Abstract
Continuous spermatogenesis in post-pubertal mammals is dependent on spermatogonial stem cells (SSCs), which balance self-renewing divisions that maintain stem cell pool with differentiating divisions that sustain continuous sperm production. Rodent stem and progenitor spermatogonia are described by their clonal arrangement in the seminiferous epithelium (e.g., Asingle, Apaired or Aaligned spermatogonia), molecular markers (e.g., ID4, GFRA1, PLZF, SALL4 and others) and most importantly by their biological potential to produce and maintain spermatogenesis when transplanted into recipient testes. In contrast, stem cells in the testes of higher primates (nonhuman and human) are defined by description of their nuclear morphology and staining with hematoxylin as Adark and Apale spermatogonia. There is limited information about how dark and pale descriptions of nuclear morphology in higher primates correspond with clone size, molecular markers or transplant potential. Do the apparent differences in stem cells and spermatogenic lineage development between rodents and primates represent true biological differences or simply differences in the volume of research and the vocabulary that has developed over the past half century? This review will provide an overview of stem, progenitor and differentiating spermatogonia that support spermatogenesis; identifying parallels between rodents and primates where they exist as well as features unique to higher primates.
Keywords: Spermatogonial stem cells, Stem cells, Adark, Apale, Spermatogenic lineage development, Testis, Male fertility
1. Introduction
Spermatogonial stem cells (SSCs) are the adult tissue stem cells in the testis that are at the foundation of spermatogenesis and essential for male fertility (Phillips et al., 2010). SSCs are defined by their dual potentials: 1) self-renew to maintain the stem cell pool and 2) differentiate to maintain continuous sperm production in post-pubertal males (de Rooij & Grootegoed, 1998). Similar to other adult tissue stem cells, SSCs are rare, comprising only 0.03% of total germ cells in mice (Tegelenbosch & de Rooij, 1993). However, numerous transit amplifying mitotic divisions in progenitor and differentiating spermatogonia, followed by two meiotic divisions give rise to millions of sperm each day (Phillips et al., 2010).
2. Spermatogonial stem cells and spermatogenic lineage development: lessons from the rodent
In the post-natal rodent testis, SSC activity is broadly believed to reside in the population of isolated (single) spermatogonia located on the basement membrane of the seminiferous tubules (Huckins, 1971; Oakberg, 1971a; de Rooij, 1973). These rare cells are called the Asingle spermatogonia (As), which divide once every three days and make up about 0.03% of the total germ cells in the mouse testis (Tegelenbosch & de Rooij, 1993; Huckins & Oakberg, 1978a). Mitotic division of As produces a pair of spermatogonia (Apaired; Apr) that will either complete cytokinesis to produce two new As (self-renewing division) or remain joined by an intracytoplasmic bridge and produce a chain of four Aaligned spermatogonia (Aal4) at the next division (Phillips et al., 2010; de Rooij & Griswold, 2012) (Fig. 1A). The Aal4 spermatogonia may undergo one or more mitotic divisions to form larger chains of 8, 16 and sometimes 32 Aal spermatogonia. Collectively, As, Apr and Aal make up the population of undifferentiated spermatogonia that comprises 0.3% of germ cells in the rodent testis; As make up 10% of undifferentiated spermatogonia (0.03% of germ cells; Fig. 2A) (Phillips et al., 2010; Huckins, 1971; Oakberg, 1971a; Valli et al., 2015; Oakberg, 1971b). Larger clones of Aal spermatogonia differentiate to A1 spermatogonia. In this context, a clone is defined as the group of interconnected cells that arise from a single As spermatogonia. In Rodents, the clones become so large that they fill entire segments of seminiferous tubule due to sequential mitotic divisions from A1 spermatogonia that produce types A2, A3, A4, Intermediate and B spermatogonia, which divide to produce primary spermatocytes. Two meiotic divisions from primary spermatocytes give rise to secondary spermatocytes and round spermatids, which undergo spermiogenesis (morphological differentiation) to produce mature sperm. Thus, through a series of transit amplifying mitotic and meiotic divisions, a relatively small pool of stem cells in the rodent testis produces 40 million sperm per gram of testis parenchyma each day (Figs. 1A and 2A) (Tegelenbosch & de Rooij, 1993; Valli et al., 2015; Thayer et al., 2001).
Fig. 1.
Clonal development in the spermatogenic lineages of rodents, monkeys and humans. Undifferentiated spermatogonia are described as As, Apr or Aal in the rodents and Adark or Apale in monkey and human. During spermatogenic development, Asingle (As) and Adark and/or Apale undergo one or more mitotic divisions to give rise to cells of larger clones (chains) of interconnected cells sizes through transit-amplifying mitotic divisions. A) Clonal development in rodents features 3–4 transit amplifying divisions in the pool of undifferentiated As, Apr and Aal spermatogonia followed by 6 amplifying divisions in the pool of differentiated spermatogonia (A1–A4, Intermediate, B), which give rise to primary spermatocytes. Two additional meiotic divisions produce round spermatids that undergo spermiogenesis to produce sperm. B) Clonal development of spermatogonia in monkeys features 0, 1 or 2 transit amplifying divisions in the pool of undifferentiated Adark/Apale spermatogonia, followed by 4 transit amplifying divisions of differented spermatogonia (B1–B4), which give rise to primare spermatocytes. C) Clonal development of spermatogonia in humans features 0, 1 or 2 transit amplifying divisions in the pool of undifferentiated Adark/Apale spermatogonia followed by a single a single transit amplyfying division in differentiated B spermatogonia that give rise to primary spermatocytes. Thus, there are typically 12 transit amplifying divisions in rodents; 8 in monkeys and 5 in humans between stem cell and sperm. The reduced number of transit amplyfing divisions in monkeys and humans is compensated in part by a larger stem cell pool (see Fig. 2).
Fig. 2.
Schematic comparison of the stem cell pools and sperm output in mice, monkeys and humans. A) The spermatogenic lineage in mice features a relatively small pool of As, Apr and Aal undifferentiated spermatogonia (0.3% of germ cells). However, with about 12 transit amplifying divisions between stem cells and sperm (see Fig. 1), the small pool of stem cells produced 40 million sperm per gram of testicular tissue per day. B) The spermatogenic lineage in monkeys features a relatively larger pool of Adark/Apale spermatogonia (4% of germ cells) and this compensates for the reduced number of transit amplying divisions between stem cell and sperm. Sperm output in monkeys is similar to mice: 41 million sperm per gram of testicular tissue per day. C) The spermatogenic lineage in humans features the largest pool of undifferentiated Adark/Apale spermatogonia (22% of germ cells). However, with only one transit amplifying division of differentiated spermatogonia, sperm output in humans is reduced to 4.4 million sperm per gram of testicular tissue per day. Thus distinguishing features of spermatogenic lineage development in mice, monkeys and men include 1) the size of the pool of undifferentiated stem/progenitor spermatogonia; 2) the number of transit amplifying divisions in differentiated spermatogonia and 3) sperm output.
