Abstract
AIM
To reveal the mechanisms of heat-shock transcription factor 4 (HSF4) mutation-induced cataract.
METHODS
GSE22362, including 3 HSF4-null lens and 3 wild-type lens, was obtained from Gene Expression Omnibus database. After data preprocessing, the differentially expressed genes (DEGs) were identified using the limma package. Based on Database for Annotation, Visualization and Integrated Discovery (DAVID) tool, functional and pathway enrichment analyses were performed for the DEGs. Followed by protein-protein interaction (PPI) network was constructed using STRING database and Cytoscape software. Furthermore, the validated microRNA (miRNA)-DEG pairs were obtained from miRWalk2.0 database, and then miRNA-DEG regulatory network was visualized by Cytoscape software.
RESULTS
A total of 176 DEGs were identified in HSF4-null lens compared with wild-type lens. In the PPI network, FBJ osteosarcoma oncogene (FOS), early growth response 1 (EGR1) and heme oxygenase (decycling) 1 (HMOX1) had higher degrees and could interact with each other. Besides, mmu-miR-15a-5p and mmu-miR-26a-5p were among the top 10 miRNAs in the miRNA-DEG regulatory network. Additionally, mmu-miR-26a-5p could target EGR1 in the regulatory network.
CONCLUSION
FOS, EGR1, HMOX1, mmu-miR-26a-5p and mmu-miR-15a-5p might function in the pathogenesis of HSF4 mutation-induced cataract.
Keywords: cataract, heat-shock transcription factor 4, differentially expressed genes, protein-protein interaction network, regulatory network
INTRODUCTION
As a clouding of the lens in the eye, cataract may result in blurry vision, faded colors, trouble with bright lights, halos around light, and trouble seeing at night[1]. Cataract-caused poor vision may further lead to an increased risk of falling into depression[2]. The risk factors for cataract include smoking tobacco, diabetes, alcohol, and prolonged exposure to sunlight[3]. Globally, cataract accounts for one-third of visual impairment and half of blindness[4]–[5]. Cataract results in blindness of about 1 to 4 per 100 000 children and 10 to 40 per 100 000 children in the developed countries and the developing countries, respectively[6]. In worldwide, cataract can induce disability in 53.8 million people, and 52.2 million of them are living in poor countries[7]. Thus, it's important to explore the mechanisms of cataract and develop novel therapies.
Previous study reports that heat-shock transcription factor 4 (HSF4) is critical for lens development and its disruption can cause cataract through reducing the expression of lens beaded filament, down-regulating γ-crystallin, and decreasing the post-translational modification of αA-crystallin[8]–[9]. HSF4 pathogenic mutations result in nuclear cataracts via abrogating the induction of expression and DNase activity of DLAD (DNase 2β)[10]. Mou et al[11] deem that vimentin is targeted by HSF4 in lens and plays a role in the aberrant lens development and cataractogenosis induced by HSF4 mutation. Based on a p53-dependent manner, HSF4 mutations may induce cataract through affecting the switch between the proliferation of lens epithelial cell (LEC) and the differentiation of secondary fiber cell[12]. In 2010, He et al[13] deposited GSE22362 and analyzed the differentially expressed genes (DEGs) in HSF4 homozygous lens, finding many genes co-regulated by HSF4 (especially the down-regulated DNase Iiβ, an enzyme for the denucleation of lens fiber cells). However, the action mechanisms of HSF4 mutations in lens development and cataract formation remaining largely unknown.
Using the microarray data deposited by He et al[13], we further analyzed the DEGs between HSF4-null lens and wild-type lens. Subsequently, the functions of the DEGs were predicted using enrichment analysis. Furthermore, the key genes co-regulated by HSF4 in lens were deeply investigated by protein-protein interaction (PPI) network and microRNA (miRNA)-DEG regulatory network analyses. This study might contribute to finding the mechanisms regarding how HSF4 mutations induce cataract formation.
SUBJECTS AND METHODS
Microarray Data
The dataset of GSE22362 was downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database, which was sequenced on the platform of GPL8321 [Mouse430A_2] Affymetrix Mouse Genome 430A 2.0 Array. The dataset included 3 HSF4-null lens and 3 wild-type lens. Lens of wild-type and day E15.5 transgenic embryos were isolated and then kept in RNA Later (Ambion, Woodlands, TX, USA). He et al[13] deposited GSE22362, and their study was performed according to the approved protocol of the Association for Research in Vision and Ophthalmology and the Albert Einstein College of Medicine Animal Institute Committee.
