Abstract
The Pax3/7 transcription factor family is integral to developmental gene networks contributing to important innovations in vertebrate evolution, including the neural crest. The basal chordate lineage of amphioxus is ideally placed to understand the dynamics of the gene regulatory network evolution that produced these novelties. We report here the discovery that the cephalochordate lineage possesses two Pax3/7 genes, Pax3/7a and Pax3/7b. The tandem duplication is ancestral to all extant amphioxus, occurring in both Asymmetron and Branchiostoma, but originated after the split from the lineage leading to vertebrates. The two paralogues are differentially expressed during embryonic development, particularly in neural and somitic tissues, suggesting distinct regulation. Our results have implications for the study of amphioxus regeneration, neural plate and crest evolution, and differential tandem paralogue evolution.
Introduction
Susumu Ohno proposed in 19701 that gene duplication might be an important evolutionary mechanism for generating diversity. The evolutionary fate of paralogues is influenced by both the mechanism of duplication and by the properties and functions of the genes involved, and various models have been developed to explain their adaptive trajectory2,3. Genes with a high degree of connectivity to regulatory regions and other gene products, and that are related to functions including development, neurogenesis, and organismal complexity, have been preferentially preserved following vertebrate whole genome duplications (WGDs). In contrast, functions primarily related to the immune response are over-represented among tandem/segmental duplications4. Because of the preferential survival of control genes such as transcription factors, duplications have had an important and complex influence on the evolution and expansion of gene regulatory networks (GRNs) (reviewed by Voordeckers et al.)5. Duplications of developmental control genes, and the opportunities for morphological novelty and complexity that they afford, are therefore important in the course of evolution.
The WGD events now thought to have occurred at the origin of vertebrates represent one such juncture, when the sudden genomic redundancy may have allowed the vertebrates to develop their synapomorphies6, specifically the head, neural crest, and neurogenic placodes7. The members of the GRN used to regulate the ontogenesis of the neural crest were mostly present in the chordate ancestor, but several were recruited from separate genetic pathways in vertebrates8, perhaps only possible because of the relaxation of genetic constraints afforded by WGD. Among the constituents of the ancestral neural patterning GRN is the neural plate border specification homeobox transcription factor Pax3/79, which is present in vertebrates as the ohnologues Pax3 and Pax7. These genes are necessary for neural crest induction10,11, and play later essential roles in neural crest cell migration, proliferation and differentiation (reviewed by Monsoro-Burq)10. Pax3/7 genes also play an important role in somitogenesis and myogenesis, specifying primitive myogenic cells12,13, and later maintaining a population of quiescent muscle satellite cells13 that reactivate to perform muscle repair and regeneration14,15. Pax3/7 is believed to have ancient neurogenic16 and possible myogenic17,18 functions.
Vertebrate Pax3 and Pax7 have retained a high degree of sequence similarity, and possess similar but also non-redundant roles in somitogenesis and neural plate, tube, and crest development that diverge as development progresses19,20. They also participate in clade-specific hierarchies of interdependent regulation21,22. Their functions diverge more clearly in both embryonic and adult muscle development, with Pax3 interfacing with 10-fold fewer transcription regulating sites than Pax7, with a comparatively much lower affinity for homeodomain motifs23, and conferring different properties to muscle satellite cells24. Thus, Pax3 and Pax7 illustrate how functional divergence in ohnologues following WGD may have contributed to the elaboration of vertebrate novelties and their diversification.
Comparative studies in cephalochordates, the invertebrate chordate sister group to Olfactores (tunicates and vertebrates), can provide important insight into the evolution of GRNs underlying vertebrate innovations. Unlike vertebrates, cephalochordates did not undergo whole genome duplication, have relatively ancestral-like genomes and possess conserved chordate morphology, including the presence of a dorsal hollow nerve cord, a notochord and segmented musculature (reviewed in Bertrand and Escriva)25. Previous research identified a single Pax3/7 in the amphioxus Branchiostoma floridae, the embryonic expression of which broadly recapitulates that of vertebrate Pax3 and Pax726. Pax3/7 expression has also been reported in adult muscle satellite-like cells in the amphioxus regenerative blastema27, a role that may be ancestral among bilaterians17.
Unexpectedly, we have discovered a divergent Pax3/7 gene, unlike the known Branchiostoma lanceolatum or B. floridae orthologue26, but closely resembling the gene described in B. belcheri28. Previous studies had identified only a single copy of Pax3/7 in cephalochordates, expressed in the neural plate border at the onset of neurulation, in somitogenesis, in the later development of the nervous system and larval musculature26,29,30 and in the adult segmental muscles31. We show here that the cephalochordate clade underwent a tandem duplication of the Pax3/7 gene before the most recent common ancestor of extant cephalochordates, and that the two paralogues are differentially regulated in amphioxus development. Our discovery is a clear example of developmental control gene duplication and evolution in the context of a chordate genome untouched by WGD events.
Results
Cephalochordates possess two Pax3/7 paralogues
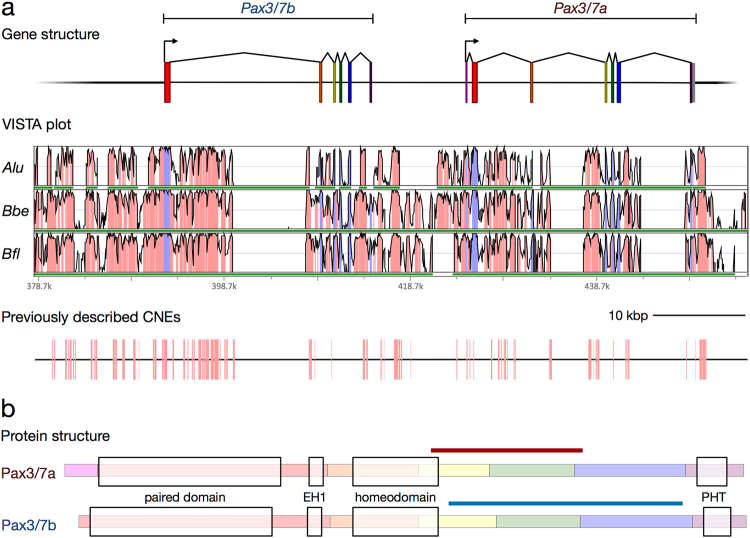
While classifying the homeobox gene complement of a regenerative transcriptome of the European amphioxus Branchiostoma lanceolatum32, we identified a transcript that much more closely resembled the previously described B. belcheri Pax3/7 homologue28 than either those of B. lanceolatum or B. floridae26. Exhaustive searches in the available Branchiostoma genomes indicated that Branchiostoma species possess two paralogues of Pax3/7, which we named in order of original discovery, Pax3/7a (described in 199926 in B. floridae, and in 200833 in B. lanceolatum) and Pax3/7b (described in 200528 in B. belcheri). Construction of gene models revealed that the genes lie adjacent to one another in the genome and are separated by approximately 10 kbs. They are also in the same orientation, and share a similar exon and domain structure (Fig. 1), indicating that the paralogues are probably the result of a tandem gene duplication. We also found Pax3/7a and Pax3/7b in the Asymmetron lucayanum transcriptome34 and genome35, suggesting that the Pax3/7 duplication event is likely to have occurred in the common ancestor of all extant cephalochordates.
