Abstract
Emodin is a natural anthraquinone derivative that is present in various herbal preparations. The pharmacological effects of emodin include anticancer, hepatoprotective, anti-inflammatory, antioxidant and even antimicrobial activities. However, emodin also has been reported to induce hepatotoxicity, nephrotoxicity, genotoxicity and reproductive toxicity. The mechanism of emodin’s adverse effects is complicated and currently not well understood. This study aimed to establish a cell metabonomic method to investigate the toxicity of emodin and explore its potential mechanism and relevant targets. In the present study, metabonomic profiles of cell extracts and cell culture media obtained using the 1H NMR technique were used to assess emodin toxicity in HepG2 cells. Multivariate statistical analyses such as partial least squares-discriminant analysis (PLS-DA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) were used to characterize the metabolites that differed between the control and emodin groups. The results indicated that emodin resulted in differences in 33 metabolites, including acetate, arginine, aspartate, creatine, isoleucine, leucine and histidine in the cell extract samples and 23 metabolites, including alanine, formate, glutamate, succinate and isoleucine, in the cell culture media samples. Approximately 8 pathways associated with these metabolites were disrupted in the emodin groups. These results demonstrated the potential for using cell metabonomics approaches to clarify the toxicological effects of emodin, the underlying mechanisms and potential biomarkers. Our findings may help with the development of novel strategies to discover targets for drug toxicity, elucidate the changes in regulatory signal networks and explore its potential mechanism of action.
Introduction
Valuable safety information in traditional Chinese medicine (TCM) comes directly from its clinical use. This information can be used as an important reference for TCM safety evaluations. Emodin, which is an active ingredient of Chinese medical herbs, such as Rheum palmatum1, Polygonum multiflorum2 and Polygonum cuspidatum3, has been used for over 2,000 years in eastern Asia and is still present in various herbal preparations4. Previous studies have reported multiple pharmacological benefits of emodin, such as anticancer, hepatoprotective, anti-inflammatory, antioxidant and antimicrobial activities5. Emodin may be an effective therapeutic agent for prophylaxis and for various diseases including constipation, asthma, atopic dermatitis, atherosclerosis, hepatopathy osteoarthritis, diabetes and its complications, Alzheimer’s disease and tumors5. However, toxic effects of emodin, such as nephrotoxicity, hepatotoxicity, genotoxicity and reproductive toxicity, have also been reported6–10. Emodin was found to induce apoptotic responses in human hepatocellular carcinoma cell (HCC) lines and HepG211. Emodin produced reactive oxygen species (ROS) in these cells, which reduced the intracellular mitochondrial transmembrane potential (ΔΨm). This activated caspase-9 and caspase-3, leading to DNA fragmentation and apoptosis. Some studies have reported that emodin has the potential to disrupt glutathione and fatty acid metabolism in human liver cells12.
Drug-induced cytotoxicity is related to cell metabolism13. As a systemic approach, metabonomics adopts a “top-down” strategy to holistically reflect the function of organisms including terminal symptoms of metabolic network and understanding systemic metabolic changes caused by interventions14. Combining novel models with molecular profiling technologies, metabonomics provides new insights into the molecular basis of toxicity and provides a rich source of biomarkers that are urgently needed in 21st century toxicology research15. The metabolic profile of a whole organism does not provide relevant information about specific cell types under different conditions, which is crucial for creating a more holistic understanding of cell functions and for drug development16. Cell cultures may provide an alternative for understanding the specific metabolism of drug candidates17. Metabolic analysis of cell cultures has many potential applications and advantages over currently used methods for cell testing18.
Nuclear magnetic resonance (NMR) is an easy and convenient research tool. NMR is a suitable analysis method for examining complex compositions in omics research, particularly in modern TCM research. Principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA) and orthogonal partial least squares-discriminant analysis (OPLS-DA) methods were applied to maximize the distinction between groups, focusing on differences in metabolic variations, and to assess the correlations between the observed NMR results.
In this study, a novel attempt was made to explore the possible mechanisms and relevant targets of emodin toxicity based on NMR non-targeted metabolomics. The objective was to establish a cell metabonomic method and identify biomarkers for investigating the toxic effects of emodin. This study may provide new ideas and methods for investigating and understanding the toxicity mechanism of TCM.
Results
Emodin inhibited viability and HepG2 cell proliferation
An MTT assay showed that emodin inhibited HepG2 cell growth in both concentration- and time-dependent manners (Fig. 1). The 50% inhibitory concentration (IC50) values of emodin at 12 h and 24 h were 41.3 μM and 32.1 μM, respectively.
Figure 1.
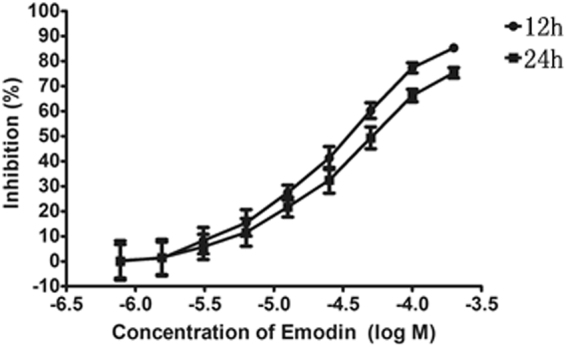
Emodin inhibited viability and proliferation in HepG2 cells. HepG2 cell were treated with different concentrations of emodin for 12 h and 24 h. Cell growth was determined using an MTT assay and was directly proportional to the absorbance at a wavelength of 570 nm. Data are expressed as the means ± S.D. for the three independent experiments.
Apoptosis caused by emodin
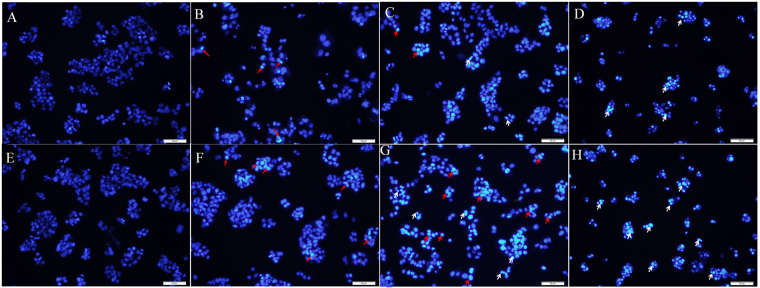
After 12 h and 24 h of exposure, emodin was found to induce HepG2 cells apoptosis using a DAPI staining assay. As shown in Fig. 2, viable normal cells exhibited an evenly distribution and deep blue fluorescence in both the nucleus and chromatin, while the apoptotic cells showed bright white fluorescence. Changes in the DAPI staining were observed in apoptotic cells in the emodin-treated group. All of the emodin concentrations tested (10, 50, and 100 μM) decreased the number of cells. In the emodin-treated groups, the cells were found to have different degrees of chromatin condensation, nuclear condensation and fragmentation, as indicated by the arrows in Fig. 2. Increasing the incubation time produced fragmented nuclei at 24 h. The apoptotic area in the 100 μM emodin sample was obviously larger than that of the 50 μM and 10 μM samples. Exposure for 24 h and at 100 μM were chosen for conducting the following metabolomic study.