Undifferentiated stem and progenitor spermatogonia in rodent testes are defined in part by clone size, as indicated above, and in part by molecular markers (e.g., ID4, PAX7, BMI1, EOMES, GFRa1, NANOS2, UTF1, ZBTB16, SALL4, LIN28, FOXO1 and others). Markers can be observed by immunohistochemistry in histological cross sections; in whole mount preparations of seminiferous tubules or by fluorescence-activated cell sorting (FACS), but clone size can only be observed in whole mount preparations of seminiferous tubules. Based on whole mount immunohistochemistry, ID4, PAX7, BMI1 and EOMES appear to have the most restricted pattern of expression, which is limited to As spermatogonia (Oatley et al., 2011; Aloisio et al., 2014; Komai et al., 2014; Braun et al., 2017). GFRa1, NANOS2 and UTF1 have expression limited to As, Apr and Aal4 (Suzuki et al., 2009; van Bragt et al., 2008; Meng et al., 2000), while ZBTB16, SALL4, LIN28, CDH1 and FOXO1 are expressed by most or all undifferentiated As, Apr and Aal spermatogonia (Costoya et al., 2004; Buaas et al., 2004; Hobbs et al., 2012; Eildermann et al., 2012a; Gassei & Orwig, 2013; Tokuda et al., 2007; Goertz et al., 2011), including some overlap with cKIT+ differentiating spermatogonia (Fig. 3A). Based on the restricted pattern of expression, some have suggested that cells expressing ID4, PAX7 and/or BMI1 might be the ultimate spermatogonial stem cells (SSCultimate) (Helsel et al., 2017; Lord & Oatley, 2017; de Rooij, 2017). Indeed, the expression of each marker on functional stem cells has been confirmed by SSC transplantation and/or lineage tracing. However, there is little information about the extent of overlap among these markers; whether any of these proteins mark the entire population of As spermatogonia or whether the entire population of functional stem cells resides in the population of As spermatogonia. In fact, molecular heterogeneity among undifferentiated spermatogonia of all clone sizes has been repeatedly documented (Suzuki et al., 2009; Gassei & Orwig, 2013; Hermann et al., 2015; Nakagawa et al., 2010; Zheng et al., 2009).
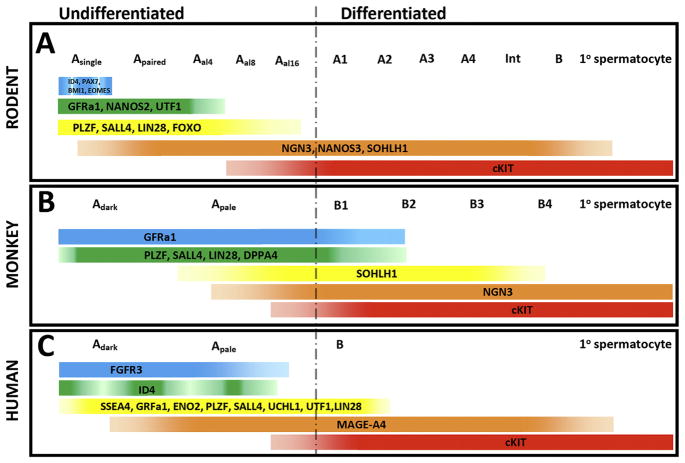
Fig. 3.
Spermatogonial markers in rodents, monkeys and humans. A) Rodents; B) Monkeys; C) Humans. Several markers are conserved from rodents to monkeys to humans, suggesting their importance in spermatogenic lineage development. GFRa1, PLZF, SALL4 and LIN28 are conserved markers of undifferentiated spermatogonia. cKIT is a conserved marker of differentiating/differentiated spermatogonia. The following references describe markers in this figure that were not referenced elsewhere in the text: NGN3 (Yoshida et al., 2004); SOHLH1 (Ballow et al., 2006).
It seems reasonable to suppose that the stem cell pool also extends to some Apr spermatogonia because As must transit through an Apr state in the process of self-renewal (see Fig. 1); this concept has been described as “false pairs” and is nicely reviewed in (de Rooij & Griswold, 2012). Furthermore, Hara and colleagues provided live video imaging of GFRa1-GFP spermatogonia data to suggest that fragmentation of larger clones (eg., Aal4 fragmenting to Aal3 + As or Apr + 2 As or 4 As) was an important contributor to maintenance of the As pool ((Hara et al., 2014); Fig. 1A). While the fate of the fragmenting clones could not be documented in that study, clones of Aal3 (possibly resulting from clone fragmentation) have been observed by others (Suzuki et al., 2009; Gassei & Orwig, 2013; Tokuda et al., 2007; Hara et al., 2014). However, one concern with the clone fragmentation model is that it is based entirely on observations of GFRa1 positive cells and does not account for the contribution of GFRa1 negative cells that are known to exist in the pool of As (Suzuki et al., 2009; Gassei & Orwig, 2013) and the pool of transplantable stem cells (Grisanti et al., 2009; Garbuzov et al., 2018). Any model that considers only a part of the stem cell pool defined by a single molecular marker is likely to be incomplete.
To date, the only way to definitively identify a spermatogonial stem cell is by observing its capacity to produce and maintain spermatogenesis long-term, by transplantation (Brinster & Zimmermann, 1994; Brinster & Avarbock, 1994) or lineage tracing (Aloisio et al., 2014; Komai et al., 2014; Nakagawa et al., 2007). These are retrospective assays. There is no evidence that the mouse SSCs can be prospectively defined completely and exclusively by a specific clone size or molecular marker (see discussion above). However, it is generally agreed that smaller clones are more undifferentiated while larger clones are more differentiated and that cKIT marks the transition to differentiated type A1 spermatogonia. Differentiated type A1 spermatogonia in rodents appear to be equivalent to type B1 in nonhuman primates and type B in humans based on appearance of heterochromatin and initiation of cKIT expression (Figs. 1 and 3). Co-staining with cKIT and a marker of un-differentiated spermatogonia (e.g., PLZF, SALL4, CDH1, UCHL1, etc) in whole mount preparations of seminiferous tubules can help to define the clone size where undifferentiated stem/progenitor spermatogonia transition to differentiated spermatogonia. In mice, this transition occurs most frequently at a clone size of 16 (Fig. 1A), but can also happen at clones sizes of 8 and less frequently at smaller clone sizes (Suzuki et al., 2009; Gassei & Orwig, 2013; Tokuda et al., 2007; Hara et al., 2014). In nonhuman primates and humans, this transition occurs at smaller clone sizes (Fig. 1B & C; see discussion below).