Data Preprocessing and Differential Expression Analysis
The raw data were preprocessed by the Robust MultiArray Averaging (RMA) method[14] in R package oligo (version 1.38.0, http://bioconductor.org/packages/release/bioc/html/oligo.html), including background correction, normalization and expression calculation. Probes were annotated based on platform annotation files, and the probes without corresponding gene symbols were removed. For the probes mapped to one and the same gene symbol, their average value was obtained as the gene expression value. Using the limma (Linear Models for Microarray Data) package[15] (version3.10.3, http://www.bioconductor.org/packages/2.9/bioc/html/limma.html) in R language, the DEGs between HSF4-null and wild-type lens were identified. Genes with |logFC (fold-change)| >1 and P-value <0.05 were identified as DEGs.
Functional and Pathway Enrichment Analysis
Gene Ontology (GO) is a database developed for describing biological process, molecular function and subcellular location of gene products[16]. Kyoto Encyclopedia of Genes and Genomes (KEGG) database, which is composed by genes and their corresponding functions, can be used for predicting potential functions of gene lists[17]. Using database for annotation, visualization and integrated discovery (DAVID) tool (version 6.8, https://david-d.ncifcrf.gov/)[18], the up-regulated and down-regulated genes were conducted with GO functional and KEGG pathway enrichment analyses, respectively. The terms with P-value <0.05 and count (the number of enriched genes) ≥2 were selected as significant terms.
Construction of Protein-protein Interaction Network
The PPI pairs among the DEGs were predicted by the Search Tool for the Retrieval of Interacting Genes (STRING, version 10, http://www.string-db.org/)[19] database, with PPI score (medium confidence) was set as 0.4. Then, PPI network was constructed using Cytoscape software (version 3.2.0, http://www.cytoscape.org)[20]. Subsequently, degree centrality of the nodes in the PPI network were calculated by the CytoNCA plug-in[21] in Cytoscape software, with parameter was set as without weight. The nodes with higher degrees were identified as the hub proteins[22].
MicroRNA-gene Regulatory Network Analysis
Using miRWalk2.0 database (validated gene-miRNA interaction information retrieval system, http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)[23], the validated miRNA-DEG pairs were obtained. Afterwards, miRNA-DEG regulatory network was visualized by Cytoscape software[20].
RESULTS
Differential Expression Analysis
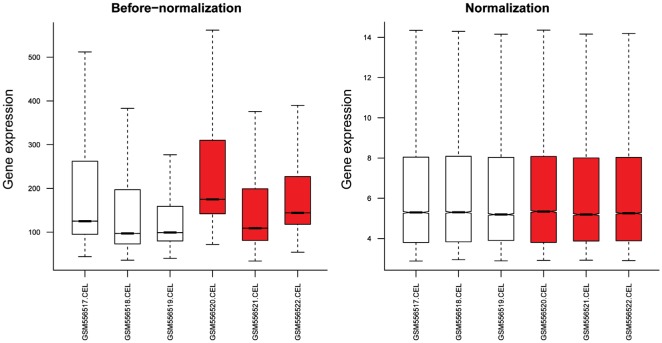
The gene expression distribution before and after normalization are shown in Figure 1.
Figure 1. The gene expression distribution before and after normalization.
Red and white boxes represent HSF4-null and wild-type lens, respectively.
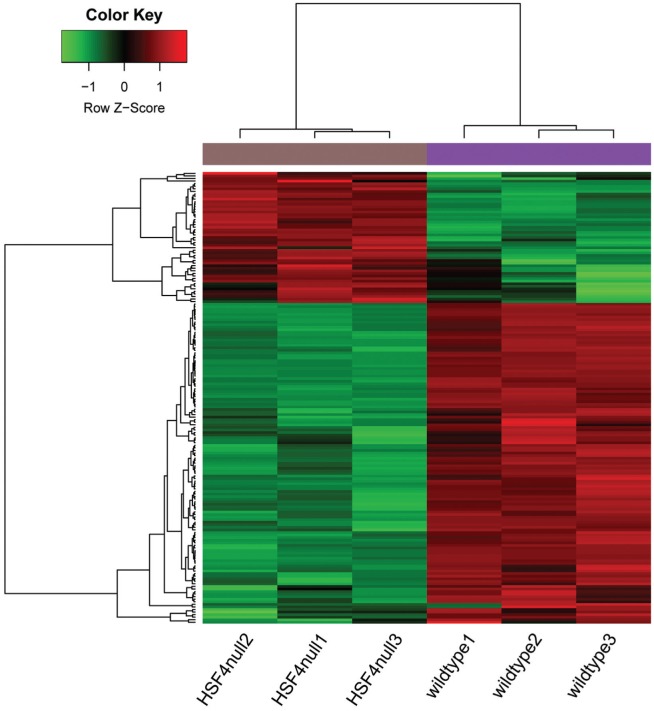
The median values were almost at the same level, indicating that the effect of data preprocessing was good. With |logFC| >1 and P-value <0.05 as the thresholds, a total of 176 DEGs (51 up-regulated and 125 down-regulated genes) were identified in HSF4-null lens compared with wild-type lens. The number of down-regulated genes exceeded that of up-regulated genes. The heatmap of the DEGs is shown in Figure 2.
