Abstract
AIM
To explore the clinical feature and genetic etiology of a Chinese Knobloch syndrome family.
METHODS
Ocular examinations and magnetic resonance imagings (MRIs) were performed on the family. Whole exome sequencing was conducted on the two patients. Sanger sequencing was utilized to validate the presence of variation in the family as well as in 100 normal controls. Real-time quantitative polymerase chain reaction (PCR) was used to detect the expression level of COL18A1 in peripheral blood lymphocytes of the patients and normal carriers.
RESULTS
The affected subjects presented with vision loss, exotropia, cataracts, retinal detachment, and other complications. A homozygous c.4759_4760delCT (p.Leu1587ValfsX72) mutation (rs398122391) in COL18A1 was identified in the two patients, cosegregating with the phenotypes, and did not be detected in 100 normal controls. This mutation caused significant decreased expression of COL18A1 mRNA in the patients.
CONCLUSION
The findings strongly indicate that this mutation is the disease-causing mutation. Moreover, this is the first Knobloch syndrome pedigree reported in the Chinese population.
Keywords: Knobloch syndrome, COL18A1, whole exome sequencing
INTRODUCTION
Knobloch syndrome [Mendelian Inheritance in Man (MIM) #267750, #608454] is a very rare autosomal recessive developmental disorder characterized by the association of vitreoretinal degeneration with or without occipital skull abnormalities[1]. Vitreoretinal degeneration usually includes recurrent retinal detachment, retinitis pigmentosa-like features, lens subluxation, congenital high myopia, macular abnormalities, and occipital skull abnormalities, which can include bone defects or cutis aplasia encephalocele[2]. The ocular features of the disease are similar to Stickler syndrome with an optically empty vitreous, severe chorioretinal degeneration, and high myopia, which is due to mutations in collagen genes[3]. Since the original report in 1971 by Knobloch and Layer, at least 90 cases of Knobloch syndrome from 47 families have been described, each with varying degrees of clinical heterogeneity[4]–[8].
COL18A1 [Online Mendelian Inheritance in Man (OMIM)# 120328] has been identified as the disease-causing gene for Knobloch syndrome. The COL18A1 gene includes 43 exons and is reported to encode two distinct isoforms in humans through the use of two promoters[9]. A homozygous or compound heterozygous mutation affects the gene product of COL18A1. Numerous mutations in COL18A1 have been identified in unrelated families who have Knobloch syndrome, thus confirming its causal relationship with the syndrome[10]. To date, more than 21 different mutations have been described in patients from various ethnicities[2],[5]–[6],[10]–[14]. However, the COL18A1 mutation and Knobloch syndrome phenotype have not been reported in Chinese patients.
In this study, we conducted thorough clinical and genetic analyses of a Chinese family with Knobloch syndrome. A homozygous c.4759_4760delCT mutation in COL18A1 was detected in the patients, and this mutation was absent in unaffected parents and 100 ethnicity-matched healthy controls. Our findings indicate that this mutation might be a candidate disease-causing mutation. Furthermore, this is the first Knobloch syndrome case found in a Chinese descent pedigree.
SUBJECTS AND METHODS
Subjects
A Knobloch syndrome pedigree from Hunan Province, China, be composed of five members (two males and three females) from two generations (Figure 1A). Ocular examinations and magnetic resonance imagings (MRIs) were made on these patients. A group of 100 controls who were ethnically matched with patients were recruited mainly from the hospital and other volunteers. This study was approved by the Second Xiangya Hospital Ethics Committee, and all of the pedigree members and controls gave their written informed consent complying with the Declaration of Helsinki principles.
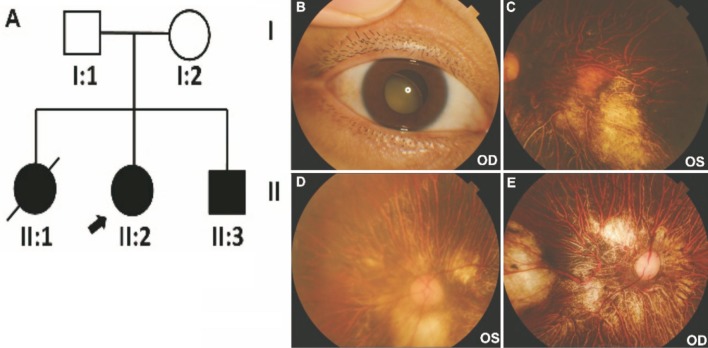
Figure 1. Ocular phenotypes of patients II:2 and II:3.
