Abstract
Inhibition of dipeptidyl peptidase IV (DPP-IV) may be exploited to maintain the incretin effect during the postprandial phase. As a result, glycemic regulation and energy homeostasis may be improved. Food protein-derived peptides have been identified as natural agents capable of inhibiting DPP-IV. Ile-Pro-Ile is the most potent DPP-IV inhibitory peptide identified to date. A minimum analog peptide set approach was used to study peptide analogs of Ile-Pro-Ile. The DPP-IV half maximal inhibitory concentration (IC50) values of the 25 peptides evaluated ranged from 3.9 ± 1.0 µM (Ile-Pro-Ile) to 247.0 ± 32.7 µM (Phe-Pro-Phe). The presence of Pro at position 2 of tripeptides was required to achieve high DPP-IV inhibition. Most peptides behaved as competitive inhibitors of DPP-IV with the exception of peptides with a N-terminal Trp, which were mixed-type inhibitors. While possessing the structure of preferred DPP-IV substrates, most peptides studied were particularly stable during 30 min incubation with DPP-IV. Molecular docking revealed that Ile-Pro-Ile and its peptide analogs interacted in a very similar manner with the active site of DPP-IV. In addition, no correlation was found between the Hydropathic INTeraction score and the DPP-IV IC50 values of the peptides studied. This outcome suggests that free energy may not be directly responsible for enzyme inhibition by the peptides. Finally, novel DPP-IV inhibitory peptides were identified using the strategy employed herein. These results may be relevant for the development of food protein-derived peptides with serum glucose lowering and food intake regulatory properties in humans.
Keywords: dipeptidyl peptidase IV inhibition, Ile-Pro-Ile (diprotin A), bioactive peptides, peptide analogs, design of experiments, quantitative structure activity relationship, molecular docking
Introduction
The inhibition of dipeptidyl peptidase IV (DPP-IV) is one of the numerous therapeutic strategies used for the management of type 2 diabetes. DPP-IV can cleave incretins such as glucagon-like peptide-1 (GLP-1) and glucose inhibitory polypeptide, resulting in a loss of their insulinotropic activity during the postprandial phase. The inhibition of DPP-IV has been shown to result in a better regulation of glycemia in type 2 diabetic subjects (1, 2). For this reason, several DPP-IV inhibitory drugs, known as gliptins, have been developed as antidiabetic agents (1). In addition, GLP-1 has been shown to modulate energy homeostasis by slowing down gastric emptying, inducing satiety, and reducing food intake (3).
Food proteins also contain peptide sequences, which are able to inhibit DPP-IV in vitro, in cell cultures and in small animals [for reviews, see Ref. (4, 5)]. Therefore, food protein-derived peptides may have an effect on the regulation of glucose metabolism in vivo. Food protein hydrolyzates have often been characterized for their DPP-IV inhibitory potency by quantifying their half maximal inhibitory concentration (IC50). Food protein-derived hydrolyzates are generally ~104 times less potent than gliptins (6). However, food and drug interventions may be complimentary in the management of type 2 diabetes (7). In fact, combinations of gliptins and hydrolyzates can have additive effects in terms of their ability to inhibit DPP-IV (6).
To date, the most potent DPP-IV inhibitory peptides reported in the literature are Ile-Pro-Ile and Val-Pro-Leu, also known as diprotin A and B, respectively. Their DPP-IV IC50 value is 3.5 and 15.8 µM, respectively (8). Both peptides were originally isolated from Bacillus cereus cultures (8). However, Ile-Pro-Ile and Val-Pro-Leu are also present in the primary sequence of several food proteins (9). The details of particularly potent DPP-IV inhibitory peptides displaying IC50 values < 100 µM have recently been reviewed (5). Ile-Pro-Ile and Val-Pro-Leu were, respectively, shown to be >12 and 2 times more potent than other food protein-derived peptides. In addition, only nine other tripeptides having DPP-IV IC50 values < 100 µM have been identified to date (Table S1 in Supplementary Material). Interestingly, all these tripeptides possess a Pro at position 2. This suggests that peptides having the structure Xaa1-Pro-Xaa3 (with Xaa an amino acid residue), may be of interest in studying those properties, which dictate the potency of DPP-IV inhibitory peptides. Furthermore, Pro-containing peptides have been reported to be particularly resistant to gastrointestinal digestion and potentially bioavailable as demonstrated by their detection in the gastrointestinal tract and in the circulation of humans [for reviews, see Ref. (10–12)].