Figure 2. The heatmap of the DEGs.
Color changes from green to red indicates that expression values range from low to high.
Functional and Pathway Enrichment Analysis
The top 10 GO terms enriched for the up-regulated genes were listed in Table 1, including cytoskeleton organization (P=2.06E-03), cellular response to fibroblast growth factor stimulus (P=2.41E-03) and cellular response to tumor necrosis factor (P=2.42E-03). Besides, 4 pathways were enriched for the up-regulated genes, including malaria (P=2.39E-04), rheumatoid arthritis (P=1.87E-02), Chagas disease (American trypanosomiasis) (P=2.86E-02) and tumor necrosis factor (TNF) signaling pathway (P=3.18E-02) (Table 1).
Table 1. The terms enriched for the up-regulated genes.
| ID | Description | Count | P | Genes |
| GO | ||||
| 0007010 | Cytoskeleton organization | 4 | 2.06E-03 | CCL12, CCL2, TAGLN, MICAL2 |
| 0044344 | Cellular response to fibroblast growth factor stimulus | 3 | 2.41E-03 | HYAL1, CCL2, COL1A1 |
| 0071356 | Cellular response to tumor necrosis factor | 4 | 2.42E-03 | CCL12, HYAL1, CCL2, COL1A1 |
| 0006935 | Chemotaxis | 4 | 2.95E-03 | CCL12, CCL2, CMTM3, CYR61 |
| 0030199 | Collagen fibril organization | 3 | 4.05E-03 | SFRP2, COL1A1, GREM1 |
| 0051591 | Response to cAMP | 3 | 6.84E-03 | FOS, PYGM, COL1A1 |
| 2000502 | Negative regulation of natural killer cell chemotaxis | 2 | 7.28E-03 | CCL12, CCL2 |
| 0001666 | Response to hypoxia | 4 | 1.13E-02 | EGR1, CCL2, PYGM, BNIP3 |
| 0006915 | Apoptotic process | 6 | 1.21E-02 | DNASE1, SGK1, CHAC1, SFRP2, BNIP3, GREM1 |
| 0048023 | Positive regulation of melanin biosynthetic process | 2 | 1.21E-02 | TYRP1, PMEL |
| KEGG | ||||
| mmu05144 | Malaria | 4 | 2.39E-04 | HBA-A1, CCL12, CCL2, HBB-B1 |
| mmu05323 | Rheumatoid arthritis | 3 | 1.87E-02 | CCL12, FOS, CCL2 |
| mmu05142 | Chagas disease (American trypanosomiasis) | 3 | 2.86E-02 | CCL12, FOS, CCL2 |
| mmu04668 | TNF signaling pathway | 3 | 3.18E-02 | CCL12, FOS, CCL2 |
ID: Identification; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; TNF: tumor necrosis factor.
Meanwhile, enrichment analysis was also performed for the down-regulated genes. For the down-regulated genes, 10 GO terms including cellular response to cisplatin (P=3.18E-04), negative regulation of inclusion body assembly (P=6.62E-04) and lipid storage (P=1.03E-02) were enriched (Table 2). Moreover, protein processing in endoplasmic reticulum (P=2.00E-02) was the only pathway enriched for the down-regulated genes (Table 2).