Figure 2.
Fluorescent images of DAPI for the HepG2 cells exposed to 10, 50, or 100 μM of emodin for 12 h and 24 h. Different concentrations (10, 50, 100 μM) of emodin caused a decrease in cell numbers. The normal cells exhibited an evenly distributed, deep blue fluorescent color in the nucleus and the chromatin, while the apoptotic cells resulting from emodin exposure showed a bright white fluorescent color. In the emodin-treated groups, cells showed different degrees of chromatin condensation, nuclear condensation and fragmentation, as indicated by the arrows. The red arrows indicate chromatin condensation, while the white arrows indicate nucleus fragmentation. (A) Normal group (12 h); (B) low emodin group (10 μM, 12 h); (C) middle emodin group (50 μM, 12 h); (D) high emodin group (100 μΜ, 12 h); (E) normal group (24 h); (F) low emodin group (10 μM, 24 h); (G) middle emodin group (50 μM, 24 h); (H) high emodin group (100 μΜ, 24 h) (Bars = 50 μm).
1H NMR analysis of cell extracts and culture media
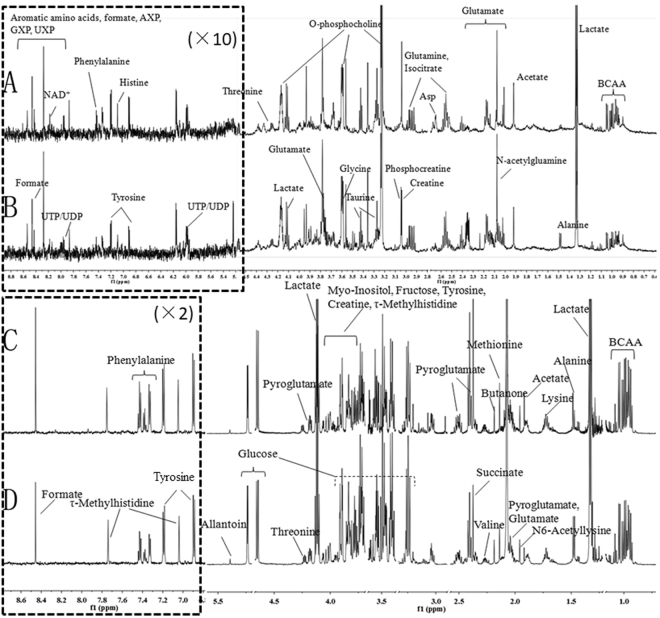
We analyzed cell extracts and culture media to investigate the potential for using metabolic fingerprinting to characterize emodin toxicological responses in liver cells. Figure 3 shows the 600 MHz 1H-NMR CPMG spectra of cell extracts and cell culture media from the high emodin group and control group. As used in previous studies19–21 and in our in-house NMR database (HMDB), the resonance assignments were performed and confirmed by a series of 2D NMR spectra including 1H-1H COSY, 1H-13C HMBC and 1H-13C HSQC. The cell extract NMR spectra were dominated by alanine (δ1.48, δ3.78), acetate (δ1.92), glutamine (δ2.13, δ2.44), leucine (δ0.96, δ1.69, δ3.74), valine (δ0.99, δ1.05, δ2.28), lactate (δ1.33, δ4.12), glucose (δ3.49, δ3.72, δ5.24), and glycine (δ3.58), among others. In the 1H NMR spectra of cell culture media, the primary changes were in signals for low-molecular-weight metabolites, such as threonine (δ1.33, δ3.56), isoleucine (δ1.02, δ1.46, δ8.09), valine (δ0.99, δ1.05, δ2.28), alanine (δ1.48, δ3.78), and glutamine (δ2.13, δ2.44). Multivariate data analysis was further performed to obtain a more detailed analysis of the metabolic differences between groups.
Figure 3.
Representative one-dimensional 600 MHz 1H NMR spectra of cell extracts and culture media from the normal and high emodin groups. (A) spectra of cell extracts from the high emodin group; (B) spectra of cell extracts from the normal group; (C) Spectra of cell culture media from the high emodin group; (D) spectra of cell culture media from the normal group. BCAA (Branched-chain amino acid); AXP (AMP, ADP, ATP); GXP (GMP, GDP; GTP); UXP (UMP, UDP, UTP).
Multivariate data analysis and the selection of potential biomarkers
PCA was performed using a mean-centered scaling approach. The data were visualized in the form of the principal component (PC) score plots to identify general metabolic trends and possible outliers. PCA from NMR data from HeLa cells and cell culture media revealed a clear dose-response (Figure S1) for emodin. The first two principal components (PC1 and PC2) explained 47.2% and 26.3% of the variation in the model in high-dose cell samples and 64.5% and 31.0% for the model and high-dose cell culture media samples. The majority of the samples were located within the 95% confidence interval. Further analyses of the NMR data from cells and cell culture media using PLS-DA showed a clear differentiation between the emodin group and control group (Figure S2).
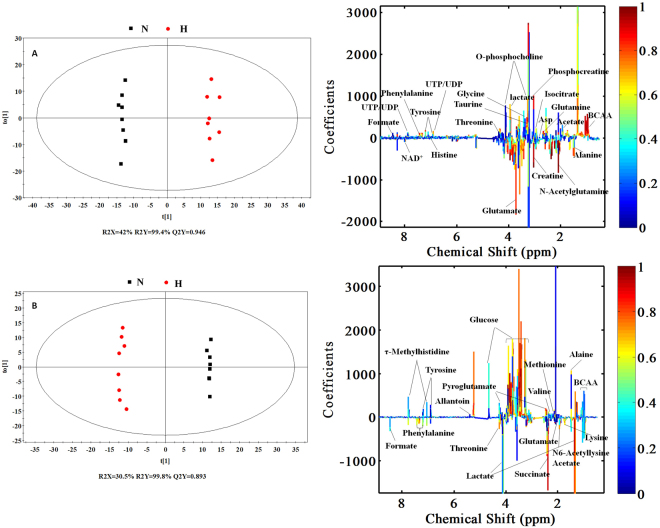
An OPLS-DA model was then performed to minimize the possible influence of between-group variability and screen metabonomic differences among the three groups. The significance of the contribution of the identified metabolites to the observed physiological changes was compared. The score plots (Figs 4 and S3) showed distinct separations between the normal group and the emodin exposure groups in the cell extract and culture media samples. A seven-fold cross-validation was applied to estimate the predictive ability of the OPLS-DA models. The parameters for the classification of control versus the high emodin group for the cell extract and cell culture media samples were respectively, R2X = 42%, Q2Y = 0.946 and R2X = 30.5%, Q2Y = 0.993, which demonstrated an acceptable goodness of fit and a high-quality predictability.