Figure 1.
The structure and conservation of the cephalochordate Pax3/7 gene pair. (a) Gene models (top), VISTA plot (middle) and map of previously described B. floridae/A. lucayanum CNEs36 (bottom) on the Branchiostoma lanceolatum scaffold. Alu = A. lucayanum; Bbe = B. belcheri; Bfl = B. floridae. In the VISTA plot, the horizontal axis indicates position on the B. lanceolatum genomic scaffold; green bars indicate coverage by the genome of the labelled species, and vertical axis indicates percent identity in a 45 bp rolling window, with a range of 50% to 100%. Pink colouration indicates regions exceeding the threshold of 90%, while blue indicates exonic sequence. Details of the scaffolds used in the VISTA analysis are reported in Supplementary Table S1. Scale bar = 10,000 base pairs. (b) Protein structure of the Pax3/7 genes in amphioxus. Each exon is highlighted with a colour corresponding with its colour in (a). Conserved domains are indicated with light boxes; the paired domain, the EH1 domain (also known as the Octapeptide motif or TN), the homeodomain, and the Paired-type Homeodomain Tail45. The positions of the paralogue-specific probes on the transcripts are marked with a coloured bar. An annotated sequence alignment is presented in Supplementary Fig. S3.
Pax3/7b has lost its first exon
Previous reconstructions of Pax3/7a seem to have inadvertently combined the 5′ end of the first Pax3/7b exon with the paralogous exon of Pax3/7a, probably due to the almost complete conservation of nucleotide identity between the two genes in the paired box domain. Also, the Paired box-containing exon in Pax3/7a does not have a start codon. However, transcriptomic data led us to identify a new Pax3/7a 5′ exon containing several potential start codons. Comparison with the exon structures of other deuterostome Pax3/7 homologues indicates that the presence of an exon before the paired box-containing exon is probably the ancestral state (Supplementary Table S4), indicating that Pax3/7b has most likely lost the ancestral first exon.
The cephalochordate Pax3/7 locus is highly conserved
We aligned the relevant genomic scaffolds for B. lanceolatum, B. floridae, B. belcheri, and A. lucayanum in mVISTA (Fig. 1a), revealing high levels of conservation of non-coding sequence near the cephalochordate Pax3/7 locus (≥90% identity over the majority of the ~74 kb window shown in Fig. 1). We identified 84 B. floridae/A. lucayanum pairs of CNEs (Conserved Non-coding Elements) previously described by Yue et al., Supplementary File 635 within 20 kbs of the B. lanceolatum Pax3/7 locus (Fig. 1a), covering about 12% of the non-coding sequence in this region. No CNE was found to reoccur in this window, implying divergence in the cis-regulatory landscape of the two paralogues.
Pax3/7a and b are not direct orthologues to Pax3 and Pax7
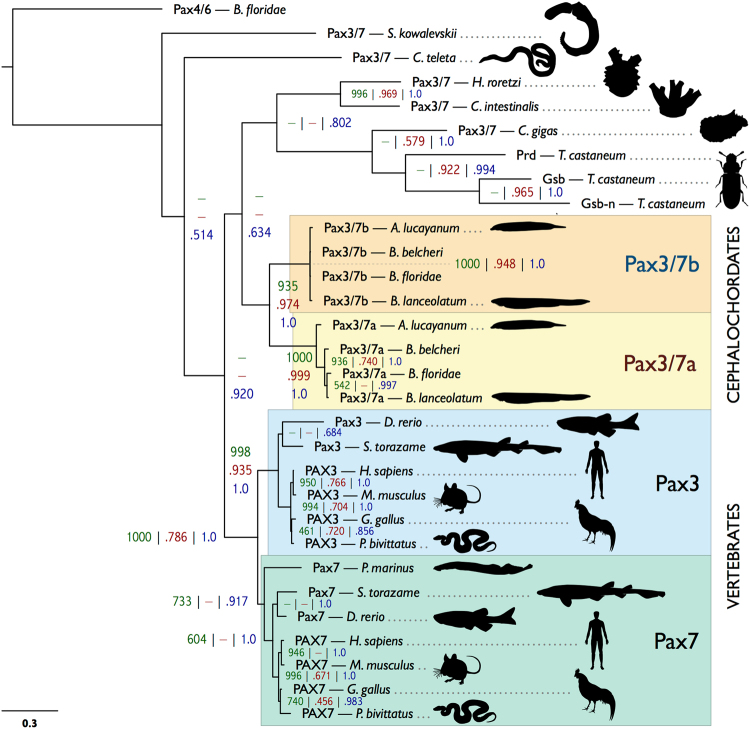
We performed a phylogenetic analysis of a selection of available Pax3/7 family sequences from vertebrates, tunicates, cephalochordates, hemichordates, annelids, molluscs and insects (Fig. 2). Support values are formatted as number of neighbour joining bootstraps out of 1000, proportion of maximum likelihood bootstraps out of 1.0, and Bayesian posterior probability out of 1.0, separated by vertical bars. Our analysis produces strongly-supported cephalochordate-only clades containing Pax3/7a (1000 | 0.999 | 1.0), Pax3/7b (1000 | 0.948 | 1.0) and Pax3/7a + Pax3/7b (935 | 0.974 | 1.0). The vertebrate sequences group similarly; Pax3 (998 | 0.935 | 1.0), Pax7 (733 | − | 0.917) and Pax3 + Pax7 (1000 | 0.736 | 1.0). Despite the more ambiguous placement of the other Pax3/7 sequences included in the analysis, these strongly-supported clades corroborate the hypothesis that the cephalochordate Pax3/7 duplication event was separate to that of the vertebrates, and that neither Pax3/7a nor Pax3/7b is a direct orthologue of either Pax3 or Pax7. The Pax3/7 sequences of tunicates (H. roretzi and C. intestinalis) and the non-chordate deuterostome sequence (S. kowalevskii) have diverged substantially, which is reflected in their phylogenetic distance from the chordate and cephalochordate genes. BLAST searches were performed in available echinoderm data, but, as in previous studies36 no Pax3/7 homologue was found.
Figure 2.
Bayesian tree of Pax3/7 genes. Support values are presented as follows: bootstraps out of 1000 from PHYLIP (dark green) | bootstraps out of 1.0 from equivalent nodes from maximum likelihood (dark red) | posterior probabilities from equivalent nodes from a Bayesian analysis (dark blue). Absence of an equivalent node in the corresponding analysis is indicated by a dash. The accession numbers of all included sequences are reported in Supplementary Table S2 (Supplementary File 1). The scale bar in the lower left corner indicates amino acid substitutions per site. B. floridae = Branchiostoma floridae; S. kowalevskii = Saccoglossus kowalevskii; C. teleta = Capitella teleta; H. roretzi = Halocynthia roretzi; C. intestinalis = Ciona intestinalis; C. gigas = Crassostrea gigas; T. castaneum = Tribolium castaneum; A. lucayanum = Asymmetron lucayanum; B. belcheri = Branchiostoma belcheri; B. lanceolatum = Branchiostoma lanceolatum; D. rerio = Danio rerio; S. torazame = Scyliorhinus torazame; H. sapiens = Homo sapiens; M. musculus = Mus musculus; G. gallus = Gallus gallus; P. bivitattus = Python bivitattus; P. marinus = Petromyzon marinus.