A: The Chinese Knobloch syndrome pedigree. Roman numerals referred to generations, and individuals within a generation were numbered from left to right, as per convention. Proband was noted with arrow, and died proband was noted with slash line. Filled symbol referred to the patient. Open symbol referred unaffected individual; B: A total cataract complicated with lens subluxation in the right eye of II:2; C: A myopic fundus with myopic macular scarring in the left eye of II:2; D, E: Fundus examination showed high myopic fundus in both eyes of II:3.
Whole Exome Sequencing and Data Analysis
Genomic DNA was extracted from the peripheral blood lymphocytes using the standard phenol-chloroform method. DNA samples of probands II-2 and II-3 were used for whole exome sequencing. Briefly, genomic DNA was sheared to 200-250 bp fragments with a Covaris LE220 Focused-ultrasonicator (Covaris, Woburn, Massachusetts, USA). Library construction was performed by the NimbleGen SeqCap EZ System (Roche NimbleGen, Madison, WI, USA) according to the manufacturer's instructions. We performed 2×150-cycles paired-end sequencing on an Illumina HiSeq2500 Analyzer (Illumina, San Diego, California, USA) following the manufacturer's instructions. Illumina Pipeline software (version 1.3.4, Illumina, San Diego, California, USA) was used to perform base calling and calculations of quality values for every base. Trim Galore! software (Babraham Bioinformatics, Babraham, Cambridgeshire, UK) was used to perform adapter and quality trimming.
Reads were aligned to the human reference genome [University of California, Santa Cruz (UCSC; http://genome.ucsc.edu) hg19 and National Center for Biotechnology Information (NCBI) GRCh37] using BWA-MEM single nucleotide variations (SNVs) and indels (insertions and deletions) were called by GATK3.4. SNVs and indels with a read depth of ≥8× and a quality of ≥30 were reserved for subsequent analysis.
The polymorphism SNVs were annotated based on the SNP database (dbSNP) and 1000 genomes. Hard filter and soft filter [Variant Quality Score Recalibration (VQSR)] methods were used to filter unreliable SNPs and indels. Gene-based and region-based annotation of functionally genetic variants were implemented by ANNOVAR software (Philadelphia, Pennsylvania, USA).
Sanger Sequencing of Implicated Mutant
Polymerase chain reaction (PCR) amplification and Sanger sequencing of the amplicons were used to validate the presence of variations in the COL18A1 gene (NM_130444) identified via exome sequencing. Primer sequences were: forward primer 5′-GCCCCTCAGTGTGTCACTTG-3′ and reverse primer 5′-CTAGGCGCCAGTGTCTGTAA-3′. PCR was performed in a 10 µL reaction mixture containing 0.1 µL TaKaRa La Taq™ (Takara Biotechnology, Dalian, China), 1 µL 10× LA Taq Buffer II, 30 ng of each primer, and 30 ng of genomic DNA. The amplification conditions consisted of an initial step of denaturation at 94°C for 1min, followed by 33 cycles of denaturation at 94°C for 30s, annealing at 60°C for 30s, and extension at 72°C for 20s. A final extension was performed at 72°C for 10min.