3. Stem cells and spermatogenic lineage development in higher primates
Nonhuman primate and human testes contain two morphologically distinct types of undifferentiated spermatogonia, identified as Adark and Apale, based on differences in nuclear morphology and staining intensity with hematoxylin (Clermont & Leblond, 1959; Clermont & Antar, 1973; Clermont, 1966). Adark spermatogonia are “relatively small, spherical or slightly ovoid” cells on the basement membrane of seminiferous tubules having dark, dense chromatin in their “uniformly stained” nuclei. Apale spermatogonia are identified as “relatively larger, oval” or almost round cells on the basement membrane of the seminiferous tubules having pale, elongated nuclei with “coarser” or more “granular chromatin”. Nucleoli may be visible in both Adark and Apale spermatogonia (Fig. 4A). B-type spermatogonia are identified by their relatively larger size, location on or close to the basement membrane of the seminiferous tubules; clear and roundish nuclei and they are differentiated from one another by the granulation and density of hetero-chromatin staining. B1 spermatogonia are least heterochromatic and B4 spermatogonia are most heterochromatic (Clermont & Leblond, 1959) Some studies have identified a “rarefraction zone” (chromatin free zone) in a subpopulation of Adark spermatogonia (Fig. 4B indicates examples of Adark with rarefaction zone and Adark without rarefaction zone). The observation of a rarefaction zone may be fixation-dependent and is used more frequently to describe Adark spermatogonia in humans than in nonhuman primates (see review from von Kopylow et al. in this special issue) (von Kopylow et al., 2010; Lim et al., 2011; Paniagua & Nistal, 1984; Schulze, 1978). There are currently few researchers with the experience or patience to use the classic Adark and Apale descriptors of primate spermatogonia. However, researchers who do so provide a valuable link between contemporary molecular readouts and the histological descriptions in the classic literature.
Fig. 4.
Histological and immunohistochemical evaluation of Adark and Apale spermatogonia in monkeys and humans. Periodic acid, Schiffs’ and Hematoxylin (PAS-H) staining in monkey (A and C) and human (B and D) testis section reveals Adark (black arrows) and Apale (red arrows) spermatogonia on the basement membrane of the seminiferous tubules. The sub-population of Adark spermatogonia with a rarefraction zone are indicated by a green arrow in (B). Colorimetric staining for UTF1 (brown color) with PAS-H staining confirms that UTF1 is a conserved marker of most, but not all Adark (black arrow) and Apale (red arrows) spermatogonia in monkey (C) and Human (D) testes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In 1959, Clermont and Leblond proposed that A1 (Adark) are the stem cells, which divide to either self–renew and maintain the stem cell pool or give rise to the A2 (Apale) progenitor cells that may undergo one or more transit-amplifying divisions before giving rise to differentiated B1 spermatogonia. Clermont revised his model 10 years later based on in vivo labeling with 3H-thymidine, which indicated that Apale, but not Adark, incorporated 3H-thymidine in Vervet monkeys (Cercopithecus aethiops). Since Adark did not appear to self-renew under steady state conditions, he proposed that Apale are the “active” stem cells that maintain spermatogenesis in the adult testis while Adark are “reserve” stem cells that regenerate spermatogenesis when it is destroyed by noxious insult (e.g., chemotherapy or radiation) (Clermont, 1969). Experimental evidence supporting this model are derived from observations in nonhuman primates and men that X-irradiation caused a striking depletion of spermatogenesis, including the entire population of Apale, which were subsequently replenished from the surviving pool of Adark spermatogonia (Clifton & Bremner, 1983; Oakberg, 1968; Oakberg, 1975; van Alphen et al., 1988). Ehmcke and Schlatt argued that low mitotic index and regenerative capacity of Adark is consistent with the characteristics of a “true stem cell” and the regular proliferation of Apale is indicative of “renewing progenitors” (Ehmcke & Schlatt, 2006). During the past 50 years, eight studies have reported on the acute labeling index of Adark and Apale spermatogonia. While four studies observed no labeling in the Adark spermatogonia (Buageaw et al., 2005; Schlatt & Weinbauer, 1994; de Rooij et al., 1986; Simorangkir et al., 2009), consistent with the results of Clermont; four studies reported a wide range (0.06% to 18%) of Adark labeling (Clermont & Antar, 1973; Fouquet & Dadoune, 1986; Ehmcke et al., 2005; Kluin et al., 1983). In all of those studies, 3H-thymidine or BrdU was administered as a single bolus, and this may not effectively label a very slow cycling population of stem cells. Chronic labeling studies are needed to determine whether Adark are indeed quiescent or whether Adark are slow-cycling, active stem cells in steady state spermatogenesis. We have proposed that Adark and at least some Apale are the same population of cells that are simply at different stages of the cell cycle (i.e., Adark: G0 versus Apale: G1/S/G2/M) (Hermann et al., 2010). The concept that Adark are in prolonged G0 is supported by observations of von Kopylow and colleagues, who found the Ki67 was expressed by Apale, but not Adark spermatogonia of the human testis (von Kopylow et al., 2012a). We believe this indicates that Adark are slow cycling (long G0 phase), not quiescent or reserve, because when we treated adult Rhesus macaques with BrdU in the drinking water for three weeks, nearly 15% of Adark incorporate label (Fayomi and Orwig, In Preparation).
4. Clonal expansion in higher primates
Three dimensional reconstruction mapping of serial cross sections and camera lucida drawings indicated that Adark and Apale spermatogonia in monkey testes are arranged in clones of 1, 2 or 4 cells, suggesting that there are only 1 or 2 transit amplifying divisions before differentiation to B1 spermatogonia (Clermont & Leblond, 1959; Clermont, 1969). This is fewer than the 3–4 transit amplifying divisions that occur in undifferentiated rodent spermatogonia before differentiation to type A1 spermatogonia (See Fig. 1A & B; Fig. 2A & B). Subsequent divisions from B1 spermatogonia in nonhuman primates produce types B2, B3 and B4 spermatogonia, primary spermatocytes, secondary spermatocytes and spermatids that undergo spermiogenesis to produce mature sperm (Clermont & Leblond, 1959). Thus, the number of transit amplifying divisions from B1 spermatogonia to spermatocyte in monkeys (four) is less than the six divisions from A1 spermatogonia to spermatocytes in rodents (Fig. 1A & B; Fig. 2A & B). Despite these differences in spermatogonial transit amplifying divisions, sperm output in rodents and monkeys is about the same (~40 million sperm per gram of testicular parenchyma per day; see Figs. 1 & 2). In contrast, men have only one generation of differentiated type B spermatogonia and sperm output is reduced to 4.4 million sperm per gram of testicular parenchyma per day (Fig. 1C; Fig. 2C; reviewed in (Valli et al., 2015)).