Figure 4.
OPLS-DA scores plots (left panel) and corresponding coefficient loading plots (right panel) derived from the 1H NMR spectra of cell extracts (A) and cell culture media (B) samples obtained from different groups. N: normal group; H: high emodin group (100 μM)
Metabonomic changes and biomarker identification
In the color-coded coefficient loading plots (Fig. 4), valuable biochemical distinctions between the normal group and the high emodin group were identified. The plots depicted considerable metabonomic differences between the control and emodin groups. Multivariate statistical analysis showed increased concentrations of 2-hydroxy-3-methylvalerate, acetate, arginine, aspartate, creatine, isoleucine, leucine, histidine, lactate, lysine, NADP+, O-phosphocholine, phenylalanine, taurine, tyrosine, valine and threonine in the cell extracts of the high emodin group and decreased concentrations of 2-hydroxybutyrate, 2-oxoglutarate, alanine, creatine phosphate, glucose, glutathione, glutamine, glycine, isocitrate, N-acetylglutamate, N-acetylglutamine, proline, UDP-glucuronate, ATP, pyroglutamate, N,N-dimethylglycine, betaine, glutamate and N-acetylglycine. The relevant data for the altered metabolites, including chemical shifts, are summarized in Table 1.
Table 1.
Quantitative comparison of metabolites (33) found in the cell extracts.
| Metabolites | Chemical Shiftc | Integrals in H and N groupa |
Integral in M and N group |
Integral in L and N group |
|||
|---|---|---|---|---|---|---|---|
| FCb | VIP | FC | VIP | FC | VIP | ||
| 2-Hydroxybutyrate | 1.64 (m) | 0.8091* | 1.2184 | 0.8772 | 0.6682 | 0.8289 | 1.3138 |
| Acetate | 1.92 (s) | 1.2581** | 1.3336 | 0.9371 | 0.6518 | 0.8917 | 0.9660 |
| Alanine | 1.48 (d), 3.78 (q) | 0.7854** | 1.4726 | 0.7198** | 1.8466 | 0.9261 | 1.0691 |
| Arginine | 1.91 (m), 3.24 (t), 3.78 (t) | 1.1801** | 1.3154 | 0.9308 | 0.8094 | 0.9198 | 1.0006 |
| Aspartate | 2.68 (m), 3.90 (m) | 2.3843** | 1.9762 | 1.3485** | 1.7590 | 1.0918 | 1.0695 |
| Creatine | 3.04 (m) | 1.4486** | 1.6717 | 1.2119* | 1.2233 | 1.1061 | 0.9868 |
| Creatine phosphate | 3.05 (s), 3.96 (s) | 0.7406** | 1.9484 | 0.9241** | 1.4916 | 1.0026 | 0.1080 |
| Glucose | 3.49 (t), 3.72 (dd), 5.24 (d) | 0.5721* | 1.1068 | 0.4758* | 1.3593 | 0.5937* | 1.5728 |
| Glutathione | 2.97 (dd), 3.77 (m) | 0.4812** | 1.9913 | 0.6360** | 2.1425 | 0.8855 | 1.8079 |
| Isoleucine | 1.02 (d), 1.46 (m) | 1.4619** | 1.9293 | 0.9565 | 0.7286 | 0.9423 | 1.0425 |
| Leucine | 0.96 (d), 1.69 (m), 3.74 (m) | 1.6182** | 1.9211 | 0.9703 | 0.5433 | 0.9528 | 0.8010 |
| Glutamine | 2.13 (m), 2.44 (m) | 0.6633** | 1.7608 | 0.8495* | 1.3052 | 0.8998* | 1.5323 |
| Glycine | 3.58 (s) | 0.7727** | 1.3490 | 0.7976* | 1.3745 | 0.8868 | 1.0789 |
| Histidine | 7.09 (s) | 1.6638** | 1.6217 | 1.2003 | 1.0026 | 1.0393 | 0.4060 |
| Isocitrate | 2.97 (m), 4.02 (d) | 0.7867** | 1.3550 | 0.7174** | 1.7326 | 0.8080* | 1.7588 |
| Lactate | 1.33(d), 4.12(q) | 1.2566* | 1.1083 | 1.3198** | 1.6731 | 1.2986** | 1.9174 |
| Lysine | 1.46 (m), 1.73 (m), 3.77 (t) | 1.1092* | 1.1748 | 1.1189* | 1.2745 | 1.0932 | 1.2703 |
| N-Acetylglutamate | 2.05 (m), 2.23 (t) | 0.5184** | 1.9286 | 0.5883** | 2.1010 | 0.5746** | 2.7253 |
| N-Acetylglutamine | 1.93 (dd), 2.12 (s) | 0.5353** | 1.9909 | 0.7079** | 2.1119 | 0.9326 | 1.3865 |
| NADP+ | 8.47(s), 8.83(d) | 1.4298** | 1.3746 | 1.0824 | 0.5863 | 1.0376 | 0.3253 |
| O-Phosphocholine | 3.24 (s), 3.60 (m), 4.15 (m) | 1.3489** | 1.3257 | 1.0051 | 0.1356 | 1.0013 | 0.2452 |
| Phenylalanine | 3.29 (dd), 3.98 (m), 7.38 (m) | 2.1359** | 1.7687 | 1.3651* | 1.3178 | 1.0465 | 0.3762 |
| Proline | 3.38 (m), 4.15 (m) | 0.7138** | 1.3935 | 0.6666** | 1.7600 | 0.7077** | 2.2799 |
| Taurine | 3.26 (t), 3.42 (t) | 1.5803** | 1.8060 | 1.4417** | 1.8751 | 1.1620 | 1.2841 |
| Tyrosine | 3.06 (dd), 3.94 (m), 6.90 (d) | 1.2952** | 1.7079 | 1.1039 | 1.0807 | 0.9324 | 0.9615 |
| Valine | 0.99 (d), 1.05 (d), 2.28 (m) | 1.4327** | 1.8617 | 1.0486 | 0.7198 | 0.9751 | 0.4531 |
| ATP | 4.22(m), 4.29(m) | 0.7931** | 1.3549 | 0.6575** | 1.8749 | 0.7501** | 2.1786 |
| Threonine | 1.33 (d), 3.56 (d) | 1.2368** | 1.5755 | 1.1438* | 1.3897 | 1.1418 | 1.4358 |
| N,N-Dimethylglycine | 2.92 (s), 3.72 (s) | 0.5694** | 1.6912 | 0.5577** | 1.9002 | 0.7364** | 1.9582 |
| Betaine | 3.24 (s), 3.89 (s) | 0.7408** | 1.6815 | 0.7418** | 1.8939 | 0.8049** | 2.1787 |
| Glutamate | 2.11 (m), 2.33(m), 3.75 (m) | 0.4799** | 2.0016 | 0.6705** | 2.1172 | 0.8733** | 2.1524 |
| 2-Oxoglutarate | 2.4 (t), 3.01 (t) | 0.8135** | 1.4803 | 0.8325** | 1.6518 | 0.8979* | 1.5029 |
| N-Acetylglycine | 2.05 (s), 3.74 (d) | 0.7879** | 1.3319 | 0.7194** | 1.7719 | 0.8957 | 1.0959 |
aThe relative integrals of metabolites were determined from the 1H NMR analysis of cell extracts of each group. N: normal group; H: high emodin group (100 μΜ); M: middle emodin group (50 μM); L: low emodin group (10 μM).
