than 5000 tumors samples encompassing 13 different types of cancers has shown that cancer associated lncRNAs identify distinct molecular signatures that can be used to identify novel cancer driver mutations in lncRNA loci (51).
Similarly, the vast majority of cancer relevant microRNAs, including let-7, miR-19, and the miR-34 family of microRNAs are found within cancer-prone genomic regions or within fragile genomic sites (52). These sites often harbor driver cancer mutations. This non-random distribution of microRNAs has also been instrumental to categorizing human cancers.
It has been further shown that tumorigenesis can result from the specific mis-regulation of single microRNAs, with oncogenic (onco-miRs) or tumor suppressive function (52). Additionally, microRNA dependent inhibition of Dicer (53), and Dicer1 germline mutations are associated with tumorigenesis (54), strongly suggesting a fundamental role of aberrant microRNA biogenesis in tumor development.
Due to their functional versatility, lncRNAs can also regulate key cellular hallmarks of cancer A non-exhaustive list of lncRNA-mediated functions includes regulation of genomic instability (55), cell proliferation/cell cycle (56,57), hormone signaling (58,59), metastasis and changes in tumor microenvironment (60,61). Mechanistically, lncRNAs act as molecular partners for proteins thereby regulating their function or stability. Multiple lncRNAs are known to regulate chromatin modifications by interacting (directly or indirectly) with PRC2 or its subunit Ezh2, a known oncogenic driver of prostate and other cancer types (59–62) (Figure 1). enhancer-RNAs (eRNAs) have also been reported to both regulate the expression of p53 dependent cell cycle arrest genes and to be transcriptionally activated by p53 itself (63).
In addition to each of these RNA species, tRNAs are emerging as important and novel regulators of tumorigenesis (64). In general, cancer cells have higher levels of tRNAs than normal cells. Importantly, preferential or selective expression of certain tRNA pools correlate with distinct cellular states (65). Strikingly, a recent paper reports the selective up-regulation of two distinct tRNAs (tRNAArg CCG and tRNAGlu UUC) in metastatic breast cells compared to normal cells and further, the authors go on to demonstrate that the specific up-regulation of these tRNAs alone is sufficient to “bias” ribosomes to translate pro-metastatic mRNAs with the corresponding cognate codons, therefore directly modulating cancer cell behavior (64). These findings implicate tRNA in the development of malignancies and other human disease (66,67).
With the discovery that cells express a multitude of circRNA, recent work has now uncovered the generation of novel, cancer specific circRNAs through chromosomal translocation. While chromosomal translocation results in the formation of chimeric proteins with acquired oncogenic properties, it has now been established that such fusion events can also lead to the formation of novel fusion circRNAs with pro-oncogenic functions, as reported for the MLL-AF9 fusion circRNA (68) (Figure 1). However, how commonly fusion circRNAs arise from the 2000 or more chromosomal translocations associate to human cancer to date, and their impact on cancer progression remains to be determined. Interestingly though, comprehensive analysis of “non-fusion circRNAs” in breast, cervical, gastric and oral carcinoma has revealed a significant and robust association between their deregulated expression and oncogenic transformation, suggesting that circRNAs may function as predictive biomarkers and perhaps cancer drivers and suppressors (69,70).
The competitive endogenous hypothesis
One unifying hypothesis that may explain, at least in part, the complex influence of non-coding genes on our understanding of biology, hitherto focused on the protein coding dimension, is the competitive endogenous hypothesis (ceRNA) (71,72) (Figure 2). The foundation of this theory is based on the principle that transcripts sharing the same microRNA response elements influence each other’s activity by competing for the same pool of microRNAs. Different types of cancers, including prostate cancer (19,73), lymphoma (20), melanoma (74) and glioblastoma (75), are affected by competition among different species of RNAs. The study of the ceRNA network for the tumor suppressor gene PTEN perfectly illustrates the diverse range by which a single transcript can have both coding and coding independent functions. The 3’UTR of the PTEN mRNA harbors multiple microRNA binding sites, while importantly, the PTENP1 pseudogene shares some of these binding sites, specifically those for miR-17; miR-19, miR-21; miR-26 and miR-214. Over-expression of the 3’UTR of PTENP1 is sufficient to increase the cellular level of PTEN mRNA and protein; and conversely down-regulation of PTENP1 triggers the opposite effects. Importantly, this phenomenon is not observed in Dicer null cells, clearly demonstrating that its biological functions are microRNA dependent and that PTENP1 acts as a molecular decoy for PTEN microRNAs. Importantly, loss of PTENP1 is observed in prostate cancer and decrease level of PTENP1 in prostate cancer cells increase their proliferation rate and results in tumor burden. A number of other transcripts (VAPA, CNOT6L, ZEB2) have also been show to modulate PTEN activity through common MREs. Therefore, predictions of ceRNA networks could help to identify novel nodes of tumorigenesis (76). The list of ceRNA that play a role in tumorigenesis includes genes such as pseudogenes of BRAF (20), KRAS (19), and key oncogenic drivers (18). Additionally, long non-coding genes can also efficiently function as ceRNAs. One of these lncRNA is lnc-MD-1, which regulates the muscle differentiation program by acting as a molecular decoy for the muscle specific myo-miR-133 and miR-135 (77).