Table 2. The terms enriched for the down-regulated genes.
| ID | Description | Count | P | Genes |
| GO | ||||
| 0072719 | Cellular response to cisplatin | 3 | 3.18E-04 | TIMELESS, HMOX1, SLC31A1 |
| 0090084 | Negative regulation of inclusion body assembly | 3 | 6.62E-04 | DNAJB2, DNAJB1, DNAJA4 |
| 0019915 | Lipid storage | 3 | 1.03E-02 | CRY2, DGAT2, BSCL2 |
| 0043029 | T cell homeostasis | 3 | 1.34E-02 | SPNS2, RAG1, FAS |
| 0002246 | Wound healing involved in inflammatory response | 2 | 1.70E-02 | CD44, HMOX1 |
| 0045766 | Positive regulation of angiogenesis | 4 | 3.19E-02 | HMOX1, HSPB1, CELA1, ANGPT2 |
| 0042752 | Regulation of circadian rhythm | 3 | 3.54E-02 | CRY2, TIMELESS, MAPK10 |
| 0006457 | Protein folding | 4 | 3.67E-02 | HSPA4L, DNAJB1, DNAJB4, DNAJA4 |
| 0008630 | Intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 3.80E-02 | PGAP2, HMOX1, EPHA2 |
| 0014066 | Regulation of phosphatidylinositol 3-kinase signaling | 2 | 4.47E-02 | 1190002N15RIK, CEACAM1 |
| KEGG | ||||
| mmu04141 | Protein processing in endoplasmic reticulum | 5 | 2.00E-02 | MAP3K5, HSPA4L, DNAJB2, DNAJB1, MAPK10 |
ID: Identification; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genome.
Protein-protein Interaction Network Analysis
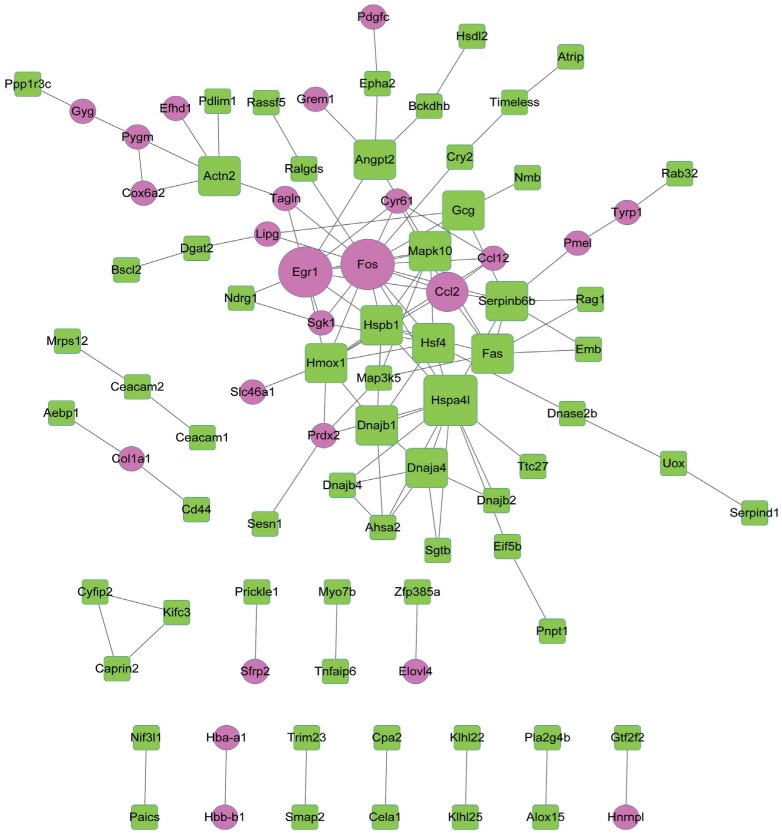
The PPI network constructed for the DEGs included 88 nodes and 121 interactions (Figure 3).
Figure 3. The PPI network constructed for the DEGs.
Red circles and green squares separately represent up-regulated and down-regulated genes. A larger size indicates a higher degree of the node.
According to the degrees of nodes, the top 20 nodes with higher degrees were identified, including FBJ osteosarcoma oncogene (FOS, degree=16), early growth response 1 (EGR1, degree=10) and heme oxygenase (decycling) 1 (HMOX1, degree=9) (Table 3).
Table 3. The top 20 nodes with higher degrees in the PPI network.
| Gene | Description | Degree |
| FOS | Up | 16 |
| HSPA4l | Down | 13 |
| EGR1 | Up | 10 |
| HMOX1 | Down | 9 |
| HSPB1 | Down | 9 |
| HSF4 | Down | 8 |
| SERPINB6B | Down | 8 |
| CCL2 | Up | 7 |
| FAS | Down | 7 |
| MAPK10 | Down | 6 |
| DNAJB1 | Down | 6 |
| DNAJA4 | Down | 6 |
| GCG | Down | 5 |
| ACTN2 | Down | 5 |
| ANGPT2 | Down | 5 |
| MAP3K5 | Down | 4 |
| CYR61 | Up | 4 |
| CCL12 | Up | 4 |
| SGK1 | Up | 4 |
| AHSA2 | Down | 4 |
Especially, FOS, EGR1 and HMOX1 had interactions with each other in the PPI network. Afterwards, pathway enrichment analysis was performed for the top 20 nodes, the enriched pathways mainly included TNF signaling pathway (P=1.37E-06), Chagas disease (American trypanosomiasis) (P=3.64E-05) and influenza A (P=2.62E-04) (Table 4).