bFC: fold change.
cThe chemical shifts in bold were used to calculate integrals and p values.
**Compared with the normal group, p < 0.01; *compared with the normal group, p < 0.05.
Metabolite differences in the cell culture media samples were also identified between the emodin-exposure groups and the control group. The levels of 2-methylglutarate, 3-methylglutarate, 4-methylglutarate, allantoin, formate, glutamate, succinate, isoleucine, lactate, N,N-dimethylglycine, N6-acetyllysine, ornithine, pyroglutamate, threonine, valine, τ-methylhistidine and phenylalanine were elevated in the emodin groups, whereas glucose and acetate levels decreased. The altered metabolites are presented in Table 2.
Table 2.
Quantitative comparison of metabolites (23) found in the cell culture media.
| Metabolites | Chemical Shiftc | Integral in H and N groupa | Integral in M and N group | Integral in L and N group | |||
|---|---|---|---|---|---|---|---|
| FCb | VIP | FC | VIP | FC | VIP | ||
| Alanine | 1.48 (d), 3.78 (q) | 0.5546** | 2.1042 | 0.7392** | 2.1955 | 0.8930* | 1.4488 |
| Allantoin | 5.41 (s) | 1.9970* | 1.2710 | 1.5914 | 0.6796 | 1.9288 | 1.3164 |
| Formate | 8.46 (s) | 1.2739** | 2.1095 | 1.3103** | 2.3911 | 1.2997** | 2.6689 |
| Fructose | 3.69 (m), 3.82 (m), 3.90 (dd) | 0.9063** | 2.0266 | 0.9555** | 1.9630 | 1.0569* | 1.7715 |
| Glucose | 3.49 (t), 3.72 (dd), 5.24 (d) | 0.6004** | 2.1696 | 0.7428** | 2.3890 | 0.9310** | 1.9516 |
| Arginine | 1.91 (m), 3.24 (t), 3.78 (t) | 0.6724** | 2.1627 | 0.8021** | 2.3940 | 0.9427** | 1.9785 |
| Methionine | 2.14 (s), 2.16 (m), 3.86 (t) | 0.6966** | 2.1016 | 0.8249** | 2.1955 | 0.9507 | 1.3813 |
| Glutamate | 2.11 (m), 2.33 (m), 3.75 (m) | 1.3120** | 1.9648 | 1.2743** | 2.1997 | 1.1584** | 2.3668 |
| Succinate | 2.41 (s) | 2.9409** | 2.1890 | 2.2460** | 2.4270 | 1.1480** | 2.3037 |
| Glutamine | 2.13 (m), 2.44 (m) | 0.2441** | 2.1028 | 0.3388** | 2.3026 | 0.5654** | 2.4428 |
| Lysine | 1.46 (m), 1.73 (m), 3.77 (t) | 0.7725** | 2.0705 | 0.8596** | 2.2625 | 0.9166** | 2.0737 |
| Isoleucine | 1.02 (d), 1.46 (m) | 1.2343** | 1.9892 | 1.1539** | 1.9826 | 1.0756* | 1.6992 |
| Lactate | 1.33 (d), 4.12 (q) | 1.2797** | 1.5165 | 1.1638* | 1.3904 | 1.0022 | 0.3572 |
| Leucine | 0.96 (d), 1.69 (m), 3.74 (m) | 0.6525** | 2.1729 | 0.7808** | 2.3874 | 0.9735 | 1.0006 |
| N,N-Dimethylglycine | 2.92 (s), 3.72 (s) | 17.1029** | 2.1976 | 9.1770** | 2.4257 | 3.1164** | 2.6144 |
| N6-Acetyllysine | 1.41 (m), 1.90 (m), 3.74 (t) | 1.3563** | 1.8345 | 1.2610** | 1.8880 | 1.0904* | 1.5103 |
| Ornithine | 1.73 (m), 3.05 (t), 3.77 (t) | 1.6490** | 1.9002 | 1.4071** | 1.8105 | 1.0372 | 0.5523 |
| Pyroglutamate | 2.39 (m), 2.50 (m), 4.17 (dd) | 1.2075** | 1.4501 | 1.1655* | 1.3869 | 0.9671 | 0.3876 |
| Threonine | 1.33 (d), 3.56 (d) | 1.4552** | 2.1414 | 1.3227** | 2.3963 | 1.1463** | 2.2457 |
| Tyrosine | 3.06 (dd), 3.94 (m), 6.90 (d) | 0.8216** | 1.3951 | 0.8603* | 1.3507 | 1.0823 | 1.3854 |
| Valine | 0.99 (d), 1.05 (d), 2.28 (m) | 1.1046* | 1.2260 | 1.0793** | 1.5845 | 1.0178 | 0.3643 |
| τ-Methylhistidine | 3.24 (dd), 3.70 (s), 7.76 (s) | 3.9045* | 1.1948 | 3.2689* | 1.4839 | 0.6081 | 0.8213 |
| Phenylalanine | 3.29 (dd), 3.98 (m), 7.38 (m) | 1.2297** | 1.9856 | 1.1529** | 2.0863 | 1.0761** | 1.8162 |
aThe relative integrals of metabolites were determined from the 1H NMR analysis of cell culture media for each group. N: normal group; H: high emodin group (100 μΜ); M: middle emodin group (50 μM); L: low emodin group (10 μM).
bFC: fold change.
cThe chemical shifts in bold were used to calculate the integrals and p values.
**Compared with the normal group, p < 0.01; *compared with the normal group, p < 0.05.
The levels of 5 metabolites (threonine, isoleucine, valine, lactate and phenylalanine) were increased in both the cell extract and the cell culture media samples of the emodin-treated groups, and the levels of alanine, glutamine and glucose were reduced. However, the concentrations of glutamate, N,N-dimethylglycine, leucine, arginine and tyrosine in the cell extracts of the emodin treated groups were reduced, while they increased in the cell culture media samples. The opposite trend was observed for the lysine concentration in the cell extract and cell culture media samples.