RNA extraction and mRNA expression analysis by real-time quantitative PCR
Total RNA was extracted from peripheral blood lymphocytes including two patients, two normal carriers, and three unrelated normal controls using Trizol reagent (Life technologies, NY, USA). cDNA was synthesized from 0.2 µg total RNA through the RevertAid First Strand cDNA Synthesis Kit (K1622, Thermo Scientific, Inc., Waltham, MA, USA) and oligo (dT) primers. Quantitative real-time PCR was carried out on an Applied Biosystems® StepOne™ Plus Real-Time PCR System (Thermo Scientific, Inc., Waltham, MA, USA) using Maxima SYBR Green qPCR Master Mixes (K0251, Thermo Scientific, Inc., Waltham, MA, USA). Data was normalized to β-actin and analyzed by the comparative CT method. Specific primer for each genes were as follows: 5′-GCACCACAGCTCCTACGTG-3′ and 5′-CTGGAAGCACTGGAAGTCG-3′ for COL18A1 whose amplification product (c.4650 to c.4809) spans the mutant cDNA site, 5′-CACGATGGAGGGGCCGGACTCATC-3′ and 5′-TAAAGACCTCTATGCCAACACAGT-3′ for β-actin.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Two-tailed Student's t-test was used to determine the significance of difference between two groups. The data is presented as mean±SEM.
RESULTS
Clinical Findings
A 33-year-old female patient (the proband, II:2) was referred to our hospital with the chief complaint of progressive vision loss. An external observation showed right exotropia. On ocular examination at presentation, the best corrected visual acuity was counting fingers at 0.1 m in the right eye and counting fingers at 0.5 m in the left eye, with accurate projection of rays (PR) in all quadrants of each eye. Intraocular pressure was 25 mm Hg in the right eye and 21 mm Hg in the left eye. A slit-lamp examination revealed a total cataract complicated with lens subluxation in the right eye (Figure 1B), and a slight posterior subcapsular cataract in the left eye. A fundus examination was not feasible due to the total cataract in the right eye. However, a fundus examination in the left eye showed a myopic fundus (Figure 1C) with vitreous detachment and myopic macular scarring.
The patient's 31-year-old brother (II:3) underwent a cataract combined with vitreoretinal surgery for a cataract and retinal detachment in the right eye at the age of 18y. He also had a nuclear and posterior subcapsular cataract in the left eye. A fundus examination showed a high myopic fundus in both eyes (Figure 1D, 1E). Both II:2 and II:3 were confirmed to have structural brain malformations. MRI showed bilateral frontal gyrus dysplasia in both patients (data not shown). Their elder sister (II:1) died from epilepsy several years earlier, she also had poor vision. Consanguinity was detected in their grandparents, who were first cousins.
Whole Exome Sequencing and Sanger Validation
We selected the probands II:2 and II:3 in the pedigree for whole exome sequencing and obtained over 69 billion clean reads each. In our sequencing results, a total of 71.55% and 69.98% of the qualified bases were mapped to a reference sequence with the mean read depth of approximately 85.5629-fold and 119.6063-fold, respectively (Table 1). The exon coverage ratio was over 99% for the two samples, and these reads covered over 94% of the reference within 200 bp of the target regions (Table 1). After alignment and variations calling, there were 4034 nonsynonymous homozygous variants in the exonic region that overlapped among the two patients. Based on the allele frequency data of 1000 genomes and Exome Sequencing Project (ESP) genomes as well as annotations in ClinVar, HGMD, and the OMIM database, a homozygous frameshift variation c.4759_4760delCT (rs398122391, chr21:46930004) in COL18A1 exon 40 was found in both probands II:2 and II:3, which might lead to a truncated COL18A1 protein (p.Leu1587ValfsX72).
Table 1. Summary of the exome sequencing mapping results.
| Sample | Total mapped reads | Reads on target regions | Target regions covered (%) | Reads on target ±200 regions (%) | Target ±200 regions covered (%) | Target bases covered (%) | Mean coverage of target bases |
| II-2 | 69096792 | 49475427 (71.55) | 99.39 | 52267144 (75.59) | 94.43 | 99.39 | 85.5629 |
| II-3 | 98753647 | 69238502 (69.98) | 99.78 | 75431309 (76.24) | 98.42 | 99.78 | 119.6063 |
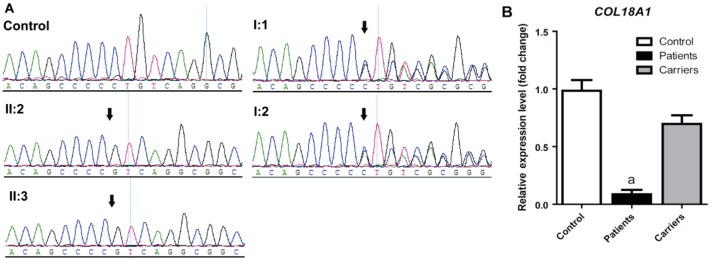
By Sanger sequencing the homozygote variant, c.4759_4760delCT in COL18A1 was found in the affected probands II:2 and II:3 (Figure 2A). Both I:1 and I:2 were carriers of the heterozygous c.4759_4760delCT variant (Figure 2A). The variant on COL18A1 was completely cosegregated with the Knobloch syndrome phenotype in this family. In addition, the variant was absent in 100 unrelated controls. Our results indicate that the variation c.4759_4760delCT in the COL18A1 gene is the disease-causing mutation in this Knobloch syndrome pedigree.
