Abstract
Objective
We characterized associations between central nervous system (CNS) adverse events and brain neurotransmitter transporter/receptor genomics among participants randomized to efavirenz-containing regimens in AIDS Clinical Trials Group studies in the United States.
Methods
Four clinical trials randomly assigned treatment-naïve participants to efavirenz-containing regimens. Genome-wide genotype and PrediXcan were used to infer gene expression levels in tissues including 10 brain regions. Multivariable regression models stratified by race/ethnicity were adjusted for CYP2B6/CYP2A6 genotypes that predict plasma efavirenz exposure, age and sex. Combined analyses also adjusted for genetic ancestry.
Results
Analyses included 167 cases with grade 2 or greater efavirenz-consistent CNS adverse events within 48 weeks of study entry, and 653 efavirenz-tolerant controls. CYP2B6/CYP2A6 genotype level was independently associated with CNS adverse events (O.R.: 1.07; p = 0.044). Predicted expression of 6 genes postulated to mediate efavirenz CNS side effects (SLC6A2, SLC6A3, PGR, HTR2A, HTR2B, HTR6) were not associated with CNS adverse events after correcting for multiple testing, the lowest P-value being for PGR in hippocampus (p=0.012), nor were polymorphisms in these genes or AR and HTR2C, the lowest P-value being for rs12393326 in HTR2C (p=6.7×10−4). As a positive control, baseline plasma bilirubin concentration was associated with predicted liver UGT1A1 expression level (p = 1.9×10−27).
Conclusions
Efavirenz-related CNS adverse events were not associated with predicted neurotransmitter transporter/receptor gene expression levels in brain or with polymorphisms in these genes. Variable susceptibility to efavirenz-related CNS adverse events may not be explained by brain neurotransmitter transporter/receptor genomics.
Keywords: HIV, efavirenz, pharmacogenomics, neurotransmitter transporter, neurotransmitter receptor
Introduction
Efavirenz is a frequently prescribed antiretroviral, with its efficacy demonstrated in multiple clinical trials [1–6]. However, central nervous system symptoms (CNS) are common with efavirenz [7–9]. Several CYP2B6 polymorphisms predict increased plasma efavirenz exposure, including CYP2B6 516G→T (rs3745274) [10–15], 983T→C (rs28399499) [15–18], and 15582C→T (rs4803419) [15]. A CYP2A6 polymorphism, -48T→G (rs28399433), also affects efavirenz pharmacokinetics [19–22] when present with CYP2B6 slow metabolizer genotypes [19, 22]. These polymorphisms explain approximately 35% of interindividual variability in plasma efavirenz exposure [15]. A possible association with efavirenz pharmacokinetics has also been reported with UGT2B7 genotype [21] but with small effect size [22].
Increased likelihood of efavirenz CNS symptoms has been attributed to CYP2B6 slow metabolizer genotypes [18, 23, 24]. In an initial analysis of AIDS Clinical Trials Group (ACTG) data [18], CYP2B6 slow metabolizer genotypes were associated with adverse events in 276 white participants (p=0.04) but not in 217 black participants (p=0.58). Similarly, among 563 patients who initiated efavirenz-containing regimens at a clinic in the Southeastern United States, slow metabolizer CYP2B6 genotypes were associated with efavirenz discontinuation for CNS symptoms in 335 white patients (p = 0.001) but not in 198 black patients (p = 0.27) [23]. Among 1833 ACTG study participants in the United States, an association between CYP2B6 genotype and suicidality was strongest among white participants but nearly null among black participants [24]. The reason for this apparent difference by race is not known.
Brain neurotransmitter transporters/receptors are postulated to mediate efavirenz CNS symptoms, including the norepinephrine transporter (encoded by SLC6A2), dopamine transporter (SLC6A3), progesterone PR-B (PGR), serotonin receptors 5-HT2A (HTR2A), 5-HT2B (HTR2B), 5-HT2C (HTR2C), and 5-HT6 (HTR6), and androgen receptor (AR) (Daria Hazuda, personal communication). Lower expression levels of neurotransmitter transporter/receptor genes in brain tissue could possibly confer increased susceptibility to efavirenz side effects. This hypothesis may be addressed indirectly using, PrediXcan, a novel computational algorithm that uses genome-wide genotype data to infer RNA expression levels for individual genes in various human organs and tissues [25].
We examined whether predicted expression levels of selected neurotransmitter transporter/receptor genes in brain were associated with risk for CNS adverse events in participants randomized to receive efavirenz-containing regimens in ACTG studies. We also examined whether individual polymorphisms in these genes were associated with CNS adverse events with efavirenz.
Methods
Study Design and Participants
Data were pooled from antiretroviral-naïve individuals who had been randomly assigned to initiate efavirenz-containing regimens in four studies: ACTG 384 (ClinicalTrials.gov: NCT00000919) [26, 27], A5095 (ClinicalTrials.gov: NCT00013520) [2, 28], A5142 (NCT00050895) [3], and A5202 (NCT00118898) [6]. Drug class components of the regimens were randomly assigned (efavirenz-based regimen vs. comparator regimen) except for nucleoside analogue choice in A5142. Genetic association testing was limited to participants who consented to genetic testing under ACTG protocol A5128 [29]. Participants self-reported race/ethnicity.
Each protocol required reporting of signs, symptoms, or diagnoses at each visit, severe and life-threatening graded signs or symptoms [30], and signs or symptoms that led to change in study regimen. Diagnoses were not graded. Further, study A5142 required report of all moderate signs or symptoms, study ACTG 384 required entry of all signs and symptoms grade 3 or greater, all signs and symptoms which resulted in dose modification regardless of grade, and all grade 2 or greater CNS symptoms, and A5095 and A5202 required report of moderate CNS symptoms. Site institutional review boards approved each study, and participants provided written informed consent.
Outcomes
The outcome of interest was new onset grade 2 or greater CNS signs or symptoms that were consistent with possible efavirenz effect. These included agitation, behavior changes, abnormal cognition, confusion, depression, difficulty concentrating, dizziness, abnormal dreams, excessive anger, hyperactivity, inappropriate behavior, insomnia, lethargy, change in level of consciousness, lightheadedness, memory loss, psychiatric mental status change, rage, or sleeping problems. Adverse event data were based on self-report, and did not involve questionnaires specifically targeting CNS events. Death by suicide was also considered a possible efavirenz effect.