Pathway analyses
Based on the target metabolites, a metabolic pathway analysis was performed using MetPA to reveal the most relevant pathways related to emodin. The potential target pathway was determined using pathway topology analysis and evaluated for an impact value above 0. For this impact value, we found 8 potential target pathways (Lysine degradation; Phenylalanine metabolism; Glycine, serine and threonine metabolism; Arginine and proline metabolism; Pyruvate metabolism; Lysine biosynthesis; Alanine, aspartate and glutamate metabolism; D-Glutamine and D-glutamate metabolism) related to 14 of the metabolites identified in this research. The 8 pathways, which included more than one target, were disrupted across the emodin treatments groups (Fig. 5).
Figure 5.
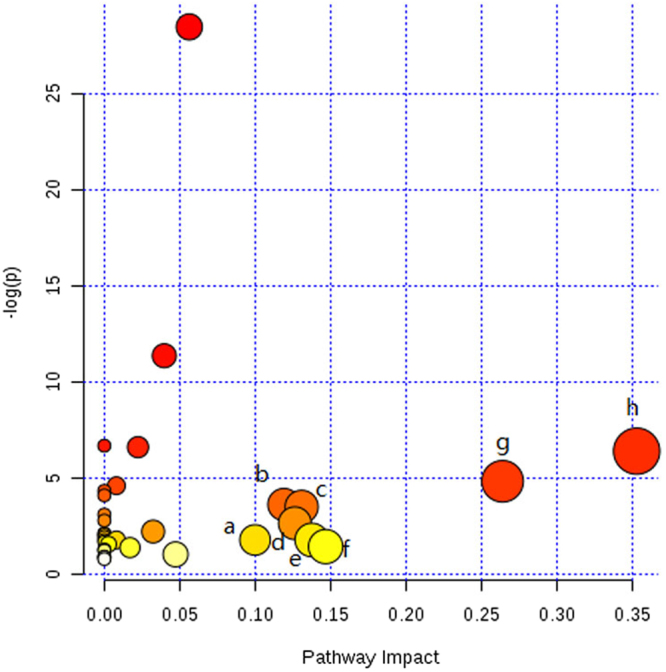
Summary of pathway analysis with MetPA. (a) Lysine degradation (b) Phenylalanine metabolism (c) Glycine, serine and threonine metabolism (d) Arginine and proline metabolism (e) Pyruvate metabolism (f) Lysine biosynthesis (g) Alanine, aspartate and glutamate metabolism (h) D-Glutamine and D-glutamate metabolism.
Discussion
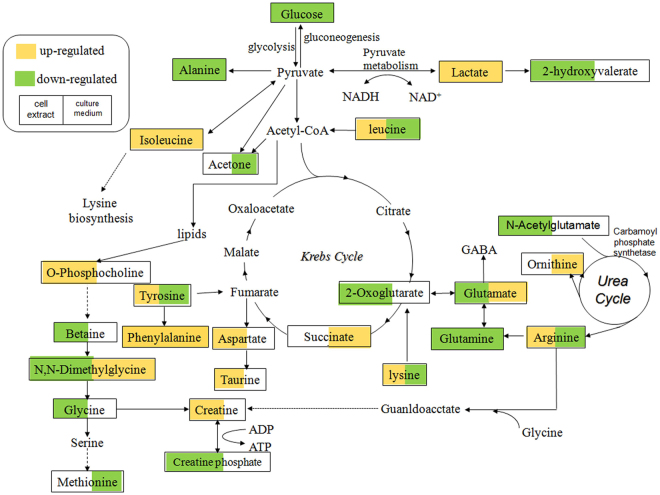
Drug-induced hepatotoxicity is a subject of interest for both the toxicology industry and in clinical research. It has been reported that emodin induces P450s, 1A1 and 1B1 in human lung adenocarcinoma CL5 cells22. Some recent studies have concluded that emodin suppresses proliferation of HepG2 cells and induces apoptosis11,23. Cytotoxicity was also observed in L-02 and BEL cells after exposure to emodin at a concentration of 50 μΜ24. There are limited reports on the cytotoxicity of emodin in HepG2 cells25. Therefore, we studied the global metabolic therapeutic effects using emodin concentrations of 10, 50 and 100 μM in cultures of HepG2 cells at an experimental exposure time of 24 h. The 100 μM dose of emodin represented the IC50 for cell viability at 24 h of treatment and caused cell apoptosis. We tried to investigate the underlying metabonomic characteristics of emodin in HepG2 cells and the influence of cellular toxicity on small-molecule metabolites. Based on 1H NMR, metabonomics were performed to screen for biomarkers of emodin toxicity in HepG2 cells. We demonstrated that the detection of emodin toxicity was possible using metabolic profiling. Finally, 45 compounds concentrations and 8 pathways were disrupted by emodin exposure. Several types of metabolites were detected in the HepG2 cell 1H NMR spectra, including some essential amino acids, non-essential amino acids (aspartate, glycine, histidine, arginine, proline, taurine, alanine and glutamate), intermediates of the tricarboxylic acid cycle (TCA) (lactate and isocitrate), ATP and carboxylic acid (2-hydroxyisobutyrate). The culture media NMR spectra were characterized by multiple metabolic intermediates and end-products including the glycolysis and TCA intermediates (lactate and succinate), the metabolic wastes (formate and allantoin) and a series of nutrient substrates, including some amino acids and glucose, which could provide all the essential elements for cell growth. The schematic representation of the metabolic network is shown in Fig. 6.
Figure 6.
Disrupted metabolic pathways detected by 1H NMR analysis. Metabolites in yellow were up-regulated both in the cell extracts and the cell culture media samples; Metabolites in green were down-regulated in both the cell extracts and the cell culture media samples.
Metabolites related to energy metabolism. The observed trends in glucose and lactate were opposite in both the cell extract and the cell culture media samples. For example, when glucose decreased, the lactate level increased. Conversely, emodin increased glycolytic activity. Reduced gluconeogenesis and glycolysis promoted acetyl-CoA production for use in the Krebs cycle. However, the acetyl-CoA demand from the Krebs cycle was exceeded by the amount of acetyl-CoA derived from glycolysis, and ketone bodies were thus produced26. Accordingly, the level of 2-hydroxybutyrate was reduced in the cell extract in the emodin group. 2-Hydroxybutyrate is a highly energetic compound that transports the energy of liver, and with the help of β-hydroxybutyrate dehydrogenase, it is converted to acetoacetic acid27,28. The release of acetate was closely associated with glucose utilization because it is the main method for pyruvate production from glycolysis in the cytoplasm. The level of alanine was closely related to pyruvate and glucose, as it is the primary energy source in the alanine-glucose cycle29. As a byproduct of choline metabolism, creatine is formed when guanidinoacetate receives a S-adenosylmethionine (SAM) methyl from guanidinoacetate methyltransferase30. Emodin may disrupt energy metabolism and lead to an increase of creatine in HepG2 cells, which was shown to be an emergency energetic regulator31.
