Abstract
Several biogeographic barriers in the Central African highlands have reduced gene flow among populations of many terrestrial species in predictable ways. Yet, a comprehensive understanding of mechanisms underlying species divergence in the Afrotropics can be obscured by unrecognized levels of cryptic diversity, particularly in widespread species. We implemented a multilocus phylogeographic approach to examine diversity within the widely distributed Central African pygmy chameleon, Rhampholeon boulengeri. Gene-tree analyses coupled with a comparative coalescent-based species delimitation framework revealed R. boulengeri as a complex of at least six genetically distinct species. The spatiotemporal speciation patterns for these cryptic species conform to general biogeographic hypotheses supporting vicariance as the main factor behind patterns of divergence in the Albertine Rift, a biodiversity hotspot in Central Africa. However, we found that parapatric species and sister species inhabited adjacent habitats, but were found in largely non-overlapping elevational ranges in the Albertine Rift, suggesting that differentiation in elevation was also an important mode of divergence. The phylogeographic patterns recovered for the genus-level phylogeny provide additional evidence for speciation by isolation in forest refugia and dating estimates indicated that the Miocene was a significant period for this diversification. Our results highlight the importance of investigating cryptic diversity in widespread species to improve understanding of diversification patterns in environmentally diverse regions such as the montane Afrotropics.
Keywords: Biodiversity, Biogeography, Burundi, Diversification, Democratic Republic of the Congo, Kenya, Molecular systematics, Phylogeography, Rwanda, Uganda
Graphical abstract
1. Introduction
The East African Rift valley system started to form in the early Oligocene from hot mantle plumes causing up-lift of the African plate resulting in rifting, the formation of horst and grabens, and associated volcanic activity (Chorowicz, 2005; Paul et al., 2014). The Albertine Rift (AR) portion in Central Africa was initiated in the late Oligocene (Roberts et al., 2012) and increased geophysical rifting in the AR occurred during the Miocene (Macgregor, 2015). Rifting oscillations influenced forest environments in the AR, largely through uplift events that altered climate and drainage patterns across the region (Sepulchre et al., 2006). Miocene volcanism has also contributed to the age and distribution of AR forests (Griffiths, 1993). The paleoclimate of the AR was generally stable through the Cretaceous (Maley, 1996), during which tropical Africa was dominated by a nearly continuous rainforest block. African rainforests began to decline in extent throughout the Cenozoic, with a pronounced increase in forest losses after the mid-Miocene (Kissling et al., 2012). Altered precipitation patterns across East Africa, driven by global cooling, contributed to the decline of the African tropical forest ecosystem in the Miocene (Zachos et al., 2001). Decreased Miocene rainfall is linked to the expansion of grass-dominated savannas across East Africa (Jacobs et al., 1999), and as grasslands expanded, forests contracted, and thereby forest connectivity was greatly reduced during this period (Kissling et al., 2012). These ancient geologically and climatically induced forest dynamics during the Miocene have left a profound legacy on the geographic distribution of genetic diversity in forest-distributed fauna in the AR (e.g., Tolley et al., 2011), and may have left a greater genetic imprint than Quaternary ice ages (Hewitt, 2000).
The proposed timing and mechanisms that underlie the remarkably high biodiversity in forests of the AR are not conclusive. One line of evidence supports recent species divergence within Pleistocene (upper limit ca. 1.8 Mya) refugial habitats (i.e., Pleistocene Forest Refuge Hypothesis [Mayr and O’Hara, 1986]), whereas another suggests that divergence occurred before Pleistocene climatic changes and species have been maintained as paleoendemics since the Miocene (ca. 5–23 Mya) (i.e., Evolutionary Museum Hypothesis, a derivative of the Montane Speciation Hypothesis [Fjeldså and Lovett, 1997]). Both of these hypotheses are based on allopatric models of speciation from isolation in forest refugia, but they differ greatly in their timing of diversification events. Speciation in forest refugia that formed in response to Pleistocene climatic changes have been implicated as biogeographic drivers among small mammals (Demos et al., 2014, 2015), land snails (Boxnick et al., 2015; Wronski and Hausdorf, 2008), and birds (Bowie et al., 2006; Voelker et al., 2013). In contrast, several other taxa, including frogs (Larson et al., 2016; Portillo et al., 2015), chameleons (Hughes et al., in press; Tolley et al., 2011), and snakes (Greenbaum et al., 2015; Menegon et al., 2014) likely diversified during pre-Pleistocene biogeographic events, such as the reduction of forests in response to global cooling in the Miocene. Afromontane forests have functioned as stable refugia during ancient climate changes and thereby promoted vicariance-driven diversification in some AR taxa (e.g., Hughes et al., in press); however, this model does not fully account for the lack of genetic structure found in some widespread AR species (e.g., Greenbaum et al., 2013, 2015). Several physical biogeographic barriers have been identified in the AR, including the Virunga volcanoes that have been active from the Plio–Pleistocene to the present (Ebinger and Furman, 2003), and the uplift of the Rwenzori mountains that occurred around the Plio–Pleistocene boundary (ca. 3–2 Mya [Kaufmann et al., 2015]). These physical features have influenced patterns of gene flow for taxa between various highland areas of the AR (e.g., Huhndorf et al., 2007). However, genetic patterns for some AR taxa are not congruent with respect to identified barriers, and thus species-specific responses have been frequently detected (e.g., Hughes et al., in press). Much of the AR is ancient and several of its prominent geological features emerged before Pleistocene aridification pulses altered African ecosystems (e.g., deMenocal, 1995), and as a result, the AR represents an ideal region to test several biogeographic hypotheses regarding the timing and environmental mechanisms of biotic evolution.
The pygmy chameleon genus Rhampholeon currently contains 19 described taxa that are largely restricted to sub-montane and montane forests distributed across West, Central, and East Africa (Uetz et al., 2017). Many species of Rhampholeon are endemic to small forest fragments that face immediate threats of deforestation, and thus nine species are currently considered Endangered or Critically Endangered (IUCN, 2017). The Eastern Arc Mountains and Southern Rift Highlands, stretching from Kenya south to Tanzania, Malawi, and Mozambique, represent the highest regional concentration of species diversity for Rhampholeon with 16 species (Tolley and Herrel, 2013). The only pygmy chameleon species in West Africa is R. spectrum and its distribution extends from Nigeria and Central African Republic south to Gabon (Tilbury, 2010). Rhampholeon hattinghi and R. boulengeri occur allopatrically in the AR highlands of Central Africa. The recently described R. hattinghi is a Critically Endangered pygmy chameleon endemic to Mount Nzawa, Democratic Republic of the Congo (DRC), a massif in the southern AR (Tolley and Tilbury, 2015). Rhampholeon boulengeri is currently assessed as Least Concern, because it has a relatively large distribution in forest habitats across the AR, west into the Congo Basin (DRC) and east to Kakamega Forest (Kenya), and much of this forest is still relatively intact (Tolley and Plumptre, 2014). In addition to having one of the largest geographic distributions of any Rhampholeon species, R. boulengeri also occurs in forests across a remarkably wide range of elevations from 500 m to nearly 2300 m (Tilbury, 2010). Rhampholeon hattinghi is similar in appearance to R. boulengeri, and thus was initially considered to be a disjunct population of the more widespread species (e.g. Tilbury, 2010); however, genetic data revealed it as an independently evolving lineage (Tilbury and Tolley, 2015). Steindachner (1911) described R. boulengeri from a series of specimens collected by Rudolf Grauer in 1908. However, the type locality was imprecisely given as “forest beyond the sand hills on the north-western shores of Lake Tanganyika”. Tilbury and Tolley (2015) considered the Itombwe Plateau as the type locality because Rudolf Grauer collected specimens in 1908 from forests of the Itombwe Plateau, which is located to the northwest of Lake Tanganyika, however, Grauer did not write books about his travels and the precise localities where he collected in the plateau are unknown (Greenbaum, in press).
In general, Rhampholeon are considered forest specialists with low vagility (Branch et al., 2014), and are thus unlikely to disperse over long distances regardless of suitable habitat corridors (Matthee et al., 2004). As a result, most pygmy chameleon species are endemic to the montane localities from where they were originally described (Uetz et al., 2017). The morphology of Rhampholeon is considered highly conservative (Branch et al., 2014), from which a potential for cryptic species results (Bickford et al., 2007), particularly in geographic regions that have received only cursory attention to the biota, such as the AR (Greenbaum, in press). Moreover, several recent accounts have drawn attention to the likelihood that R. boulengeri represents a species complex (Tilbury, 2010; Tolley and Plumptre, 2014; Tilbury and Tolley, 2015). Therefore, R. boulengeri is an excellent model for investigating how diverse landscapes with complex histories of geomorphological and climatic changes have influenced the distribution of genetic diversity. In this study, we investigated the evolutionary history of R. boulengeri with a statistical framework to test three hypotheses related to cryptic diversity. We use a multilocus gene-tree and a comparative approach with four coalescent-based species-tree estimations to test whether R. boulengeri represents a single widespread species in the AR or a complex of genetically distinct species. We assess phylogeographic patterns and compare species distributions to test whether allopatric speciation driven by forest fragmentation underlies the diversification in the R. boulengeri species complex and for the genus in general. We implement fossil-calibrated Bayesian methods on a large-scale phylogeny to determine whether the timing of diversification in Rhampholeon follows a single break-up of African forests followed by isolation, or multiple forest fragmentations and reconnections over time.
2. Materials and methods
2.1. Taxon sampling and DNA sequencing
Forty-six samples of R. boulengeri were collected during field surveys in various forests across four Central African countries of the AR from 2008–2016, including Burundi, DRC, Rwanda, and Uganda (Table 1). Two additional samples were collected from the Yala Nature Reserve, Kakamega Forest, western Kenya. We also included additional sequences in our analyses that were not generated for this study: ND2 fragments and one RAG1 fragment for two individuals from Bwindi Impenetrable National Park, Uganda (CAS 2016 81–82); a 16S fragment for an individual from Irangi (near Kahuzi-Biega National Park), DRC (ZFMK 47571); and a 16S fragment for an individual from Cyamudongo Forest, Rwanda (ZFMK 55104) (Fisseha et al., 2013). For phylogenetic analyses, we included 18 of the 19 currently recognized Rhampholeon species and three species of Rieppeleon as outgroups (Branch et al., 2014). We excluded the species R. beraduccii from phylogenetic analyses because only a single sequence for the mitochondrial fragment 16S is available on GenBank. Rhampholeon beraduccii, from the Mahenge Mountains in southern Tanzania, is a member of the subgenus Rhinodigitum with close affinities to R. acuminatus; however, this phylogenetic placement was based on analyses of the 16S gene only (Mariaux and Tilbury, 2006; Fisseha et al., 2013).
Table 1.
Species identifications, specimen catalog numbers, GenBank accession numbers, and collecting localities for pygmy chameleons (Rhampholeon) analyzed in this study.