Pax3/7 paralogues are differentially expressed during development
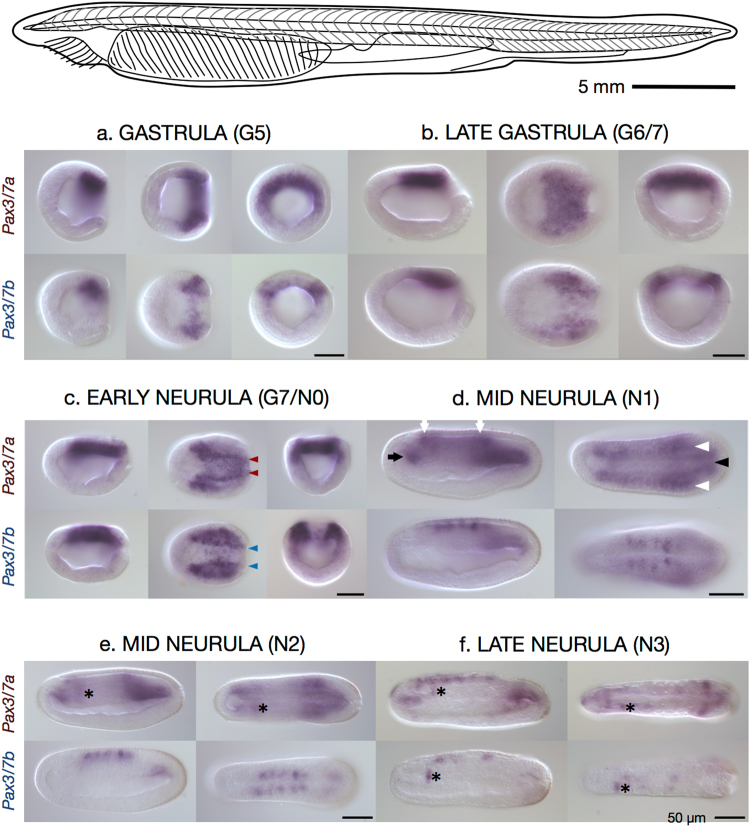
To visualise the expression of the two Pax3/7 paralogues in early development, we performed whole mount in situ hybridisation on a time-course of B. lanceolatum embryos from mid-gastrula (G5) to L2 larvae (Fig. 3, Supplementary Fig. S1) and A. lucayanum embryos (Supplementary Fig. S2) using probes designed to target the divergent 3′ end of Pax3/7a and Pax3/7b transcripts (see Fig. 1, Supplementary Fig. S3). The Pax3/7a probe covered a region with 54.5% similarity (with 33 gaps) when aligned with MAFFT to Pax3/7b, and the Pax3/7b probe covered a region with 51.1% similarity (with 72 gaps) when aligned to Pax3/7a. Our results indicate that the paralogues are differentially expressed during embryonic development.
Figure 3.
Expression of Pax3/7a and Pax3/7b in a B. lanceolatum early developmental time course. Top: Illustrative line drawing of adult B. lanceolatum. Scale bar ≈5 mm. Below: Whole mount in situ hybridisation images of Pax3/7a-specific probe (top row of each block) and Pax3/7b-specific probe (bottom row of each block) in B. lanceolatum embryos. Views are presented, in left-to-right order: lateral, dorsal, and blastoporal (gastrula and early neurula only). Lateral and dorsal views are oriented with the anterior to the left. (a) Gastrula, G5, 10 hours post fertilisation (hpf). (b) Late gastrula, G6/7, 12 hpf. (c) Early neurula, G7/N0, 14 hpf. (d,e) Mid neurulae: N1, 16 hpf and N2, 21 hpf. (f) Late neurula, N3, 24 hpf. Domains of expression are marked throughout as follows: coloured arrowheads — differentially patterned neural plate border expression; black arrow — expression in the anterior mesodermal tissue; white arrows — in the anterior end and posterior of the neural tube; white arrowheads — in the postero-lateral somitic tissue; black arrowhead — in the postero-medial notochord tissue; asterisk — (placed immediately posteriorly to) the sinistral domain of expression found in both paralogues and in A. lucayanum. Scale bars = 50 micrometres.
Expression of Pax3/7a
In mid-gastrulae (G5, Fig. 3a), Pax3/7a is expressed in a semicircular band in the dorsal endoderm of the blastoporal lip. This expression pattern remains relatively diffuse in the late gastrula (G6/7, Fig. 3b), but by the early neurula (N0, Fig. 3c) the expression domain has become condensed into lines running symmetrically either side of the midline (Fig. 3c, red arrowheads) with enlarged anterior patches, though weak expression persists throughout the posterior. By the hatchling neurula (N1, Fig. 3d), Pax3/7a has diffuse expression with greater concentration in five indistinct, bilaterally symmetrical areas; the anterior mesodermal tissue (black arrow), the anterior end and posterior of the neural tube (white arrows), the postero-lateral somitic tissue (white arrowheads), and the postero-medial notochord tissue (black arrowhead). These expression domains continue with little change through to the mid neurula (N2, Fig. 3e), except for the appearance of a distinct domain of asymmetrical Pax3/7a expression in the anterior (marked throughout by an asterisk placed just posteriorly), which is consistently absent or very weak on the right side. In the late neurula (N3, Fig. 3f), Pax3/7a expression has become condensed into the anterior and posterior mesodermal regions, and into the left anterior somite. Patchy and granular neural regions of expression have also appeared. The asymmetrical domain persists into the early larva (L1, Supplementary Fig. S1a) while the other domains of expression are substantially reduced such that only a few anterior neural and the posterior mesodermal domains are present. This pattern continues in the L2 and L3 larvae (Supplementary Fig. S1b and c), with faint, patchy neural expression reappearing in the latter stage.