Table 4. The pathways enriched for the top 20 nodes in the PPI network.
| ID | Description | Count | P | Genes |
| mmu04668 | TNF signaling pathway | 6 | 1.37E-06 | CCL12, FOS, MAP3K5, CCL2, FAS, MAPK10 |
| mmu05142 | Chagas disease (American trypanosomiasis) | 5 | 3.64E-05 | CCL12, FOS, CCL2, FAS, MAPK10 |
| mmu05164 | Influenza A | 5 | 2.62E-04 | CCL12, CCL2, FAS, DNAJB1, MAPK10 |
| mmu05168 | Herpes simplex infection | 5 | 5.53E-04 | CCL12, FOS, CCL2, FAS, MAPK10 |
| mmu04010 | MAPK signaling pathway | 5 | 1.16E-03 | FOS, MAP3K5, HSPB1, FAS, MAPK10 |
| mmu04141 | Protein processing in endoplasmic reticulum | 4 | 3.80E-03 | MAP3K5, HSPA4L, DNAJB1, MAPK10 |
| mmu04621 | NOD-like receptor signaling pathway | 3 | 5.11E-03 | CCL12, CCL2, MAPK10 |
| mmu05323 | Rheumatoid arthritis | 3 | 1.07E-02 | CCL12, FOS, CCL2 |
| mmu05146 | Amoebiasis | 3 | 2.10E-02 | SERPINB6B, HSPB1, ACTN2 |
| mmu05161 | Hepatitis B | 3 | 3.17E-02 | FOS, FAS, MAPK10 |
| mmu04932 | Non-alcoholic fatty liver disease | 3 | 3.63E-02 | MAP3K5, FAS, MAPK10 |
ID: Identification.
MicroRNA-gene Regulatory Network Analysis
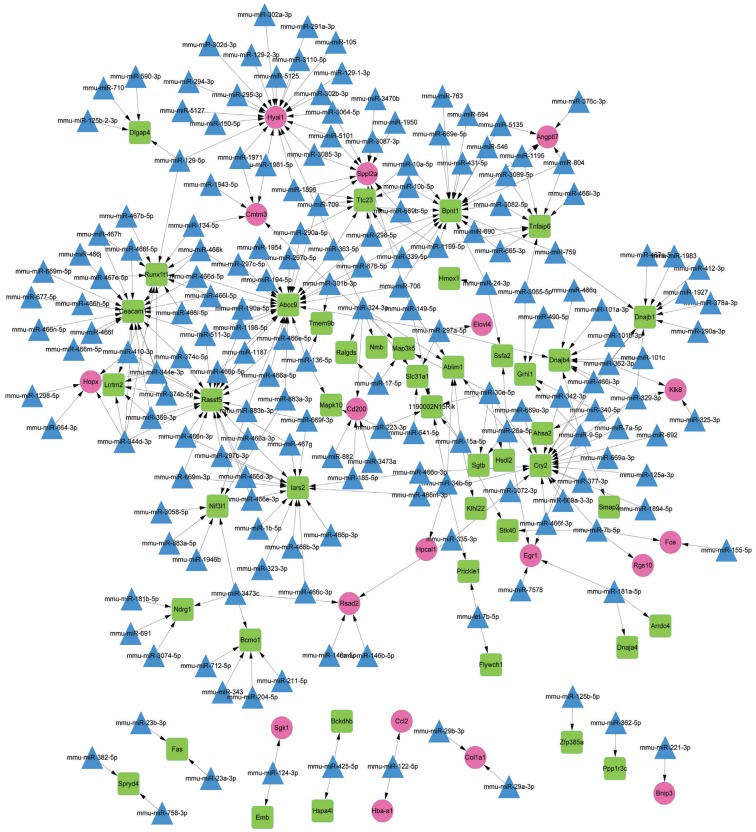
The constructed miRNA-DEG regulatory network had 243 nodes (including 18 up-regulated genes, 44 down-regulated genes and 181 miRNAs) (Figure 4). Among the nodes involved in the regulatory network, the top 10 genes and miRNAs (including mmu-miR-15a-5p and mmu-miR-26a-5p) with higher degrees were listed in Table 5. In addition, mmu-miR-26a-5p could target EGR1 in the regulatory network.