| Species | Catalog No. | 16S | ND2 | RAG1 | Locality |
|---|---|---|---|---|---|
| Rhampholeon sp. 1 | JS 41688 | Kenya: Kakamega Forest, Yala Nature Reserve | |||
| Rhampholeon sp. 1 | JS 41690 | Kenya: Kakamega Forest, Yala Nature Reserve | |||
| Rhampholeon sp. 1 | CAS 201681 | * | AY524916 | Uganda: Kabale District, Ihihizo River, Bwindi Impenetrable National Park | |
| Rhampholeon sp. 1 | CAS 201682 | * | AY524915 | N/A | Uganda: Kabale District, Ihihizo River, Bwindi Impenetrable National Park |
| Rhampholeon sp. 1 | ELI 2764 | Uganda: Kabale District, Ihihizo River, Bwindi Impenetrable National Park | |||
| Rhampholeon sp. 1 | ELI 2816 | Uganda: Kabale District, Ihihizo River, Bwindi Impenetrable National Park | |||
| Rhampholeon sp. 1 | CFS 1025 | DRC: North Kivu Province, Bunyantenge village | |||
| Rhampholeon sp. 1 | CFS 1026 | DRC: North Kivu Province, Mount Vibende | |||
| Rhampholeon sp. 1 | CFS 1013 | DRC: North Kivu Province, Kaunzo village | |||
| Rhampholeon sp. 1 | MUSE 10193 | DRC: South Kivu Province, Kahuzi-Biega National Park, Luyuyu | |||
| Rhampholeon sp. 1 | MUSE 10226 | DRC: Maniema Province, Kahuzi-Biega National Park, Nkumwa | |||
| Rhampholeon sp. 1 | MTSN 6898 | DRC: South Kivu Province, Kahuzi-Biega National Park, Madiriri | |||
| Rhampholeon sp. 1 | MTSN 6899 | DRC: South Kivu Province, Kahuzi-Biega National Park, Madiriri | |||
| Rhampholeon sp. 2 | CRSN 2833 | DRC: Ituri Province, Bongobongo village | |||
| Rhampholeon sp. 2 | CRSN 2837 | DRC: Ituri Province, Lodjo village | |||
| Rhampholeon sp. 2 | DFH 558 | Uganda: Western Region, Budongo Central Forest Reserve, Sonso River | |||
| Rhampholeon sp. 2 | DFH 559 | Uganda: Western Region, Budongo Central Forest Reserve, Sonso River | |||
| Rhampholeon sp. 2 | ELI 464 | DRC: South Kivu Province, Bizombo village | |||
| Rhampholeon sp. 2 | ELI 465 | DRC: South Kivu Province, Bizombo village | |||
| Rhampholeon sp. 2 | EBG 2737 | DRC: South Kivu Province, Bizombo village | |||
| Rhampholeon sp. 2 | MUSE 10178 | DRC: South Kivu Province, Kahuzi-Biega National Park, Ikundwe | |||
| Rhampholeon sp. 2 | MUSE 10589 | DRC: Maniema Province, Kahuzi-Biega National Park, Kyasa | |||
| Rhampholeon sp. 2 | ZFMK 47571 | AM055647 | N/A | N/A | DRC: South Kivu Province, Irangi village |
| Rhampholeon sp. 2 | DFH 1101 | Uganda: Western Region, Kibale Forest National Park, Ngogo Research Center | |||
| Rhampholeon sp. 3 | ELI 602 | DRC: South Kivu Province, Kalundu village | |||
| Rhampholeon sp. 3 | ELI 617 | DRC: South Kivu Province, Kalundu village | |||
| Rhampholeon sp. 3 | CRSN 2984 | DRC: South Kivu Province, Mwana village | |||
| Rhampholeon sp. 3 | EBG 1286 | DRC: South Kivu Province, in the vicinity of Irangi village | |||
| Rhampholeon sp. 3 | EBG 1346 | DRC: South Kivu Province, in the vicinity of Irangi village | |||
| Rhampholeon sp. 3 | ELI 739 | DRC: South Kivu Province, Tumungu village | |||
| Rhampholeon sp. 3 | MUSE 10146 | DRC: South Kivu Province, Mabwe village | |||
| Rhampholeon sp. 3 | MTSN 7123 | Rwanda: Nyungwe National Park, Cyamudongo Forest | |||
| Rhampholeon sp. 3 | MUSE 10146 | DRC: South Kivu Province, Tshibati village | |||
| Rhampholeon sp. 3 | JMD 2014-53 | Rwanda: Nyungwe National Park, Kamiranzovu Swamp | |||
| Rhampholeon sp. 3 | JMD 2014-101 | Rwanda: Nyungwe National Park, Kamiranzovu Swamp | |||
| Rhampholeon sp. 3 | ZFMK 55104 | AM055645 | N/A | N/A | Rwanda: Nyungwe National Park, Cyamudongo Forest |
| Rhampholeon sp. 4 | ELI 1159 | Burundi: Bubanza District, Kibira National Park | |||
| Rhampholeon sp. 4 | MTSN 7213 | Rwanda: Nyungwe National Park, Mount Bigugu | |||
| Rhampholeon sp. 5 | ELI 906 | Burundi: Bururi District, Bururi Forest Nature Reserve | |||
| Rhampholeon sp. 5 | ELI 907 | Burundi: Bururi District, Bururi Forest Nature Reserve | |||
| Rhampholeon sp. 5 | ELI 911 | Burundi: Bururi District, Bururi Forest Nature Reserve | |||
| Rhampholeon sp. 5 | CFS 1599 | Burundi: Bururi District, Bururi Forest Nature Reserve Uganda: Western Region, Rwenzori |
|||
| Rhampholeon sp. 5 | ELI 2851 | Mountains National Park, near Nyakalengisa entrance | |||
| Rhampholeon sp. 5 | CT 347 | Uganda: Western Region, Rwenzori Mountains National Park Uganda: Western Region, Rwenzori |
|||
| Rhampholeon sp. 5 | ELI 2838 | Mountains National Park, near Nyakalengisa entrance | |||
| Rhampholeon sp. Itombwe | EBG 1613 | KM589410 | KM589405 | KM589416 | DRC: South Kivu Province, Bichaka village, Itombwe Plateau |
| Rhampholeon sp. Itombwe | EBG 1700 | DRC: South Kivu Province, Bichaka village, Itombwe Plateau | |||
| Rhampholeon sp. Itombwe | EBG 1701 | DRC: South Kivu Province, Bichaka village, Itombwe Plateau | |||
| Rhampholeon sp. Itombwe | EBG 1702 | KM589411 | KM589406 | KM589417 | DRC: South Kivu Province, Bichaka village, Itombwe Plateau |
| Rhampholeon acuminatus (T) | CT 153 | HF570459 | N/A | N/A | Tanzania: Nguru Mountains |
| Rhampholeon bruessoworum (T) | PEM-R 20374 | HG798975 | HG798989 | HG798999 | Mozambique: Mount Inago |
| Rhampholeon bruessoworum (T) | PEM-R 20375 | HG798976 | HG798990 | HG799000 | Mozambique: Mount Inago |
| Rhampholeon chapmanorum (T) | PEM-R 16245 | AY524881 | AY524919 | AY524956 | Malawi: Malawi Hill |
| Rhampholeon gorongosae (T) | PEM-R 16252 | AY524873 | AY524911 | AY524949 | Mozambique: Mount Gorongosa |
| Rhampholeon gorongosae (T) | PEM-R 16253 | AY524874 | AY524912 | AY524950 | Mozambique: Mount Gorongosa |
| Rhampholeon hattinghi (P) | PEM-R 19196 | KM589414 | KM589408 | N/A | DRC: Tanganyika Province, Mount Nzawa |
| Rhampholeon hattinghi (P) | PEM-R 19197 | KM589415 | KM589409 | N/A | DRC: Tanganyika Province, Mount Nzawa |
| Rhampholeon maspictus (T) | PEM-R 17911 | HG798971 | HG798984 | HG798997 | Mozambique: Mount Mabu |
| Rhampholeon maspictus (T) | PEM-R 17912 | HG798972 | HG798985 | HG798998 | Mozambique: Mount Mabu |
| Rhampholeon marshalli | PEM-R 16243 | AY524870 | AY524908 | AY524946 | Zimbabwe: Vumba Mountains |
| Rhampholeon marshalli | PEM-R 16244 | AY524871 | AY524909 | AY524947 | Zimbabwe: Vumba Mountains |
| Rhampholeon moyeri (P) | MTSN 001TA | AY524876 | AY524914 | AY524952 | Tanzania: Udzungwa Mountains |
| Rhampholeon moyeri (P) | MTSN 002TA | N/A | AY524913 | AY524951 | Tanzania: Udzungwa Mountains |
| Rhampholeon nchisiensis (T) | PEM-R 16242 | AY524883 | AY524921 | AY524958 | Malawi: Nchisi Mountain |
| Rhampholeon nchisiensis | PEM-R 16249 | AY524886 | AY524924 | AY524961 | Zambia: Nyika Plateau |
| Rhampholeon nebulauctor (P) | PEM-R 17280 | HG798973 | HG798987 | N/A | Mozambique: Mount Chiperone |
| Rhampholeon nebulauctor (A) | PEM-R 17281 | HG798974 | HG798988 | N/A | Mozambique: Mount Chiperone |
| Rhampholeon platyceps (T) | PEM-R 16250 | AY524880 | AY524918 | AY524955 | Malawi: Mount Mlanje |
| Rhampholeon platyceps (T) | PEM-R 16251 | AY524879 | AY524917 | AY524954 | Malawi: Mount Mlanje |
| Rhampholeon spectrum | CAS 207682 | AY524864 | AY524901 | AY524939 | Equatorial Guinea: Bioko Island |
| Rhampholeon spectrum | CAS 207683 | AY524863 | AY524900 | AY524938 | Equatorial Guinea: Bioko Island |
| Rhampholeon spinosus | CT 118 | HF570460 | HF570510 | HF570779 | Tanzania: West Usambara Mountains |
| Rhampholeon temporalis (T) | PEM-R 16254 | AY524866 | AY524904 | AY524942 | Tanzania: East Usambara Mountains |
| Rhampholeon temporalis (T) | PEM-R 16255 | AY524867 | AY524905 | AY524943 | Tanzania: East Usambara Mountains |
| Rhampholeon tilburyi (T) | PEM-R 17134 | EF114322 | EF114330 | EF114338 | Mozambique: Namuli Massif |
| Rhampholeon tilburyi (T) | PEM-R 17135 | EF114323 | EF114331 | EF114339 | Mozambique: Namuli Massif |
| Rhampholeon uluguruensis (T) | ZMB 48421 | AY524896 | AY524934 | N/A | Tanzania: Uluguru Mountains |
| Rhampholeon uluguruensis (T) | ZMB 48431 | AY524897 | AY524935 | N/A | Tanzania: Uluguru Mountains |
| Rhampholeon viridis | CT 204 | HF570461 | HF570511 | HF570780 | Tanzania: North Pare Mountains |
| Rhampholeon viridis (T) | PEM-R 16259 | AY524869 | AY524907 | AY524945 | Tanzania: South Pare Mountains |
| Rhampholeon viridis (T) | PEM-R 16260 | AY524868 | AY524906 | AY524944 | Tanzania: South Pare Mountains |
| Rieppeleon brachyurus | PEM-R 16263 | AY524898 | AY524936 | AY524968 | Tanzania: Near Tamota |
| Rieppeleon brachyurus | PEM-R 16264 | AY524899 | AY524937 | AY524969 | Tanzania: Near Tamota |
| Rieppeleon brevicaudatus (T) | PEM-R 16256 | AY524887 | AY524925 | AY524962 | Tanzania: East Usambara Mountains |
| Rieppeleon brevicaudatus (T) | PEM-R 16257 | AY524888 | AY524926 | AY524963 | Tanzania: East Usambara Mountains |
| Rieppeleon kerstenii | CAS 169939 | AY524890 | AY524928 | AY524965 | Kenya: Kilifi |
| Rieppeleon kerstenii | N/A | AY524892 | AY524930 | AY524967 | N/A |
Institutional abbreviations follow Sabaj (2016). DRC = Democratic Republic of the Congo;
= data not used due to poor quality; A = allotype; H = holotype; P = paratype; T = topotype.
We harvested tissues from the liver or hind limb muscle of chameleons before formalin fixation and preserved these tissues in 2-ml vials containing 99% ethanol. Genomic DNA was isolated from tissue samples with the Qiagen DNeasy tissue kit (Qiagen Inc., Valencia, CA, USA). PCR amplification and cycle sequencing of two mitochondrial gene fragments were carried out with the following primers for ND2: L4347 (Macey et al., 1997a) and H5934 (Macey et al., 1997b), and 16S: L2510 and H3080 (Palumbi, 1996). A fragment of the nuclear gene RAG1 was sequenced using primers G396 (R13) and G397 (R18) (Groth and Barrowclough, 1999). Although RAG1 is a relatively slowly evolving nuclear gene (Groth and Barrowclough, 1999), it has been demonstrated to be a useful marker for studying deep divergences among vertebrates (San Mauro et al., 2004; Sullivan et al., 2006) and it also has been used extensively in pygmy chameleon systematics (e.g., Tilbury and Tolley, 2015). We used 25 μL PCR reactions with an initial denaturation step of 95 °C for 2 min, followed by denaturation at 95 °C for 35 s, annealing at 50 °C for 35 s, and extension at 72 °C for 95 s, with 4 s added to the extension per cycle for 32 (mitochondrial genes) or 34 (nuclear gene) cycles. Amplification products were visualized on a 1.5% agarose gel stained with Invitrogen SYBR Safe DNA gel stain (Thermo Fisher Scientific, Waltham, MA, USA). Sequencing reactions were purified with Agencourt CleanSEQ magnetic bead solution (Beckman Coulter Inc., Brea, CA, USA) and sequenced with an ABI 3130xl automated sequencer at the University of Texas at El Paso (UTEP) Border Biomedical Research Center (BBRC) Genomic Analysis Core Facility.
2.2. Gene trees and species trees
We interpreted chromatograph data using the program SeqMan Pro (Swindell and Plasterer, 1997) and made alignments for each gene using MUSCLE v. 3.6 (Edgar, 2004) in the program Mesquite v. 3.04 (Maddison and Maddison, 2015). We conservatively trimmed sequences and made other minor manual adjustments in the program MacClade v. 4.08 (Maddison and Maddison, 2005). We used PHASE v. 2.1.1 (Stephens and Donnelly, 2003) in the program DnaSP v. 5.1 (Librado and Rozas, 2009) to phase haplotypes for the nuclear fragment (RAG1). We transformed alignments using SeqPHASE (Flot, 2010) and excluded haplotypes with probabilities lower than 0.7 (Harrigan et al., 2008). In addition, we note that most double peaks in the nuclear marker were remedied in SeqMan Pro because one allele exhibited a much stronger signal than the other did (i.e., unequal heights) (Fontaneto et al., 2015). Phased RAG1 sequences were used for all species-delimitation analyses (see below).
Phylogenetic analyses were initially conducted on mitochondrial and nuclear data sets that revealed similar topologies, and thus we used the concatenated data set for all analyses. Maximum likelihood (ML) analyses were conducted with the GTRGAMMA model in RAxML v. 8.2.2 (Stamatakis, 2006; Stamatakis, 2014). All parameters were estimated and a random starting tree was used. Support values for clades inferred by ML analyses were assessed with the rapid bootstrap algorithm with 1,000 replicates (Stamatakis et al., 2008). Bayesian inference (BI) analyses were conducted in MrBayes v. 3.2.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003). Our model included seven data partitions, including a single partition for 16S and three independent partitions for each codon position for the protein-coding genes ND2 and RAG1. Concatenated data sets were partitioned identically for ML and BI analyses and were run on the CIPRES Science Gateway v. 3.3 (http://www.phylo.org/). The Akaike Information Criterion (AIC) and greedy search algorithm in PartitionFinder v. 1.1 (Lanfear et al., 2012) were used to establish the best model of evolution for each marker. The selected models of evolution were used for all BI analyses, but in cases where the model selected in PartitionFinder was not available in MrBayes, we set the number of rate categories and other parameters to match the best model. Bayesian analyses were conducted with random starting trees, run for 20 million generations, and Markov chains were sampled every 1,000 generations. To verify that multiple runs converged, AWTY (Nylander et al., 2008) was used. Burn-in was set at 25%, and thus the initial 5,000 trees were discarded. Phylogenies were visualized using FigTree v. 1.3.1 (Rambaut and Drummond, 2009). Bayesian posterior probabilities ≥ 0.95 (Hillis and Bull, 1993; Alfaro et al., 2003) and bootstrap values ≥ 70% (Felsenstein, 1981, 1985) were considered as strong support. Net sequence divergences (uncorrected p-distances) between Rhampholeon lineages for each marker were estimated using MEGA v. 7.0 (Kumar et al., 2016).