Figure 2. Sanger sequencing validation and RNA expression of the mutation COL18A1 c.4759_4760delCT in the Chinese Knobloch syndrome pedigree.
A: The figure shows the DNA sequence of COL18A1 in the Chinese autosomal recessive pedigree and normal control. The arrow referred to mutant base; B: mRNA expression of COL18A1 in lymphocytes of affected individuals, carriers, and controls. Mean expression (±SEM) of COL18A1 in affected individuals (n=2), carriers (n=2), and controls (n=3) measured by quantitive real-time PCR. aP<0.0001.
Decreased COL18A1 mRNA expression level in Patients
To comfirm that the homozygous frameshift variation influences the mRNA expression level of COL18A1, we performed quantitative PCR using lymphocytes from two cases (II:2 and II:3), two carriers (I:1 and I:2) within the family and three controls outside the family. As is shown in Figure 2B, the patients revealed decreased COL18A1 expression (8.67%±3.84%; P=0.0009) significantly compared with controls, while these carriers only displayed moderate declined COL18A1 expression (69.67%±7.42%; P=0.0731).
DISCUSSION
Knobloch syndrome is an autosomal recessive developmental disorder characterized by typical eye abnormalities including high myopia, vitreoretinal degeneration (often with retinal detachment), cataract, and dislocated lens, with various occipital skull defects ranging from scalp defects to encephalocele[4]. Additional clinical phenotypes, such as renal anomalies, lung hypoplasia, central nervous system malformations, refractory seizures, and intellectual impairment of varying severity with a Lennox-Gastaut phenotype, have also occasionally been reported[5],[10],[15]. Our patients revealed similar ocular abnormalities as well as structural brain malformations, which were comparable to phenotypes reported in other studies and populations.
In our Knobloch syndrome pedigree, whole exome sequencing was utilized to identify the c.4759_4760delCT (p.Leu1587ValfsX72) variant of COL18A1 in the pedigree, which was validated by Sanger sequencing and completely cosegregated within the pedigree. This variant has also been reported in the probands of two Saudi families[6]. One proband was a 6-month-old boy who had a retinal detachment, high myopia, and occipital cutis aplasia. The other proband was an 11-year-old boy with various ocular anomalies, high myopia, and epilepsy[6]. Ocular abnormalities of our patients were similar to the two Saudi probands. Our results directly implicated the COL18A1 mutation c.4759_4760delCT as the genetic etiology of the Chinese Knobloch syndrome pedigree, which was the first Knobloch syndrome mutation reported in the Chinese population.
COL18A1 encodes collagen XVIII, a widely expressed, non-fibrillar collagen that is an essential component of the extracellular matrix (ECM). Collagen XVIII is expressed in vascular and basement membranes (BM), and has multiple functions in ocular and neurologic development, including angiogenesis, BM maintenance, and in the Wnt/β-catenin signaling pathway[16]. It is present in almost all ocular structures of human eyes, including Bruch's membrane, the lens capsule, the BM of the iris, the aqueous humor, vitreous, and retina[17]. Our homozygote frameshift mutation is predicted to create premature stop codons, and our quantitative PCR analysis showed that the expression level of mutated COL18A1 was reduced significantly compared with controls. These results indicated that our mutant COL18A1 led to insufficient of collagen XVIII protein, which in turn resulted in severe ocular defects in our patients[5]. Collagen XVIII is expressed in embryonic brain[18] as well, and it plays a critical role in brain development[19]. Therefore, patients often present with brain malformations, such as mental retardation, seizures, and structural brain malformations[20], which are similar to our patients' brain phenotype.