Figure 4. The microRNA-DEG regulatory network.
Red circles, green squares and blue triangles represent up-regulated genes, down-regulated genes and miRNAs, respectively.
Table 5. The top 10 genes and microRNAs in the regulatory network.
| Gene | Description | Degree | miRNA | Degree |
| ABCC9 | Down | 26 | mmu-miR-324-3p | 6 |
| HYAL1 | Up | 21 | mmu-miR-344d-3p | 5 |
| BPNT1 | Down | 21 | mmu-miR-410-3p | 5 |
| CEACAM1 | Down | 21 | mmu-miR-344e-3p | 5 |
| CRY2 | Down | 20 | mmu-miR-30e-5p | 5 |
| RASSF5 | Down | 20 | mmu-miR-15a-5p | 5 |
| IARS2 | Down | 15 | mmu-miR-26a-5p | 4 |
| RUNXLT1 | Down | 12 | mmu-miR-9-5p | 4 |
| SPPL2A | Up | 11 | mmu-miR-297a-5p | 4 |
| DNAJB1 | Down | 10 | mmu-miR-7b-5p | 4 |
DISCUSSION
This study was aimed to reveal the genes and miRNAs involved in HSF4 mutation-induced cataract, which provided targets for the further experimental researches and the clinical therapy of this disease. In this study, a total of 176 DEGs were identified in HSF4-null lens, including 51 up-regulated and 125 down-regulated genes. FOS, EGR1 and HMOX1 had relatively higher degrees in the PPI network. Besides, the pathways of TNF signaling pathway, Chagas disease (American trypanosomiasis) and influenza A were enriched for the top 20 nodes in the PPI network. Moreover, mmu-miR-15a-5p and mmu-miR-26a-5p also were among the top 10 miRNAs in the miRNA-DEG regulatory network.
FOS protein was temporarily expressed in equatorial LECs following anterior capsule rubbing, indicating that equatorial LECs are transcriptionally activated after the anterior lens surface experience minor mechanical stimuli[24]. FOS, UBC (E2 ubiquitin-conjugating protein UBC), EGR1 and PTGS2 (prostaglandin-endoperoxide synthase 2) expression levels are mediated by HSF4 mutations in cataract and can serve as therapeutic targets for the disease[25]. The expression of FOS and JUN (jun proto-oncogene) are dysregulated in the terminal differentiation process of lens fiber cells[26]. FOS can interact with JUN to constitute the AP-1 (activating protein 1) transcription factor, and AP-1 may act in mediating the behavior of lens cells during lens wound repair[27]. These declared that FOS might function in the pathogenesis of HSF4 mutation-induced cataract.
EGR1 may function in the embryonic fibrotic phenotype in the β1MLR10 (β1 F/F and homozygous for MLR10-Cre) lens, and may also be involved in the pathogenesis of posterior capsular opacification (PCO) and other lens diseases[28]. The mRNA levels of EGR1 in a mammalian retina exhibiting a biphasic response to adversing ocular growth stimuli, indicating that retinal EGR1 may serve as a signal for directing ocular growth in various species[29]. Nakajima et al[30] demonstrate that increased activity of EGR1 may play a role in selenite-induced LEC death. Ma et al[31] find that HMOX1 protects LECs from oxidant stress induced by hydrogen peroxide (H2O2) through decreasing the generation of reactive oxygen species (ROS) and enhancing the activity of antioxidant enzyme, and therefore restraining caspase family-dependent apoptosis. Therefore, EGR1 and HMOX1 might play roles in formation of HSF4 mutation-induced cataract. FOS, EGR1 and HMOX1 could interactions with each other in the PPI network, indicating that FOS, EGR1 and HMOX1 might also act in HSF4 mutation-induced cataract through interacting with other genes.
Hsa-miR-15a-5p, hsa-miR-15a-3p, and hsa-miR-16-1-5p are overexpressed in the LECs of age-related cataract, and may promote the progression of age-related cataract through inhibiting the expression levels of the anti-apoptotic genes myeloid cell leukemia sequence 1 (MCL1) and B-cell CLL/lymphoma 2 (BCL2)[32]. Previous studies report that miR-26b plays an important role in growth and proliferation of LECs in cataract rat[33]–[34]. Downregulation of miR-26b can postpone the progression of oxidative cataract via mediating cell proliferation, and suppressing LEC apoptosis, nuclear factor kappa B (NF-κB) expression and inflammatory factors[35]. mmu-miR-26a-5p could target EGR1 in the miRNA-DEG regulatory network, suggesting that mmu-miR-26a-5p targeting EGR1 and mmu-miR-15a-5p might also be involved in the procession of HSF4 mutation-induced cataract.