When inferring species limits from multilocus data, two issues are widely recognized: the underlying species tree can be different from individual gene trees (Maddison, 1997) and that simply increasing the number of loci does not necessarily improve the delimitation of species (Degnan and Rosenberg, 2009). To account for these uncertainties, we implemented a comparative coalescent framework to estimate species trees for the AR Rhampholeon clade and we conservatively interpreted species across four separate species-tree approaches. For approach 1, we used the Bayesian *BEAST (Heled and Drummond, 2010) in the program BEAST v. 1.8.4 (Drummond et al., 2012) to estimate a species tree for the focal taxa. *BEAST necessitates the prior assignment of individuals to presumed species, so we based our initial species assignments on reciprocally monophyletic clades recovered in the concatenated gene-tree analyses. As additional evidence for species assignments, we compared uncorrected p-distances of the ND2 locus among these clades to described species of Rhampholeon. Models of sequence evolution were chosen using the AIC in PartitionFinder. We specified unlinked site, clock, and tree models, and implemented a Yule process tree prior as this analysis investigates interspecific relationships. We estimated species trees with five concurrent runs of 200 million generations that totaled 2 billion generations sampled every 20,000 generations. Each run produced 10,000 trees and all runs were combined using LogCombiner (Drummond et al., 2012) for a total of 50,000 trees. We discarded the initial 10% of trees as burn-in with the program TreeAnnotator (Drummond et al., 2012), and used the program Tracer v. 1.5 (Rambaut and Drummond, 2007) to asses Effective Sample Size (ESS) values, which were > 200 for all parameters. For approach 2, we used the maximum pseudo-likelihood function in the program MP-EST (Liu et al., 2010) on the web-server STRAW (Shaw et al., 2013) to estimate a species tree from our collection of three ML gene trees (16S, ND2, RAG1). Individual gene trees were generated with RAxML under the GTRGAMMA model with 1,000 rapid bootstrap inferences on each genetic data set with three Rieppeleon species as the outgroup. The pseudo-likelihood function is derived from coalescent theory, assumes no gene flow, and can be validated with bootstrap support (Liu et al., 2010). For approach 3, we used the Bayesian program bPTP (Zhang et al., 2013) on the web-server (http://species.h-its.org/) to estimate a species tree using our ML tree of the concatenated data. The MCMC analysis was run for 500,000 generations with thinning set to 100 and burn in at 10%. We used a rooted tree and did not exclude the Rieppeleon outgroup. Species representation was examined using trace plots to check for convergence of the maximum likelihood’s value of each node. For approach 4, we used the coalescent model GMYC (Pons et al., 2006; Fujisawa and Barraclough, 2013) on the web-server (http://species.h-its.org/gmyc/) to estimate a species tree for the focal taxa using both single and multi-threshold models for the dated phylogeny based on the concatenated data set obtained in BEAST (see below).
2.3. Divergence dating
We estimated divergence times for the comprehensive Rhampholeon phylogeny using a fossil-calibrated Bayesian approach in the program BEAST v. 1.8.4 (Drummond et al., 2012) run on the CIPRES Science Gateway. We implemented an uncorrelated log-normal relaxed clock model with an estimated clock rate to allow for rate heterogeneity among lineages (Drummond et al., 2006). We estimated tree shape under the Yule prior (pure birth) on our multilocus data set because this prior is best suited for phylogenies describing the relationships between different species and assumes a constant speciation rate. In order to maximize calibration points, and because inadequate outgroup selections can produce misleading dating estimates (Sauquet, 2013), we included at least three species per chameleon genus (when available on GenBank) to provide a robust representation of Chamaeleonidae in these dating analyses. Furthermore, to utilize as many fossil calibrations as possible, we included several major representatives of the superorder Lepidosauria totaling 22 squamate taxa plus Sphenodon punctatus (Table S1). In some cases, chimeric sequences were constructed using more than one species from the same genus for these more distantly related groups (Pyron et al., 2013). We enforced monophyly for six chameleon genera based on the relationships recovered in the family level phylogeny of Tolley et al. (2013) (i.e., Calumma + Furcifer; Bradypodion + Nadzikambia; Trioceros + Kinyongia), because initial runs produced topologies inconsistent with interspecific relationships among chameleon genera, most likely due to incomplete taxon sampling at the family level in our phylogeny. Fossil calibrations were placed on nine nodes that correspond to some of the oldest known fossils of Lepidosauria (Table 2). Secondary calibrations were placed on five nodes to achieve temporal congruence with the most comprehensive time-calibrated chameleon phylogeny published to date—dating analyses that were based on 12 genetic markers and included over 90% of all named chameleon species (Tolley et al., 2013). For each calibration, we used a translated log-normal distribution, with an offset equal to the age of the fossil or node split. The treatment of date estimates from independent molecular analyses as point calibrations without consideration of associated error can increase the probability of type I errors (Ho and Phillips, 2009). We attempted to mitigate this potential pitfall by accounting for calibration uncertainty in our use of dates from Tolley et al. (2013) and including multiple primary calibrations rather than relying on a single point calibration (Graur and Martin, 2004). We estimated phylogenetic relationships from five concurrent runs of 100 million generations each and we sampled trees every 5,000 generations. We used the program LogCombiner to combine the trees produced from the five runs, which resulted in 100,000 trees. We discarded 10% of the trees (10,000 trees) as burn-in and summarized parameter values from the posterior probabilities on the maximum clade credibility tree using the program TreeAnnotator. The program Tracer was used to confirm stationarity and adequate ESS of the posterior probabilities (> 200 for each estimated parameter). Bayesian posterior probabilities ≥ 95% were considered as strong support.
Table 2.
Divergence-date priors for primary (top) and secondary (bottom) calibrations. Node numbers correspond to those indicated in Fig. S2. The translated log-normal (TL) zero-offset is presented in millions of years ago (Mya), parameter values (mean and standard deviation) follow in parentheses, and posterior (calculated) ages are presented as median with 95% confidence interval in parentheses.
| PRIMARY CALIBRATIONS
| |||
|---|---|---|---|
| Node | TL zero-offset (mean, SD) | Median (95% CI) | Source |
| 1 | 238 (1.4, 0.7) | 242 (239.4–249.6) | Fossil rhynchocephalian from Middle Triassic (Jones et al., 2013) |
| 2 | 161 (1.8, 1.0) | 167 (162.2–192.3) | Stem scincomorph Balnealacerta from Middle Jurassic (Evans, 1998) |
| 3 | 110 (1.8, 1.3) | 116 (110.7–161.3) | Stem teiids (e.g. Ptilodon) from Early Cretaceous (Nydam and Cifelli, 2002; Winkler et al., 1990) |
| 4 | 61 (1.6, 0.8) | 66.1 (62.4–80) | Fossil amphisbaenian Plesiorhineura tsentasi from Middle Paleocene (Sullivan, 1985) |
| 5 | 128 (1.0, 0.5) | 130.7 (129.2–134.2) | Fossil lizard Dalinghosaurus longidigitus from Early Cretaceous (Evans and Wang, 2005) |
| 6 | 70 (1.8, 1.0) | 76.1 (71.2–101.3) | Fossil anguid Odaxosaurus from Late Cretaceous (Sullivan and Lucas, 1996) |
| 7 | 70 (1.8, 1.0) | 76.1 (71.2–101.3) | Stem acrodont iguanian clade Priscagaminae from Late Cretaceous (Keqin and Norell, 2000) |
| 8 | 70 (1.2, 1.9) | 73.3 (70.2–145.6) | Fossil pleurodont iguanian Saichangurvel from Late Cretaceous (Conrad and Norell, 2007) |
| 9 | 99 (1.0, 0.5) | 101.7 (100.2–105.2) | Stem chameleon from Albian-Cenomanian boundary, Cretaceous (Daza et al., 2016) |
|
SECONDARY CALIBRATIONS | |||
| Node | TL zero-offset (mean, SD) | Median (95% CI) | Source |
|
| |||
| 10 | 62.8 (1.0, 0.5) | 64.8 (63.3–68.3) | Node 1 by codon in Table S4 (Tolley et al., 2013) |
| 11 | 51.2 (1.0, 0.5) | 53.9 (52.4–57.4) | Node 2 by codon in Table S4 (Tolley et al., 2013) |
| 12 | 47.5 (1.0, 0.5) | 50.2 (48.7–53.7) | Node 3 by codon in Table S4 (Tolley et al., 2013) |
| 13 | 45.6 (1.0, 0.5) | 48.3 (46.8–51.8) | Node 4 by codon in Table S4 (Tolley et al., 2013) |
| 14 | 33.4 (1.0, 0.5) | 36.1 (34.6–39.6) | Node 5 by codon in Table S4 (Tolley et al., 2013) |
3. RESULTS
3.1. Sampling, sequencing, and evolutionary models
We generated 138 new sequences of three genetic markers for a multilocus data set consisting of 2045 bp per sample (16S: 450 bp; ND2: 690 bp; RAG1: 905 bp). There were no gaps in the alignments of any of the three loci after we conservatively omitted a hypervariable region consisting of 16 bp from the 16S ribosomal gene. The most appropriate substitution models estimated for each locus were GTR+G for 16S; GTR+G for ND2 1st codon position, HKY+G for ND2 2nd codon position, TrN+G for ND2 3rd codon position; and HKY+G for RAG1 1st and 2nd codon positions, K80 for RAG1 3rd codon position.
3.2. Gene trees and genetic distances
Phylogenetic relationships among Rhampholeon species, reconstructed from the concatenated data set, were similar and with comparable node support using BI and ML methods. Six distinct clades representing R. boulengeri were recovered (Fig. 1). The recently described species R. hattinghi, endemic to Mount Nzawa in the southern AR, was recovered with strong support as sister to the R. boulengeri clade. A population of R. cf. boulengeri, collected from high-elevation forests of the Itombwe Plateau in eastern DRC (R. sp. Itombwe), formed a distinct clade that was weakly supported as sister to R. sp. 3. Rhampholeon sp. 1 contained samples from populations occupying mid to high-elevation forests in eastern DRC, southwestern Uganda, and the Yala Nature Reserve near Kakamega Forest Reserve in western Kenya. Rhampholeon sp. 1 was recovered as sister to R. sp. 2 (with strong support in BI analyses), which comprised widely distributed populations from low and mid-elevation forests in eastern DRC and western Uganda (Figs. 1–3). Rhampholeon sp. 3 contained populations from mostly mid-elevation forests of the Itombwe Plateau and some high-elevation forests around the southern and western sides of Lake Kivu, including Nyungwe National Park in Rwanda and Kahuzi-Biega National Park in DRC (Figs. 1–3). Rhampholeon sp. 4 contained two samples collected from high-elevation forests of the Rugege Highlands, including Mount Bigugu of Nyungwe National Park and Mpishi village near Kibira National Park in Burundi, and was found to be closely related to R. sp. 5, which contained populations from high-elevation forests of Bururi Forest Reserve in southern Burundi and Rwenzori Mountains National Park in western Uganda (Figs. 1–3).
Figure 1.
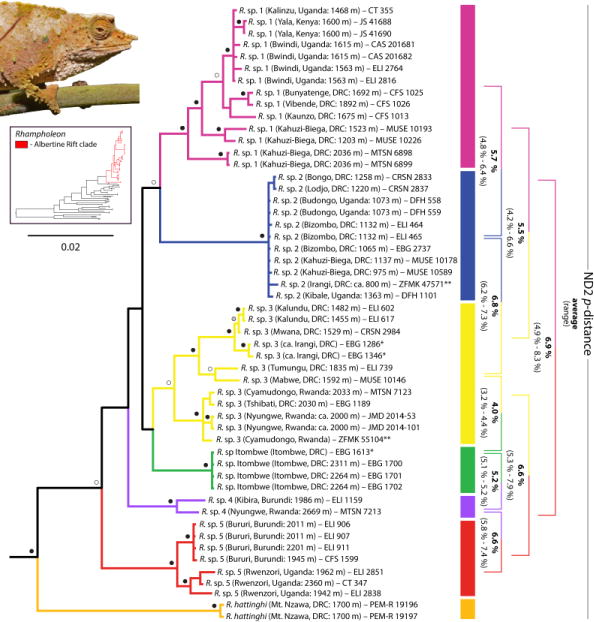
Bayesian phylogeny of pygmy chameleon species (genus Rhampholeon) from the Albertine Rift, Central Africa, and western Kenya. Color-coded rectangles correspond to the Albertine Rift/western Kenya species as follows (from top of phylogeny down): pink – R sp. 1; blue – R. sp. 2; yellow – R. sp. 3; green – R. sp. Itombwe; purple – R. sp. 4; red – R. sp. 5; orange – R. hattinghi. This color scheme is retained throughout all figures. Uncorrected p-distances for the ND2 marker are given as a range for selected species on the right. Nodes supported by both ML (≥ 70% bootstrap) and BI (≥ 0.95 posterior probabilities) are denoted with black circles, nodes supported by BI only are denoted with white circles, and nodes supported by ML only are denoted with grey circles. * = specific locality not available. ** = only 16S data were available.
Figure 3.
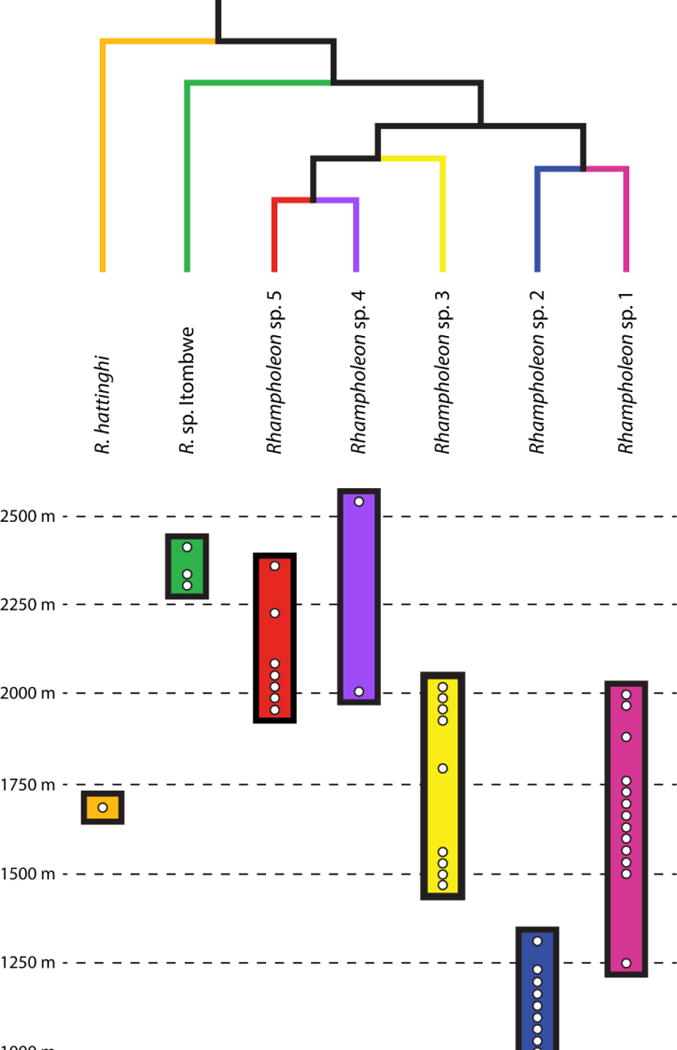
Elevational zonation of seven pygmy chameleon species (genus Rhampholeon) from the Albertine Rift, Central Africa, and western Kenya. The upper and lower known elevational limits of species distributions are indicated by colored rectangles. White circles within the colored rectangles represent samples used in the phylogenetic analyses. The topology is based on the complete phylogeny in Fig. 4.