In conclusion, our results suggest that c.4759_4760delCT (p.Leu1587ValfsX72) of COL18A1 is a pathogenic mutation in the Knobloch syndrome family as determined by whole exome sequencing. Further functional studies and animal studies of p.Leu1587ValfsX72 will be required to interpret its role in the etiopathogenesis of Knobloch syndrome.
Acknowledgments
We are grateful to our patients and their family and to the normal controls who participated in this study. We thank Cong Wang from Hunan Clinical Research Center of Ophthalmic Disease, Hunan, China for DNA extraction.
Authors' contributions: Ding C and Zhang LS conceived and designed the experiments; Zhang LS and Li HB performed the experiments; Zeng J and Yang Y helped to analyze the data; Ding C and Zhang LS wrote the paper. Correspondence and requests for materials should be addressed to Ding C.
Foundations: Supported by the National Natural Science Foundation of China (No.81300758, No.81700837); Department of Science and Technology, Hunan (No.2015TP2007).
Conflicts of Interest: Zhang LS, None; Li HB, None; Zeng J, None; Yang Y, None; Ding C, None.
REFERENCES
- 1.Hull S, Arno G, Ku CA, Ge Z, Waseem N, Chandra A, Webster AR, Robson AG, Michaelides M, Weleber RG, Davagnanam I, Chen R, Holder GE, Pennesi ME, Moore AT. Molecular and clinical findings in patients with Knobloch syndrome. JAMA Ophthalmol. 2016;134(7):753–762. doi: 10.1001/jamaophthalmol.2016.1073. [DOI] [PubMed] [Google Scholar]
- 2.Haghighi A, Tiwari A, Piri N, Nürnberg G, Saleh-Gohari N, Haghighi A, Neidhardt J, Nürnberg P, Berger W. Homozygosity mapping and whole exome sequencing reveal a novel homozygous COL18A1 mutation causing Knobloch syndrome. PLoS One. 2014;9(11):e112747. doi: 10.1371/journal.pone.0112747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snead MP, McNinch AM, Poulson AV, Bearcroft P, Silverman B, Gomersall P, Parfect V, Richards AJ. Stickler syndrome, ocular-only variants and a key diagnostic role for the ophthalmologist. Eye (Lond) 2011;25(11):1389–1400. doi: 10.1038/eye.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan AO, Aldahmesh MA, Mohamed JY, Al-Mesfer S, Alkuraya FS. The distinct ophthalmic phenotype of Knobloch syndrome in children. Br J Ophthalmol. 2012;96(6):890–895. doi: 10.1136/bjophthalmol-2011-301396. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki O, Kague E, Bagatini K, Tu H, Heljasvaara R, Carvalhaes L, Gava E, de Oliveira G, Godoi P, Oliva G, Kitten G, Pihlajaniemi T, Passos-Bueno MR. Novel pathogenic mutations and skin biopsy analysis in Knobloch syndrome. Mol Vis. 2009;15:801–809. [PMC free article] [PubMed] [Google Scholar]
- 6.Aldahmesh MA, Khan AO, Mohamed JY, Levin AV, Wuthisiri W, Lynch S, McCreery K, Alkuraya FS. No evidence for locus heterogeneity in Knobloch syndrome. J Med Genet. 2013;50(8):565–566. doi: 10.1136/jmedgenet-2013-101755. [DOI] [PubMed] [Google Scholar]
- 7.Gradstein L, Hansen RM, Cox GF, Altschwager P, Fulton AB. Progressive retinal degeneration in a girl with Knobloch syndrome who presented with signs of ocular albinism. Doc Ophthalmol. 2017;134(2):135–140. doi: 10.1007/s10633-017-9574-1. [DOI] [PubMed] [Google Scholar]
- 8.AlBakri A, Ghazi NG, Khan AO. Biometry, optical coherence tomography, and further clinical observations in Knobloch syndrome. Ophthalmic Genet. 2017;38(2):138–142. doi: 10.3109/13816810.2016.1164197. [DOI] [PubMed] [Google Scholar]