Expression of Pax3/7b
In mid-gastrulae (G5, Fig. 3a), Pax3/7b is expressed in smaller lateral patches in the dorsum in both germ layers. This lateral expression pattern continues in the late gastrula (G6/7, Fig. 3b); by the early neurula (N0, Fig. 3c) the interior lateral borders of the expression domain have become strongly resolved (Fig. 3c, blue arrowheads), though weak medial expression continues. Pax3/7b expression overlaps with Pax3/7a in the posterior regions but with a much weaker signal. By the N1 stage (Fig. 3d), in contrast to Pax3/7a, there are five distinct, symmetrical domains of Pax3/7b expression in the dorsolateral neural tube. These spots are flanked at their anterior and posterior limits by the weaker, more diffuse regions of Pax3/7a expression (white arrows). This expression pattern continues with little change through to the mid neurula (N2, Fig. 3e). By stage N3 (Fig. 3f), the neural regions of expression are reduced in size and number, retaining only the two anterior-most and posterior-most spots, while the strong asymmetrical domain of expression in the left anterior somite previously distinguished by Pax3/7a expression is now also labeled by Pax3/7b (asterisks). This domain persists with strong expression into the early larva (L1, Supplementary Fig. S1a) while expression ceases elsewhere in the L2 and L3 larvae (Supplementary Fig. S1b and c).
Discussion
Gene duplication is an important mechanism in evolution, providing a potent source of new genetic material on which evolution can act outside the constraints on single-copy genes. Transcription factors stand out as a particularly important subset of retained and adapted paralogous genes. Paralogue divergence includes subfunctionalisation and neofunctionalisation of binding specificity and motif recognition, upstream regulatory control, and cofactor interaction, which all provide opportunities for more intricate spatiotemporal expression control and the potential for the generation of novel gene regulatory networks and morphology5.
The two rounds of whole genome duplication (2R-WGD) at the base of the vertebrate lineage37 provided an ample source of stoichiometrically-balanced raw genetic material, possibly facilitating the elaboration of vertebrate novelties including the head, neural crest, and neurogenic placodes6,7. In contrast, cephalochordate genomes bear no indications of paleopolyploidy events37–39, and share more similarities in terms of architecture and gene content with the chordate ancestral genome than other extant chordate clades40. Cephalochordates therefore have many fewer paralogues than vertebrates, though both RNA-mediated and DNA-mediated duplications have been described. Among the latter, homeobox genes are most numerous; paralogues have been found in Evx41,42, Emx41,43,44, Mnx, Vent, Nk1, Nedx, Uncx, Lhx2/9, Irx, Pou344, and Hox9-1539,45, many of which are the result of small-scale tandem duplications. Of these, only Vent1 and Vent2 have been the subject of detailed functional assays, which established their cis- and trans-regulation in the amphioxus dorsoventral patterning regulatory network46 and their expression in pharmacologically manipulated embryos47.
Our data from three species of Branchiostoma and Asymmetron lucayanum, a representative of the earliest branching of the extant amphioxus genera, support the idea that tandem gene duplication may have been an important mechanism for generating cell type diversity in the cephalochordate ancestor. We report that amphioxus possess two paralogues of Pax3/7, a gene notable for its functions in neural plate border specification, its vertebrate roles in neural crest and placode specification, and for its involvement in somitogenesis, myogenesis and the population of regenerative muscle satellite cells possibly common to all bilaterians17. We confirm that this duplication predates the modern cephalochordate radiation but post-dates the divergence from other chordates, implying that the chordate ancestor had a single copy.
One of our key findings is that Pax3/7a and Pax3/7b diverged symmetrically but heterogeneously between duplication and the cephalochordate radiation (Fig. 2). They share very strong nucleotide sequence conservation and 100% amino acid sequence identity in the paired domain, EH1/Octapeptide motif and homeodomain, possibly the result of gene conversion. In contrast, they have diverged substantially in the linker regions, the N-terminus (where Pax3/7b seems to have lost an exon) and the four exons of the C-terminus. The paralogues have changed little since their divergence, both in coding sequence and local CNEs; of the pair, Pax3/7a has changed more since the Asymmetron/Branchiostoma speciation events, indicating it might be under slightly relaxed selection, but has a more prototypical PHT domain48 (Supplementary Fig. S3; Supplementary File 3, residues 447–467 at positions 801–829), while Pax3/7b is more conserved among species. Pronounced evolutionary asymmetry is common amongst tandem paralogues (reviewed by Holland et al.)49, for instance, in AmphiEvx; however, examples in which asymmetry is not observed have also been documented (AmphiEmx).
Although cephalochordates are considered to be slow-evolving, the pattern we observe in paralogue divergence is also consistent with the recent estimate that the crown cephalochordate node dates to only 38.8–46.0 million years ago (MYA), in contrast to previous results placing it ~120–250 MYA (see Igawa et al.50 and references therein). Based on their calibration date of the cephalochordate/Olfactores split approximately 550 MYA, the duplication, fixation, fate-determination and preservation phases of paralogue evolution (see Innan and Kondrashov)51 all occurred in the ~500 MYA interval during which no evident radiation occurred. Comparatively rapid change and quicker preservation is considered typical of tandem duplications5, although as Pax3/7 genes are transcription factors involved in development, and specifically neurogenesis52, their sequence and expression domain change may have been severely constrained.
Symmetry of sequence evolution rate between paralogues is considered indicative of subfunctionalisation53. The evolutionary trajectory of cephalochordate Pax3/7 duplicates, based on the symmetry of sequence change evident in Fig. 2, seems to accord with the duplication-degeneration-complementation (DDC) model of Force et al.3 or the specialisation model of Hughes2. According to these models, the duplicated pair, under relaxed purifying selection, accumulates either mutations that complementarily degrade (DDC) or improve (specialisation) their capacity to perform subsets of their pre-duplication function, until the loss of either paralogue is deleterious. The only non-duplicated chordate or deuterostome outgroups for ancestral Pax3/7 function are found in the tunicates and hemichordates. However, both groups have a highly divergent Pax3/7 sequence, and the former of which has a very derived genome and morphology. Consequently, it is difficult to determine the exact set of ancestral functions of the Pax3/7 pro-orthologue in the chordate ancestor. Nevertheless, a conserved role in neural border specification is highly probable, given enrichment of Pax3/7 in lateral neuroblasts in a number of bilaterians9.
Although the DNA-binding domains of Pax3/7a and Pax3/7b are identical, it is likely that the differences in the C-terminus and in the linker regions between the conserved domains are sufficient to alter their functionality. Amino-terminal sequence changes have been shown to affect the binding specificity of DNA-binding domains and homeodomains in general18,54 and Pax genes specifically (reviewed by Mayran et al.)52.
Small sequence changes have the potential to differentially modify the binding affinity of the paired domain and homeodomain, the binding modality of the paired subdomains, and subnuclear localisation56. The modest differences between Pax3 and Pax7 sequence, located mostly in the C-terminus, are enough to produce substantial differences in target activation in myogenesis23 (Mayran et al.55 and references therein). The extent of these substantial functional effects caused by the minor differences in mutants, splice variants and between vertebrate Pax3 and Pax7 is an indication that Pax3/7a and Pax3/7b, which have diverged more than Pax3/Pax7, probably behave differently with regard to target recognition and interaction with cofactors. Such sequence change has been highlighted as an important but under-appreciated mechanism in the evolution of developmental GRNs57.