Pairwise sequence divergences (uncorrected p-distances) between the undescribed lineages were generally high and comparable to currently recognized Rhampholeon species (Table S2). For the ND2 locus, p-distances ranged from 4.8–6.4% between R. sp. 1 and R. sp. 2; 6.2–7.3% between R. sp. 2 and R. sp. 3; 4.1–6.6% between R. sp. 3 and R. sp. 4; and 5.8–7.4% between R. sp. 4 and R. sp. 5. Moreover, p-distance ranges for this locus between undescribed clades and R. sp. Itombwe from the Itombwe Plateau were also relatively high: 4.2–5.9% for R. sp. 1; 5.9–6.2% for R. sp. 2; 3.2–4.4% for R. sp. 3; 5.1–5.2% for R. sp. 4; and 5.1–6.4% for R. sp. 5. Lastly, intraspecific p-distances among these species were comparatively low (see Table S2).
3.3. Divergence dating
Results from the calibrated dating analyses indicate that the genus Rhampholeon diverged from other chameleons in the early Eocene at 53.59 Mya (51.92–56.19 Mya, 95% highest posterior densities [HPD]), and initial branching within the genus occurred in the mid-Eocene at 44.99 Mya (39.29–50.44 Mya, HPD) (Fig. 4). The majority of species-level diversification occurred in the Miocene, although several lineages arose earlier (Eocene and Oligocene), and a few originated in the Pliocene. The only West African species, R. spectrum, diverged in the early Eocene at 41.31 Mya (33.56–47.33 Mya, HPD) from its sister clade, which includes species from the Eastern Arc Mountains of Tanzania. The most southerly species, R. gorongosae and R. marshalli, also diverged in the Eocene from a sister clade containing primarily East African species. The AR clade diverged in the mid-Miocene at 18.06 Mya (12.59–23.27 Mya, HPD) from its closest relative, R. acuminatus from the Nguru Mountains in eastern Tanzania. The earliest divergence of AR Rhampholeon (i.e., split between R. hattinghi and the R. boulengeri clade) occurred near the start of the late Miocene at 11.14 Mya (7.68–14.68 Mya, HPD) (Fig. 4). The divergence of R. sp. Itombwe from the other five R. cf. boulengeri species (R. sp. 1, R. sp. 2, R. sp. 3, R. sp. 4, and R. sp. 5) was dated in the late Miocene around 7.16 Mya (5.34–9.01 Mya, HPD). The initial divergence within the remaining R. boulengeri clade was estimated in the late Miocene at 6.25 Mya (4.78–7.72 Mya, HPD), with most of the species-level divergence at the Miocene–Pliocene boundary around 4–5 Mya (Fig. 4). For example, estimated splits between R. sp. 1 and R. sp. 2 occurred at 5.33 Mya (3.84–6.81 Mya, HPD); R. sp. 3 was estimated to have diverged from the clade containing R. sp. 4 and R. sp. 5 at 5.74 Mya (4.27–7.08 Mya, HPD); and the divergence between R. sp. 4 and R. sp. 5 was estimated to have occurred at 5.54 Mya (3.69–6.61 Mya, HPD).
Figure 4.
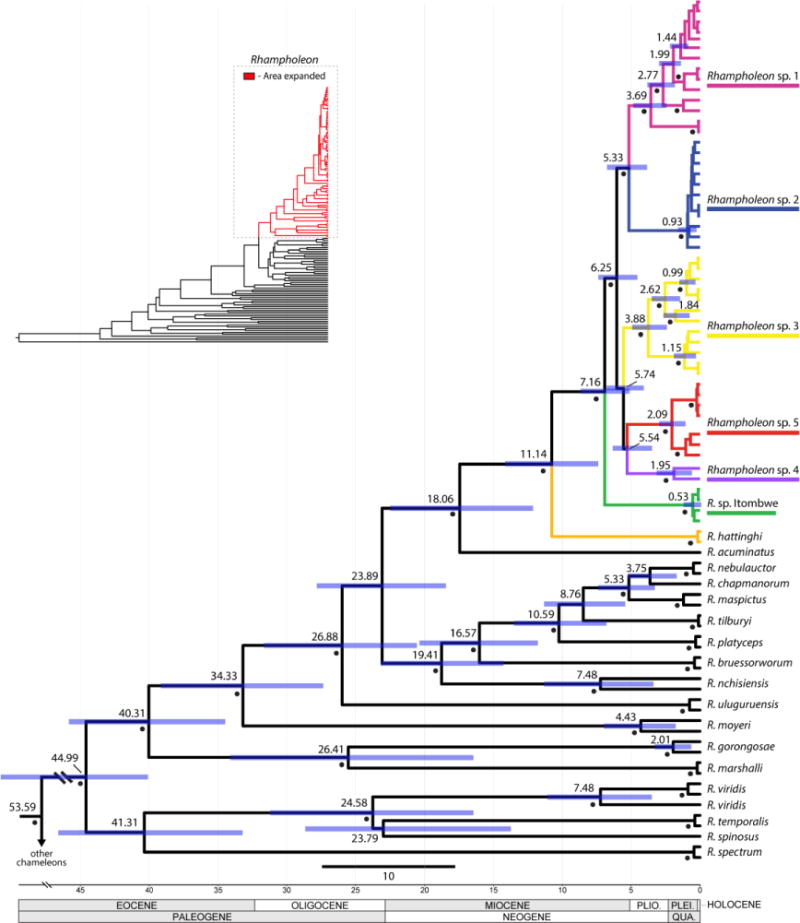
Bayesian chronogram of the pygmy chameleon genus Rhampholeon. Posterior probabilities ≥ 95% are denoted by filled circles adjacent to nodes. Numbers near nodes denote mean highest posterior densities (HPD) and blue bars at nodes represent 95% HPD.
3.4. Species delimitation
The four species-tree analyses recovered similar clade topology and node support values to our concatenated gene-tree analyses. The nodes representing R. hattinghi and R. sp. Itombwe were generally well-supported across analyses (Fig. 5). Similarly, node support values for R. sp. 2 were high in the species trees derived from MP-EST and bPTP, yet weaker support for this species was found in the *BEAST species tree. The results from GMYC and bPTP differed from *BEAST and MP-EST in that additional species were delimited beyond the five cryptic species recognized in the gene-tree analyses of the R. boulengeri clade. For example, GMYC recovered four additional species and bPTP six additional species, whereas MP-EST and *BEAST both recovered the more conservative estimate of five species (Fig. 5). Evaluating the congruent evidence of these four coalescent-based species-tree inferences leads us to recognize five cryptic species within the R. boulengeri clade.
Figure 5.
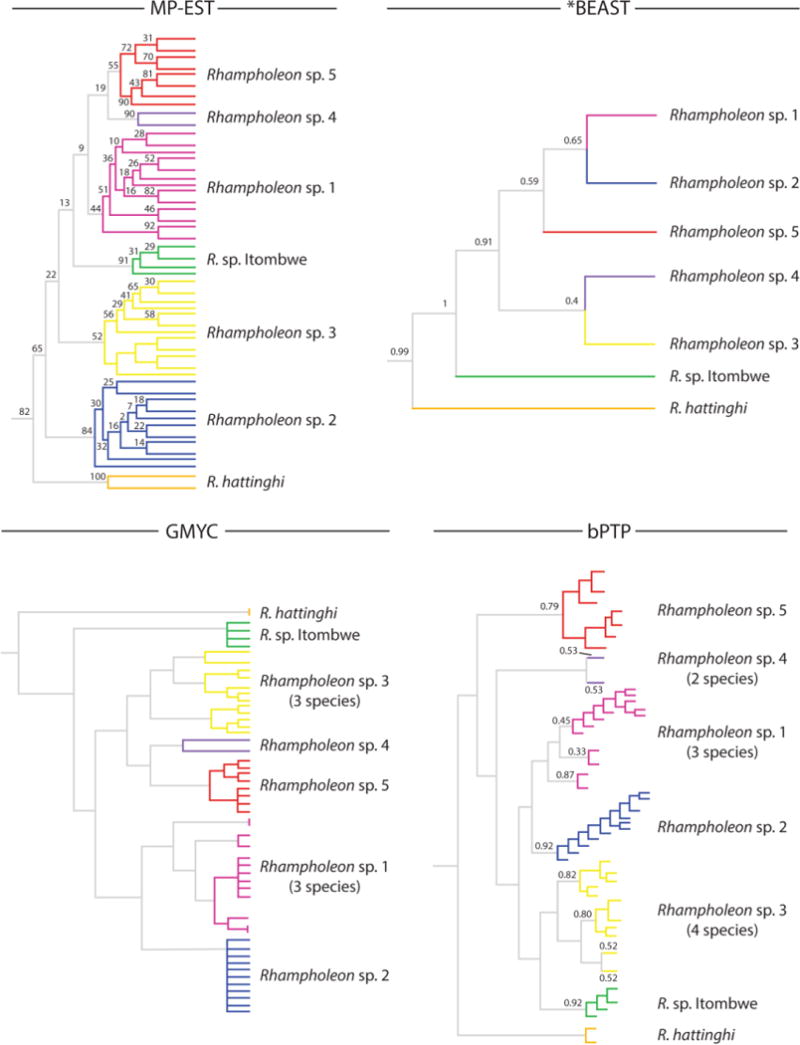
Comparative species-tree estimations for seven pygmy chameleon species (genus Rhampholeon) from the Albertine Rift, Central Africa, and western Kenya. Numbers above nodes denote posterior probabilities for the Bayesian analyses *BEAST and bPTP, and bootstrap values for MP-EST.
4. Discussion
4.1. Molecular systematics and species delimitation
While our phylogenetic analyses are consistent with the evolutionary relationships across the genus Rhampholeon recovered in previous studies (Matthee et al., 2004; Mariaux and Tilbury 2006; Fisseha et al., 2013; Tolley et al., 2013; Branch et al., 2014; Tilbury and Tolley, 2015), the additional populations sampled from diverse forest environments of the Albertine Rift (AR) represent five previously unrecognized species. All of the AR species were recovered in the same clade, and importantly, none of the new species were conspecific with described Rhampholeon from the AR (i.e., R. hattinghi). We found three pygmy chameleon species associated with the Itombwe Plateau, which is likely to be the type locality for R. boulengeri (see commentary in Introduction). Because the type specimens were not consulted, we did not assign the nominal R. boulengeri to any of the three lineages with populations from the Itombwe Plateau, South Kivu Province, eastern DRC. Nevertheless, one lineage (R. sp. Itombwe) is likely to be the bona fide R. boulengeri, because it is currently endemic to the plateau, but an integrative taxonomic assessment is required before names can be appropriately assigned within this clade. The recognition of widespread cryptic diversity in R. boulengeri was not entirely unexpected given the extensive range of this species and previous accounts that pointed to the potential for cryptic species. Indeed, a recent phylogeny by Tilbury and Tolley (2015) showed three distinct clades of R. boulengeri across three localities in the AR (samples EBG 1613, 1702 from the Itombwe Plateau, eastern DRC; two samples CAS 2016 81–82 from Bwindi Impenetrable National Park, southwestern Uganda; and one sample CT 347 from Rwenzori Mountains National Park, western Uganda). The comprehensive sampling in our present study that included those samples and multiple populations across the range of R. boulengeri in the AR and Kenya, allowed for much greater resolution for detecting species, and thus provided a far greater understanding of diversity in R. boulengeri across its geographic distribution. In particular, we can confirm the two cryptic species recovered by Tilbury and Tolley (2015) plus the recognition of at least six cryptic species currently recognized under R. boulengeri. The species-level phylogeographical patterns we recovered were indicative of diversification in isolation, either due to forest fragmentation (allopatric), or in some cases, elevational zonation (parapatric). Nevertheless, we found that one new species is actually widespread (R. sp. 1), extending from eastern DRC to western Kenya, a finding that supports the strong affinities of herpetofauna between these highlands (Bwong et al., 2009; Lötters et al., 2007; Wagner and Böhme 2007; Wagner et al., 2008). Based on our findings, we speculate that observations of R. boulengeri from other localities in the AR and western Kenya likely represent one of the new species, or potentially additional novel lineages (e.g., de Witte, 1965; Spawls et al., 2002; Tilbury, 2010; Stipala, 2014).
Across all four approaches to reconstruct species trees, we consistently found support for the recognition of at least six species within R. boulengeri sensu lato (including the nominal species). Of the four species-tree reconstruction methods, two directly estimated phylogenies based on the genetic data (*BEAST and MP-EST), and these produced a speciation scenario most similar to the gene-tree analyses. The other two methods (GMYC and bPTP) assessed tree topologies based on the gene trees generated from ML and BI analyses, and these produced inflated speciation scenarios relative to the other approaches. We considered that the additional lineages delimited by GMYC (four) and bPTP (six) to be artifacts of the model and disregarded them as over-split species. The recognition of six species within R. boulengeri is based on our conservative interpretation of the results across species-tree and gene-tree estimates, which has improved our understanding of diversity in this group at a greater resolution than prior studies. These species are morphologically conservative (i.e., cryptic), and thus morphological characters reported in historical accounts on the species (e.g., Schmidt, 1919) are insufficient to delimit species boundaries in this group. The long-standing recognition of R. boulengeri as a single species was due to limited sampling across the geographic range of the species, lack of topotypic material in previous molecular studies, and absence of species-tree approaches to delimit species, which have been shown to accurately recover species in empirical tests (e.g., Camargo et al., 2012).
A caveat to the MP-EST results is that the branch lengths in this analysis represent coalescent units and not sequence divergences (Liu et al., 2010). It is likely that incomplete lineage sorting in the deeper branches of the gene tree led to the recovery of low support for some nodes in the species-tree produced by *BEAST (Drummond et al., 2012). Although GMYC has been demonstrated to be a useful tool for species-delimitation (e.g., Talavera et al., 2013), concerns have been raised regarding spatially induced increases in intraspecific genetic variation leading to over-splitting (Bergsten et al., 2012). It has also been demonstrated that GMYC can over-estimate the number of species when compared with other evidence commonly used in taxonomic studies (e.g., morphology) (Miralles and Vences, 2013). Incomplete lineage sorting and loci with different evolutionary rates cannot be ruled out as sources of topological incongruence across species-tree analyses (Sistrom et al., 2014; Xi et al., 2014).