Cases were participants with grade 2 or greater efavirenz-consistent CNS signs or symptoms, or with death due to suicide, documented within 48 weeks after study entry while still being prescribed efavirenz, or within 2 weeks after efavirenz was discontinued. Controls were participants with no documented efavirenz-consistent CNS signs or symptoms regardless of any grade after study entry while being prescribed efavirenz for at least 96 weeks. Cases and controls were excluded for any neuropsychological signs or symptoms of any grade documented at study entry.
Covariates
Baseline covariates included in multivariable models included age, sex and CYP2B6/CYP2A6 genotype. The first two principal components generated from genome-wide genotype data were also included to minimize confounding by unrecognized population stratification. Analyses performed separately among self-identified white, black, and Hispanic participants also adjusted for age, sex and CYP2B6/CYP2A6 genotype, but not principal components.
Genetic assays and data
Genotypes for CYP2B6 516G→T, 983T→C, 15582C→T and CYP2A6 -48T→G were largely available from a MassARRAY® iPLEX Gold (Sequenom, Inc.) assay, generated by Vanderbilt Technologies for Advanced Genomics (VANTAGE) [15]. Genome-wide genotype data largely available from a previous immunogenomics project [31] were generated by Illumina HumanHap 650Y array for A5095 and by Illumina 1M duo array for A5142 and A5202. Quality control and imputation of genome-wide data was performed as described elsewhere [32]. The PLINK program and R statistical programming language were used for QC procedures [33, 34]. Polymorphisms were censored for call rates <99%. We excluded 18 samples where genetically inferred sex differed from clinical data, or missing sex status that could not be inferred, 105 samples with overall genotyping call rates <99%, and 17 samples with cryptic relatedness based on identity by descent (IBD) estimates >0.3 from approximately 100,000 pruned SNPs.
Post QC data were imputed to 1000 Genomes [35] after converting to genome build 37 using liftOver [36] and stratifying by chromosome to parallelize imputation processing. ShapeIt2 [37] was used to check strand alignment and to phase data. The IMPUTE2 algorithm [38] was used to impute additional genotypes that were available in the 1000 Genomes reference panel, but not directly genotyped. Each chromosome was segmented into 6 MB regions with at least 3500 reference variants in each region. Imputed genotypes were included if posterior probabilities exceeded 0.9.
Quality of imputed data was assessed following the Electronic Medical Records and Genomics (eMERGE) protocol [39]. Each chromosome from each phase was checked for 100% concordance with genotyped data. We dropped imputed SNPs with imputation scores <0.7, genotyping call rates <99% and minor allele frequencies (MAF) <0.01.
Twelve composite CYP2B6/CYP2A6 genotype levels that predict progressively greater plasma efavirenz exposure were defined by combinations of three CYP2B6 and one CYP2A6 polymorphisms [15, 22] as described elsewhere [22]. Each CYP2B6/CYP2A6 polymorphism (rs3745274, rs28399499, rs4803419, and rs28399433) was in Hardy-Weinberg equilibrium in white, black, and Hispanic participants analyzed separately except rs4803419 in white participants (multiple testing-corrected P = 0.045). Consent for genetic analysis was obtained under ACTG protocol A5128 [29], and the ACTG approved this use of DNA.
PrediXcan
PrediXcan was used to infer, from genome-wide genotype data, the heritable component of RNA expression levels in 43 available reference tissues [25], using the 2015 PrediXcan models. PrediXcan was able to infer expression of approximately 10,000 genes in each tissue. The 10 reference brain tissues analyzed included anterior cingulate cortex, caudate, cerebellar hemisphere, cerebellum, cortex, frontal cortex, hippocampus, hypothalamus, nucleus accumbens, and putamen. The additional 33 reference tissues included adipose subcutaneous, adrenal gland, aorta, coronary artery, tibial artery, breast, EBV-transformed lymphocytes, transformed fibroblasts, sigmoid colon, transverse colon, gastroesophageal junction, esophagus mucosa, esophagus muscularis, heart atrial appendage, heart left ventricle, liver, lung, skeletal muscle, tibial nerve, ovary, pancreas, pituitary gland, skin suprapubic not sun exposed, skin lower leg sun exposed, terminal ileum, terminal ileum (Elastic Net), spleen, stomach, testis, thyroid, whole blood unscaled, whole blood, and cross tissue.
Statistical analysis
We characterized associations between predicted expression of six autosomal brain neurotransmitter transporter/receptor genes (SLC6A2, SLC6A3, PGR, HTR2A, HTR2B, HTR6) and efavirenz CNS adverse events. We also characterized associations between polymorphisms in these six genes as well as AR and HTR2C and efavirenz CNS adverse events. Because AR and HTR2C are on the X chromosome, PrediXcan cannot infer their expression. Analyses controlled for CYP2B6/CYP2A6 genotype level as a linear covariate, age, and sex. The first two principal components, calculated using EIGENSOFT [40] were used to adjust for global ancestry in analyses that pooled all race/ethnicity groups. Associations with CNS adverse events were evaluated using logistic regression models, stratified by race/ethnicity. As a positive control, associations between baseline total plasma bilirubin concentration and hepatic UGT1A1 expression were similarly evaluated, controlling for CYP2B6/CYP2A6 genotype level, age, sex, and the first two principal components. The Bonferroni method was used to adjust for multiple testing.
Results
Study participants
Of 4,742 participants from ACTG studies ACTG 384, A5095, A5142, and A5202, a total of 2,171 who had been randomly assigned to initiate efavirenz-containing regimens consented to genetic testing at research sites in the United States. Of these participants, 1,425 were successfully genotyped for CYP2B6/CYP2A6 and had imputed genome-wide genotype and principal component data. Of these participants, 820 met definitions as either cases with documented grade 2 or greater efavirenz-consistent CNS adverse events by week 48 (n = 167) or efavirenz-tolerant controls that continued efavirenz for at least 96 weeks without documented CNS adverse events (n = 653). Participant disposition is presented in Figure 1. Baseline characteristics of study participants are shown in Table 1. Females were underrepresented among cases versus controls.