Regardless of putative differences in downstream activity, Pax3/7a and Pax3/7b are expressed differently during gastrulation and neurulation in B. lanceolatum, demonstrating that the paralogues have diverged in their cis-regulation. Pax3/7a and Pax3/7b are expressed in partially overlapping but distinct domains in the neural plate (G5 to N0, Fig. 3a–c, red and blue arrowheads), presumably as the result of modification of an ancestral neural plate domain. Pax3/7a is expressed throughout the dorso-posterior mesoderm prior to neurulation (G5 and G6/7, Fig. 3a,b) while Pax3/7b is restricted to smaller, bilaterally symmetrical dorso-posterior regions in both the mesoderm and ectoderm, consistent with a role in the initial specification of the neural plate border. Distinct lateral lines of expression do appear in Pax3/7a in the late gastrula/early neurula (G7/N0, Fig. 3c), but diffuse expression remains throughout the posterior. By the mid-neurula, the paralogues seem to have switched to a different expression programme, one in which their expression patterns have the least overlap. Particularly notable are the tight, defined neural spots of Pax3/7b and the appearance of the asymmetrical, sinistral domain (the anterior somite26) of expression that first appears in Pax3/7a (left of asterisk throughout, N2, Fig. 3e) and later appears in Pax3/7b (N3, Fig. 3f). As the embryo becomes a larva, the two expression patterns converge until both expression patterns are largely restricted to the asymmetrical domain (L1 and 2, Supplementary Fig. S1a–c). Thus, divergence between duplicate expression patterns increases during gastrulation and early neurulation, peaking at mid-neurula stages, consistent with function partitioning. Although we still know very little about Asymmetron lucayanum developmental gene expression, our data indicate similar results for Pax3/7 paralogues in this species (Supplementary Fig. S2). This is currently the only example in cephalochordates in which a gene duplication event has been shown to predate the divergence of extant lineages and for which expression data exist in more than one genus.
Our results broadly recapitulate previous Pax3/7 expression data from B. lanceolatum (Fig. 3H,I and J of Somorjai et al.)33, considering that the latter used a probe with probable cross-reactivity between the 5′ conserved region of Pax3/7a and Pax3/7b. In contrast, the B. lanceolatum expression patterns are not a perfect subset of the Pax3/7(a) domains reported for B. floridae (Fig. 5 of Holland et al.)26, who used a similarly cross-reactive probe. Potentially missing from our patterns are the anterior somitic and mesodermal expression (Fig. 5F,G,I and K of Holland et al.)26, the distinct anterior neural spot (arrow, Fig. 5K,M,P and Q of Holland et al.)26 and the larval axial musculature and notochord expression (Fig. 5M,P, and Q of Holland et al.)26. Minor discrepancies are not unusual, but significant differences among Branchiostoma species are rare33. It is possible that these differences are caused by the general variability between probes for the same target, Pax gene probe cross-reactivity, or experimental sensitivity. The probes we used were by necessity relatively short in order to limit possible cross-reaction of highly conserved regions, but the expression patterns we observed are highly specific and reproducible, suggesting they reflect the core domains of Pax3/7a and Pax3/7b.
In contrast to what we see in amphioxus, differences between vertebrate Pax3 and Pax7 early developmental expression are much less pronounced, to the extent that they have ‘swapped’ expression profiles during evolution (see Monsoro-Burq10, and references therein). Pax3/Pax7 appear in the neural plate border during neural induction in the early gastrula, and intensify at the lateral edges to mark the dorsal edge of the closing neural tube, a pattern comparable to late gastrula/early neurula expression in amphioxus. Pax3 and/or Pax7 are also expressed throughout the posterior dorsal neuraxis, an approximate analogue of the neural spots in Pax3/7b and later Pax3/7a, though these spots are more spatiotemporally restricted.
While Pax3 and Pax7 appear to play semi-redundant roles in neural development, they diverge in function in vertebrate myogenesis (reviewed by Buckingham & Relaix)58. Pax3 acts broadly from the onset of myogenesis in the presomitic mesoderm to the dermomyotome, while Pax7 expression is later and restricted to a dermomyotomal subdomain. These PAX3/PAX7 positive cells form a proliferative muscle progenitor population that eventually positions itself underneath the basal lamina on the muscle fibres. In the adult, these cells become a heterogenous population of quiescent satellite cells; all are maintained by Pax7 expression, but some also expresses Pax3, which is known in this context to be an inadequate substitute, binding 10-fold fewer targets, most of which are also targets of PAX7. During myogenesis, Pax3 and Pax7 seem to be responsible for maintaining the cells in a proliferative/quiescent but undifferentiated state. Lack or cessation of Pax3 or Pax7 expression in a cell can lead to apoptosis or cell cycle exit and muscle differentiation via MyoD, depending on the precise context.
Although the later myogenic roles of amphioxus Pax3/7 genes are yet to be thoroughly characterised, at least one of the paralogues is known to be expressed in adult muscle, as Pax3/7b has been amplified from adult B. belcheri segmental muscle31. Whether both paralogues are involved in adult muscle development redundantly, or rather show temporal or tissue-specific patterns of expression (similarly to Pax3 and Pax7 in post-embryonic muscle development and regeneration in mice) is still unclear. Our initial identification of Pax3/7b transcripts in a tail blastema transcriptome clearly identifies a role in the adult regeneration process. However, previous characterization of Pax3/7 in a population of satellite-like cells and the nerve cord during tail regeneration utilized a cross-reactive in situ hybridisation probe27. We therefore cannot currently rule out changes in paralogue function during postembryonic processes in amphioxus. Future studies are required to determine to what extent divergence has occurred in expression, downstream targets, and interaction with co-factors in both myogenic and neural contexts.
Amphioxus Pax3/7 has been considered a useful proxy for understanding the properties and deployment of the chordate proto-Pax3/7. Our findings showing independent vertebrate and cephalochordate Pax3/7 duplications – and the resulting functional and regulatory divergence – offer new insight into genomic constraint/plasticity, and evolvability of gene duplicates and GRNs in different duplication contexts. In amphioxus, tandem duplication and divergence of Pax3/7 has resulted in a subfunctionalisation (and possibly neofunctionalisation) of ancestral neural plate border9 and muscle-related17,18 functions, many of which parallel those seen in vertebrate Pax3 and Pax7 following WGD. Dissecting the regulatory landscape of Pax3/7 genes in amphioxus, including the function of the CNEs partitioned between paralogues, should shed further light on genome architecture evolution in chordates.
Conclusions
We show that cephalochordates, which are considered to be a significant outgroup to vertebrates in the study of the evolution of the neural crest GRN, have two Pax3/7 paralogues where it was previously thought that this family was represented by a single-copy gene in these animals. This discovery has implications both for previous and future studies of amphioxus development and regeneration, and for vertebrate studies in which cephalochordates are used as an outgroup. The amphioxus Pax3/7 gene pair also offers a tantalising and tractable example of cis-regulatory and sequence subfunctionalisation after tandem duplication of a developmental transcription factor involved in the development of key chordate features.