To summarize, there are a total of 12 transit amplifying divisions from the isolated As spermatogonia in rodents to the terminally differentiated sperm, which should yield 4096 sperm per stem cell that commits to differentiate (12 doublings = 212 = 4096) (Russell et al., 1990). The actual yield is considerably less due to massive apoptosis (~50%) that occurs in the differentiated type A2-A4 spermatogonia (de Rooij, 1973; Huckins & Oakberg, 1978b; Huckins, 1978). By comparison, there appear to be only 8 transit amplifying divisions in non-human primates and 5 transit amplifying divisions in humans between the isolated Adark/Apale undifferentiated spermatogonia and terminally differentiated sperm (Fig. 1). Assuming similar stem cell pool sizes and spermatogenic lineage development dynamics, one might expect that sperm output in nonhuman primates (28 = 256) and humans (25 = 32) to be reduced 16-fold and 128-fold, respectively, compared with mice. However, as indicated above and in Figs. 1 and 2, this is not the case. Sperm output in monkeys is equivalent to rodents and sperm output in humans is reduced by only 10-fold compared with rodents. The contribution of apoptosis to sperm output in higher primates is not known, but the size of the stem cell pool is likely to be major contributor to differences in sperm output among species.
5. The pool of stem/progenitor spermatogonia in higher primates is larger than rodents
As described above, the precise molecular or clone size definition of functional stem cells in the rodent testis is subject to debate. However, the broader pool of stem & transit amplifying progenitors in rodents is understood to include As, Apr and Aal spermatogonia with a cKIT negative phenotype. Similarly, the precise definition of functional stem cells in primate testes are subject to debate (Valli et al., 2015; Hermann et al., 2009), but the broader pool of stem/progenitor spermatogonia resides in the population of Adark and Apale spermatogonia with a cKIT negative phenotype. In rodents, the population of As, Apr and Aal un-differentiated spermatogonia comprises 0.3% of germ cells in the testis (Fig. 2A). In nonhuman primates, Adark and Apale spermatogonia are present in equal numbers and comprise 4% of germ cells in the testis (Marshall & Plant, 1996). Like nonhuman primates, Adark and Apale spermatogonia are present in equal numbers in the human testis (Clermont, 1966; Schulze, 1978; Paniagua et al., 1987) and constitute 22% of germ cells in the testis (Paniagua et al., 1987). Thus, the larger pool of stem/progenitor cells in the testes of higher primates compensates, in part, for the reduced number of transit amplifying divisions (Figs. 1 & 2). The large pool of stem/progenitor cells in higher primates may also be a mechanism to reduce the replicative demand on each individual stem cells in longer lived species.
6. Molecular description of spermatogonia in higher primates
Based on expression of conserved molecular markers, Adark and some Apale spermatogonia in nonhuman primates and humans exhibit an undifferentiated phenotype, similar to As, Apr and some Aal rodent spermatogonia (GFRa1+, PLZF+, SALL4+, cKIT−). Some Apale have a transition phenotype similar to larger chain Aal spermatogonia in rodents (e.g., GFRa1+/SOHLH1+/NGN3+/cKIT+) (Hermann et al., 2009; Ramaswamy et al., 2014). Markers of undifferentiated spermatogonia that are conserved from rodents to nonhuman primates to humans include GFRa1, UTF1, PLZF, SALL4 and LIN28 (van Bragt et al., 2008; Meng et al., 2000; Costoya et al., 2004; Buaas et al., 2004; Hobbs et al., 2012; Eildermann et al., 2012a; Gassei & Orwig, 2013; Zheng et al., 2009; Hermann et al., 2009; Ramaswamy et al., 2014; Aeckerle et al., 2012; Lin et al., 2015; Di Persio et al., 2017; Valli et al., 2014; Zheng et al., 2014; Sachs et al., 2014) (See UTF1 staining of monkey and human testis cross sections in Fig. 4C and D). ID4 is conserved in the undifferentiated spermatogonia of rodents (Oatley et al., 2011) and humans (Sachs et al., 2014), but has not been described in nonhuman primates. cKIT appears to be a conserved marker of differentiated spermatogonia, marking the transition to A1 spermatogonia in rodents; B1 spermatogonia in nonhuman primates and B spermatogonia in humans (Hermann et al., 2009; Valli et al., 2014). There are no molecular markers that distinguish the entire population of Adark from the entire population of Apale, perhaps because both are elements of the same stem cell pool that are in different stages of the cell cycle (Hermann et al., 2010). However, a few markers have been identified that are restricted to the subpopulation of Adark with a rarefaction zone (EXOSC10, FGFR3, OCT2) (Lim et al., 2011; von Kopylow et al., 2012a; von Kopylow et al., 2012b) and a few markers are restricted to Apale (DMRT1, Ki67, SSX2-4) or a subpopulation of Apale (NGN3, cKIT) (von Kopylow et al., 2012a; Hermann et al., 2009).