Although our species-tree analyses provide a suitable scenario for speciation among lineages of R. boulengeri, multi-species coalescent models can be misleading, recovering high posterior probabilities at the population level rather than the species level (Sukumaran and Knowles, 2017). This can be exacerbated by improper parameter selections (Olave et al., 2014) leading to overestimation of species delimitation (Niemiller et al., 2012; Carstens et al., 2013). Regardless, our comparative approach to species-delimitation with a conservative interpretation leads to the same general outcome across the methods, and we are therefore confident in our overall interpretation.
Based on monophyly in phylogenetic analyses and supported by morphological characters (Klaver and Böhme, 1986; Loveridge, 1956; Matthee et al., 2004), Rhampholeon are divided into three sub-genera: Rhampholeon (Rhampholeon) with four species (spectrum, spinosus, temporalis, and viridis); Rhampholeon (Rhinodigitum) with 13 species (acuminatus, beraduccii, boulengeri, bruessorworum, chapmanorum, hattinghi, maspictus, moyeri, nchisiensis, nebulauctor, platyceps, tilburyi, and uluguruensis); and Rhampholeon (Bicuspis) with two species (gorongosae and marshalli). These sub-generic allocations have been upheld in recent taxonomic investigations of the genus (e.g., Branch et al., 2014). Although our the divisions of Matthee et al. (2004), the utility of sub-generic molecular phylogenetic results support classifications in systematics is equivocal and thus its usage has become generally uncommon in herpetological classifications (e.g., Frost et al., 2009; Frost and Hillis, 1990), with some notable exceptions (e.g., Wallach et al., 2009).
4.2. Historical biogeography
The estimated divergence dates we recovered for the genus Rhampholeon closely resembled analyses by Townsend et al. (2011) and Tolley et al. (2013), and were roughly similar to those by Matthee et al. (2004). The similarity of our dates to the independent ones of Tolley et al. (2013) was not entirely unexpected given our co-opting of some dates as secondary calibrations. Taxon sampling for Rhampholeon varied across these studies: Townsend et al. (2011) included five species; Matthee et al. (2004) included 12; Tolley et al. (2013) included 13; and we included 18. Our dates were also consistent with the estimates provided by Branch et al. (2014) that were derived from a general rate of evolutionary change of the ND2 marker for several Mozambican Rhampholeon species. The 95% HPD intervals overlapped across these studies indicating that multiple independent approaches to divergence dating converged on similar estimates for the timing of lineage diversification in Rhampholeon.
In contrast to other studies on East African taxa that found genetic legacies left by Quaternary climatic changes (e.g., Cox et al., 2014; Demos et al., 2014; Roy et al., 2014), there was no evidence of Pleistocene radiations in Rhampholeon (i.e., no support for the Pleistocene Forest Refuge Hypothesis [Mayr and O’Hara, 1986]). Rather, we found evidence that paleoendemic lineages persisted in montane forest refugia since the Eocene (i.e., support for aspects of the Evolutionary Museum Hypothesis, although in montane regions, whereas the hypothesis originally identified lowlands as refugia [Fjeldså and Lovett, 1997]). These ancient lineages were maintained in small montane forest refugia during the increasingly arid climate of the Pliocene and Pleistocene (e.g., deMenocal, 1995, 2004). Diversification dates for forest-dependent Rhampholeon species in our genus-level phylogeny generally overlap with those of Couvreur et al. (2008) for diversification of African tropical rainforests since the Oligocene and follow general patterns of African forest declines during the Miocene presented by Kissling et al. (2012). The high number of ancient species confined to small forest fragments suggests that Rhampholeon lineages did not immigrate during the early Pliocene when forest connectivity likely increased across East Africa (e.g., Maley, 1996; Zachos et al., 2001).
The estimated divergence dates recovered within Rhampholeon suggest an initial split around 45 Mya (± 10 Mya) (this study; Matthee et al., 2004; Townsend et al., 2011; Tolley et al., 2013). This divergence does not correspond to two spatially disparate clades, but rather, consists of two deeply divergent clades that are generally sympatric at present. One of the major clades includes the West African R. spectrum, which split from an East African group (R. spinosus, R. temporalis, and R. viridis) around 40 Mya (± 10 Mya), generally coincident with the break-up of West/Central and East African forests (Couvreur et al., 2008). The other major Rhampholeon clade, which includes all the remaining species in the genus, diverged in the Eocene and has remained on the eastern side of sub-Saharan Africa. The Eastern Arc Mountains and Southern Rift Highlands harbor a particularly species-rich Rhampholeon clade, which first diversified in the Eocene and then underwent extensive diversification events during the Miocene, especially in the highlands of Malawi and Mozambique. The Central African clade that is restricted to high-elevation forests of the AR diverged from a common ancestor with the East African R. acuminatus in the Miocene around 18 Mya (± 5 Mya) (this study; Matthee et al., 2004; Tolley et al., 2013). This period correlates with some of the initial uplift (Wichura et al., 2010) and subsequent drying of East Africa’s climate (Sepulchre et al., 2006), which caused the break-up of forest habitats between Central and East Africa.
We found that the Miocene epoch was an important period for diversification of East African Rhampholeon species. In the Miocene, the environment of the AR was experiencing dramatic changes from both climatological and geological factors, which derive from arid conditions induced by a combination of reduced atmospheric CO2 concentrations globally (Cerling et al., 1997) and tectonic uplifts that altered climatic patterns in East Africa (Sepulchre et al., 2006). Global cooling trends that began after the late Oligocene warming heightened during the Miocene, and these drops in temperature altered precipitation patterns (Jacobs, 2004; Sepulchre et al., 2006), which increased aridity across the African continent (Böhme, 2003; Werdelin and Sanders, 2010; Wichura et al., 2015). A global cooling trend in the Miocene is evident from sedimentation records (Pickford et al., 1993), and supported by climate-driven faunal turnovers from the fossil record of East Africa (Leakey et al., 1996) and the AR (Senut and Pickford, 1994). Rainfall vicissitudes throughout the Neogene across Central and East Africa (Pickford, 1992; Wynn, 2003) resulted in drastically transformed vegetation patterns (Cerling et al., 1997; Feakins et al., 2005), which manifested in the extensive development of savannas (Cerling, 1992; Meadows and Linder, 1993; Jacobs et al., 1999; Jacobs, 2004) and the interrelated fragmentation of forests (Couvreur et al., 2008; Kissling et al., 2012). Not only was the Miocene climate a significant influence, but tectonic activities were also drivers of major environmental change in the AR. Initial rifting of the AR began around the Oligocene–Miocene boundary (Roberts et al., 2012), and most of the geophysical rifting and volcanism in East Africa occurred during the Miocene (e.g., Wichura et al., 2010), with increased activity from 10–5 Mya around the Miocene–Pliocene boundary (Paul et al., 2014; Macgregor, 2015). These geological changes also shifted climatic patterns towards increased aridity and thus reinforced the global weather trends, which were linked to substantial decreases in the extent of tropical rainforest across sub-Saharan Africa in the Miocene (Kissling et al., 2012). General trends of forest fragmentation during the Miocene likely underlie most of the species-level diversification patterns in Rhampholeon, a finding that is similar to East African thicket rats (Bryja et al., 2017) and forest chameleons (Hughes et al., in press).
Among the AR Rhampholeon species, diversification events since the mid-Miocene were most common, a finding that is similar to several other forest-adapted taxa in the region (e.g., Tolley et al., 2011; Greenbaum et al., 2015; Portillo et al., 2015; Larson et al., 2016; Hughes et al., in press). Short internodes and low support among the species in the R. boulengeri complex are consistent with a rapid radiation event across the Miocene–Pliocene boundary (ca. 6–4 Mya). Diversification patterns for the entire genus Rhampholeon are consistent with vicariance-driven speciation via forest fragmentation during the Miocene; however, this pattern does not fully explain the diversification within the R. boulengeri complex. For example, the distribution of R. sp. 5 is extensive yet disjunct, ranging from the Rwenzori Mountains to the highlands of southern Burundi. The geologically young Rwenzori Mountains (Kaufmann et al., 2015) were surrounded by bodies of water since the mid-Pleistocene (Beadle, 1981), which were likely barriers to dispersal. In fact, many vertebrate taxa are endemic to this massif (Butynski and Kalina, 1993), including two forest chameleon species (Tilbury, 2010). However, we found that R. sp. 5 was present in high-elevation forests of the Rwenzori Mountains and Bururi Nature Reserve of southern Burundi. These two localities are separated by a great distance (> 450 km), and although samples from these locales formed distinct clades, we detected minimal intraspecific genetic distance between these clades (Fig. S2). Furthermore, we estimated that R. sp. 5 diverged prior to the uplift of the Rwenzori Mountains (ca. 3–2 Mya), and thus geological uplift alone cannot explain the diversification of this species. It is possible that R. sp. 5 occurs at other localities between these sites, but potential populations have yet to be sampled. If the allopatric distribution between populations of R. sp. 5 is real, perhaps there were forest connections that are now gone, or fluctuations in the historical water levels of the AR crater lakes (Salzburger et al., 2014) isolated ancestral populations, and subsequent population-level extinctions between these sites produced its present-day distribution. Nevertheless, there are several endemic species that have widespread, yet disjunct distributions in the AR, including one bird (Bradypterus graueri [Kahindo et al., 2017]), three tree frogs (Hyperolius castaneus, H. discodactylus, and Leptopelis karissimbensis [Greenbaum et al., 2013; Liedtke et al., 2014; Portillo et al., 2015]), and three small mammals (Hylomyscus vulcanorum, Lophuromys woosnami, and Sylvisorex vulcanorum [Huhndorf et al., 2007; Demos et al., 2014, 2015]).
4.3. Elevational zonation and cryptic diversity
A model of allopatric speciation driven by forest fragmentation throughout the Cenozoic has been proposed for forest-dependent chameleons (Tolley et al. 2011; Matthee et al., 2004; Townsend et al., 2011; Tolley et al., 2013; Branch et al., 2014; Ceccarelli et al., 2014). This model fits the pattern of cladogenesis for pygmy chameleons from our genus level phylogeny, but is a poor fit within the R. boulengeri complex because we found evidence that sister species occur in near sympatry yet occupy distinct elevational zones. The complex patterns of cryptic diversity we found in the AR suggest that traditional biogeographic barriers for the region may be inadequate to explain the diversity in R. boulengeri, and thus we contend that elevational zonation and parapatric speciation have played more significant roles in generating diversity within this group than previously recognized. Elevational zonation as a mechanism of speciation has been well characterized in the Americas for amphibians and reptiles (e.g., Arteaga et al., 2016; Kozak and Wiens, 2010; Hutter et al., 2013; Wake and Lynch, 1976), yet its role in the divergence of African taxa is not well understood. Fuchs et al. (2011) found species-level genetic differentiation between montane and lowland forms of the bird species Phyllastrephus debilis from the Eastern Arc Mountains of Tanzania. However, Cox et al. (2014) found mixed support for an elevational gradient speciation model in their study on the songbird genus Zosterops from the Eastern Afromontane highlands of Kenya.
In contrast to the Montane Speciation Hypothesis (Fjeldså and Lovett, 1997) that predicts geographical isolation will result from the inability of species to adapt to new environmental conditions (i.e., allopatric speciation via niche conservatism [Kozak and Wiens, 2010]), we argue that the Gradient Speciation Hypothesis (Moritz et al., 2000) is more appropriate to explain diversification in the R. boulengeri complex. This hypothesis predicts that sister taxa will occupy distinct but adjacent habitats, because new species formation occurred via adaptation to different climatic regimes along an altitudinal gradient (i.e., parapatric speciation via niche differentiation [Moritz et al. 2000]). The six species of the R. boulengeri species complex, five of which are endemic to the region, occur along an elevational gradient (800–2,600 m) in the AR and they might be expected to show little genetic differentiation across a relatively small area of continuous habitat, yet they exhibit an extraordinary degree of genetic diversification. This is significant considering that all of the species are members of an ancient clade that diversified in the late Miocene. Furthermore, the elevated levels of genetic variation were not accompanied by pronounced morphological variation, which is likely because the selection pressures exerted upon the phenotype to occupy the leaf-litter ecological niche have been the same across species. Several species in the R. boulengeri complex are partly sympatric (e.g., R. sp. 1, R. sp. 2, R. sp. 3, and R. sp. Itombwe), but not syntopic because they occur in largely non-overlapping elevational zones (Figs. 2–3). These patterns of elevational zonation were likely promoted by parapatric speciation, in which adaptation to physical factors such as temperature or differentiation in climatic niches initiated processes that lead to species formation. Specifically, pulses of forest expansion and contraction throughout the Miocene–Pliocene boundary could be invoked to explain a parapatric pattern of high-elevation species (R. sp. 1) as sister to a lower-elevation species (R. sp. 2). As historical forests migrated up in elevation, ancestral populations may have adapted to novel lower thermal limits and thus physiological thresholds changed, and during periods of greater forest connectivity, dispersal to warmer, low-elevation forests may have been hampered. A similar scenario may also explain the divergence of R. sp. 3 to the clade containing R. sp. 4 and R. sp. 5, and perhaps explain the divergence between R. hattinghi and R. sp. Itombwe.
Figure 2.
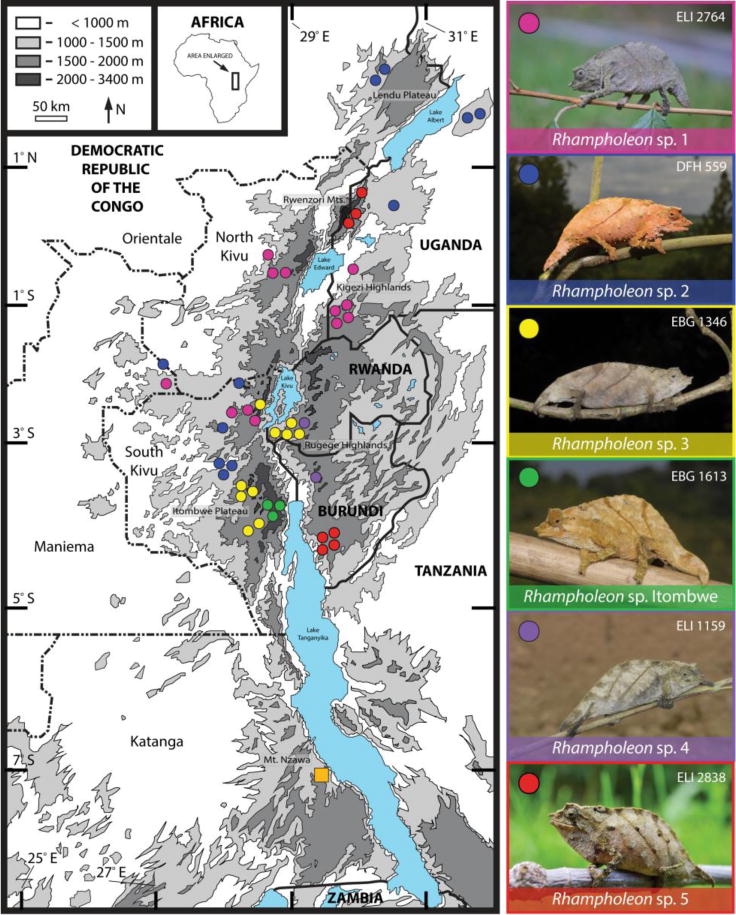
Elevation map of the Albertine Rift, Central Africa showing sampling localities of pygmy chameleons (genus Rhampholeon) used in this study. Two samples from western Kenya are not shown. Photographs of representative individuals for the new species are displayed on the right. Orange square represents the species R. hattinghi.