Methods
Genomic & transcriptomic analysis
A tBLASTn59 search of a transcriptome, generated from the pre-amputation and blastemal tissues of a regenerating B. lanceolatum post-anal tail (14 dpa/stage 2 sensu Somorjai et al.)27 assembled with developmental transcriptomic data from Oulion et al.60, for homeodomains selected from HomeoDB61 retrieved a partial Pax3/7b sequence. Subsequent identification and comparison was done by alignment in Jalview 2.x62. The exon structures of Pax3/7a and Pax3/7b were manually predicted with reference to tBLASTn searches of the known sequences against the available genomes: the B. lanceolatum draft assembly (Bl71nemr) (European Amphioxus Genome Consortium), the B. floridae reference genome version 2.037, the B. belcheri draft assembly38 (HapV2), and the A. lucayanum draft assembly35, and used to manually produce diagrams of the gene and protein structure, in reference to domains predicted by the Conserved Domain Database63 and the Pax gene conserved regions identified by Vorobyov & Horst48.
Transcriptomic support for both cephalochordate Pax3/7 paralogues was obtained using tBLASTn and MEGABLAST searches of the A. lucayanum transcriptome34, B. floridae cDNA library29, a B. lanceolatum SRA (BioProject: PRJNA285432) and the unpublished regenerative transcriptome.
Visualisation
Curated genomic sequences from B. lanceolatum, B. floridae, B. belcheri and A. lucayanum were uploaded to the web interface for mVISTA64 along with manually predicted annotations of the B. lanceolatum scaffold. These were aligned using the AVID alignment algorithm65 and the alignment was visualised with 45 bp calculation window, 45 bp minimum conserved width, and 90% conserved identity threshold parameters. Full details of the scaffolds and curation are presented in Supplementary Table S1.
The region of the B. lanceolatum genome represented in the VISTA plot was used as a query for a BLASTn search against the Conserved Non-Coding Elements database presented in Supplementary File 6 of Yue et al.35. Matching sequences were retrieved from the CNE database, and the sequences aligned back to the query using MAFFT–addfragments mode66. Spreadsheet tools were used to extract positional information and to generate the visualisation of distribution.
Exon positions for various deuterostome Pax3/7 genes were extracted from their NCBI records listed under the accession numbers in Supplementary Table S4; protein domain positions were predicted using the Conserved Domain Database63 and manually corrected where homeodomain prediction was too short.
Phylogenetic analysis
Protein sequences were predicted from the genomes of B. lanceolatum, B. floridae, B. belcheri, and A. lucayanum with reference to the published B. floridae Pax3/7a (EEN66816.1) and B. belcheri Pax3/7b (ABK54280.1) sequences. Gaps in the A. lucayanum gene models due to incomplete coverage were partially filled by manual assembly of the results of a tBLASTn search of the B. lanceolatum Pax3/7a and b protein sequences against the A. lucayanum SRA archive (SRR1138336). Complete coverage of A. lucayanum Pax3/7b was not possible.
Protein sequences for Homo sapiens, Mus musculus, Gallus gallus, Python bivittatus, Danio rerio, and Scyliorhinus torazame Pax3 and Pax7; Petromyzon marinus Pax7; Halocynthia roretzi, Saccoglossus kowalevskii, Crassostrea gigas, and Capitella teleta Pax3/7; and Tribolium castaneum Paired, Gooseberry, and Gooseberry-neuro were retrieved from the NCBI; all accession numbers for the phylogeny are reported in Supplementary Table S3. Sequences were aligned in Jalview using the MAFFT alignment algorithm with default settings66 and manually corrected.
Model selection was performed in ModelGenerator v0.8567 using default settings and 4 gamma categories. The model recommended (JTT + G + F) or its closest possible equivalent was selected in all subsequent phylogenetic analyses. A neighbour-joining analysis was performed in PHYLIP 3.6968, a maximum-likelihood analysis in MEGA-CC69, and a Bayesian analysis on the CIPRES Science Gateway70, using MrBayes 3.2.671 on XSEDE. Full details of settings used in the analysis are presented in the Supplementary Note. The support values for equivalent nodes between analyses were mapped using the Python script in Supplementary File 3 onto the Bayesian tree and the consensus tree output was visualised in FigTree 1.4.2.
Embryo collection
Adult European amphioxus (Branchiostoma lanceolatum) were collected from Argeles-sur-mer (France), kept in a semi-closed circulating system at 16.5 °C, and induced to spawn as described previously72. Populations of A. lucayanum were collected from Bimini (Bahamas), kept in filtered seawater at 25 °C, and induced to spawn as described previously35. Embryos for in situ hybridisation were fixed at relevant time points in fresh 4% PFA in MOPS salts (0.1 M MOPS, 2 mM MgSO4, 1 mM EGTA, & 0.5 M NaCl), transferred into 70% Ethanol and stored at −20 °C. Branchiostoma lanceolatum embryos were staged according to modifications suggested by Zhang et al.73.
Cloning and probe synthesis
RNA was extracted from B. lanceolatum embryos fixed at a selection of developmental stages using TRIsure (Bioline) using the supplier’s protocol. A. lucayanum embryos fixed in RNAlater (Invitrogen) were transferred to TRIsure and treated similarly. cDNA libraries were produced using the Tetro cDNA Synthesis kit (Bioline). Gene fragments for probe synthesis were amplified by PCR using gene-specific primers (Supplementary Table S2) designed using B. lanceolatum transcriptomic (see above) and genomic sequences from B. lanceolatum and A. lucayanum cDNA. The amplicons were ligated into pGEM-T Easy vector (Promega) and transformed into the XL10-Gold (Stratagene) competent E. coli cell strain using standard heat shock protocols. Selected clones were cultured and extracted using peqGOLD (Peqlab) or Promega plasmid miniprep kits and sequenced for verification using Universal M13F (5′-GTAAACGACGGCCAGT-3′) and M13R (5′-AACAGCTATGACCATG-3′) primers at the University of Oxford Zoology Sequencing Service. Probe template was produced using PCR with M13 primers. Bands were verified using agarose electrophoresis and precipitated using sodium acetate (3 M, pH 5.2) and ethanol. DIG-labelled (Roche) antisense probes were transcribed in vitro using T3 and SP6 enzymes as appropriate, following standard protocols.
In situ hybridisation
Whole mount in situ hybridisation was performed as previously reported33. In brief, embryos fixed in 4% paraformaldehyde and stored at −20 °C were rehydrated in NaPBS + 0.1% Tween and permeabilised with proteinase K, followed by post-fixation and acetic anhydride treatment to reduce background. A paralogue- and species-specific DIG-labelled probe was hybridised overnight at 65 °C to target mRNA in the embryos. Excess probe was washed back out through decreasing concentrations of formamide before being treated with RNAses to reduce background. The embryos were blocked against non-specific antibody binding and exposed to alkaline phosphatase-associated anti-DIG antibodies overnight at 4 °C. The embryos were finally stepped into buffer and NBT and BCIP (alkaline phosphatase substrates) introduced.