Figure 1. Derivation of the study sample.
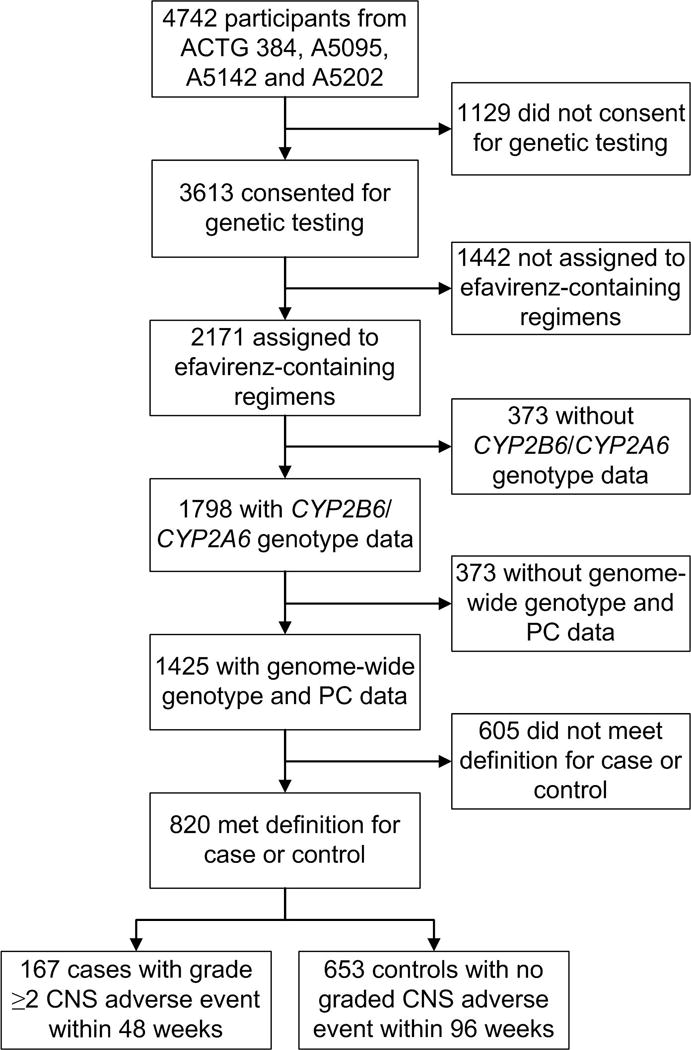
Derivation cases who developed grade 2 or greater efavirenz-consistent CNS adverse events by week 48, and efavirenz-tolerant controls who continued efavirenz for at least 96 weeks without efavirenz-consistent CNS adverse events, during participation in ACTG 384, A5095, A5142 or A5202 is shown. Abbreviation: PC, principal components.
Table 1.
Baseline characteristics of participants included in analyses of grade 2 or greater CNS adverse events after being randomly assigned to efavirenz-containing regimens.
| Cases (n=167) |
Controls (n=653) |
Total (N=820) |
||
|---|---|---|---|---|
| Parent study, n (%) | ACTG384 | 28 (13.9) | 173 (86.1) | 201 |
| A5095 | 63 (34.4) | 120 (65.6) | 183 | |
| A5142 | 15 (10.1) | 133 (89.9) | 148 | |
| A5202 | 61 (21.2) | 227 (78.8) | 288 | |
| Sex, n (%) | Male | 156 (16.4) | 539 (77.6) | 695 |
| Female | 11 (8.8) | 114 (91.2) | 125 | |
| Race or ethnic group, n (%) | White Non-Hispanic | 81 (22.8) | 274 (77.2) | 355 |
| Black Non-Hispanic | 53 (20.1) | 211 (79.9) | 264 | |
| Hispanic | 26 (14.1) | 158 (85.9) | 184 | |
| Asian | 2 (25.0) | 6 (75.0) | 8 | |
| Native American | 3 (42.9) | 4 (57.1) | 7 | |
| Multiracial | 1 (100) | 0 (0) | 1 | |
| Unknown | 1 (100) | 0 (0) | 1 | |
| Age, Median (IQR) | 38 (31, 45) | 37 (31, 45) | 37 (31, 45) |
CYP2B6/CYP2A6 genotype and CNS adverse events
In logistical regression analyses, among the 820 participants who were evaluable as either cases who developed grade 2 or greater CNS adverse events by week 48 (n = 167), or efavirenz-tolerant controls (n = 653), and controlling for age, sex, and the first 2 principal components, CYP2B6/CYP2A6 genotype level was associated with grade 2 or greater CNS adverse events within 48 weeks (O.R.: 1.07; 95% C.I.: 1.00 to 1.15; p = 0.044). In analyses performed separately among 335 white, 264 black, and 184 Hispanic participants, and adjusted for age and sex but not principal components, odds ratios for association between CYP2B6/CYP2A6 genotype level and grade 2 or greater CNS event by week 48 were similar to the total group (1.10 for white, 1.07 for black, and 1.08 for Hispanic participants), but none were statically significant (P>0.10 for each). In the above multivariable models, female sex was associated with fewer grade 2 or greater CNS adverse events within 48 weeks among all participants (O.R.: 0.33; 95% C.I.: 0.17 to 0.63; p = 0.001), and among white participants (O.R.: 0.14; 95% C.I.: 0.18 to 1.04; p = 0.055) and black participants (O.R.: 0.43; 95% C.I.: 0.19 to 0.96; p = 0.040) analyzed separately.