Metabolites related to the urea cycle and the TCA cycle. Characterized by a decreased utilization of succinate, emodin-induced metabolic changes were determined using the OPLS-DA, which may be related to the low activity of succinate dehydrogenase induced by emodin hepatotoxicity. Arginine, which is produced when arginine-succinate is split, is an important intermediate product in the urea cycle. Catalyzed by arginase in the liver, arginine produces urea and ornithine via hydrolysis, which initiates a new cycle of urea cycle. In the emodin groups, the splitting of arginine-succinate may have been affected by emodin. Arginine release was abnormal and eventually affected the urea cycle and TCA.
Metabolites related to oxidative stress and immune response. As a derivative of glycine, N,N-dimethylglycine is an important intermediate in the metabolism of choline to glycine. Choline is oxidized to betaine, which is then demethylated to form N,N-dimethylglycine. Dimethylglycine is oxidatively demethylated to form sarcosine32. It has been reported that N,N-dimethylglycine decreases oxidative stress33 and improves immune responses34. Compared with the control group, emodin significantly increased the levels of N,N-dimethyglycine and betaine in the cell media samples and decreased the level of N,N-dimethyglycine in the cell extracts. This finding indicates that emodin might influence the biological processes of oxidative stress and immune response.
Metabolism of other amino acids. Due to hepatocellular injury or hepatic ATP consumption, the levels of some amino acids (leucine and Isoleucine) were found to be increased in the HepG2 cell extracts. The concentrations of amino acids in HepG2 cells may vary according to the degree of cell damage. Elevated urinary taurine has long been identified as a specific marker of liver impairments35–37, including necrosis and steatosis, which correlate with the hepatocyte necrosis observed by histopathology. Here, the elevated taurine levels in the HepG2 cells may support a disruption of hepatic function. Increased levels of some amino acids including glutamate, threonine, isoleucine, phenylalanine and valine were observed, which suggests that emodin may facilitate the protein catabolism and reduce protein synthesis.
Two pathways (alanine, aspartate and glutamate metabolism and D-glutamine and D-glutamate metabolism) were mainly disrupted by emodin. The results showed a decrease in the levels of alanine, glutamate and glutamine, and an increase in the level of aspartate in the cell extract samples. The level of glutamate increased in the cell culture media samples, while alanine and glutamine decreased. The biosynthesis of these amino acids is connected to intermediates of the citrate cycle. A significant disruption of the citrate cycle was also observed in the emodin exposed cells. Emodin disrupted glutathione metabolism in HepG2 cells, marked by a decreased level of glutathione, which is consistent with results reported by Liu et al.12. Glutathione is a cellular antioxidant. Therefore, using glutathione to modulate drug metabolism is an important mechanism for drug detoxification13. As a substrate, aspartate can be used to produce glutamate and oxaloacetic acid, and glutamate is one of the substrates used to synthesize glutathione. With the depletion of glutathione, the toxicity of emodin is increased, resulting in oxidative stress and peroxidation reactions.
The liver is an essential organ for drug and xenobiotic metabolism. Drug-induced hepatotoxicity may result from the direct toxicity of compound or from indirect toxicity due to its active metabolites. One limitation of our study is that we were only able to detect and conduct qualitative analysis on metabolites that were disrupted by emodin exposure in HepG2 cells. We were thus not able to directly measure the pathways and mechanisms affected. Future studies should identify accurate biomarkers, the mechanism of action for emodin and how to intervene to reduce emodin’s toxic effects.
Conclusions
In this study, we explored emodin toxicity using a combination of NMR and pattern recognition data to establish cell metabonomics from samples of cell extracts and cell culture media. Emodin disrupted several classes of metabolites and disrupted biological processes including the Krebs cycle, amino acid metabolism and purine metabolism. Moreover, this study may contribute to the development of new strategies to elucidate TCM toxicity information, identify potential targets for candidate drugs and clarify the relevant signal network.
Materials and Methods
Cell culture and viability
The HepG2 cells were purchased from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and used within twenty passages. HepG2 cells were maintained in high-glucose Dulbecco’s-modified eagle medium (DMEM) with 10% fetal calf serum. Cells were cultured in a humidified atmosphere at 37 °C and with 5% CO2.
Cell viability was evaluated using an MTT assay38. HepG2 cells were seeded in a 96-well microtiter plate with 1 × 104 cells/well left to adhere overnight before treatment. Emodin (purchased from the National Institute for Food and Drug Control) was diluted in dimethylsulfoxide (DMSO) and high-glucose DMEM and added to the cells for 12 h or 24 h incubations. The final concentration was 0.78 to 200 μM. The cell media, which contained only DMSO, was used as a control. Absorbance was determined at 570 nm using an ELISA reader. Three replicates were performed for each cell group.
DAPI staining
HepG2 cells (approximately 1 × 106 cells/well) were seeded in 6-well plates. Three different concentrations of emodin (10, 50, 100 μM) were added to the adherent cells and incubated for 24 h. At room temperature, HepG2 cells were stained by DAPI (5 μg/mL) for 10 min. The DAPI dye was then rinsed out. The changes in the cells were observed and the stained cells were photographed.
Sample preparation
There were four study groups: a high emodin group (100 μM, HG), a middle emodin group (50 μM, MG), a low emodin group (10 μM, LG) and control group (normal cultured HepG2 cells). Approximately 1 × 106 cells/well of HepG2 cells were seeded in 6-well plates and left to adhere overnight. Three different concentrations of emodin (10, 50, 100 μM) were then added to the cells and incubated for 24 h.
Cell extract preparation: Cell extracts were prepared using a precooled methanol-chloroform-water system (2:2:3). After removing the culture media from the culture dish, cells were quickly washed three times with ice-cold PBS (pH 7.4) to remove media components. Cells were then quenched with 1.4 ml of methanol and 0.7 ml of ultrapure H2O and homogenized. Next, 1.4 ml of chloroform and 1.4 ml of ultrapure H2O were added to the homogenate and mixed for extraction. The solution was centrifuged at 10,000 g for 15 min at 4 °C. The aqueous phase was lyophilized for the next metabonomic analysis. Lyophilized cell extracts were resuspended with D2O and a sodium phosphate buffer. Samples were mixed uniformly and centrifuged at 10,000 g for 10 min at 4 °C. The supernatant was collected for further assessment.