In conclusion, a total of 176 DEGs were identified in HSF4-null lens through a series of bioinformatics analyses. Besides, FOS, EGR1, HMOX1, mmu-miR-26a-5p and mmu-miR-15a-5p might play roles in the pathogenesis of HSF4 mutation-induced cataract. However, these results were obtained from bioinformatics analyses and in-depth experimental researches should be done in the future.
Acknowledgments
Foundation: Supported by the Scientific and Technological Developing Scheme of Jilin Province (No.20150414038GH).
Conflicts of Interest: Tian R, None; Xu Y, None; Dou WW, None; Zhang H, None.
REFERENCES
- 1.Allen D, Vasavada A. Cataract and surgery for cataract. BMJ. 2006;333(7559):128–132. doi: 10.1136/bmj.333.7559.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gimbel HV, Dardzhikova AA. Consequences of waiting for cataract surgery. Curr Opin Ophthalmol. 2011;22(1):28–30. doi: 10.1097/ICU.0b013e328341425d. [DOI] [PubMed] [Google Scholar]
- 3.de Jong PTVM. Cataract, age-related macular degeneration, and primary open-angle glaucoma: risk factors. Ophthalmology and the Ageing Society Springer Benlin Beidelberg. 2013:33–55. [Google Scholar]
- 4.Bourne R, Price H, Stevens G, GBD Vision Loss Expert Group Global burden of visual impairment and blindness. Arch Ophthalmol. 2012;130(5):645–647. doi: 10.1001/archophthalmol.2012.1032. [DOI] [PubMed] [Google Scholar]
- 5.Organization WH. Global data on visual impairment. 2012.
- 6.Rogers GL. Pediatric cataract surgery: techniques, complications, and management. Lippincott Williams & Wilkins. Ophthalmic Surgery, Lasers and Imaging Retina. 2005;36(6):526. doi: 10.3928/1542-8877-20051101-18. [DOI] [PubMed] [Google Scholar]
- 7.Lopez AD, Murray CJL. The global burden of disease, 1990-2020. Available at: https://www.nature.com/articles/nm1198_1241. [DOI] [PubMed]
- 8.Shi X, Cui B, Wang Z, Wend L, Xu Z, Ma J, Xu G, Kong X, Hu L. Removal of Hsf4 leads to cataract development in mice through down-regulation of gamma S-crystallin and Bfsp expression. BMC Mol Biol. 2009;10:10. doi: 10.1186/1471-2199-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv H, Huang C, Zhang J, Liu Z, Zhang Z, Xu H, You Y, Hu J, Li X, Wang W. A novel hsf4 gene mutation causes autosomal-dominant cataracts in a chinese family. G3-Genes Genomes Genetics. 2014;4(5):823–828. doi: 10.1534/g3.113.009860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui X, Wang L, Zhang J, Du R, Liao S, Li D, Li C, Ke T, Li DW, Huang H, Yin Z, Tang Z, Liu M. Hsf4 regulates dlad expression and promotes lens de-nucleation. Biochim Biophys Acta. 2013;1832(8):1167–1172. doi: 10.1016/j.bbadis.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Mou L, Xu JY, Lei X, Wu Y, Xu G, Kong X, Li W, Xu GT. Identification of vimentin as a novel target of HSF4 in lens development and cataract. Invest Ophthalmol Vis Sci. 2010;51(1):369–404. doi: 10.1167/iovs.09-3772. [DOI] [PubMed] [Google Scholar]
- 12.Huang M, Li D, Huang Y, Cui X, Liao S, Wang J, Liu F, Li C, Gao M, Chen J, Tang Z, Li DW, Liu M. Hsf4 promotes G1/S arrest in human lens epithelial cells by stabilizing p53. Biochim Biophys Acta. 2015;1853(8):1808–1817. doi: 10.1016/j.bbamcr.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 13.He S, Pirity MK, Wang WL, Wolf L, Chauhan BK, Cveklova K, Tamm ER, Ashery-Padan R, Metzger D, Nakai A, Chambon P, Zavadil J, Cvekl A. Chromatin remodeling enzyme Brg1 is required for mouse lens fiber cell terminal differentiation and its denucleation. Epigenetics Chromatin. 2010;3(1):21. doi: 10.1186/1756-8935-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 15.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using r and bioconductor. New York, NY: Springer New York; 2005. pp. 397–420. Available at: https://link.springer.com/chapter/10.1007/0-387-29362-0_23. [Google Scholar]