Predicted expression levels of neurotransmitter transporter/receptor genes and CNS adverse events
For each participant, genome-wide genotype data was used to predict the heritable component of gene expression for 43 tissues, including multiple reference regions of the brain. Primary analyses characterized associations between 6 neurotransmitter transporter/receptor genes postulated to mediate efavirenz effects in brain (SLC6A2, SLC6A3, PGR, HTR2A, HTR2B, HTR6), and grade 2 or greater CNS adverse events within 48 weeks. Among all subjects, and controlling for CYP2B6/CYP2A6 genotype level, age, sex, and 2 principal components, the lowest nominal P-value was for PGR in hippocampus (p=0.012). The two lowest nominal P-value results for each gene and associated brain regions are presented in Table 2. None were significant after correcting for multiple testing. In analyses performed separately among white, black, and Hispanic participants, and adjusted for age and sex but not principal components, there were no significant associations. The lowest P-value in white participants was for HTR2A in cerebellar hemisphere (p=0.12), in black participants was for SLC6A2 in caudate (p=0.17), and in Hispanics participants was for HTR2A in cortex (p=0.082). The two lowest nominal P-value results for each gene within race /ethnicity group, and associated brain regions are presented in Supplemental Table 1, supplemental Digital Content 1, http://links.lww.com/NMC/A133.
Table 2.
Associations between predicted expression levels of six autosomal neurotransmitter receptor/transporter genes and grade 2 or greater CNS adverse events within 48 weeks of starting efavirenz-containing regimens.
| Gene | Chromosome | Brain Region | Beta | P value |
|---|---|---|---|---|
| HTR2A | 13 | Cerebellar hemisphere | −0.61 | 0.086 |
| Frontal cortex | −1.24 | 0.677 | ||
| HTR2B | 2 | Frontal cortex | −0.77 | 0.331 |
| Hippocampus | −0.31 | 0.601 | ||
| HTR6 | 1 | Nucleus accumbens | 0.45 | 0.258 |
| Caudate | 0.53 | 0.587 | ||
| PGR | 11 | Hippocampus | −0.91 | 0.012 |
| Cortex | −1.00 | 0.204 | ||
| SLC6A2 | 16 | Cortex | 1.85 | 0.405 |
| Caudate | 0.69 | 0.454 | ||
| SLC6A3 | 5 | Nucleus accumbens | 1.59 | 0.116 |
| Hypothalamus | 0.65 | 0.192 |
Logistic regression analysis involved 820 total participants, which included 167 grade 2 or greater CNS event cases and 653 efavirenz-tolerant controls. The analysis controlled for CYP2B6/CYP2A6 genotype level, age, sex, and the first 2 principal components. The analyses included all evaluable participants without stratification for race/ancestry. The two lowest P-value results are shown for six genes postulated to mediate efavirenz CNS side effects. The positive or negative beta indicates directionality of the relationship.
Following the approach of Li et al [41], we were able to assess correlations between predicted and observed gene expression in reference datasets in brain tissue for SLC6A3, HTR2B, and HTR6. Among Caucasians, the strongest correlations were with SLC6A3 (r2 = 0.038) and HTR2B in hypothalamus (r2 = 0.028). Among Africans the strongest correlations were with SLC6A3 in hypothalamus (r2 = 0.040) and HTR6 in cortex (r2 = 0.029). These would not be considered well-predicted genes.
Predicted expression levels for all evaluable genes in brain and CNS adverse events
In secondary analyses we explored associations with expression across all evaluable autosomal genes in the brain. Among all subjects, and controlling for CYP2B6/CYP2A6 genotype level, age, sex, and 2 principal components, the lowest nominal p-value was for RCE1 (a metalloproteinase) in cerebellar hemisphere (p = 5.3×10−8). The two lowest nominal P-value results for each brain region are presented in Table 3. In analyses performed separately among white, black, and Hispanic participants, and adjusted for age and sex but not principal components, there were no significant associations after adjusting for multiple comparisons. The lowest P-value in white participants was for ACSF3 in anterior cingulate cortex (p=9.5×10−6), in black participants was for TRPC3 in cerebellar hemisphere (p=1.9×10−5), and in Hispanics participants was for KLK5 in cerebellar hemisphere (p=3.0×10−5). The two lowest nominal P-value genes for each brain region each gene within race /ethnicity group are in Supplemental Table 2, Supplemental Digital Content 1, http://links.lww.com/NMC/A133.
Table 3.
Associations between predicted expression levels of all genes in brain and grade 2 or greater CNS adverse events within 48 weeks of starting efavirenz-containing regimens.
| Brain region | Gene | Chromosome | Beta | P value |
|---|---|---|---|---|
| Anterior cingulate cortex | PAXBP1 | 21 | −6.73 | 1.7×10−6 |
| SEMA3G | 3 | 2.80 | 2.8×10−6 | |
| Caudate | EMC10 | 19 | 2.79 | 1.0×10−5 |
| SIRPB1 | 20 | 1.52 | 1.9×10−5 | |
| Cerebellar hemisphere | RCE1 | 11 | −2.64 | 5.3×10−8 |
| RABEPK | 9 | −1.21 | 4.6×10−6 | |
| Cerebellum | CDK10 | 16 | −0.73 | 1.8×10−5 |
| C4orf48 | 4 | −4.90 | 4.9×10−5 | |
| Cortex | TFAP2D | 6 | −6.61 | 1.2×10−6 |
| EHMT1 | 9 | −1.44 | 1.1×10−4 | |
| Frontal cortex | ZNF76 | 6 | −59.49 | 2.2×10−6 |
| HLA-G | 6 | 0.59 | 1.5×10−4 | |
| Hippocampus | C11orf68 | 11 | −2.45 | 5.3×10−8 |
| ZNF555 | 19 | 42.89 | 7.6×10−6 | |
| Hypothalamus | IKBKE | 1 | 0.10 | 2.7×10−7 |
| ADRBK1 | 11 | 4.63 | 2.4×10−6 | |
| Nucleus accumbens | SHANK3 | 22 | 4.80 | 2.9×10−7 |
| MSX2 | 5 | −4.92 | 1.2×10−4 | |
| Putamen | UBL4B | 1 | 5.62 | 1.0×10−5 |
| SMIM6 | 17 | −1.49 | 1.0×10−5 |
Logistic regression analysis involved 820 total participants, which included 167 grade 2 or greater CNS event cases and 653 efavirenz-tolerant controls. The analysis controlled for CYP2B6/CYP2A6 genotype level, age, sex, and the first 2 principal components. The analyses included all evaluable participants without stratification for race/ancestry. The two lowest P-value results are shown for each brain region.