Analogs of lead drug compounds are often used in drug discovery to design molecules with higher activity, improved bioavailability, or reduced side effects (13). DPP-IV inhibitory peptide analogs possessing the structure Trp-Arg-Xaa were studied using a peptide library of the 20 possible peptides (14). The most potent peptide discovered using this approach was Trp-Arg-Glu having a DPP-IV IC50 value of 350 µM. However, the utilization of a peptide library strategy to study Xaa1-Pro-Xaa3 would involve the analysis of 400 peptides (20 × 20 amino acids for Xaa1 and Xaa3). Therefore, in order to reduce the number of peptides for analysis, the concept of minimum analog peptide set (MAPS) has been introduced (15, 16). MAPS has been described during the study of peptide analogs employed for the development of meaningful quantitative structure activity relationship (QSAR) models for angiotensin converting enzyme (ACE) inhibition. MAPS correspond to specific peptide sets containing a minimum number of sequences, which are chosen by factorial or fractional factorial design to incorporate physicochemical properties relevant for a target bioactivity. To date, MAPS has been employed in the prediction of oxytocin (oxytocic and pressor activity), pepstatin (inhibition of porcine pepsin), and bradykinin (bradykinin potentiating) analogs (16). MAPS approaches do not appear to have been widely employed for the study of food protein-derived bioactive peptides.
The aim of this study was, therefore, to apply a MAPS-based approach to design peptide analogs of Ile-Pro-Ile. This was achieved by developing a QSAR model for DPP-IV inhibitory peptides to better understand the physicochemical characteristics, which are relevant to DPP-IV inhibition. In a second stage, a factorial design was employed to design the peptide analogs. The peptides were then assessed for their in vitro DPP-IV inhibitory properties and their mode of inhibition. Finally, molecular docking studies were conducted to better understand how those peptides may interact with the active site of DPP-IV.
Materials and Methods
Reagents
High-performance liquid chromatography (HPLC) grade water was from VWR (Dublin, Ireland). Trifluoroacetic acid, formic acid (FA), tris(hydroxymethyl)aminomethane (Tris), Gly-Pro-pNA, diprotin A (Ile-Pro-Ile), porcine DPP-IV (≥10 U mg−1 protein), mass spectrometry (MS) grade water, and acetonitrile were purchased from Sigma Aldrich (Dublin, Ireland). Synthetic peptides (Ala-Ile-Pro, Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ala-Arg-Pro, Ile-Phe-Lys, Ile-Ile-Phe, Ile-Pro-Ala, Ile-Arg-Phe, Ile-Arg-Lys, Lys-Pro-Ala, Lys-Arg-Ile, Arg-Ile-Phe, Arg-Ile-Arg, Arg-Lys-Arg, Phe-Pro-Phe, Phe-Pro-Ile, Phe-Pro-Trp, Ile-Ala-Ile, Ile-Pro-Phe, Ile-Pro-Trp, Trp-Pro-Phe, Trp-Pro-Ile, Trp-Pro-Trp) with a purity > 95% (w/w) were obtained from Genscript (Piscataway, NJ, USA).
QSAR Modeling
The QSAR analysis linking the DPP-IV IC50 value of peptides to their physicochemical properties was conducted using a partial least squares regression (PLSR) model (Eq. 1) as previously described (17). The peptides used (Table 1) were all competitive inhibitors of DPP-IV, and their IC50 values were obtained using the same DPP-IV inhibition assay (18). These conditions were selected given that it was previously demonstrated that this was required in order to obtain a significant PLSR model (17). The structural v-scale [v1 (van der Waals volume), v2 (net charge index), and v3 (hydrophobic parameter of side chains)] from Lin et al. (19) was employed for the amino acid descriptors. Peptide descriptors for the two N- and C-terminal amino acids of the sequences were generated to take account of peptides with various lengths (2–9 amino acids, Table 1) as described earlier (20). The PLSR model employed is described in Eq. 1
| (1) |
with Y: the Log10(DPP-IV IC50); i: the N- or C- terminal position of the amino acid (1 or 2); j: the amino acid descriptor number (varying between 1 and 3); a and b: the coefficients of the model; c, and ε the constant and residual of the model, respectively.
Table 1.
Dipeptidyl peptidase IV (DPP-IV) half maximal inhibitory concentration (IC50) of competitive inhibitory peptides used in the quantitative structure activity relationship model.