The highest regional concentration of diversity in the R. boulengeri complex (four species) is found in the South Kivu Province of eastern DRC. In Kahuzi-Biega National Park, three species (R. sp. 1, R. sp. 2, and R. sp. 3) occur along an elevational gradient (800–2030 m), and similarly in association with the Itombwe Plateau, three species (R. sp. 2, R. sp. 3, and R. sp. Itombwe) occur along a slightly greater elevational gradient (1060–2311 m). The extensive volcanism and orogeny in this province (e.g., Pasteels et al., 1989; Kampunzu et al., 1998; Ebinger and Furman, 2003), especially during the Miocene, likely contributed to the emergence of new ecological conditions, and thus may account for some of the patterns of elevated genetic diversity in pygmy chameleons along elevational gradients in these particular highlands.
4.3. Conservation implications
Although pygmy chameleons are considered less threatened by the illegal wildlife trade than by habitat loss, several African countries supply large numbers of chameleon exports to satisfy international demand (Carpenter et al., 2004; Robinson et al., 2015). In an effort to reduce pressure on natural populations for species in the legal trade, all pygmy chameleon species were added to CITES Appendix II in 2016, which represents an important first step towards the sustainable trade in pygmy chameleons. For these reasons and more detailed below, we wish to call attention to several serious threats to the biological integrity of the AR that pose challenges to pygmy chameleon conservation. Although the AR is extremely biologically diverse (see Plumptre et al., 2007), the amount of unrecognized diversity in pygmy chameleons in the AR suggests that the discovery of cryptic taxa in this region is still in its initial stages (see Bickford et al., 2007). Moreover, a recent study demonstrated that Central Africa is one of the three most under-sampled regions on the continent with respect to its herpetofauna (Tolley et al., 2016). The forests that harbor elevated levels of pygmy chameleon diversity in the AR face severe challenges from an extremely dense human population (Burgess et al., 2007) and international demand for petroleum products (von Einsiedel, 2014). Near ubiquitous occupation of land by humans across the AR has imposed unprecedented pressures upon its natural environments, especially the forest habitats that are being converted to agriculture at an alarming rate (Barnes, 1990; Butsic et al., 2015). Political disputes and armed conflict among the various countries of the AR has also irreparably damaged many of its natural environments (Glew and Hudson, 2007; Hanson et al., 2009; Kanyamibwa, 1998). Adding to these problems is the threat posed by predicted climate change (Carr et al., 2013), and a recent model indicated that by 2070, over 40% of the AR region will be unsuitable for most of its current ecosystems (Ponce-Reyes et al., 2017). Unfortunately, these interwoven threats in the AR (Brooks et al., 2004) are rampant across tropical biodiversity hotspots worldwide (Mittermeier et al., 2011; Myers et al., 2000) and, in part, they underlie the current global extinction crisis (Kolbert, 2014). Exacerbating these issues is the rate of species discovery, which is thought to be so slow (Fontaine et al., 2012) that numerous species will be lost before they are known to science (Costello et al., 2013). Moreover, declines in biodiversity not only affect ecosystem function (see Loreau et al., 2001); they also induce losses to our understanding of character variation via direct losses in data. Our understanding of variation across space and through time is what evolutionary biology and biogeography are founded upon. To that end, no single type of data should be excluded as we endeavor to decipher the history of life on Earth and thus we must increase the rate of rescue for as many types of data as possible (e.g., Hughes et al., 2016). Lastly, we have identified a significant gap between the taxonomy and the diversity of AR pygmy chameleons and because specific names are critical to species conservation, we plan to do a follow-up study, using an integrative taxonomic approach, to describe these distinct populations as new species.
Supplementary Material
Table S1. GenBank accession numbers for outgroup samples analyzed in Fig. S2. Alternative species used to create chimeric sequences are shown in each cell above the accession number.
Table S2. Uncorrected p-distances within and among species in the genus Rhampholeon for three molecular markers (16S, ND2, and RAG1).
Figure S1. Bayesian (A) and maximum likelihood (B) phylogenies of the genus Rhampholeon with support values adjacent to nodes.
Figure S2. Entire Bayesian chronogram of Chamaeleonidae and outgroup. Numbers near nodes denote mean highest posterior densities (HPD). Fossil-calibrated nodes are indicated with encircled red numbers and secondary calibrated nodes with encircled light blue numbers. Diversification dates within the genus Rhampholeon are presented in Fig. 4.
Highlights.
Species diversity of pygmy chameleons in the Albertine Rift is underestimated
Rhampholeon boulengeri is a complex of at least 6 species
Diversification via allopatry underlies genetic patterns in genus-level phylogeny
The R. boulengeri complex exhibits signatures of parapatric speciation
We discuss the importance of investigating cryptic diversity in the montane Afrotropics
Acknowledgments
Dodson Graduate Research Grants and Student Travel Grants from UTEP, and the Dr. Keelung Hong Graduate Research Fellowship funded fieldwork by DFH. Fieldwork by EG was supported by the Percy Sladen Memorial Fund, an IUCN/SSC Amphibian Specialist Group Seed Grant, K. Reed (MD), Villanova University, UTEP, National Geographic (Research and Exploration Grant no. 8556-08), and the National Science Foundation (DEB-1145459). We thank our field companions M. M. Aristote, W. M. Muninga, and J. -P. Mokanse. The Institutional Animal Care and Use Committee (IACUC) at UTEP approved this research (A-200902-1). Baluku Bajope of the Centre de Recherche en Sciences Naturelles (CRSN) provided project support and permits, and the Institut Congolais pour la Conservation de la Nature (ICCN) kindly granted permits to work in protected areas in DRC. We are grateful for the Wildlife Conservation Society (WCS) teams who collected specimens in eastern DRC, particularly Guillain Mitamba and Emmanuel Muhindo, and the financing by USAID, US Fish and Wildlife Service, and Arcus Foundation that enabled the surveys to be made. We thank the Uganda National Council of Science and Technology (UNCST) for granting us the research permit to conduct our ongoing Herpetofaunal Conservation Assessment of Uganda. We also thank the Uganda Wildlife Authority (UWA) and the National Forest Authority (NFA) for the necessary permits to work in the protected areas under their jurisdiction and their field staff that assisted us with access, protection, and excellent companionship to carry out our research. We are indebted to Mr. James Lutalo, Commissioner of Wildlife Conservation and CITES Authority for Uganda, and Mr. Aggrey Rwetsiba, Senior Monitoring and Research Coordinator for UWA, and other staff under their supervision, who tirelessly worked hard to make sure we received the correct documents in a timely manner. We extend our gratitude to John C. Mitani, David P. Watts, and members of the Ngogo Chimpanzee Project for their assistance in Kibale National Park. We thank Léonidas Nzigiyimpa of the Institut National pour l’Environnement et la Conservation de la Nature (INECN) of Burundi for logistical support and permit negotiations. We thank Aaron M. Bauer for insightful comments on an early draft. We would like to thank the staff of the Border Biomedical Research Center (BBRC) Genomics Analysis Core Facility for services and facilities provided. This work was supported by grant G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD), a component of the National Institutes of Health (NIH). Lastly, we acknowledge F. Portillo for his efforts on the map and technical advice on analyses.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfaro ME, Zoller S, Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- Arteaga A, Pyron RA, Peñafiel N, Romero-Barreto P, Culebras J, Bustamante L, Yánez-Muñoz MH, Guayasamin JM. Comparative phylogeography reveals cryptic diversity and repeated patterns of cladogenesis for amphibians and reptiles in northwestern Ecuador. PLoS ONE. 2016;11:e0151746. doi: 10.1371/journal.pone.0151746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RFW. Deforestation trends in tropical Africa. Afr J Ecol. 1990;28:161–173. [Google Scholar]
- Beadle LC. The Inland Waters of Tropical Africa: An Introduction to Tropical Limnology. 2nd. Longman Group, Ltd.; London, UK: 1981. [Google Scholar]
- Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, Hendrich L, Geijer J, Herrmann J, Foster GN, Ribera I. The effect of geographical scale of sampling on DNA barcoding. Syst Biol. 2012;61:851–869. doi: 10.1093/sysbio/sys037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Böhme M. The Miocene climatic optimum: evidence from ectothermic vertebrates of Central Europe. Palaeogeogr Palaeoclimatol Palaeoecol. 2003;195:389–401. [Google Scholar]
- Bowie RCK, Fjeldså J, Hackett SJ, Bates JM, Crowe TM. Coalescent models reveal the relative roles of ancestral polymorphism, vicariance, and dispersal in shaping phylogeographical structure of an African montane forest robin. Mol Phylogenet Evol. 2006;38:171–188. doi: 10.1016/j.ympev.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Boxnick A, Apio A, Wronski T, Hausdorf B. Diversity patterns of the terrestrial snail fauna of Nyungwe Forest National Park (Rwanda), a Pleistocene refugium in the heart of Africa. Biol J Linn Soc. 2015;114:363–375. [Google Scholar]
- Branch WR, Bayliss J, Tolley KA. Pygmy chameleons of the Rhampholeon platyceps complex (Squamata: Chamaeleonidae): description of four new species from isolated ‘sky islands’ of northern Mozambique. Zootaxa. 2014;3814:1–36. doi: 10.11646/zootaxa.3814.1.1. [DOI] [PubMed] [Google Scholar]
- Brooks T, Hoffmann M, Burgess N, Plumptre AJ, Williams S, Gereau RE, Mittermeier RA, Stuart S. Eastern afromontane. In: Mittermeier RA, Robles-Gil P, Hoffmann M, Pilgrim JD, Brooks TM, Mittermeier CG, Lamoreux JL, Fonseca G, editors. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Ecoregions. 2nd. CEMEX; Mexico City: 2004. pp. 241–242. [Google Scholar]
- Bryja J, Šumbera R, Peterhans K, Julian C, Aghová T, Bryjová A, Mikula O, Nicolas V, Denys C, Verheyen E. Evolutionary history of the thicket rats (genus Grammomys) mirrors the evolution of African forests since late Miocene. J Biogeogr. 2017;44:182–194. [Google Scholar]
- Burgess ND, Balmford A, Cordeiro NJ, Fjeldså J, Küper W, Rahbek C, Sanderson EW, Scharlemann JP, Sommer JH, Williams PH. Correlations among species distributions, human density and human infrastructure across the high biodiversity tropical mountains of Africa. Biol Cons. 2007;134:164–177. [Google Scholar]
- Butsic V, Baumann M, Shortland A, Walker S, Kuemmerle T. Conservation and conflict in the Democratic Republic of Congo: the impacts of warfare, mining, and protected areas on deforestation. Biol Cons. 2015;191:266–273. [Google Scholar]
- Butynski TM, Kalina J. Three new mountain national parks for Uganda. Oryx. 1993;27:214–224. [Google Scholar]
- Bwong BA, Chira R, Schick S, Veith M, Lötters S. Diversity of ridged frogs (Ptychadenidae: Ptychadena) in the easternmost remnant of the Guineo-Congolian rain forest: an analysis using morphology, bioacoustics and molecular genetics. Salamandra. 2009;45:129–146. [Google Scholar]
- Camargo A, Morando M, Avila LJ, Sites JW. Species delimitation with abc and other coalescent‐based methods: a test of accuracy with simulations and an empirical example with lizards of the Liolaemus darwinii complex (Squamata: Liolaemidae) Evolution. 2012;66:2834–2849. doi: 10.1111/j.1558-5646.2012.01640.x. [DOI] [PubMed] [Google Scholar]
- Carpenter AI, Rowcliffe JM, Watkinson AR. The dynamics of the global trade in chameleons. Biol Cons. 2004;120:291–301. [Google Scholar]
- Carr JA, Outhwaite WE, Goodman GL, Oldfield TEE, Foden WB. Vital but vulnerable: climate change vulnerability and human use of wildlife in Africa’s Albertine Rift. Gland, Switzerland, and Cambridge, UK: 2013. (Occasional Paper of the International Union for Conservation of Nature (IUCN) Species Survival Commission No. 48). [Google Scholar]
- Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- Ceccarelli FS, Menegon M, Tolley KA, Tilbury CR, Gower DJ, Laserna MH, Kasahun R, Rodriguez-Prieto A, Hagmann R, Loader SP. Evolutionary relationships, species delimitation and biogeography of Eastern Afromontane horned chameleons (Chamaeleonidae: Trioceros) Mol Phylogenet Evol. 2014;80:125–136. doi: 10.1016/j.ympev.2014.07.023. [DOI] [PubMed] [Google Scholar]
- Cerling TE. Development of grasslands and savannas in East Africa during the Neogene. Palaeogeogr Palaeoclimatol Palaeoecol. 1992;97:241–247. [Google Scholar]