Predicted UGT1A1 expression in all tissues and baseline plasma bilirubin concentration
As a positive control we considered total plasma bilirubin concentration at baseline, which should correlate inversely with UGT1A1 expression, especially in liver. By linear regression analysis involving 1354 participants evaluable for baseline plasma bilirubin, and controlling for CYP2B6/CYP2A6 genotype, age, sex, and 2 principal components, hepatic UGT1A1 expression was associated with bilirubin concentration (p = 3.4×10−27). This association was also present among white participants (p = 4.3×10−15), black participants (p = 4.2×10−12), and Hispanic participants (p = 1.5×10−6) analyzed separately without adjusting for principal components (Table 4). In contrast, for 17 non-liver tissues for which PrediXcan could predict UGT1A1 expression levels, only three had nominal p-values less than 0.05 for association of UGT1A1 expression with baseline total bilirubin - skeletal muscle (p = 0.002), non-sun-exposed skin (p = 0.005), and putamen (p = 0.046). P-values exceeded 0.10 for the other 14 tissues. This established that PrediXcan could identify a true gene expression-phenotype association in our dataset, and could do so in a tissue-specific manner.
Table 4.
Associations between baseline plasma total bilirubin concentration and predicted gene expression levels in liver.
| Race/ethnicity | Gene | Chromosome | Participants | Beta | P value |
|---|---|---|---|---|---|
| All | UGT1A1 | 14 | 1354 | 0.30 | 3.4×10−27 |
| GGACT | 2 | 1354 | −0.88 | 7.2×10−6 | |
| White | UGT1A1 | 2 | 645 | 0.32 | 4.28×10−15 |
| CSN1S1 | 4 | 645 | −0.37 | 8.72×10−5 | |
| Black | UGT1A1 | 2 | 458 | 0.30 | 4.2×10−12 |
| PGBD2 | 1 | 458 | −1.96 | 3.57×10−5 | |
| Hispanic | CLDN12 | 7 | 282 | −4.17 | 3.69×10−7 |
| UGT1A1 | 2 | 282 | 0.33 | 1.45×10−6 |
Linear regression analysis involved numbers of participants shown, for baseline plasma total bilirubin concentration. Analysis among all participants controlled for CYP2B6/CYP2A6 genotype level, age, and sex. Analysis among white, black, and Hispanic participants controlled for the same covariates but did not control for principal components. The two lowest P-value results are shown for each population group.
Neurotransmitter transporter/receptor gene polymorphisms and CNS adverse events
Primary analyses characterized associations between polymorphisms in the eight neurotransmitter transporter/receptor genes (± 100 kB) noted above (SLC6A2, SLC6A3, NR3C3, HTR2A, HTR2B, HTR2C, HTR6, NR3C4) and grade 2 or greater CNS adverse events within 48 weeks. Analysis among all participants, and controlling for CYP2B6/CYP2A6 genotype level, age, sex, and 2 principal components, the lowest nominal P-value was for rs12393326 in HTR2C (p=6.7×10−4). The two lowest nominal P-value polymorphisms among all participants and among white, black, and Hispanic participants analyzed separately are presented in Table 5. None were significant after correcting for multiple testing.
Table 5.
Associations between polymorphisms in eight neurotransmitter/transporter genes and grade 2 or greater CNS adverse events within 48 weeks of starting efavirenz.
| Race/ethnicity | Polymorphism | Gene | Chromosome | MAF | Cases | Controls | Beta | SE | P value |
|---|---|---|---|---|---|---|---|---|---|
| All | rs12393326 | HTR2C | X | G:0.07 | 164 | 815 | 0.53 | 0.16 | 6.7×10−4 |
| rs5988149 | HTR2C | X | A:0.12 | 165 | 816 | 0.38 | 0.13 | 3.6×10−3 | |
| White | rs660541 | PGR | 11 | A:0.51 | 83 | 281 | 0.41 | 0.18 | 2.2×10−2 |
| rs511484 | PGR | 11 | C:0.49 | 83 | 281 | −0.36 | 0.16 | 2.3×10−2 | |
| Black | rs2497509 | HTR2C | X | C:0.43 | 49 | 211 | 0.59 | 0.18 | 8.8×10−4 |
| rs2497510 | HTR2C | X | A:0.43 | 49 | 211 | 0.59 | 0.18 | 8.8×10−4 | |
| Hispanic | rs3785152 | SLC6A2 | 16 | T:0.08 | 26 | 158 | 1.09 | 0.46 | 1.8×10−2 |
| rs2016711 | HTR2A | 13 | G:0.03 | 26 | 158 | 1.34 | 0.61 | 2.8×10−2 |
Logistic regression analysis involved the indicated numbers of grade 2 or greater CNS event cases and efavirenz-tolerant controls. We considered polymorphisms in SLC6A2, SLC6A3, NR3C3, HTR2A, HTR2B, HTR2C, HTR6, and NR3C4 ± 100 kB. The analysis controlled for CYP2B6/CYP2A6 genotype level, age, sex, and, for the analysis involving all participants, the first 2 principal components. The two lowest P-value results are shown for each race/ancestry group.
Genome-wide polymorphisms and CNS adverse events
To complement the above analyses focused on eight neurotransmitter transporter/receptor gene polymorphisms, secondary analyses explored polymorphism associations genome-wide. Analysis among all participants, and controlling for CYP2B6/CYP2A6 genotype level, age, sex, and 2 principal components, the lowest nominal P-value was for rs7143465 in SLC8A3, which encodes solute carrier family 8 member A3 (p=2.2×10−9). The two lowest nominal P-value polymorphisms among all participants and among white, black, and Hispanic participants analyzed separately are presented in Table 6.
Table 6.