Figure 2. The ceRNA cross-talk in normal and cancer cells. A.
Competitive endogenous RNAs are transcripts that share the same microRNA response elements (MREs). ceRNAs compete with each other to bind to the same pool of microRNAs which essentially allows them to regulate each other’s expression. This mechanism of gene regulation is coding independent since it is mediated by the 3-UTR of mRNAs or by non-coding genes and represents an additional layer of gene regulation. The ceRNA effect is not observed in Dicer knock out cells. B-C. Schematics of ceRNAs role in tumorigenesis. Increasing expression of oncogenic ceRNAs (whether lncRNAs, cirRNAs, pseudogenes or mRNAs) will result in the concomitant increase in the ceRNA target protein level, due to the release of microRNA inhibition. If the ceRNA target is an oncogene, the final output will be an oncogenic transformation. Tumorigenesis may also results from a decrease in tumor suppressive ceRNAs. This will in turn lead to an increase in microRNA inhibition of tumor suppressor genes.
While these examples of ceRNAs are gene centered, looking forward, we expect to be able to identify networks of ceRNAs that overall regulate different disease states or control the transition of cells into distinct states. Given the proposed role of some circ-RNAs as microRNA sponges (e.g. cirS-7) (12) we also expect to be able to identify additional circ-centered ceRNA networks in the near future.
ceRNAs are competitive endogenous transcripts whose function is dependent on microRNA binding. However the mechanism of competition among non-coding genes can also be observed in other contexts. An interesting example of RNA/RNA crosstalk involves tRNA fragments (tRFs) and was recently shown to drive breast cancer metastasis (78). tRFs are small non coding transcripts derived from the endonucleolytic cleavage of tRNA molecules. tRFs up-regulated in low oxygen conditions modulate the cell response to stress by out competing other mRNAs for the binding to the RNA binding protein YBX1. YBX1 has been shown to promote cancer progression by favoring the translation and stability of oncogenes through preferentially binding their 3’UTR. The up-regulation of distinct hypoxia induced tRFs induces the displacement of YBX1 from its targets therefore acting as a tumor suppressor. Importantly metastatic cells overcome hypoxic stress by down-regulating the expression of hypoxia-induced tRFs and enabling YBX1 dependent up-regulation of oncogenes.
Thus, the ceRNA hypothesis provides the framework to identify and study RNA/RNA crosstalk and to predict which of these interactions may have a role in disease progression by conveniently looking at their shared MREs. Still under study are aspects of this network that evaluate cellular abundance of ceRNAs, microRNAs, and other requirements or cellular constraints that may affect the ability of any given transcript to function as a ceRNA. On the other hand, this hypothesis has propelled a renewed excitement in the field of non-coding biology with many new tools aimed at predicting ceRNA networks for genes of interest, stimulating insightful discussions on the dynamics of such RNA/RNA crosstalk, and initiating the search for novel, microRNA-independent mechanisms of cross-talk among non-coding transcripts.
Conclusions
The existence of a vast genomic space of non-coding RNAs has only recently been fully recognized and systematically explored. High throughput studies have been instrumental in highlighting the pervasive nature of non-coding genes, and an overwhelming body of evidence now indicates that non-coding genes represent critical regulators of normal cellular homeostasis, with aberrant expression of these genes in vivo contributing to the development of human diseases, including cancer.
The development of RNA based medicine represents an exciting and rapidly growing field of research. Because microRNAs have been implicated in the regulation of many diseases, conspicuous research efforts have been aimed at targeting miRNAs. Approaches to modulate miRNA activity (either restoration or inhibition) are diverse, include the development of both non-viral and viral methods and are aimed at increasing cellular uptake to limit toxicity and enhance the pharmacokinetics of the “RNA drug “overall. Clinical studies have rapidly moved from mice to non-human primates (79) and human clinical trials are currently ongoing for the treatment of cancer and non-neoplastic conditions (80, 81). Additionally, the list of genes used as cancer biomarkers is becoming increasingly populated with other classes of non-coding RNAs.
The development of novel cancer therapies and the ability to deliver more effectively and selectively these RNA drugs are contingent on the further progressive advancements in RNA medicine. Our hope is that continuous efforts to understanding the biology of RNA at the basic science level will foster and lead to the development of effective RNA based therapies for cancer and other diseases.