- Cerling TE, Harris JM, MacFadden BJ, Leakey MG, Quade J, Eisenmann V, Ehleringer JR. Global vegetation change through the Miocene/Pliocene boundary. Nature (London) 1997;389:153–158. [Google Scholar]
- Chorowicz J. The East African Rift system. J Afr Earth Sci. 2005;43:379–410. [Google Scholar]
- Conrad JL, Norell MA. A complete Late Cretaceous iguanian (Squamata, Reptilia) from the Gobi and identification of a new iguanian clade. Am Mus Novit. 2007;3584:1–47. [Google Scholar]
- Costello MJ, May RM, Stork NE. Can we name Earth’s species before they go extinct? Science (Washington) 2013;339:413–416. doi: 10.1126/science.1230318. [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Chatrou LW, Sosef MSM, Richardson JE. Molecular phylogenetics reveal multiple tertiary vicariance origins of the African rain forest trees. BMC Biol. 2008;6:54. doi: 10.1186/1741-7007-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SC, Prys‐Jones RP, Habel JC, Amakobe BA, Day JJ. Niche divergence promotes rapid diversification of East African sky island white‐eyes (Aves: Zosteropidae) Mol Ecol. 2014;23:4103–4118. doi: 10.1111/mec.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza JD, Stanley EL, Wagner P, Bauer AM, Grimaldi DA. Mid-Cretaceous amber fossils illuminate the past diversity of tropical lizards. Sci Adv. 2016;2:e1501080. doi: 10.1126/sciadv.1501080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- de Witte GF. Les caméléons de l’Afrique Centrale (République Démocratique du Congo, République du Rwanda et Royaume du Burundi). Annales Musée Royal de l’Afrique Centrale. Sciences Zoologiques. 1965;142:1–215. + pl. I–XII. [Google Scholar]
- deMenocal PB. Plio-Pleistocene African climate. Science (Washington) 1995;270:53–59. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- deMenocal PB. African climate change and faunal evolution during the Pliocene–Pleistocene. Earth Planet Sci Lett. 2004;220:3–24. [Google Scholar]
- Demos TC, Peterhans JCK, Agwanda B, Hickerson MJ. Uncovering cryptic diversity and refugial persistence among small mammal lineages across the Eastern Afromontane biodiversity hotspot. Mol Phylogenet Evol. 2014;71:41–54. doi: 10.1016/j.ympev.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Demos TC, Peterhans JCK, Joseph TA, Robinson JD, Agwanda B, Hickerson MJ. Comparative population genomics of African montane forest mammals support population persistence across a climatic gradient and quaternary climatic cycles. PLoS ONE. 2015;10:e0131800. doi: 10.1371/journal.pone.0131800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinger C, Furman T. Geodynamical setting of the Virunga volcanic province, East Africa. Acta Vulcanologica. 2003;14:1–8. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SE. Crown-group lizards from the Middle Jurassic of Britain. Palaeontogr Am. 1998;250:1–32. [Google Scholar]
- Evans SE, Wang Y. The Early Cretaceous lizard Dalinghosaurus from China. Acta Palaeontol Pol. 2005;50:725–742. [Google Scholar]
- Feakins SJ, deMenocal PB, Eglinton TI. Biomarker records of late Neogene changes in northeast African vegetation. Geology. 2005;33:977–980. [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fisseha M, Mariaux J, Menegon M. The “Rhampholeon uluguruensis complex” (Squamata: Chamaeleonidae) and the taxonomic status of the pygmy chameleons in Tanzania. Zootaxa. 2013;3746:439–453. doi: 10.11646/zootaxa.3746.3.3. [DOI] [PubMed] [Google Scholar]
- Fjeldså J, Lovett JC. Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodivers Conserv. 1997;6:325–346. [Google Scholar]
- Flot JF. SeqPHASE: a web tool for interconverting PHASE input/output files and FASTA sequence alignments. Mol Ecol Resour. 2010;10:162–166. doi: 10.1111/j.1755-0998.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- Fontaine B, Perrard A, Bouchet P. 21 years of shelf life between discovery and description of new species. Curr Biol. 2012;22:R943–R944. doi: 10.1016/j.cub.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Fontaneto D, Flot JF, Tang CQ. Guidelines for DNA taxonomy, with a focus on the meiofauna. Mar Biodiv. 2015;45:433–451. [Google Scholar]
- Frost DR, Hillis DM. Species in concept and practice: herpetological applications. Herpetologica. 1990;46:86–104. [Google Scholar]
- Frost DR, McDiarmid RW, Mendelson JR., III Response to the Point of View of Gregory B. Pauly, David M. Hillis, and David C. Cannatella, by the anuran subcommittee of the SSAR/HL/ASIH scientific and standard English names list. Herpetologica. 2009;65:136–153. [Google Scholar]
- Fuchs J, Fjeldså J, Bowie RCK. Diversification across an altitudinal gradient in the Tiny Greenbul (Phyllastrephus debilis) from the Eastern Arc Mountains of Africa. BMC Evol Biol. 2011;11:117. doi: 10.1186/1471-2148-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Barraclough TG. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glew L, Hudson MD. Gorillas in the midst: the impact of armed conflict on the conservation of protected areas in sub-Saharan Africa. Oryx. 2007;41:140–150. [Google Scholar]
- Graur D, Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Greenbaum E, Sinsch U, Lehr E, Valdez F, Kusamba C. Phylogeography of the tree frog Hyperolius castaneus (Anura: Hyperoliidae) from the Albertine Rift of Central Africa: implications for taxonomy, biogeography and conservation. Zootaxa. 2013;3731:473–494. doi: 10.11646/zootaxa.3731.4.3. [DOI] [PubMed] [Google Scholar]
- Greenbaum E, Portillo F, Jackson K, Kusamba C. A phylogeny of Central African Boaedon (Serpentes: Lamprophiidae), with the description of a new species from the Albertine Rift. Afr J Herpetol. 2015;64:18–38. [Google Scholar]
- Greenbaum E. Emerald Labyrinth: A Scientist’s Adventures in Jungles of the Congo. ForeEdge: Lebanon, NH; In press. [Google Scholar]
- Griffiths CJ. The geological evolution of East Africa. In: Lovett JC, Wasser SK, editors. Biogeography and Ecology of the Rain Forests of Eastern Africa. Cambridge University Press; Cambridge, UK: 1993. pp. 9–21. [Google Scholar]
- Groth JG, Barrowclough GF. Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Mol Phylogenet Evol. 1999;12:115–123. doi: 10.1006/mpev.1998.0603. [DOI] [PubMed] [Google Scholar]
- Hanson T, Brooks TM, Da Fonseca GAB, Hoffmann M, Lamoreux JF, Machlis G, Mittermeier CG, Mittermeier RA, Pilgrim JD. Warfare in biodiversity hotspots. Conserv Biol. 2009;23:578–587. doi: 10.1111/j.1523-1739.2009.01166.x. [DOI] [PubMed] [Google Scholar]
- Harrigan RJ, Mazza ME, Sorenson MD. Computation vs. cloning: evaluation of two methods for haplotype determination. Mol Ecol Resour. 2008;8:1239–1248. doi: 10.1111/j.1755-0998.2008.02241.x. [DOI] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature (London) 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Ho SY, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hughes DF, Walker EM, Gignac PM, Martinez A, Negishi K, Lieb CS, Greenbaum E, Khan AM. Rescuing perishable neuroanatomical information from a threatened biodiversity hotspot: remote field methods for brain tissue preservation validated by cytoarchitectonic analysis, immunohistochemistry, and x-ray microcomputed tomography. PLoS ONE. 2016;11:e0155824. doi: 10.1371/journal.pone.0155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DF, Kusamba C, Behangana M, Greenbaum E. Integrative taxonomy of the Central African forest chameleon Kinyongia adolfifriderici (Sauria: Chamaeleonidae), reveals underestimated species diversity in the Albertine Rift. Zool J Linn Soc. In press. Available at: https://doi.org/10.1093/zoolinnean/zlx005.
- Huhndorf MH, Peterhans JCK, Loew SS. Comparative phylogeography of three endemic rodents from the Albertine Rift, east central Africa. Mol Ecol. 2007;16:663–674. doi: 10.1111/j.1365-294X.2007.03153.x. [DOI] [PubMed] [Google Scholar]
- Hutter CR, Guayasamin JM, Wiens JJ. Explaining Andean megadiversity: the evolutionary and ecological causes of glassfrog elevational richness patterns. Ecol Lett. 2013;16:1135–1144. doi: 10.1111/ele.12148. [DOI] [PubMed] [Google Scholar]
- IUCN. The IUCN Red List of Threatened Species Version 2016-2. 2017 Accessed on 27 April 2017. Available at: www.iucnredlist.org.
- Jacobs BF, Kingston JD, Jacobs LL. The origin of grass-dominated ecosystems. Ann Missouri Bot. 1999;86:590–643. [Google Scholar]
- Jacobs BF. Palaeobotanical studies from tropical Africa: relevance to the evolution of forest, woodland and savannah biomes. Phil Trans R Soc B. 2004;359:1573–1583. doi: 10.1098/rstb.2004.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MEH, Anderson CL, Hipsley CA, Müller J, Evans SE, Schoch RR. Integration of molecules and new fossils supports a Triassic origin for Lepidosauria (lizards, snakes, and tuatara) BMC Evol Biol. 2013;13:208. doi: 10.1186/1471-2148-13-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahindo CM, Bates JM, Bowie RCK. Population genetic structure of Grauer’s Swamp Warbler Bradypterus graueri, an Albertine Rift endemic. Ibis. 2017;159:415–429. [Google Scholar]
- Kampunzu AB, Bonhomme MG, Kanika M. Geochronology of volcanic rocks and evolution of the Cenozoic Western Branch of the East African Rift System. J Afr Earth Sci. 1998;26:441–461. [Google Scholar]
- Kanyamibwa S. Impact of war on conservation: Rwandan environment and wildlife in agony. Biodivers Conserv. 1998;7:1399–1406. [Google Scholar]
- Kaufmann G, Hinderer M, Romanov D. Shaping the Rwenzoris: balancing uplift, erosion, and glaciation. Int J Earth Sci. 2015;104:1–18. [Google Scholar]
- Keqin G, Norell MA. Taxonomic composition and systematics of Late Cretaceous lizard assemblages from Ukhaa Tolgod and adjacent localities, Mongolian Gobi Desert. Bull Am Mus Nat Hist. 2000;249:1–118. [Google Scholar]
- Kissling WD, Eiserhardt WL, Baker WJ, Borchsenius F, Couvreur TL, Balslev H, Svenning JC. Cenozoic imprints on the phylogenetic structure of palm species assemblages worldwide. Proc Natl Acad Sci USA. 2012;109:7379–7384. doi: 10.1073/pnas.1120467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver CJJ, Böhme W. Phylogeny and classification of the Chamaeleonidae (Sauria) with special reference to hemipenis morphology. Bonn zool Monogr. 1986;22:1–64. [Google Scholar]
- Kolbert E. The Sixth Extinction: An Unnatural History. Bloomsbury Publishing; London, UK: 2014. [Google Scholar]
- Kozak KH, Wiens JJ. Niche conservatism drives elevational diversity patterns in Appalachian salamanders. Am Nat. 2010;176:40–54. doi: 10.1086/653031. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Larson TR, Castro D, Behangana M, Greenbaum E. Evolutionary history of the river frog genus Amietia (Anura: Pyxicephalidae) reveals extensive diversification in Central African highlands. Mol Phylogenet Evol. 2016;99:168–181. doi: 10.1016/j.ympev.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leakey MG, Feibel CS, Bernor RL, Harris JM, Cerling TE, Stewart KM, Storrs GW, Walker A, Werdelin L, Winkler AJ. Lothagam: a record of faunal change in the Late Miocene of East Africa. J Vert Paleontol. 1996;16:556–570. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liedtke HC, Hügli D, Dehling JM, Pupin F, Menegon M, Plumptre AJ, Kujirakwinja D, Loader SP. One or two species? On the case of Hyperolius discodactylus Ahl, 1931 and H alticola Ahl, 1931 (Anura: Hyperoliidae) Zootaxa. 2014;3768:253–290. doi: 10.11646/zootaxa.3768.3.2. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Edwards SV. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 2010;10:302. doi: 10.1186/1471-2148-10-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreau M, Naeem S, Inchausti P, Bengtsson J, Grime JP, Hector A, Hooper DU, Huston MA, Raffaelli D, Schmid B, Tilman D. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science (Washington) 2001;294:804–808. doi: 10.1126/science.1064088. [DOI] [PubMed] [Google Scholar]
- Lötters S, Wagner P, Bwong BA, Schick S, Malonza PK, Muchai V, Wasonga DV, Veith M. A Fieldguide to the Amphibians and Reptiles of the Kakamega Forest. National Museums of Kenya and University of Mainz; Frankfurt am Main, Germany: 2007. [Google Scholar]
- Loveridge A. A new subgenus of Chamaeleo from Rhodesia and a new race of Mabuya from Kenya Colony. Breviora. 1956;59:1–4. [Google Scholar]
- Macey JR, Larson A, Ananjeva NB, Papenfuss TJ. Evolutionary shifts in three major structural features of the mitochondrial genome among iguanian lizards. J Mol Evol. 1997a;44:660–674. doi: 10.1007/pl00006190. [DOI] [PubMed] [Google Scholar]
- Macey JR, Larson A, Ananjeva NB, Fang ZL, Papenfuss TJ. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol. 1997b;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- Macgregor D. History of the development of the East African Rift System: a series of interpreted maps through time. J Afr Earth Sci. 2015;101:232–252. [Google Scholar]
- Maddison WP. Gene trees in species trees. Syst Biol. 1997;46:523–536. [Google Scholar]
- Maddison DR, Maddison WP. MacClade: analysis of phylogeny and character evolution Version 4-08. 2005 doi: 10.1159/000156416. Available at: http://macclade.org/index.html. [DOI] [PubMed]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis Version 3-04. 2015 Available at: http://mesquiteproject.org.