Associations between genome-wide polymorphisms and grade 2 or greater CNS adverse events within 48 weeks of starting efavirenz.
| Race/ethnicity | Polymorphism | Gene | Chromosome | MAF | Cases | Controls | Beta | SE | P value |
|---|---|---|---|---|---|---|---|---|---|
| All | rs7143465 | SLC8A3 | 14 | C:0.45 | 167 | 653 | −0.69 | 0.12 | 2.2×10−9 |
| rs1045920 | CFAP36 | 2 | T:0.45 | 166 | 653 | −0.74 | 0.13 | 2.6×10−9 | |
| Black | rs2292493 | BBS12 | 4 | T:0.13 | 52 | 211 | 1.60 | 0.31 | 2.9×10−7 |
| rs72671080 | BBS12 | 4 | A:0.13 | 52 | 211 | 1.60 | 0.31 | 3.0×10−7 | |
| Hispanic | rs62520409 | LOC105375706 | 8 | A:0.04 | 26 | 158 | 3.34 | 0.68 | 9.8×10−7 |
| rs1305010 | Intergenic | 2 | C:0.06 | 26 | 157 | 2.31 | 0.54 | 1.9×10−5 | |
| White | rs10823547 | Intergenic | 10 | G:0.50 | 82 | 281 | −0.88 | 0.18 | 1.2×10−6 |
| rs144103499 | ACSF3 | 16 | CCT:0.21 | 83 | 280 | 1.04 | 0.22 | 1.3×10−6 |
Logistic regression analysis involved the indicated numbers of grade 2 or greater CNS event cases and efavirenz-tolerant controls. The analysis controlled for CYP2B6/CYP2A6 genotype level, age, sex, and, for the analysis involving all participants, the first 2 principal components. The two lowest P-value results are shown for each race/ancestry group.
For the four genes in Table 6, we attempted to examine whether predicted expression in brain was associated with CNS events. Of these, CFAP36 was not represented in PrediXcan. Among white participants, but not among black or Hispanic participants, predicted ACSF3 expression in brain tissues tended to be associated with grade 2 or greater CNS adverse events within 48 weeks, including in anterior cingulate cortex (P = 9.46×10−6), frontal cortex P = 1.08×10−5), cortex (1.24 ×10−5), and caudate (P = 8.53×10−5). Of note, ACSF3 rs144103499 (Table 6) is not a known expression quantitative trait locus (eQTL) for ACSF3 in any tissue [42]. There were not associations with BBS12 or SLC8A3. The lowest P-value for BBS12 was in cerebellum among black participants (P = 0.092), and for SLC8A3 was in hypothalamus among all participants (P = 0.014).
Discussion
Among individuals who were randomly assigned to initial treatment with efavirenz-containing regimens in four ACTG studies, and with correction for multiple comparisons, we found no significant association between predicted expression of 6 neurotransmitter transporter and receptor genes postulated to mediate efavirenz effects in brain (SLC6A2, SLC6A3, PGR, HTR2A, HTR2B, HTR6) and grade 2 or greater CNS adverse events, both among all participants, and in analyses performed separately among white, black, and Hispanic participants. The lowest nominal P-value among all participants was for PGR in hippocampus (p=0.012). Similarly, after correction for multiple comparisons we found no significant association between individuals SNPs in these genes or AR and HTR2C and grade 2 or greater CNS adverse events, both among all participants, and in analyses performed separately among white, black, and Hispanic participants. The lowest nominal P-value was for rs12393326 in HTR2C (p=6.7×10−4).
Analyses were controlled for CYP2B6/CYP2A6 genotype, age, and sex, and in analyses that pooled all race/ethnicity groups, also the first 2 principal components. We demonstrated significant, though weak, associations between CYP2B6/CYP2A6 genotype level and grade 2 or greater CNS adverse events, which is generally consistent with several previous reports [18, 23, 24]. It was therefore important that we adjust for CYP2B6/CYP2A6 genotype level in analyses for associations with predicted neurotransmitter transporter/receptor gene expression and polymorphisms. In addition, controlling for principal components in analyses involving all participants reduced the likelihood for false associations due to unrecognized population stratification.
PrediXcan is a relatively new computational algorithm that allows the heritable component of RNA expression levels for individual genes in different tissues to be inferred from genome-wide genotype data [25]. It was developed to exploit genotype-tissue expression (GTEx) data, and evaluates aggregate effects of cis-regulatory variants (within 1MB upstream or downstream) on gene expression by an elastic net regression method, and generates potential eQTLs and their weights for each gene in each GTEx tissue type. By considering genes rather than individual polymorphisms, PrediXcan should have a greatly reduced multiple testing burden versus single-variant-single-trait association tests. PrediXcan may therefore identify loci with modest to weak effect sizes that are not significant in genome-wide association studies. To assess the performance of PrediXcan we tested for associations with baseline plasma total bilirubin concentration. It is reassuring that we found significant associations between predicted UGT1A1 expression in liver and bilirubin concentrations among all participants, and separately among white, black, and Hispanic participants. Thus PrediXcan detected a known association in a tissue-specific manner in our dataset.
By analyzing all participants pooled, as well as racial/ethnic groups of white, black, and Hispanic participants separately, we had the potential to identify consistent genetic associations across groups. While true genetic associations need not be present in all populations, finding the same association in all participants and in each race/ethnicity group increases the likelihood that the association is not by chance. For example, associations between CYP2B6 genotype level and plasma efavirenz concentrations were previously demonstrated in pooled analyses and among white, black, and Hispanic participants analyzed separately [15]. In the present analyses, neither predicted gene expression levels or polymorphisms with the lowest P-values were consistent across populations.
We cannot explain the apparent association between female sex and fewer grade 2 or greater CNS adverse events among all participants, and among white participants and black participants analyzed separately. Previous reports have been inconsistent in this regard, with studies showing no difference by sex [43, 44], increased efavirenz CNS adverse events in males [45], and increased CNS adverse events in females [46].