Cell culture media preparation: culture medium samples were prepared using 200 μL of cell media mixed with 400 μL of D2O19. All samples were mixed and then centrifuged at 10,000 g for 10 min at 4 °C. The supernatants were collected for further assessment.
1H NMR spectroscopy
All samples were analyzed by 1H NMR spectroscopy using a VARIAN VNMRS 600 MHz NMR spectrometer (Varian Inc, Palo Alto, Calif) operating at 298 K and 599.871 MHz using a 5-mm inverse-proton triple resonance probe. For intracellular metabolite extracts, the standard NOESYPR 1D pulse sequence (RD–90°–t1–90°–tm–90°–ACQ) was used with the irradiations at the water frequency during a recycle delay of 2 s and a mixing time of 100 ms to suppress the residual HOD signal.
The 1H NMR spectra of the cell culture media were obtained by the water-suppressed standard 1D Carr-Purcell-Meiboom-Gill pulse sequence (RD-90°-(τ−180°-τ) n-ACQ). The free induction decays (FIDs) were recorded by 64 K data points with a spectral width of 12000 Hz and 128 scans with a relaxation delay of 2.0 s and an acquisition time of 1.36 s. The FIDs were weighted by an exponential function with a 0.5 Hz line-broadening factor prior to Fourier transformation.
Data processing
The 1H NMR spectra were processed using MestReNova 7.1.2 software (Mestrelab Research, Spain). All 1H-NMR spectra were manually corrected for baseline and phase and referenced to the TSP signal (δ0.00) to assess the changes in the endogenous metabolites related to the emodin toxicity. In the cell extract NMR spectra, the regions of δ 5.2–4.6 and δ2.8-2.69 were removed to eliminate the water suppression and DMSO signals. For the NMR spectra of the media, the regions of δ5.19-4.68 and δ3.37-3.33, δ2.89-2.58 and δ3.69-3.61, δ1.23-1.15 were excluded to eliminate the effect of residual water, methanol, DMSO and ethanol signals, respectively.
Multivariate analysis was performed with SIMCA-P + 12.0 (Umetrics, Sweden). Principal component analysis (PCA) was applied to mean-centered data to identify outliers and to acquire the data distribution profiles. To improve the separation caused by variations among the groups and minimize other biological analytical variations, sample classes were modeled using the orthogonal partial least squares discriminant analysis (OPLS-DA) algorithm with a unit variance-scaled approach. Coefficient plots were generated with MATLAB scripts (http://www.mathworks.com) with some in-house modifications and color-coded by the absolute value of coefficients39.
To determine which variables contributed to the assignment of spectra among the groups, we analyzed the variable importance in the projection (VIP) values of each peak from the OPLS-DA models in this study. To detect significant differences in the signals between the two groups, an independent T-test was conducted in SPSS Statistics 17.0 (SPSS Inc, USA). Differences in metabolites were identified as having statistically significant results for both the multivariate analyses with VIP > 1 and the univariate analyses with p < 0.05. Additionally, the significance of the metabolites was assessed based on the unpaired Student’s T-test of chemical shifts. Most of the metabolites were identified by comparing them with the HMDB (http://www.hmdb.ca/) and the previous literature, which will be listed in the results and references.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic supplementary material
Acknowledgements
This study was funded by the Beijing Municipal Science & Technology Commission (No. Z171100001717029), the Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2018ZX09201009-002-003) and the National Natural Science Foundation of China (No. 81603346, 81573832 and 81703945).
Author Contributions
Z.F.L., A.L., J.G. and C.C. planned and performed the experiments, collected and analyzed the data, and wrote the paper. J.G. analyzed the results. All the other authors including T.S.W., C.G., Y.J.Y., C.Y.M., L.W.G., Y.Y. and J.G. discussed the results and commented on the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Chang Chen and Jian Gao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27359-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhong-Feng Li, Email: lizf@cnu.edu.cn.
An Liu, Email: aliu@icmm.ac.cn.
References
- 1.Wang JB, et al. Hepatotoxicity or hepatoprotection? Pattern recognition for the paradoxical effect of the Chinese herb Rheumpalmatum L. in treating rat liver injury. PLoS One. 2011;6:e24498. doi: 10.1371/journal.pone.0024498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee MH, Kao L, Lin C. Comparison of the antioxidant and transmembrane permeative activities of the different Polygonum cuspidatum extracts in phospholipid-based microemulsions. J Agr Food Chem. 2011;59:9135–9141. doi: 10.1021/jf201577f. [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Zhao R, Wang W, Mao X, Yu J. Lipid regulation effects of Polygoni Multiflori Radix, its processed products and its major substances on steatosis human liver cell line L02. J Ethnopharmacol. 2012;139:287–293. doi: 10.1016/j.jep.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Dong X, et al. Emodin: A Review of its Pharmacology, Toxicity and Pharmacokinetics. Phytother Res. 2016;30:1207–1218. doi: 10.1002/ptr.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo C, Capasso R. Constipation and botanical medicines: an overview. Phytother Res. 2015;29:1488–1493. doi: 10.1002/ptr.5410. [DOI] [PubMed] [Google Scholar]
- 6.Wang C. Emodin induces apoptosis through caspase 3-dependent pathway in HK-2 cells. Toxicology. 2007;231:120–128. doi: 10.1016/j.tox.2006.11.064. [DOI] [PubMed] [Google Scholar]
- 7.Wang QX, et al. Cytotoxicity of free anthraquinone from Radix et Rhizoma Rhei to HK-2 Cells. Chinese Journal of New Drugs. 2007;16:189–199. [Google Scholar]
- 8.Li CL, Ma J, Zheng L, Li HJ, Li P. Determination of emodin in L-02 cells and cell culture media with liquid chromatography-mass spectrometry: application to a cellular toxicokinetic study. J Pharmaceut Biomed. 2012;71:71–78. doi: 10.1016/j.jpba.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 9.Peng C, Wang Y, Li Y. Toxicity mechanism of emodin on interstitial cells of Cajal. Pharmacol Pharm. 2013;4:331–339. doi: 10.4236/pp.2013.43048. [DOI] [Google Scholar]