7. Cell surface markers of undifferentiated spermatogonia in higher primates
To date, no cell surface marker has been identified in any species with expression restricted to functional spermatogonial stem cells. GFRa1 is a conserved marker that appears to be most restricted to undifferentiated spermatogonia (i.e., As, Apr, Aal4 in rodents and Adark and Apale in higher primates). This marker has been used to isolate and enrich undifferentiated spermatogonia (Garbuzov et al., 2018; Buageaw et al., 2005; Gassei et al., 2010; He et al., 2012), but many investigators have reported difficulty sorting SSCs using GFRa1 antibodies (personal communications and unpublished data). It is also now clear that half of functional stem cells in the adult mouse testis are in the GFRa1 negative fraction (Garbuzov et al., 2018). This may indicate that stem cells oscillate between GFRa1+ and GFRa1− states depending on cell cycle status, signals from the SSC niche, density of germ cells on the basement membrane or other circumstances. In contrast, GFRa1 appears to be expressed by all Adark and Apale spermatogonia in the Rhesus macaques (Hermann et al., 2009), which presumably include the entire population of functional stem cells. ITGA6 is another robust and conserved marker that can be used to isolate and enrich SSCs from rodent, monkey and human testis cell suspensions (Valli et al., 2014; Shinohara et al., 2000; Maki et al., 2009). ITGA6 expression is not restricted to SSCs or even germ cells, but the entire population of functional SSCs can be recovered and are significantly enriched in the ITGA6+ fraction of rodent (Shinohara et al., 2000) and human testis cells (Valli et al., 2014). SSEA4 has not been used to isolate mouse SSCs, but is expressed by undifferentiated spermatogonia in monkey and human testes and has been used effectively to isolate transplantable SSCs (Zheng et al., 2014; Muller et al., 2008; Izadyar et al., 2011; Eildermann et al., 2012b; Smith et al., 2014). CD9 is expressed by a subpopulation of MAGEA4+ spermatogonia in human testes and can be used to isolate transplantable stem cells (Zohni et al., 2012). ITGA6, SSEA4 and CD9 are effective single markers for isolating primate spermatogonia because they clearly segregate the heterogeneous testis cell suspension into positive and negative fractions and have been tested functionally by xeno-transplantation into infertile mouse recipients. Thus, the majority of functional SSCs are captured in the positive fractions with limited loss to the negative fractions. Other cell surface markers that have been used to isolate and enrich functional SSCs, alone or in combination with other markers, include CD90, EpCAM and GPR125 (Hermann et al., 2009; Valli et al., 2014; Kubota et al., 2003; Ryu et al., 2004; Nickkholgh et al., 2014).
8. SSC transplantation bioassay in higher primates
Similar to rodents, transplantation is the established method to quantify functional stem cells in higher primates. Of course, homologous species transplantation is not possible in humans. Homologous species SSC transplantation is possible in primates (Hermann et al., 2012; Jahnukainen et al., 2011), but not practical as a routine biological assay. Therefore, xenotransplantation to the testes of infertile, immune deficient mice has emerged as the gold standard to quantify functional stem cells from monkey or human cells populations. Human and monkey SSCs do not regenerate complete spermatogenesis when transplanted into mouse testes. However, they do migrate to the seminiferous tubule basement membrane and produce chains or networks of spermatogonia that persist for many months after transplantation (Hermann et al., 2009; Valli et al., 2014; Izadyar et al., 2011; Zohni et al., 2012; Nagano et al., 2001; Nagano et al., 2002; Hermann et al., 2007; Wu et al., 2009; Sadri-Ardekani et al., 2009; Sadri-Ardekani et al., 2011; Dovey et al., 2013; Clark et al., 2017; Durruthy Durruthy et al., 2014; Ramathal et al., 2014). It is not currently possible to recapitulate complete spermatogenesis from monkey or human cells using the xenotransplantation assays. Perhaps one day this challenge will be overcome by transplantation to more closely related species and/or using an organ culture system similar to that described for producing eggs or sperm from primordial germ cell-like cells (PGCLCs) in mice (Hayashi et al., 2012; Zhou et al., 2016).
9. Concluding remarks
Although different vocabularies have evolved to describe spermatogonial stem cells and spermatogenic lineage development in rodents, monkeys and humans, many features are conserved between species. For example, spermatogenesis emerges from isolated spermatogonia that give rise to clones of interconnected chains or networks of cells that become progressively more differentiated with each successive transit amplifying division. In all species, smaller clones are the more un-differentiated elements while larger clones are the more differentiated elements. Many markers are conserved from rodents to primates to humans as well as their association with undifferentiated versus transition versus differentiated spermatogonia. Some markers appear to be more species specific, but in some cases, this may be an artifact of antibody quality or availability for different species. There are important differences between rodents and higher primates. Rodents have more transit amplifying divisions in the pool of undifferentiated and differentiated spermatogonia than nonhuman primates or humans. Based on sperm output data, the difference in transit amplifying divisions appears to be fully compensated by a much larger pool of stem/progenitor spermatogonia in nonhuman primates. In contrast, the large pool of stem/progenitor spermatogonia in humans does not compensate for the reduced number of transit amplifying divisions and consequently, sperm output is reduced. Understanding similarities and differences between species will help to explain challenges in translating technologies such as SSC culture and SSC transplantation to higher primates and ultimately to the human clinic.
Acknowledgments
KEO is supported by grants from the Eunice Kennedy Shriver National Institute for Child Health and Human Development grants HD076412, HD075795 and HD092084; the US-Israel Binational Science Foundation grant 2011111 and Magee-Womens Research Institute and Foundation discretionary fund 9931. Figures in this review were created by AF for his PhD thesis, defended March 23, 2018. AF was supported by a diversity supplement to HD076412.