This study had limitations. Because providers may not have referred patients perceived to be at increased risk for CNS adverse events to studies of efavirenz, risk may be underestimated. Analyses largely involved white, black, and Hispanic participants in the United States, so findings may not translate to other countries or race/ethnicity groups. The open-label design of 3 of the 4 studies might have biased investigators into reporting CNS adverse events in patients randomized to efavirenz. While PrediXcan readily identified an association between predicted UGT1A1 expression in liver and bilirubin concentrations, this does not prove that we could detect associations of efavirenz CNS adverse events with predicted gene expression levels in brain. To our knowledge, there is no brain gene-phenotype pair in our dataset that could serve as a positive control. A larger sample size would increase power to identify associations. The present study was not designed to address rare polymorphisms, epigenetics, inducibility of gene expression, and trans regulatory elements. In addition, factors not evaluated in this study such as nicotine and alcohol use may affect expression of CYP2B6, CYP2A6, and other genes, and could conceivably differ by ancestry.
In summary, it is important to identify genetic factors that affect susceptibility to antiretroviral toxicities. The present study suggests that interindividual differences in brain neurotransmitter transporter/receptor genomics may not explain variable susceptibility to efavirenz CNS adverse events.
Supplementary Material
Acknowledgments
The authors are grateful to the many persons with HIV infection who volunteered for ACTG384, A5095, A5142, A5202 and A5128. In addition, we acknowledge the contributions of study teams and site staff for these protocols.
Joseph J. Eron has received research support from grants awarded to UNC from BMS, AbbVie, Janssen, ViiV Healthcare and Gilead. He is a consultant to BMS, AbbVie, Janssen, ViiV Healthcare, Merck and Gilead
Paul E. Sax has received research support from grants awarded to Brigham and Women’s Hospital from BMS, ViiV, and Gilead. He is a consultant to BMS, AbbVie, Janssen, ViiV, Merck and Gilead
Eric S. Daar has received research support from grants awarded to Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center from Gilead, Merck and ViiV. He is a consultant for Bristol Myers Squibb, Janssen, Merck, Teva and ViiV.
Grant support included:
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1 AI068634, UM1 AI068636 and UM1 AI106701. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This work was supported by the AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases (AI068636, AI038858, AI068634, AI038855). Grant support included AI069439, TR000445 (DWH), AI077505 (DWH), and AI50410 (University of North Carolina at Chapel Hill Center for AIDS Research). Clinical Research Sites that participated in ACTG protocols ACTG 384, A5095, A5142 and A5202, and collected DNA under protocol A5128, were supported by the following grants from NIAID: AI069477, AI027675, AI073961, AI069474, AI069432, AI069513, AI069423, AI050410, AI069452, AI69450, AI054907, AI069428, AI045008, AI069495, AI069415, AI069556, AI069484, AI069424, AI069532, AI069419, AI069471, AI025859, AI069418, AI050409, AI069501, AI069502, AI069511, AI069434, AI069465, AI069494, AI069472, AI069470, AI046376, AI072626, AI027661, AI034853, AI069447, AI032782, AI027658, AI27666, AI058740, AI046370, and by the following grants from the National Center for Research Resources (NCRR): RR00051, RR00046, RR025747, RR025777, RR024160, RR024996, and RR024156. Abbott Laboratories, Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, and GlaxoSmithKline provided study medications.
Footnotes
Potential conflicts:
No conflicts: David W. Haas, Roy M. Gulick, Yuki Bradford, Anurag Verma, Shefali S. Verma, Sarah A. Pendergrass, Gene D. Morse, Sharon Riddler, Edward P. Acosta, Marylyn D. Ritchie.
Previous presentation:
None
References
- 1.van Leth F, Phanuphak P, Ruxrungtham K, Baraldi E, Miller S, Gazzard B, et al. Comparison of first-line antiretroviral therapy with regimens including nevirapine, efavirenz, or both drugs, plus stavudine and lamivudine: a randomised open-label trial, the 2NN Study. Lancet. 2004;363(9417):1253–63. doi: 10.1016/S0140-6736(04)15997-7. [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM, Ribaudo HJ, Shikuma CM, Lustgarten S, Squires KE, Meyer WA, III, et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med. 2004;350(18):1850–61. doi: 10.1056/NEJMoa031772. [DOI] [PubMed] [Google Scholar]
- 3.Riddler SA, Haubrich R, DiRienzo AG, Peeples L, Powderly WG, Klingman KL, et al. Class-sparing regimens for initial treatment of HIV-1 infection. N Engl J Med. 2008;358(20):2095–106. doi: 10.1056/NEJMoa074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, et al. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet. 2009;374(9692):796–806. doi: 10.1016/S0140-6736(09)60918-1. [DOI] [PubMed] [Google Scholar]
- 5.Cooper DA, Heera J, Goodrich J, Tawadrous M, Saag M, Dejesus E, et al. Maraviroc versus Efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J Infect Dis. 2010;201(6):803–13. doi: 10.1086/650697. [DOI] [PubMed] [Google Scholar]
- 6.Daar ES, Tierney C, Fischl MA, Sax PE, Mollan K, Budhathoki C, et al. Atazanavir plus ritonavir or efavirenz as part of a 3-drug regimen for initial treatment of HIV-1. Ann Intern Med. 2011;154(7):445–56. doi: 10.1059/0003-4819-154-7-201104050-00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clifford DB, Evans S, Yang Y, Acosta EP, Goodkin K, Tashima K, et al. Impact of efavirenz on neuropsychological performance and symptoms in HIV-infected individuals. Ann Intern Med. 2005;143(10):714–21. doi: 10.7326/0003-4819-143-10-200511150-00008. [DOI] [PubMed] [Google Scholar]
- 8.Mollan KR, Smurzynski M, Eron JJ, Daar ES, Campbell TB, Sax PE, et al. Association between efavirenz as initial therapy for HIV-1 infection and increased risk for suicidal ideation or attempted or completed suicide: an analysis of trial data. Ann Intern Med. 2014;161(1):1–10. doi: 10.7326/M14-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arenas-Pinto A, Grund B, Sharma S, Martinez E, Cummins N, Fox J, et al. Risk of Suicidal Behavior With Use of Efavirenz: Results from the Strategic Timing of Antiretroviral Treatment Trial. Clin Infect Dis. 2018 doi: 10.1093/cid/ciy051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18(18):2391–400. [PubMed] [Google Scholar]