Acknowledgments
We would like to thank current members of the Pandolfi laboratory for critical discussion, and Lauren Southwood for insightful editing. This work is funded by grants from HFSP [GP000912014] and from IH/NCI [R35CA197529-01].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shabalina SA, Spiridonov NA. The mammalian transcriptome and the function of non-coding DNA sequences. Genome Biol. 2004;5(4):105. doi: 10.1186/gb-2004-5-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;1:R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 3.Frith MC, Pheasant M, Mattick JS. The amazing complexity of the human transcriptome. Eur J Hum Genet. 2005;13:894–897. doi: 10.1038/sj.ejhg.5201459. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;1:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 7.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noller HF. RNA structure: reading the ribosome. Science. 2005;309(5740):1508–14. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 9.McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA. 2015;6(2):173–89. doi: 10.1002/wrna.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 11.Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16(2):98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- 12.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. doi: 10.1038/nature11993. Together with ref 16, this paper is the first to show that circular RNAs are functional non-coding transcripts with a regulatory role. [DOI] [PubMed] [Google Scholar]
- 13.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 17.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression vai forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, Asangani AI, Kothari V, Prensner JR, Lonigro RJ, et al. Expressed Pseudogenes in the Transcriptional Landscape of Human Cancers. Cell. 2012;149:1622–1634. doi: 10.1016/j.cell.2012.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–32. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 22.Andi T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl C, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme Dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105(6):2111–2116. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Rourke JR, Geroges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for dicer during skeletal muscle development. Dev Biol. 2007;311(2):359–68. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fatica A, Bozzoni I. Long noncodingRNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 27.Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. Role of non-coding sequence variants in cancer. Nat Rev Genet. 2016;17(2):93–108. doi: 10.1038/nrg.2015.17. [DOI] [PubMed] [Google Scholar]
- 28.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Ann Rev Pathol. 9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nature Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 30.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25(3):137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 32.Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17(6):1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. This article provides a systematic and comprehensive analysis of human non-coding RNAs with respect to their molecular features, conservation, expression and function across different species and different human tissues. To our knowledge, this represents one of the most valuable resources in the study of lncRNAs to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng S-Y, Lin L, Soh BS, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29(8):461–8. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Wutz A, Gribnau J. X inactivation Xplained. Curr Opin Genet Dev. 2007;17(5):387–93. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 38.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P, Shao W, et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Plurip otent Cells. Cell Stem Cell. 2016;18(5):637–52. doi: 10.1016/j.stem.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–17. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemerling M, Goldman JA, Black BL, Poss KD. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532(7598):201–6. doi: 10.1038/nature17644. This research article is the first to demonstrate the existence of functional enhancer elements that are specifically activated by regenerative stimuli. Most importantly, the functionality of these non-coding elements is shown, in vivo, in a Zebrafish model of fin regeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351(6270):271–5. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson DM, Anderson KM, Chang CL, Makerewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. A micropeptie encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160(4):595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–9. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome wide analysis of noncoding regulatory mutations in cancer. Nat Genet. 2014;46(11):1160–5. doi: 10.1038/ng.3101. This paper, together with ref 51, identifies putative cancer drivers mutations in non-coding regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28(4):529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 54.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokmap KA, Suarez BK, Whelan AJ, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325(5943):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO P rotein. Cell. 2016;164(1–2):69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong Z, Wang LP, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26(3):344–57. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Banduru S, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 58.Chakravarty D, Sboner A, Nair SS, Giannopuolou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Comm. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K, Takahashi S, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32(12):1665–80. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 61.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varambally S, DHhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 63.Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell. 2013;49(3):524–35. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 64.Goodarzi H, Nguyen HC, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165(6):1416–27. doi: 10.1016/j.cell.2016.05.046. This paper reports a functional role of tRNAs in cancer progression. By looking at the expression of tRNAs in cancer cells, the authors have discovered that highly metastatic cells up-regulate certain tRNAs. This, in turn, favors the translation of oncogenic proteins that are encoded by the corresponding cognate codons. Overall, this paper describes a novel mechanism by which non-coding genes regulate tumorigenesis. Importantly, modulation of the abundance of tRNAs may be used to revert highly metastatic cells into normal-like cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gingold H, Tehler D, Christoffersen NR, Nielsen NM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–92. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. Human CLP1 Mutations alter tRNA biogenesis affecting both peripheral and central nervous system function. Cell. 2014;157(3):636–50. doi: 10.1016/j.cell.2014.02.058. This paper, together with ref 67, is the first to report a novel role of tRNAs in tissue development that is relevant for human disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, et al. CLP1 fuonder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157(3):651–63. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. This research article provides the first evidence of de novo circular RNAs, specifically originated from the aberrant fusion of their corresponding linear transcripts in a subset of tumors. [DOI] [PubMed] [Google Scholar]
- 69.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 71.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;46(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressive PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–95. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147(2):370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Léopold V, Tay Y, Provero P, Zecchina R, Pandolfi PP. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A. 2013;110(18):7154–9. doi: 10.1073/pnas.1222509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tarmontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competitive endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hildebrandt-Eriksen ES, Aarup V, Persson R, Hansen HF, Munk ME, Ørum H. A locked nucleic acid oligonucleotide targeting microRNA 122 is well-tolerated in cynomolgus monkeys. Nucleic Acid Ther. 2012;22(3):152–61. doi: 10.1089/nat.2011.0332. [DOI] [PubMed] [Google Scholar]
- 80.Jansee HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 81.Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31(7):577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]