- Maley J. The African forest – main characteristics of changes in vegetation and climate from Upper Cretaceous to the Quaternary. Proc R Soc Edinb. 1996;104:31–73. [Google Scholar]
- Mariaux J, Tilbury CR. The pygmy chameleons of the Eastern Arc Range (Tanzania): evolutionary relationships and the description of three new species of Rhampholeon (Sauria: Chamaeleonidae) Herpetol J. 2006;16:315–331. [Google Scholar]
- Matthee CA, Tilbury CR, Townsend T. A phylogenetic review of the African leaf chameleons: genus Rhampholeon (Chamaeleonidae): the role of vicariance and climate change in speciation. Proc R Soc Lond Biol. 2004;271:1967–1975. doi: 10.1098/rspb.2004.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E, O’Hara RJ. The biogeographic evidence supporting the Pleistocene forest refuge hypothesis. Evolution. 1986;40:55–67. doi: 10.1111/j.1558-5646.1986.tb05717.x. [DOI] [PubMed] [Google Scholar]
- Meadows ME, Linder HP. A palaeoecological perspective on the origin of Afromontane grasslands. J Biogeogr. 1993;20:345–355. [Google Scholar]
- Menegon M, Loader SP, Marsden SJ, Branch WR, Davenport TRB, Ursenbacher S. The genus Atheris (Serpentes: Viperidae) in East Africa: phylogeny and the role of rifting and climate in shaping the current pattern of species diversity. Mol Phylogenet Evol. 2014;79:12–22. doi: 10.1016/j.ympev.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Mittermeier RA, Turner WA, Larsen FW, Brooks TM, Gascon C. Global biodiversity conservation: the critical role of hotspots. In: Zachos FE, Habel JC, editors. Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas. Springer-Verlag; Berlin, Germany: 2011. pp. 3–22. [Google Scholar]
- Miralles A, Vences M. New metrics for comparison of taxonomies reveal striking discrepancies among species delimitation methods in Madascincus lizards. PLoS ONE. 2013;8:e68242. doi: 10.1371/journal.pone.0068242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Evol Syst. 2000;31:533–563. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, Da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature (London) 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Niemiller ML, Near TJ, Fitzpatrick BM. Delimiting species using multilocus data: diagnosing cryptic diversity in the southern cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae) Evolution. 2012;66:846–866. doi: 10.1111/j.1558-5646.2011.01480.x. [DOI] [PubMed] [Google Scholar]
- Nydam RL, Cifelli RL. Lizards from the Lower Cretaceous (Aptian–Albian) Antlers and Cloverly Formations. J Vert Paleontol. 2002;22:286–98. [Google Scholar]
- Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY are we there yet? A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Olave M, Solà E, Knowles LL. Upstream analyses create problems with DNA-based species delimitation. Syst Biol. 2014;63:263–271. doi: 10.1093/sysbio/syt106. [DOI] [PubMed] [Google Scholar]
- Palumbi S. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. 2nd. Sinauer Associates; Sunderland, MA: 1996. pp. 205–247. [Google Scholar]
- Pasteels P, Villeneuve M, De Paepe P, Klerkx J. Timing of the volcanism of the southern Kivu province: implications for the evolution of the western branch of the East African Rift system. Earth Planet Sci Lett. 1989;94:353–363. [Google Scholar]
- Paul JD, Roberts GG, White N. The African landscape through space and time. Tectonics. 2014;33:898–935. [Google Scholar]
- Pickford M. Evidence for an arid climate in western Uganda during the Middle Miocene. Comptes Rendus de l’Académie des Sciences. Série 2, Mécanique, Physique, Chimie, Sciences de l’univers. Sciences de la Terre. 1992;315:1419–1424. [Google Scholar]
- Pickford M, Senut B, Hadoto DPM. Geology and palaeobiology of the Albertine Rift valley, Uganda-Zaire: geology Vol I Publication Occasionnelle. Centre International Pour la Formation et les Echanges Geologiques, CIFEG; Orléans, France: 1993. [Google Scholar]
- Plumptre AJ, Davenport TRB, Behangana M, Kityo R, Eilu G, Ssegawa P, Ewango C, Meirte D, Kahindo C, Herremans M, Peterhans JCK, Pilgrim JD, Wilson M, Languy M, Moyer D. The biodiversity of the Albertine Rift. Biol Cons. 2007;134:178–194. [Google Scholar]
- Ponce-Reyes R, Plumptre AJ, Segan D, Ayebare S, Fuller RA, Possingham HP, Watson JEM. Forecasting ecosystem responses to climate change across Africa’s Albertine Rift. Biol Cons. 2017;209:464–472. [Google Scholar]
- Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- Portillo F, Greenbaum E, Menegon M, Kusamba C, Dehling JM. Phylogeography and species boundaries of Leptopelis (Anura: Arthroleptidae) from the Albertine Rift. Mol Phylogenet Evol. 2015;82:75–86. doi: 10.1016/j.ympev.2014.09.024. [DOI] [PubMed] [Google Scholar]
- Pyron RA, Burbrink FT, Wiens JJ. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 2013;13:93. doi: 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.5. 2007 Available at: http://tree.bio.ed.ac.uk/software/tracer/
- Rambaut A, Drummond AJ. FigTree v1.3.1. 2009 Available at: http://tree.bio.ed.ac.uk/software/figtree/
- Roberts EM, Stevens NJ, O’Connor PM, Dirks PHGM, Gottfried MD, Clyde WC, Armstrong RA, Kemp AIS, Hemming S. Initiation of the western branch of the East African Rift coeval with the eastern branch. Nat Geosci. 2012;5:289–294. [Google Scholar]
- Robinson JE, Griffiths RA, John FAS, Roberts DL. Dynamics of the global trade in live reptiles: shifting trends in production and consequences for sustainability. Biol Cons. 2015;184:42–50. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Roy J, Arandjelovic M, Bradley BJ, Guschanski K, Stephens CR, Bucknell D, Cirhuza H, Kusamba C, Kyungu JC, Smith V, Robbins MM. Recent divergences and size decreases of eastern gorilla populations. Biol Lett. 2014;10:20140811. doi: 10.1098/rsbl.2014.0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaj MH. Standard symbolic codes for institutional resource collections in herpetology and ichthyology: an online reference. Version 6.5 (16 August 2016) Washington, DC: American Society of Ichthyologists and Herpetologists; 2016. Available at: http://www.asih.org/ [Google Scholar]
- Salzburger W, Van Bocxlaer B, Cohen AS. Ecology and evolution of the African Great Lakes and their faunas. Ann Rev Ecol Evol Syst. 2014;45:519–545. [Google Scholar]
- San Mauro D, Gower DJ, Oommen OV, Wilkinson M, Zardoya R. Phylogeny of caecilian amphibians (Gymnophiona) based on complete mitochondrial genomes and nuclear RAG1. Mol Phylogenet Evol. 2004;33:413–427. doi: 10.1016/j.ympev.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Sauquet H. A practical guide to molecular dating. C R Palevol. 2013;12:355–367. [Google Scholar]
- Schmidt KP. Contributions to the herpetology of the Belgian Congo based on the collection of the American Museum Congo Expedition, 1909–1915. Part I: Turtles, crocodiles, lizards, and chamaeleons. Bull Am Mus Nat Hist. 1919;39:385–624. [Google Scholar]
- Senut B, Pickford M. Geology and palaeobiology of the Albertine Rift valley, Uganda-Zaire: palaeobiology, Vol II Publication Occasionnelle. Centre International Pour la Formation et les Echanges Geologiques, CIFEG; Orléans, France: 1994. [Google Scholar]
- Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin JJ, Brunet M. Tectonic uplift and Eastern Africa aridification. Science (Washington) 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- Shaw TI, Ruan Z, Glenn TC, Liu L. STRAW: Species TRee Analysis Web server. Nucl Acids Res. 2013;41:W238–W241. doi: 10.1093/nar/gkt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistrom M, Hutchinson M, Bertozzi T, Donnellan S. Evaluating evolutionary history in the face of high gene tree discordance in Australian Gehyra (Reptilia: Gekkonidae) Heredity. 2014;113:52–63. doi: 10.1038/hdy.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spawls S, Howell K, Drewes RC, Ashe J. A Field Guide to the Reptiles of East Africa: Kenya, Tanzania, Rwanda and Burundi. Academic Press; London, UK: 2002. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Steindachner F. Vorläufiger Bericht über drei neue Arten aus der Familie Chamaeleontidae. Anz Akad Wiss Wien. 1911;10:177–179. [Google Scholar]
- Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipala J. Mountain Dragons: In Search of Chameleons in the Highlands of Kenya. Tien Wah Press; Singapore: 2014. [Google Scholar]
- Sukumaran J, Knowles LL. Multispecies coalescent delimits structure, not species. Proc Natl Acad Sci USA. 2017;114:1607–1612. doi: 10.1073/pnas.1607921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RM. A new Middle Paleocene (Torrejonian) rhineurid amphisbaenian, Plesiorhineura tsentasi new genus, new species, from the San Juan Basin, New Mexico. J Paleo. 1985;59:1481–1485. [Google Scholar]
- Sullivan RM, Lucas SG. Palaeoscincosaurus middletoni, new genus and species (Squamata: Scincidae) from the Early Paleocene (Puercan) Denver Formation, Colorado. J Vert Paleontol. 1996;16:666–672. [Google Scholar]
- Sullivan JP, Lundberg JG, Hardman M. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using rag1 and rag2 nuclear gene sequences. Mol Phylogenet Evol. 2006;41:636–662. doi: 10.1016/j.ympev.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Swindell SR, Plasterer TN. SEQMAN. In: Swindell SR, editor. Sequence Data Analysis Guidebook Methods in Molecular Medicine. Vol. 70. Humana Press; Totowa, NJ: 1997. pp. 75–89. [Google Scholar]
- Talavera G, Dincă V, Vila R. Factors affecting species delimitations with the GMYC model: insights from a butterfly survey. Methods Ecol Evol. 2013;4:1101–1110. [Google Scholar]
- Tilbury CR. Chameleons of Africa: An Atlas Including the Chameleons of Europe, the Middle East and Asia. Edition Chimaira: Frankfurt am Main, Germany; 2010. [Google Scholar]
- Tilbury CR, Tolley KA. Contributions to the herpetofauna of the Albertine Rift: two new species of chameleon (Sauria: Chamaeleonidae) from an isolated montane forest, south eastern Democratic Republic of Congo. Zootaxa. 2015;3905:345–364. doi: 10.11646/zootaxa.3905.3.2. [DOI] [PubMed] [Google Scholar]
- Tolley KA, Tilbury CR, Measey GJ, Menegon M, Branch WR, Matthee CA. Ancient forest fragmentation or recent radiation? Testing refugial speciation models in chameleons within an African biodiversity hotspot. J Biogeogr. 2011;38:1748–1760. [Google Scholar]
- Tolley KA, Herrel A, editors. The Biology of Chameleons. University of California Press; Berkeley, CA: 2013. [Google Scholar]
- Tolley KA, Townsend TM, Vences M. Large-scale phylogeny of chameleons suggests African origins and Eocene diversification. Proc R Soc Lond Biol. 2013;280:20130184. doi: 10.1098/rspb.2013.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley KA, Plumptre AJ. Rhampholeon boulengeri The IUCN Red List of Threatened Species 2014: e.T172523A1344185. 2014 Accessed on 26 March 2017. Available at: http://www.iucnredlist.org/details/172523/0.
- Tolley KA, Tilbury CR. Rhampholeon hattinghi The IUCN Red List of Threatened Species 2015 e.T75976755A75976771. 2015 Accessed on 26 March 2017. Available at: http://www.iucnredlist.org/details/75976755/0.
- Tolley KA, Alexander GJ, Branch WR, Bowles P, Maritz B. Conservation status and threats for African reptiles. Biol Cons. 2016;204:63–71. [Google Scholar]
- Townsend TM, Tolley KA, Glaw F, Böhme W, Vences M. Eastward from Africa: palaeocurrent-mediated chameleon dispersal to the Seychelles islands. Biol Lett. 2011;7:225–228. doi: 10.1098/rsbl.2010.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Freed P, Hošek J, editors. The Reptile Database. 2017 Accessed on 17 March 2017. Available at: http://www.reptile-database.org.
- Voelker G, Marks BD, Kahindo C, A’genonga U, Bapeamoni F, Duffie LE, Huntley JW, Mulotwa E, Rosenbaum SA, Light JE. River barriers and cryptic biodiversity in an evolutionary museum. Ecol Evol. 2013;3:536–545. doi: 10.1002/ece3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Einsiedel O. Virunga [movie] Grain Media Ltd.; London, UK: 2014. dir. [Google Scholar]
- Wagner P, Böhme W. Herpetofauna Kakamegensis–the amphibians and reptiles of Kakamega Forest, western Kenya. Bonn zool Beitr. 2007;55:123–150. [Google Scholar]
- Wagner P, Köhler J, Schmitz A, Böhme W. The biogeographical assignment of a west Kenyan rain forest remnant: further evidence from analysis of its reptile fauna. J Biogeogr. 2008;35:1349–1361. doi: 10.1111/j.1365-2699.2008.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake DB, Lynch JF. The distribution, ecology, and evolutionary history of plethodontid salamanders in tropical America. Nat Hist Mus Los Ang Cty Sci Bull. 1976;25:1–65. [Google Scholar]
- Wallach V, Wüster W, Broadley DG. In praise of subgenera: taxonomic status of cobras of the genus Naja Laurenti (Serpentes: Elapidae) Zootaxa. 2009;2236:26–36. [Google Scholar]
- Werdelin L, Sanders WJ, editors. Cenozoic Mammals of Africa. University of California Press; Berkeley, CA: 2010. [Google Scholar]
- Wichura H, Bousquet R, Oberhänsli R, Strecker MR, Trauth MH. Evidence for middle Miocene uplift of the East African Plateau. Geology. 2010;38:543–546. [Google Scholar]
- Wichura H, Jacobs LL, Lin A, Polcyn MJ, Manthi FK, Winkler DA, Strecker MR, Clemens M. A 17-My-old whale constrains onset of uplift and climate change in East Africa. Proc Natl Acad Sci USA. 2015;112:3910–3915. doi: 10.1073/pnas.1421502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler DA, Murry PA, Jacobs LL. Early Cretaceous (Comanchean) vertebrates of central Texas. J Vert Paleontol. 1990;10:95–116. [Google Scholar]
- Wronski T, Hausdorf B. Distribution patterns of landsnails in Ugandan rain forests support the existence of Pleistocene forest refugia. J Biogeogr. 2008;35:1759–1768. [Google Scholar]
- Wynn J. Miocene and Pliocene paleosols of Lothagam. In: Leakey MG, Harris J, editors. Lothagam: The Dawn of Humanity in Eastern Africa. Columbia University Press; New York City, NY: 2003. pp. 31–38. [Google Scholar]
- Xi Z, Liu L, Rest JS, Davis CC. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst Biol. 2014;63:919–932. doi: 10.1093/sysbio/syu055. [DOI] [PubMed] [Google Scholar]
- Zachos JC, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science (Washington) 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. GenBank accession numbers for outgroup samples analyzed in Fig. S2. Alternative species used to create chimeric sequences are shown in each cell above the accession number.
Table S2. Uncorrected p-distances within and among species in the genus Rhampholeon for three molecular markers (16S, ND2, and RAG1).
Figure S1. Bayesian (A) and maximum likelihood (B) phylogenies of the genus Rhampholeon with support values adjacent to nodes.
Figure S2. Entire Bayesian chronogram of Chamaeleonidae and outgroup. Numbers near nodes denote mean highest posterior densities (HPD). Fossil-calibrated nodes are indicated with encircled red numbers and secondary calibrated nodes with encircled light blue numbers. Diversification dates within the genus Rhampholeon are presented in Fig. 4.