- 11.Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319(4):1322–6. doi: 10.1016/j.bbrc.2004.05.116. [DOI] [PubMed] [Google Scholar]
- 12.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genom. 2005;15(1):1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult AIDS Clinical Trials Group Study. J Infect Dis. 2005;192(11):1931–42. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Novoa S, Barreiro P, Rendon A, Jimenez-Nacher I, Gonzalez-Lahoz J, Soriano V. Influence of 516G>T polymorphisms at the gene encoding the CYP450-2B6 isoenzyme on efavirenz plasma concentrations in HIV-infected subjects. Clin Infect Dis. 2005;40(9):1358–61. doi: 10.1086/429327. [DOI] [PubMed] [Google Scholar]
- 15.Holzinger ER, Grady B, Ritchie MD, Ribaudo HJ, Acosta EP, Morse GD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genom. 2012;22(12):858–67. doi: 10.1097/FPC.0b013e32835a450b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemo. 2008;61(4):914–8. doi: 10.1093/jac/dkn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Sonnerborg A, Rane A, Josephson F, Lundgren S, Stahle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genom. 2006;16(3):191–8. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 18.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202(5):717–22. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genom. 2009;19(4):300–9. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 20.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6 (c.516G–>T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67(4):427–36. doi: 10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS. 2009;23(16):2101–6. doi: 10.1097/QAD.0b013e3283319908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas DW, Kwara A, Richardson DM, Baker P, Papageorgiou I, Acosta EP, et al. Secondary metabolism pathway polymorphisms and plasma efavirenz concentrations in HIV-infected adults with CYP2B6 slow metabolizer genotypes. J Antimicrob Chemother. 2014;69(8):2175–82. doi: 10.1093/jac/dku110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leger P, Chirwa S, Turner M, Richardson DM, Baker P, Leonard M, et al. Pharmacogenetics of efavirenz discontinuation for reported central nervous system symptoms appears to differ by race. Pharmacogenet Genomics. 2016;26(10):473–80. doi: 10.1097/FPC.0000000000000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollan KR, Tierney C, Hellwege JN, Eron JJ, Hudgens MG, Gulick RM, et al. Race/Ethnicity and the Pharmacogenetics of Reported Suicidality With Efavirenz Among Clinical Trials Participants. J Infect Dis. 2017;216(5):554–64. doi: 10.1093/infdis/jix248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins GK, De GV, Shafer RW, Smeaton LM, Snyder SW, Pettinelli C, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349(24):2293–303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafer RW, Smeaton LM, Robbins GK, De GV, Snyder SW, D’Aquila RT, et al. Comparison of four-drug regimens and pairs of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349(24):2304–15. doi: 10.1056/NEJMoa030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulick RM, Ribaudo HJ, Shikuma CM, Lalama C, Schackman BR, Meyer WA, III, et al. Three- vs four-drug antiretroviral regimens for the initial treatment of HIV-1 infection: a randomized controlled trial. JAMA. 2006;296(7):769–81. doi: 10.1001/jama.296.7.769. [DOI] [PubMed] [Google Scholar]
- 29.Haas DW, Wilkinson GR, Kuritzkes DR, Richman DD, Nicotera J, Mahon LF, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4(5):287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 30.U.S. Department of Health and Human Services, National Institutes of Health, National Institute of Allergy and Infectious Diseases, Division of AIDS. Division of AIDS Table for Grading the Severity of Adult and Pediatric Adverse Events, Version 1.0. 2009 Updated August 2009. Available at: http://rsc.tech-res.com/docs/default-source/safety/table_for_grading_severity_of_adult_pediatric_adverse_events.pdf. Accessed April 28, 2018.
- 31.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore CB, Verma A, Pendergrass S, Verma SS, Johnson DH, Daar ES, et al. Phenome-wide Association Study Relating Pretreatment Laboratory Parameters With Human Genetic Variants in AIDS Clinical Trials Group Protocols. Open Forum Infect Dis. 2015;2(1):ofu113. doi: 10.1093/ofid/ofu113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 35.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.liftOver [Internet] Available from: http://genome.ucsc.edu/cgi-bin/hgLiftOver. Accessed April 28, 2018.
- 37.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 38.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma SS, de Andrade M, Tromp G, Kuivaniemi H, Pugh E, Namjou B, et al. Imputation and quality control steps for combining multiple genome-wide datasets. doi: 10.3389/fgene.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price A EIGENSOFT [Internet] Available from: http://www.hsph.harvard.edu/alkes-price/software/. Accessed April 28, 2018.
- 41.Li B, Verma SS, Veturi YC, Verma A, Bradford Y, Haas DW, et al. Evaluation of PrediXcan for prioritizing GWAS associations and predicting gene expression. Pac Symp Biocomput. 2018;23:448–59. [PMC free article] [PubMed] [Google Scholar]
- 42.The GTEx Project. 2018 Available at: https://www.gtexportal.org/home/documentationPage. Accessed April 22, 2018.
- 43.Wynberg E, Williams E, Tudor-Williams G, Lyall H, Foster C. Discontinuation of Efavirenz in Paediatric Patients: Why do Children Switch? Clin Drug Investig. 2017 doi: 10.1007/s40261-017-0605-1. [DOI] [PubMed] [Google Scholar]
- 44.Smith KY, Tierney C, Mollan K, Venuto CS, Budhathoki C, Ma Q, et al. Outcomes by sex following treatment initiation with atazanavir plus ritonavir or efavirenz with abacavir/lamivudine or tenofovir/emtricitabine. Clin Infect Dis. 2014;58(4):555–63. doi: 10.1093/cid/cit747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhoro M, Ngara B, Kadzirange G, Nhachi C, Masimirembwa C. Genetic variants of drug metabolizing enzymes and drug transporter (ABCB1) as possible biomarkers for adverse drug reactions in an HIV/AIDS cohort in Zimbabwe. Curr HIV Res. 2013;11(6):481–90. doi: 10.2174/1570162x113119990048. [DOI] [PubMed] [Google Scholar]
- 46.Bisaso KR, Mukonzo JK, Ette EI. Markov model for characterizing neuropsychologic impairment and Monte Carlo simulation for optimizing efavirenz therapy. J Clin Pharmacol. 2015;55(11):1229–35. doi: 10.1002/jcph.533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