References
- Aeckerle N, Eildermann K, Drummer C, Ehmcke J, Schweyer S, Lerchl A, Bergmann M, Kliesch S, Gromoll J, Schlatt S, Behr R. The pluripotency factor LIN28 in monkey and human testes: a marker for spermatogonial stem cells? Mol Hum Reprod. 2012;18:477–488. doi: 10.1093/molehr/gas025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloisio GM, Nakada Y, Saatcioglu HD, Pena CG, Baker MD, Tarnawa ED, Mukherjee J, Manjunath H, Bugde A, Sengupta AL, Amatruda JF, Cuevas I, Hamra FK, Castrillon DH. PAX7 expression defines germline stem cells in the adult testis. J Clin Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294:161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- Braun RE, Sharma M, Srivastave A, Fairfield HE, Bergstrom DE. Identification of Slow-Cycling Long-Term Spermatogonial Stem Cells and their Regulation by PLZF. Society for the Study of Reproduction; Washington DC: 2017. p. 279. [Google Scholar]
- Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci U S A. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci U S A. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Buageaw A, Sukhwani M, Ben-Yehudah A, Ehmcke J, Rawe VY, Pholpramool C, Orwig KE, Schlatt S. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- Clark AT, Gkountela S, Chen D, Liu W, Sosa E, Sukhwani M, Hennebold JD, Orwig KE. Primate primordial germ cells acquire transplantation potential by Carnegie stage 23. Stem Cell Rep. 2017;9:329–341. doi: 10.1016/j.stemcr.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont Y. Renewal of spermatogonia in man. Am J Anat. 1966;118:509–524. doi: 10.1002/aja.1001180211. [DOI] [PubMed] [Google Scholar]
- Clermont Y. Two classes of spermatogonial stem cells in the monkey (Cercopithecus aethiops) Am J Anat. 1969;126:57–71. doi: 10.1002/aja.1001260106. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Antar M. Duration of the cycle of the seminiferous epithelium and the spermatogonial renewal in the monkey Macaca arctoides. Am J Anat. 1973;136:153–165. doi: 10.1002/aja.1001360204. [DOI] [PubMed] [Google Scholar]
- Clermont Y, Leblond CP. Differentiation and renewal of spermatogonia in the monkey, Macacus rhesus. Am J Anat. 1959;104:237–273. doi: 10.1002/aja.1001040204. [DOI] [PubMed] [Google Scholar]
- Clifton DK, Bremner WJ. The effect of testicular x-irradiation on spermatogenesis in man. A comparison with the mouse. J Androl. 1983;4:387–392. doi: 10.1002/j.1939-4640.1983.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. Spermatogonial stem cell renewal in the mouse. I Normal situation. Cell Tissue Kinet. 1973;6:281–287. doi: 10.1111/j.1365-2184.1973.tb01617.x. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J Androl. 2012;33:1085–1095. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr Opin Cell Biol. 1998;10:694–701. doi: 10.1016/s0955-0674(98)80109-9. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, van Alphen MM, van de Kant HJ. Duration of the cycle of the seminiferous epithelium and its stages in the rhesus monkey (Macaca mulatta) Biol Reprod. 1986;35:587–591. doi: 10.1095/biolreprod35.3.587. [DOI] [PubMed] [Google Scholar]
- Di Persio S, Saracino R, Fera S, Muciaccia B, Esposito V, Boitani C, Berloco BP, Nudo F, Spadetta G, Stefanini M, de Rooij DG, Vicini E. Spermatogonial kinetics in humans. Development. 2017;144:3430–3439. doi: 10.1242/dev.150284. [DOI] [PubMed] [Google Scholar]
- Dovey SL, Valli H, Hermann BP, Sukhwani M, Donohue J, Castro CA, Chu T, Sanfilippo JS, Orwig KE. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123:1833–1843. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durruthy Durruthy J, Ramathal C, Sukhwani M, Fang F, Cui J, Orwig KE, Reijo Pera RA. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum Mol Genet. 2014;23:3071–3084. doi: 10.1093/hmg/ddu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmcke J, Schlatt S. A revised model for spermatogonial expansion in man: lessons from non-human primates. Reproduction. 2006;132:673–680. doi: 10.1530/rep.1.01081. [DOI] [PubMed] [Google Scholar]
- Ehmcke J, Simorangkir DR, Schlatt S. Identification of the starting point for spermatogenesis and characterization of the testicular stem cell in adult male rhesus monkeys. Hum Reprod. 2005;20:1185–1193. doi: 10.1093/humrep/deh766. [DOI] [PubMed] [Google Scholar]
- Eildermann K, Aeckerle N, Debowski K, Godmann M, Christiansen H, Heistermann M, Schweyer S, Bergmann M, Kliesch S, Gromoll J, Ehmcke J, Schlatt S, Behr R. Developmental expression of the pluripotency factor Sal-like protein 4 in the monkey, human and mouse testis: restriction to premeiotic germ cells. Cells Tissues Organs. 2012a;196:206–220. doi: 10.1159/000335031. [DOI] [PubMed] [Google Scholar]
- Eildermann K, Gromoll J, Behr R. Misleading and reliable markers to differentiate between primate testis-derived multipotent stromal cells and spermatogonia in culture. Hum Reprod. 2012b;27:1754–1767. doi: 10.1093/humrep/des091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet JP, Dadoune JP. Renewal of spermatogonia in the monkey (Macaca fascicularis) Biol Reprod. 1986;35:199–207. doi: 10.1095/biolreprod35.1.199. [DOI] [PubMed] [Google Scholar]
- Garbuzov A, Pech MF, Hasegawa K, Sukhwani M, Zhang RJ, Orwig KE, Artandi SE. Purification of GFRα1+ and GFRα1– spermatogonial stem cells reveals a niche-dependent mechanism for fate determination. Stem Cell Rep. 2018 doi: 10.1016/j.stemcr.2017.12.009. http://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0053976&type=printable. [DOI] [PMC free article] [PubMed]
- Gassei K, Orwig KE. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One. 2013;8:e53976. doi: 10.1371/journal.pone.0053976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K, Ehmcke J, Dhir R, Schlatt S. Magnetic activated cell sorting allows isolation of spermatogonia from adult primate testes and reveals distinct GFRa1-positive subpopulations in men. J Med Primatol. 2010;39:83–91. doi: 10.1111/j.1600-0684.2009.00397.x. [DOI] [PubMed] [Google Scholar]
- Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanti L, Falciatori I, Grasso M, Dovere L, Fera S, Muciaccia B, Fuso A, Berno V, Boitani C, Stefanini M, Vicini E. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells. 2009;27:3043–3052. doi: 10.1002/stem.206. [DOI] [PubMed] [Google Scholar]
- Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, Simons BD, Yoshida S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell. 2014;14:658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M. Offspring from oocytes derived from in vitro primordial germ cell–like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- He Z, Kokkinaki M, Jiang J, Zeng W, Dobrinski I, Dym M. Isolation of human male germline stem cells using enzymatic digestion and magnetic-activated cell sorting. In: Chan W-Y, Blomberg LA, editors. Germline Development: Methods and Protocols. Springer; New York, New York, NY: 2012. pp. 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel AR, Yang QE, Oatley MJ, Lord T, Sablitzky F, Oatley JM. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Lin CC, Sheng Y, Tomko J, Rodriguez M, Shuttleworth JJ, McFarland D, Hobbs RM, Pandolfi PP, Schatten GP, Orwig KE. Characterization, cryopreservation, and ablation of spermatogonial stem cells in adult rhesus macaques. Stem Cells. 2007;25:2330–2338. doi: 10.1634/stemcells.2007-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Simorangkir DR, Chu T, Plant TM, Orwig KE. Molecular dissection of the male germ cell lineage identifies putative spermatogonial stem cells in rhesus macaques. Hum Reprod. 2009;24:1704–1716. doi: 10.1093/humrep/dep073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Hansel MC, Orwig KE. Spermatogonial stem cells in higher primates: are there differences from those in rodents? Reproduction. 2010;139:479–493. doi: 10.1530/REP-09-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G, Peterson K, Masterson K, Ramsey C, Ward T, Lienesch M, Volk A, Cooper DK, Thomson AW, Kiss JE, Penedo MC, Schatten GP, Mitalipov S, Orwig KE. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB, McCarrey JR. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod. 2015;92:54. doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs RM, Fagoonee S, Papa A, Webster K, Altruda F, Nishinakamura R, Chai L, Pandolfi PP. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell. 2012;10:284–298. doi: 10.1016/j.stem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I Their morphology, proliferation and maturation. Anat Rec. 1971;169:533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- Huckins C. The morphology and kinetics of spermatogonial degeneration in normal adult rats: an analysis using a simplified classification of the germinal epithelium. Anat Rec. 1978;190:905–926. doi: 10.1002/ar.1091900410. [DOI] [PubMed] [Google Scholar]
- Huckins C, Oakberg EF. Morphological and quantitative analysis of spermatogonia in mouse testes using whole mounted seminiferous tubules. II The irradiated testes. Anat Rec. 1978a;192:529–542. doi: 10.1002/ar.1091920407. [DOI] [PubMed] [Google Scholar]
- Huckins C, Oakberg EF. Morphological and quantitative analysis of spermatogonia in mouse testes using whole mounted seminiferous tubules, I. The normal testes. Anat Rec. 1978b;192:519–528. doi: 10.1002/ar.1091920406. [DOI] [PubMed] [Google Scholar]
- Izadyar F, Wong J, Maki C, Pacchiarotti J, Ramos T, Howerton K, Yuen C, Greilach S, Zhao HH, Chow M, Chow YC, Rao J, Barritt J, Bar-Chama N, Copperman A. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26:1296–1306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- Jahnukainen K, Ehmcke J, Quader MA, Saiful Huq M, Epperly MW, Hergenrother S, Nurmio M, Schlatt S. Testicular recovery after irradiation differs in prepubertal and pubertal non-human primates, and can be enhanced by autologous germ cell transplantation. Hum Reprod. 2011;26:1945–1954. doi: 10.1093/humrep/der160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin PM, Kramer MF, de Rooij DG. Testicular development in Macaca irus after birth. Int J Androl. 1983;6:25–43. doi: 10.1111/j.1365-2605.1983.tb00321.x. [DOI] [PubMed] [Google Scholar]
- Komai Y, Tanaka T, Tokuyama Y, Yanai H, Ohe S, Omachi T, Atsumi N, Yoshida N, Kumano K, Hisha H, Matsuda T, Ueno H. Bmi1 expression in long-term germ stem cells. Sci Rep. 2014;4:6175. doi: 10.1038/srep06175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H, Avarbock MR, Brinster RL. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc Natl Acad Sci U S A. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Goriely A, Turner GD, Ewen KA, Jacobsen GK, Graem N, Wilkie AO, Rajpert-De Meyts E. OCT2, SSX and SAGE1 reveal the phenotypic heterogeneity of spermatocytic seminoma reflecting distinct subpopulations of spermatogonia. J Pathol. 2011;224:473–483. doi: 10.1002/path.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZYC, Hirano T, Shibata S, Seki NM, Kitajima R, Sedohara A, Siomi MC, Sasaki E, Siomi H, Imamura M, Okano H. Gene expression ontogeny of spermatogenesis in the marmoset uncovers primate characteristics during testicular development. Dev Biol. 2015;400:43–58. doi: 10.1016/j.ydbio.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Lord T, Oatley JM. A revised Asingle model to explain stem cell dynamics in the mouse male germline. Reproduction. 2017;154:R55–R64. doi: 10.1530/REP-17-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CB, Pacchiarotti J, Ramos T, Pascual M, Pham J, Kinjo J, Anorve S, Izadyar F. Phenotypic and molecular characterization of spermatogonial stem cells in adult primate testes. Hum Reprod. 2009;24:1480–1491. doi: 10.1093/humrep/dep033. [DOI] [PubMed] [Google Scholar]
- Marshall GR, Plant TM. Puberty occurring either spontaneously or induced precociously in rhesus monkey (Macaca mulatta) is associated with a marked proliferation of Sertoli cells. Biol Reprod. 1996;54:1192–1199. doi: 10.1095/biolreprod54.6.1192. [DOI] [PubMed] [Google Scholar]
- Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H, Saarma M, Sariola H. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Muller T, Eildermann K, Dhir R, Schlatt S, Behr R. Glycan stem-cell markers are specifically expressed by spermatogonia in the adult non-human primate testis. Hum Reprod. 2008;23:2292–2298. doi: 10.1093/humrep/den253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, McCarrey JR, Brinster RL. Primate spermatogonial stem cells colonize mouse testes. Biol Reprod. 2001;64:1409–1416. doi: 10.1095/biolreprod64.5.1409. [DOI] [PubMed] [Google Scholar]
- Nagano M, Patrizio P, Brinster RL. Long-term survival of human spermatogonial stem cells in mouse testes. Fertil Steril. 2002;78:1225–1233. doi: 10.1016/s0015-0282(02)04345-5. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Nabeshima Y, Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickkholgh B, Mizrak SC, Korver CM, van Daalen SK, Meissner A, Repping S, van Pelt AM. Enrichment of spermatogonial stem cells from long-term cultured human testicular cells. Fertil Steril. 2014;102:558–565 e555. doi: 10.1016/j.fertnstert.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Mammalian Gametogenesis and Species Comparisons in Radiation Response of the Gonads. Effects of Radiation on Meiotic Systems. International Atomic Energy Agency; Vienna. 1968. pp. 3–15. [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec. 1971a;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Spermatogonial stem-cell renewal in the mouse. Anat Rec. 1971b;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Oakberg EF. Effects of radiation on the testis. In: Hamilton DW, Greep RO, editors. Handbook of Physiology. Am. Physiol. Soc; Washington DC: 1975. (Section 7) [Google Scholar]
- Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85:347–356. doi: 10.1095/biolreprod.111.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, Nistal M. Morphological and histometric study of human spermatogonia from birth to the onset of puberty. J Anat. 1984;139(Pt 3):535–552. [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, Codesal J, Nistal M, Rodriguez MC, Santamaria L. Quantification of cell types throughout the cycle of the human seminiferous epithelium and their DNA content. A new approach to the spermatogonial stem cell in man. Anat Embryol. 1987;176:225–230. doi: 10.1007/BF00310055. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond Ser B Biol Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Razack BS, Roslund RM, Suzuki H, Marshall GR, Rajkovic A, Plant TM. Spermatogonial SOHLH1 nucleocytoplasmic shuttling associates with initiation of spermatogenesis in the rhesus monkey (Macaca mulatta) Mol Hum Reprod. 2014;20:350–357. doi: 10.1093/molehr/gat093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramathal C, Durruthy-Durruthy J, Sukhwani M, Arakaki JE, Turek PJ, Orwig KE, Reijo Pera RA. Fate of iPSCs derived from azoospermic and fertile men following xenotransplantation to murine seminiferous tubules. Cell Rep. 2014;7:1284–1297. doi: 10.1016/j.celrep.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, SinhaHikim AP, Clegg ED. Histological and Histopathological Evaluation of the Testis. Cache River Press; Clearwater, FL: 1990. [Google Scholar]
- Ryu BY, Orwig KE, Kubota H, Avarbock MR, Brinster RL. Phenotypic and functional characteristics of spermatogonial stem cells in rats. Dev Biol. 2004;274:158–170. doi: 10.1016/j.ydbio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Sachs C, Robinson BD, Andres Martin L, Webster T, Gilbert M, Lo HY, Rafii S, Ng CK, Seandel M. Evaluation of candidate spermatogonial markers ID4 and GPR125 in testes of adult human cadaveric organ donors. Andrology. 2014;2:607–614. doi: 10.1111/j.2047-2927.2014.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Mizrak SC, van Daalen SK, Korver CM, Roepers-Gajadien HL, Koruji M, Hovingh S, de Reijke TM, de la Rosette JJ, van der Veen F, de Rooij DG, Repping S, van Pelt AM. Propagation of human spermatogonial stem cells in vitro. J Am Med Assoc. 2009;302:2127–2134. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- Sadri-Ardekani H, Akhondi MA, van der Veen F, Repping S, van Pelt AM. In vitro propagation of human prepubertal spermatogonial stem cells. J Am Med Assoc. 2011;305:2416–2418. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- Schlatt S, Weinbauer GF. Immunohistochemical localization of proliferating cell nuclear antigen as a tool to study cell proliferation in rodent and primate testes. Int J Androl. 1994;17:214–222. doi: 10.1111/j.1365-2605.1994.tb01245.x. [DOI] [PubMed] [Google Scholar]
- Schulze W. Light and electron microscope studies of the morphology of A spermatogonia in men with normal spermatogenesis and in patients treated with anti-androgens. Andrologia. 1978;10:307–320. [PubMed] [Google Scholar]
- Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci U S A. 2000;97:8346–8351. doi: 10.1073/pnas.97.15.8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simorangkir DR, Marshall GR, Plant TM. A re-examination of proliferation and differentiation of type A spermatogonia in the adult rhesus monkey (Macaca mulatta) Hum Reprod. 2009;24:1596–1604. doi: 10.1093/humrep/dep051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JF, Yango P, Altman E, Choudhry S, Poelzl A, Zamah AM, Rosen M, Klatsky PC, Tran ND. Testicular niche required for human spermatogonial stem cell expansion. Stem Cells Transl Med. 2014;3:1043–1054. doi: 10.5966/sctm.2014-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009;336:222–231. doi: 10.1016/j.ydbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Tegelenbosch RA, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- Thayer KA, Ruhlen RL, Howdeshell KL, Buchanan DL, Cooke PS, Preziosi D, Welshons WV, Haseman J, vom Saal FS. Altered prostate growth and daily sperm production in male mice exposed prenatally to subclinical doses of 17α-ethinyl oestradiol. Hum Reprod. 2001;16:988–996. doi: 10.1093/humrep/16.5.988. [DOI] [PubMed] [Google Scholar]
- Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- Valli H, Sukhwani M, Dovey SL, Peters KA, Donohue J, Castro CA, Chu T, Marshall GR, Orwig KE. Fluorescence- and magnetic-activated cell sorting strategies to isolate and enrich human spermatogonial stem cells. Fertil Steril. 2014;102:566–580. doi: 10.1016/j.fertnstert.2014.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli H, Phillips BT, Gassei K, Nagano MC, Orwig KE. Spermatogonial stem cells and spermatogenesis. In: Plant TM, Zeleznik AJ, editors. Knobil and Neill’s Physiology of Reproduction. Elsevier; San Diego: 2015. pp. 595–635. [Google Scholar]
- van Alphen MM, van de Kant HJ, de Rooij DG. Depletion of the spermatogonia from the seminiferous epithelium of the rhesus monkey after X irradiation. Radiat Res. 1988;113:473–486. [PubMed] [Google Scholar]
- van Bragt MP, Roepers-Gajadien HL, Korver CM, Bogerd J, Okuda A, Eggen BJ, de Rooij DG, van Pelt AM. Expression of the pluripotency marker UTF1 is restricted to a subpopulation of early A spermatogonia in rat testis. Reproduction. 2008;136:33–40. doi: 10.1530/REP-07-0536. [DOI] [PubMed] [Google Scholar]
- von Kopylow K, Kirchhoff C, Jezek D, Schulze W, Feig C, Primig M, Steinkraus V, Spiess AN. Screening for biomarkers of spermatogonia within the human testis: a whole genome approach. Hum Reprod. 2010;25:1104–1112. doi: 10.1093/humrep/deq053. [DOI] [PubMed] [Google Scholar]
- von Kopylow K, Staege H, Spiess AN, Schulze W, Will H, Primig M, Kirchhoff C. Differential marker protein expression specifies rarefaction zone-containing human Adark spermatogonia. Reproduction. 2012a;143:45–57. doi: 10.1530/REP-11-0290. [DOI] [PubMed] [Google Scholar]
- von Kopylow K, Staege H, Schulze W, Will H, Kirchhoff C. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A spermatogonia. Histochem Cell Biol. 2012b;138:759–772. doi: 10.1007/s00418-012-0991-7. [DOI] [PubMed] [Google Scholar]
- Wu X, Schmidt JA, Avarbock MR, Tobias JW, Carlson CA, Kolon TF, Ginsberg JP, Brinster RL. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci U S A. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Takakura A, Ohbo K, Abe K, Wakabayashi J, Yamamoto M, Suda T, Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol. 2004;269:447–458. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner K, Wang P. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev Biol. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Thomas A, Schmidt CM, Dann CT. Quantitative detection of human spermatogonia for optimization of spermatogonial stem cell culture. Hum Reprod. 2014;29:2497–2511. doi: 10.1093/humrep/deu232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang M, Yuan Y, Wang X, Fu R, Wan H, Xie M, Liu M, Guo X, Zheng Y, Feng G, Shi Q, Zhao XY, Sha J, Zhou Q. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18:330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
- Zohni K, Zhang X, Tan SL, Chan P, Nagano M. CD9 is expressed on human male germ cells that have a long-term repopulation potential after transplantation into mouse testes. Biol Reprod. 2012;87:27. doi: 10.1095/biolreprod.112.098913. [DOI] [PubMed] [Google Scholar]