- 16.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H, FlyBase Consortium Flybase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altermann E, Klaenhammer TR. Pathwayvoyager: pathway mapping using the kyoto encyclopedia of genes and genomes (kegg) database. BMC genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. David bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. String v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9(11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of biological networks. BioSystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 22.He X, Zhang J. Why do hubs tend to be essential in protein networks? PLoS Genet. 2006;2(6):e88. doi: 10.1371/journal.pgen.0020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dweep H, Gretz N. miRwalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697–697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 24.Shirai K, Saika S, Okada Y, Senba E, Ohnishi Y. Transcription activation in lens epithelial cells after anterior capsule rubbing in rats: expression of c-Fos. J Cataract Refract Surg. 2003;29(8):1601–1604. doi: 10.1016/s0886-3350(02)02043-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W, Zhao W, Zhao J, Wang D, Li J. Screening of potential target genes for cataract by analyzing mRNA expression profile of mouse Hsf4-null lens. BMC Ophthalmol. 2015;15:76. doi: 10.1186/s12886-015-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rinaudo JA, Zelenka PS. Expression of c-fos and c-jun mRNA in the developing chicken lens: relationship to cell proliferation, quiescence, and differentiation. Exp Cell Res. 1992;199(1):147–153. doi: 10.1016/0014-4827(92)90472-k. [DOI] [PubMed] [Google Scholar]
- 27.Shirai K, Okada Y, Saika S, Senba E, Ohnishi Y. Expression of transcription factor AP-1 in rat lens epithelial cells during wound repair. Exp Eye Res. 2001;73(4):461–468. doi: 10.1006/exer.2001.1052. [DOI] [PubMed] [Google Scholar]
- 28.Terrell A. Β1-integrin may regulate egr1 (early growth response 1) within the lens. University of Delaware; 2013. Avilabel at: http://dspace.udel.edu/handle/19716/12965. [Google Scholar]
- 29.Ashby RS, Zeng G, Leotta AJ, Tse DY, McFadden SA. Egr-1 mRNA expression is a marker for the direction of mammalian ocular growth. Invest Ophthalmol Vis Sci. 2014;55(9):5911–5921. doi: 10.1167/iovs.13-11708. [DOI] [PubMed] [Google Scholar]
- 30.Nakajima T, Belusko PB, Walkup RD, Azuma M, Shearer TR. Involvement of Egr-1 in lens epithelial cell death induced by selenite. Exp Eye Res. 2006;82:874–878. doi: 10.1016/j.exer.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Ma T, Chen T, Li P, Ye Z, Zhai W, Jia L, Chen W, Sun A, Huang Y, Wei S, Li Z. Heme oxygenase-1 (HO-1) protects human lens epithelial cells (SRA01/04) against hydrogen peroxide (H2O2)-induced oxidative stress and apoptosis. Exp Eye Res. 2016;146:318–329. doi: 10.1016/j.exer.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Liu S, Zhang F, Jiang P, Wu X, Liang Y. Expression of the microRNAs hsa-miR-15a and hsa-miR-16-1 in lens epithelial cells of patients with age-related cataract. Int J Clin Exp Med. 2014;8(2):2405–2410. [PMC free article] [PubMed] [Google Scholar]
- 33.Liang N, Zhou X, Zhao M, Zhao D, Zhu Z, Li S, Yang H. Down-regulation of microRNA-26b modulates non-small cell lung cancer cells chemoresistance and migration through the association of PTEN. Acta Biochim Biophys Sin (Shanghai) 2015;47(7):530–538. doi: 10.1093/abbs/gmv046. [DOI] [PubMed] [Google Scholar]
- 34.Dong N, Xu B, Benya SR, Tang X. miRNA-26b inhibits the proliferation, migration, and epithelial-mesenchymal transition of lens epithelial cells. Mol Cell Biochem. 2014;396(1-2):229–238. doi: 10.1007/s11010-014-2158-4. [DOI] [PubMed] [Google Scholar]
- 35.Liu C, Han H. Downregulation of mir-26b impact on lens epithelial cells in cataract rat. Int J Clin Pathol. 2016;9(9):8784–8790. [Google Scholar]