Equipment and settings
To capture the images used in Fig. 3 and Supplementary Figs S1 and S2, embryos were mounted in 95% glycerol/5% PBS and examined under a Leitz DMRB microscope (Leica Microsystems) with Nomarski optics. Images were captured using a Retiga 2000R camera in the QCapture software suite (QImaging) and processed in the GNU Image Manipulation Package (GIMP) and Inkscape.
Ethics statement
No specific permits were required for collection of animals used in this study. All procedures were in compliance with regulations for the experimental use of non-cephalopod invertebrates in the UK and EU (DIRECTIVE 2010/63/EU OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 22 September 2010 on the protection of animals used for scientific purposes).
Statement of data availability
Full sequences for the genes described in the current study are available from the NCBI database (accession numbers in Supplementary Table S3).
Electronic supplementary material
Acknowledgements
We thank Simon Dailey for support with in situ hybridizations, past and present members of the Somorjai and Ferrier laboratories for discussion, and Linda and Nick Holland of the Scripps Institution of Oceanography (U.C. San Diego) for their help in collecting Asymmetron material. Funding for this research was provided by the European Union’s Horizon 2020 research and innovation programme under grant agreement number 654428 (“CORBEL”) and a MASTS (Marine Alliance for Science and Technology Scotland) PECRE young investigator award to IMLS. This work was supported by the Branchiostoma lanceolatum genome consortium, which provided access to the Branchiostoma lanceolatum genome sequence.
Author Contributions
I.M.L.S. designed and funded the study. I.M.L.S. and D.E.K.F. supervised T.B.O. T.B.O. conducted bioinformatics analyses and expression experiments. I.M.L.S. and T.B.O. collected embryonic material. I.M.L.S. and T.B.O. wrote the paper based on initial drafts by T.B.O. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-27700-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ohno, S. Evolution by gene duplication. (Springer Science & Business Media, 1970).
- 2.Hughes AL. The evolution of functionally novel proteins after gene duplication. Proceedings of the Royal Society of London. Series B. Biological Sciences. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- 3.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huminiecki L, Heldin CH. 2R and remodeling of vertebrate signal transduction engine. BMC Biology. 2010;8:146. doi: 10.1186/1741-7007-8-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voordeckers K, Pougach K, Verstrepen KJ. How do regulatory networks evolve and expand throughout evolution? Systems biology Nanobiotechnology. 2015;34:180–188. doi: 10.1016/j.copbio.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: Comparative analyses in vertebrates. Genome Research. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: A new head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 8.Meulemans D, Bronner-Fraser M. Central role of gene cooption in neural crest evolution. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution. 2005;304:298–303. doi: 10.1002/jez.b.21047. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, et al. Conserved gene regulatory module specifies lateral neural borders across bilaterians. Proceedings of the National Academy of Sciences. 2017;114:E6352–E6360. doi: 10.1073/pnas.1704194114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monsoro-Burq AH. PAX transcription factors in neural crest development. Paramutation & Pax Transcription Factors. 2015;44:87–96. doi: 10.1016/j.semcdb.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Basch ML, Bronner-Fraser M, García-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–222. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- 12.Kassar-Duchossoy L, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes & Development. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Lin G, Slack JMW. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development. 2006;133:2303–2313. doi: 10.1242/dev.02397. [DOI] [PubMed] [Google Scholar]
- 15.Morrison JI, Lööf S, He P, Simon A. Salamander limb regeneration involves the activation of a multipotent skeletal muscle satellite cell population. J Cell Biol. 2006;172:433–440. doi: 10.1083/jcb.200509011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navet S, et al. The Pax gene family: Highlights from cephalopods. PLOS ONE. 2017;12:e0172719. doi: 10.1371/journal.pone.0172719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konstantinides N, Averof M. A common cellular basis for muscle regeneration in arthropods and vertebrates. Science (New York, N.Y.) 2014;343:788–791. doi: 10.1126/science.1243529. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Matthews KS, Bondos SE. Internal regulatory interactions determine DNA binding specificity by a Hox transcription factor. Journal of Molecular Biology. 2009;390:760–774. doi: 10.1016/j.jmb.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relaix F, Rocancourt D, Mansouri A, Buckingham M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes & Development. 2004;18:1088–1105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JA, Zembrzycki A, Mansouri A, Ziman M. Pax7 is requisite for maintenance of a subpopulation of superior collicular neurons and shows a diverging expression pattern to Pax3 during superior collicular development. BMC Developmental Biology. 2008;8:62. doi: 10.1186/1471-213X-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maczkowiak F, et al. The Pax3 and Pax7 paralogs cooperate in neural and neural crest patterning using distinct molecular mechanisms in Xenopus laevis embryos. Developmental Biology. 2010;340:381–396. doi: 10.1016/j.ydbio.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agoston Z, Li N, Haslinger A, Wizenmann A, Schulte D. Genetic and physical interaction of Meis2, Pax3 and Pax7 during dorsal midbrain development. BMC Developmental Biology. 2012;12:10. doi: 10.1186/1471-213X-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soleimani VD, et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Developmental Cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q, et al. PAX3 + skeletal muscle satellite cells retain long-term self-renewal and proliferation. Muscle & Nerve. 2016;54:943–951. doi: 10.1002/mus.25117. [DOI] [PubMed] [Google Scholar]
- 25.Bertrand S, Escriva H. Evolutionary crossroads in developmental biology: amphioxus. Development. 2011;138:4819–4830. doi: 10.1242/dev.066720. [DOI] [PubMed] [Google Scholar]
- 26.Holland LZ, Schubert M, Kozmik Z, Holland ND. AmphiPax3/7, an amphioxus paired box gene: Insights into chordate myogenesis, neurogenesis, and the possible evolutionary precursor of definitive vertebrate neural crest. Evolution & Development. 1999;1:153–165. doi: 10.1046/j.1525-142x.1999.99019.x. [DOI] [PubMed] [Google Scholar]
- 27.Somorjai IML, Somorjai RL, Garcia-Fernàndez J, Escrivà H. Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:517–522. doi: 10.1073/pnas.1100045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang W, Xu H-L, Lin L-P, Su B, Wang Y-Q. Construction of a BAC library for Chinese amphioxus Branchiostoma belcheri and identification of clones containing Amphi-Pax genes. Genes & Genetic Systems. 2005;80:233–236. doi: 10.1266/ggs.80.233. [DOI] [PubMed] [Google Scholar]
- 29.Yu J-K, et al. A cDNA resource for the cephalochordate amphioxus Branchiostoma floridae. Development Genes and Evolution. 2008;218:723–727. doi: 10.1007/s00427-008-0228-x. [DOI] [PubMed] [Google Scholar]
- 30.Kozmik Z, et al. Pax–Six–Eya–Dach network during amphioxus development: Conservation in vitro but context specificity in vivo. Developmental Biology. 2007;306:143–159. doi: 10.1016/j.ydbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Zhang Q, Wang W, Wang Y. Spatiotemporal expression of Pax genes in amphioxus: Insights into Pax-related organogenesis and evolution. Science China Life Sciences. 2010;53:1031–1040. doi: 10.1007/s11427-010-4040-2. [DOI] [PubMed] [Google Scholar]
- 32.Dailey, S. C. Evolutionary developmental and genomic insights from a tail regeneration transcriptome of the cephalochordate Branchiostoma lanceolatum, PhD thesis, University of St Andrews (2017).