- 10.Luo T, et al. Emodin inhibits human sperm functions by reducing sperm [Ca2+] i and tyrosine phosphorylation. Reprod Toxicol. 2014;51:14–21. doi: 10.1016/j.reprotox.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Jing XB, Ueki N, Cheng JD, Imanishi H, Hada T. Induction of apoptosis in hepatocellular carcinoma cell lines by emodin. Jpn J Cancer Res. 2002;93:874–882. doi: 10.1111/j.1349-7006.2002.tb01332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Liu Y, Qu Y, Cheng M, Xiao H. Metabolomic profiling of emodin induced cytotoxicity in human liver cells and mechanistic study. Toxicol Res. 2015;4:948–955. doi: 10.1039/C4TX00246F. [DOI] [Google Scholar]
- 13.Liu J, et al. Metabolite profiling and identification of triptolide in rats. J Chromatogr B-Analyt Technol Biomed Life Sci. 2013;939:51–58. doi: 10.1016/j.jchromb.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol. 2006;2:52–57. doi: 10.1038/msb4100095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang A, Sun H, Xu H, Qiu S, Wang X. Cell Metabolomics. OMICS. 2013;17:495–501. doi: 10.1089/omi.2012.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuperlović-Culf M, Barnett DA, Culf AS, Chute I. Cell culture metabolomics: applications and future directions. Drug Discov Today. 2010;15:610–621. doi: 10.1016/j.drudis.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Aihua Z, Hui S, Hongying X, Shi Q, Xijun W. Cell Metabolomics. OMICS. 2013;17:495–501. doi: 10.1089/omi.2012.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.du Preez I, du Loots. T. Altered fatty acid metabolism due to rifampicin-resistance conferring mutations in the rpoB gene of Mycobacterium tuberculosis: Mapping the potential of pharmaco-metabolomics for global health and personalized medicine. OMICS. 2012;16:596–603. doi: 10.1089/omi.2012.0028. [DOI] [PubMed] [Google Scholar]
- 19.Ouattara DA, et al. Metabolomics-on-a-chip and metabolic flux analysis for label-free modeling of the internal metabolism of HepG2/C3A cells. Mol Biosyst. 2012;8:1908–1920. doi: 10.1039/c2mb25049g. [DOI] [PubMed] [Google Scholar]
- 20.Massimi M, et al. Effects of resveratrol on HepG2 cells as revealed by 1H-NMR based metabolic profiling. Biochim Biophys Acta. 2012;1820:1–8. doi: 10.1016/j.bbagen.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. The antitumor effect of formosanin C on HepG2 cell as revealed by 1H-NMR based metabolic profiling. Chem Biol Interact. 2014;220:193–199. doi: 10.1016/j.cbi.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Wang HW, Chen TL, Yang PC, Ueng TH. Induction of cytochromes P450 1A1 and 1B1 by emodin in human lung adenocarcinoma cell line CL5. Drug Metab Dispos. 2001;29:1229–1235. [PubMed] [Google Scholar]
- 23.Hsu CM, et al. Emodin inhibits the growth of hepatoma cells: finding the common anti-cancer pathway using Huh7, Hep3B, and HepG2cells. Biochem Bioph Res. 2010;392:473–478. doi: 10.1016/j.bbrc.2009.10.153. [DOI] [PubMed] [Google Scholar]
- 24.Qin B, et al. Chemical Reactivity of Emodin and Its Oxidative Metabolites to Thiols. Chem Res Toxicol. 2016;29:2114–2124. doi: 10.1021/acs.chemrestox.6b00191. [DOI] [PubMed] [Google Scholar]
- 25.Cui YT, et al. Involvement of PI3K/Akt, ERK and p38 signaling pathways in emodinmediated extrinsic and intrinsic human hepatoblastoma cell apoptosis. Food and Chemical Toxicology. 2016;92:26–37. doi: 10.1016/j.fct.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 26.McGarry JD, Foster DW. Regulation of hepatic fatty acid oxidation and ketone body production. Annual Review of Biochemistry. 1980;49:395–420. doi: 10.1146/annurev.bi.49.070180.002143. [DOI] [PubMed] [Google Scholar]
- 27.Boulagnon C, Garnotel R, Fornes P, Gillery P. Post-mortem biochemistry of vitreous humor and glucose metabolism: anupdate. ClinChem Lab Med. 2011;49:1265–1270. doi: 10.1515/CCLM.2011.638. [DOI] [PubMed] [Google Scholar]
- 28.Hockenhull J, Dhillo W, Andrews R, Paterson S. Investigation of markers to indicate and distinguish death due to alcoholic ketoacidosis, diabetic ketoacidosis and hyperosmolar hyperglycemic state using post-mortem samples. Forensic Science International. 2012;214:142–147. doi: 10.1016/j.forsciint.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 29.Mazurek S, Eigenbroadt E. The tumor metabolome. Anticancer Research. 2003;23:1149–1154. [PubMed] [Google Scholar]
- 30.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiological Reviews. 2000;80:1107–1213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 31.Morvan D, Demidem A. Metabolomics by proton nuclear magnetic resonance spectroscopy of the response to chloroethylnitrosourea reveals drug efficacy and tumor adaptive metabolic pathways. Cancer Research. 2007;67:2150–2159. doi: 10.1158/0008-5472.CAN-06-2346. [DOI] [PubMed] [Google Scholar]
- 32.Lever, M., McEntyre, C. J., George, P. M. & Chambers, S. T. Is N,N-dimethylglycine N-oxide a choline and betaine metabolite? Biol Chem 2017, Jan 13 (2017). [DOI] [PubMed]
- 33.Takahashi T, et al. N-Dimethylglycine decreases oxidative stress and improves in vitro development of bovine embryos. J Reprod Dev. 2016;62:209–212. doi: 10.1262/jrd.2015-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graber CD, Goust JM, Glassman AD, Kendall R, Loadholt CB. Immunomodulating properties of dimethylglycine in humans. J Infect Dis. 1981;143:101–105. doi: 10.1093/infdis/143.1.101. [DOI] [PubMed] [Google Scholar]
- 35.Beckwith-Hall BM, et al. Nuclear magnetic resonance spectroscopic and principal components analysis investigations into biochemical effects of three model hepatotoxins. Chemical Research in Toxicology. 1998;11:260–272. doi: 10.1021/tx9700679. [DOI] [PubMed] [Google Scholar]
- 36.Bollard ME, et al. NMR-Based metabolic profiling identifies biomarkers of liver regeneration following partial hepatectomy in the rat. Journal of Proteome Research. 2010;9:59–69. doi: 10.1021/pr900200v. [DOI] [PubMed] [Google Scholar]
- 37.Waterfield CJ, Turton JA, Scales MD, Timbrell JA. Investigations into the effects of various hepatotoxic compounds on urinary and liver taurine levels in rats. Archives of Toxicology. 1993;67:244–254. doi: 10.1007/BF01974343. [DOI] [PubMed] [Google Scholar]
- 38.Yan LL, Zhang YJ, Gao WY, Man SL, Wang Y. In vitro and in vivo anticancer activity of steroid saponins of Paris polyphylla var. yunnanensis. Experimental Oncology. 2009;31:27–32. [PubMed] [Google Scholar]
- 39.Feng J, Liu H, Zhang L, Bhakoo K, Lu L. An insight into the metabolic responses of ultra-small superparamagnetic particles of iron oxide using metabonomic analysis of biofluids. Nanotechnology. 2010;21:395101. doi: 10.1088/0957-4484/21/39/395101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.