- 9.Paisán-Ruiz C, Scopes G, Lee P, Houlden H. Homozygosity mapping through whole genome analysis identifies a COL18A1 mutation in an Indian family presenting with an autosomal recessive neurological disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(7):993–997. doi: 10.1002/ajmg.b.30929. [DOI] [PubMed] [Google Scholar]
- 10.Aldahmesh MA, Khan AO, Mohamed JY, Alkuraya H, Ahmed H, Bobis S, Al-Mesfer S, Alkuraya FS. Identification of ADAMTS18 as a gene mutated in Knobloch syndrome. J Med Genet. 2011;48(9):597–601. doi: 10.1136/jmedgenet-2011-100306. [DOI] [PubMed] [Google Scholar]
- 11.Mahajan VB, Olney AH, Garrett P, Chary A, Dragan E, Lerner G, Murray J, Bassuk AG. Collagen XVIII mutation in Knobloch syndrome with acute lymphoblastic leukemia. Am J Med Genet A. 2010;152A(11):2875–2879. doi: 10.1002/ajmg.a.33621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joyce S, Tee L, Abid A, Khaliq S, Mehdi SQ, Maher ER. Locus heterogeneity and Knobloch syndrome. Am J Med Genet A. 2010;152A(11):2880–2881. doi: 10.1002/ajmg.a.33619. [DOI] [PubMed] [Google Scholar]
- 13.Keren B, Suzuki OT, Gerard-Blanluet M, Bremond-Gignac D, Elmaleh M, Titomanlio L, Delezoide AL, Passos-Bueno MR, Verloes A. CNS malformations in Knobloch syndrome with splice mutation in COL18A1 gene. Am J Med Genet A. 2007;143A(13):1514–1518. doi: 10.1002/ajmg.a.31784. [DOI] [PubMed] [Google Scholar]
- 14.O'Connell AC, Toner M. Murphy S. Knobloch syndrome: novel intra-oral findings. Int J Paediatr Dent. 2009;19(3):213–215. doi: 10.1111/j.1365-263X.2008.00923.x. [DOI] [PubMed] [Google Scholar]
- 15.Corbett MA, Turner SJ, Gardner A, Silver J, Stankovich J, Leventer RJ, Derry CP, Carroll R, Ha T, Scheffer IE, Bahlo M, Jackson GD, Mackey DA, Berkovic SF, Gecz J. Familial epilepsy with anterior polymicrogyria as a presentation of COL18A1 mutations. Eur J Med Genet. 2017;60(8):437–443. doi: 10.1016/j.ejmg.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Seppinen L, Pihlajaniemi T. The multiple functions of collagen XVIII in development and disease. Matrix Biol. 2011;30(2):83–92. doi: 10.1016/j.matbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Heljasvaara R, Aikio M, Ruotsalainen H, Pihlajaniemi T. Collagen XVIII in tissue homeostasis and dysregulation-Lessons learned from model organisms and human patients. Matrix Biol. 2017;57-58:55–75. doi: 10.1016/j.matbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Kague E, Bessling SL, Lee J, Hu G, Passos-Bueno MR, Fisher S. Functionally conserved cis-regulatory elements of COL18A1 identified through zebrafish transgenesis. Dev Biol. 2010;337(2):496–505. doi: 10.1016/j.ydbio.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 19.Fox MA. Novel roles for collagens in wiring the vertebrate nervous system. Curr Opin Cell Biol. 2008;20(5):508–513. doi: 10.1016/j.ceb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Caglayan AO, Baranoski JF, Aktar F, Han W, Tuysuz B, Guzel A, Guclu B, Kaymakcalan H, Aktekin B, Akgumus GT, Murray PB, Erson-Omay EZ, Caglar C, Bakircioglu M, Sakalar YB, Guzel E, Demir N, Tuncer O, Senturk S, Ekici B, Minja FJ, Sestan N, Yasuno K, Bilguvar K, Caksen H, Gunel M. Brain malformations associated with Knobloch syndrome--review of literature, expanding clinical spectrum, and identification of novel mutations. Pediatr Neurol. 2014;51(6):806–813. doi: 10.1016/j.pediatrneurol.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]