- 33.Somorjai I, Bertrand S, Camasses A, Haguenauer A, Escriva H. Evidence for stasis and not genetic piracy in developmental expression patterns of Branchiostoma lanceolatum and Branchiostoma floridae, two amphioxus species that have evolved independently over the course of 200 Myr. Development Genes and Evolution. 2008;218:703–713. doi: 10.1007/s00427-008-0256-6. [DOI] [PubMed] [Google Scholar]
- 34.Yue J-X, Yu J-K, Putnam NH, Holland LZ. The transcriptome of an amphioxus, Asymmetron lucayanum, from the Bahamas: A window into chordate evolution. Genome Biology and Evolution. 2014;6:2681–2696. doi: 10.1093/gbe/evu212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue J-X, et al. Conserved noncoding elements in the most distant genera of cephalochordates: The Goldilocks principle. Genome Biology and Evolution. 2016;8:2387–2405. doi: 10.1093/gbe/evw158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howard-Ashby M, et al. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Developmental Biology. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 37.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 38.Huang S, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nature Communications. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Research. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louis A, Roest Crollius H, Robinson-Rechavi M. How much does the amphioxus genome represent the ancestor of chordates? Briefings in Functional Genomics. 2012;11:89–95. doi: 10.1093/bfgp/els003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minguillón C, Ferrier DEK, Cebrián C, Garcia-Fernàndez J. Gene duplications in the prototypical cephalochordate amphioxus. Gene. 2002;287:121–128. doi: 10.1016/S0378-1119(01)00828-9. [DOI] [PubMed] [Google Scholar]
- 42.Ferrier DE, Minguillón C, Cebrián C, Garcia-Fernàndez J. Amphioxus Evx genes: Implications for the evolution of the midbrain-hindbrain boundary and the chordate tailbud. Developmental Biology. 2001;237:270–281. doi: 10.1006/dbio.2001.0375. [DOI] [PubMed] [Google Scholar]
- 43.Williams NA, Holland PWH. An amphioxus Emx homeobox gene reveals duplication during vertebrate evolution. Mol Biol Evol. 2000;17:1520–1528. doi: 10.1093/oxfordjournals.molbev.a026251. [DOI] [PubMed] [Google Scholar]
- 44.Takatori N, et al. Comprehensive survey and classification of homeobox genes in the genome of amphioxus. Branchiostoma floridae. Development Genes and Evolution. 2008;218:579–590. doi: 10.1007/s00427-008-0245-9. [DOI] [PubMed] [Google Scholar]
- 45.Feiner N, Ericsson R, Meyer A, Kuraku S. Revisiting the origin of the vertebrate Hox14 by including its relict sarcopterygian members. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2011;316B:515–525. doi: 10.1002/jez.b.21426. [DOI] [PubMed] [Google Scholar]
- 46.Kozmikova I, Smolikova J, Vlcek C, Kozmik Z. Conservation and Diversification of an Ancestral Chordate Gene Regulatory Network for Dorsoventral Patterning. PLOS ONE. 2011;6:e14650. doi: 10.1371/journal.pone.0014650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozmikova I, Candiani S, Fabian P, Gurska D, Kozmik Z. Essential role of Bmp signaling and its positive feedback loop in the early cell fate evolution of chordates. Developmental Biology. 2013;382:538–554. doi: 10.1016/j.ydbio.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Vorobyov E, Horst J. Getting the proto-Pax by the tail. Journal of Molecular Evolution. 2006;63:153–164. doi: 10.1007/s00239-005-0163-7. [DOI] [PubMed] [Google Scholar]
- 49.Holland, P. W. H., Marlétaz, F., Maeso, I., Dunwell, T. L. & Paps, J. New genes from old: Asymmetric divergence of gene duplicates and the evolution of development. Phil. Trans. R. Soc. B372 (2017). [DOI] [PMC free article] [PubMed]
- 50.Igawa T, et al. Evolutionary history of the extant amphioxus lineage with shallow-branching diversification. Scientific Reports. 2017;7:1157. doi: 10.1038/s41598-017-00786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Innan H, Kondrashov F. The evolution of gene duplications: Classifying and distinguishing between models. Nature Reviews Genetics. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 52.Roux J, Liu J, Robinson-Rechavi M. Selective constraints on coding sequences of nervous system genes are a major determinant of duplicate gene retention in vertebrates. Molecular Biology and Evolution. 2017;34:2773–2791. doi: 10.1093/molbev/msx199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yampolsky LY, Bouzinier MA. Faster evolving Drosophila paralogs lose expression rate and ubiquity and accumulate more non-synonymous SNPs. Biology Direct. 2014;9:2. doi: 10.1186/1745-6150-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzeng S-R, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 55.Mayran A, Pelletier A, Drouin J. Pax factors in transcription and epigenetic remodelling. Paramutation & Pax Transcription Factors. 2015;44:135–144. doi: 10.1016/j.semcdb.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Corry GN, et al. The PAX3 paired domain and homeodomain function as a single binding module in vivo to regulate subnuclear localization and mobility by a mechanism that requires base-specific recognition. Journal of Molecular Biology. 2010;402:178–193. doi: 10.1016/j.jmb.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 57.Cheatle Jarvela, A. M. & Hinman, V. F. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. EvoDevo6, (2015). [DOI] [PMC free article] [PubMed]
- 58.Buckingham M, Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Paramutation & Pax Transcription Factors. 2015;44:115–125. doi: 10.1016/j.semcdb.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oulion S, Bertrand S, Belgacem MR, Le Petillon Y, Escriva H. Sequencing and analysis of the Mediterranean amphioxus (Branchiostoma lanceolatum) transcriptome. PLoS ONE. 2012;7:e36554. doi: 10.1371/journal.pone.0036554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong Y, Holland PW. HomeoDB2: Functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evolution & Development. 2011;13:567–568. doi: 10.1111/j.1525-142X.2011.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research. 2015;43:D222–226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Research. 2004;32:W273–279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bray N, Dubchak I, Pachter L. AVID: A global alignment program. Genome Research. 2003;13:97–102. doi: 10.1101/gr.789803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evolutionary Biology. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Felsenstein J. PHYLIP - phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 69.Kumar S, Stecher G, Peterson D, Tamura K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–2686. doi: 10.1093/bioinformatics/bts507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. in Proceedings of the Gateway Computing Environments Workshop (GCE) 1–8 (2010).
- 71.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 72.Fuentes M, et al. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum) Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2007;308B:484–493. doi: 10.1002/jez.b.21179. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Q-J, Luo Y-J, Wu H-R, Chen Y-T, Yu J-K. Expression of germline markers in three species of amphioxus supports a preformation mechanism of germ cell development in cephalochordates. EvoDevo. 2013;4:17. doi: 10.1186/2041-9139-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.