| Peptide sequencea | Length | DPP-IV IC50 (μM) | Reference |
|---|---|---|---|
| IPI | 3 | 3.5 | (21) |
| IPIQY | 5 | 35.2 | (21) |
| WL | 2 | 43.6 | (22) |
| LPVPQ | 5 | 43.7 | (17) |
| FLQP | 4 | 65.3 | (23) |
| IPM | 3 | 79.4 | (17) |
| LPVP | 4 | 87.0 | (24) |
| MPVQA | 5 | 93.3 | (24) |
| LPYPY | 5 | 108.3 | (21) |
| HL | 2 | 143.2 | (18) |
| IP | 2 | 149.6 | (21) |
| VA | 2 | 168.2 | (18) |
| LLQLEAIR | 8 | 177.8 | (24) |
| YPYY | 4 | 194.4 | (21) |
| LPQNIPPLT | 9 | 205.2 | (17) |
| VPGEIVE | 7 | 224.5 | (17) |
| LPL | 3 | 241.4 | (21) |
| YPY | 3 | 243.7 | (21) |
| LPLPL | 5 | 325.0 | (21) |
| LPLPLL | 6 | 371.5 | (17) |
| FL | 2 | 399.6 | (18) |
| IPSK | 4 | 406.8 | (17) |
| VLGP | 4 | 580.4 | (23) |
| RP | 2 | 657.2 | (17) |
| YP | 2 | 658.1 | (21) |
| LP | 2 | 712.5 | (21) |
| ILELA | 5 | 721.1 | (24) |
| AL | 2 | 882.1 | (18) |
| LW | 2 | 993.4 | (22) |
| QPLPPT | 6 | 1,013.8 | (17) |
| SL | 2 | 2,517.1 | (18) |
| GL | 2 | 2,615.0 | (18) |
| EK | 2 | 3,216.8 | (18) |
aPeptide sequences abbreviated with the one letter amino acid code. Peptides are ordered by decreasing DPP-IV inhibitory potency.
Generation of the peptide descriptors and PLSR model was carried out using Matlab (version R2015b, MathWorks, Inc., Natick, MA, USA). Training and test sets comprising 28 and 5 peptides, respectively, were randomly chosen (100 times) by the Matlab algorithm in order to cross validate the PLSR model.
Experimental Design for the Selection of Peptide Analogs of Ile-Pro-Ile
All the Ile-Pro-Ile analogs used had a fixed length of three amino acids. In addition, the QSAR model used herein, as well as a previous QSAR model (17), highlighted the importance of the amino acid hydrophobicity in the inhibition of DPP-IV. Therefore, a factorial design consisting of 3 factors at 2 levels was employed to design peptide analogs of Ile-Pro-Ile. Each factor represented a position within the peptide sequence (Xaa1, Xaa2, and Xaa3, where Xaai designates the amino acid located at position i of the peptide) and each level corresponded to a low (−1) or high (+1) hydrophobicity parameter of side chains (v3) for the amino acid. The 15 conditions of the factorial design corresponding to the hydrophobicity of the selected peptides (Ala-Ile-Pro, Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ala-Arg-Pro, Ile-Phe-Lys, Ile-Ile-Phe, Ile-Pro-Ala, Ile-Arg-Phe, Ile-Arg-Lys, Lys-Pro-Ala, Lys-Arg-Ile, Arg-Ile-Phe, Arg-Ile-Arg, and Arg-Lys-Arg), are presented in Table 2.
Table 2.
Dipeptidyl peptidase IV (DPP-IV) half maximal inhibitory concentration (IC50), z-centered value of the hydrophobic parameter of side chains [v3 in the scale developed by Lin et al. (19)] of the amino acids constitutive of the peptides, and mode of inhibition of peptide analog of Ile-Pro-Ile.
| Set number | Peptide sequencea | z-Centered values ofv3 of the amino acids within the tripeptide | Predicted DPP-IV IC50 (μM) | Experimental DPP-IV IC50 (μM) | Mode of inhibition | Peptide hydrolyzed by DPP-IV | ||
|---|---|---|---|---|---|---|---|---|
| Xaa1 | Xaa2 | Xaa3 | ||||||
| 1 | IPI | 0.725 | 0.059 | 0.725 | 238 | 3.9 ± 1.0 | Competitive | NO |
| AIP | −0.194 | 0.725 | 0.059 | 103 | nd | nd | nd | |
| APA | −0.194 | 0.059 | −0.194 | 988 | 44.3 ± 3.2 | Competitive | NO | |
| APF | −0.194 | 0.059 | 0.716 | 168 | 65.8 ± 5.5 | Competitive | NO | |
| APR | −0.194 | 0.059 | −1.000 | 15 | 119.7 ± 3.5 | Competitive | NO | |
| ARP | −0.194 | −1.000 | 0.059 | 687 | nd | nd | nd | |
| IFK | 0.725 | 0.716 | −0.991 | 34 | nd | nd | nd | |
| IIF | 0.725 | 0.725 | 0.716 | 71 | nd | nd | nd | |
| IPA | 0.725 | 0.059 | −0.194 | 472 | 28.3 ± 3.4 | Competitive | NO | |
| IRF | 0.725 | −1.000 | 0.716 | 476 | nd | nd | nd | |
| IRK | 0.725 | −1.000 | −0.991 | 107 | nd | nd | nd | |
| KPA | −0.991 | 0.059 | −0.194 | 22,690 | 74.5 ± 7.0 | Competitive | NO | |
| KRI | −0.991 | −1.000 | 0.725 | 67,699 | nd | nd | nd | |
| RIF | −1.000 | 0.725 | 0.716 | 7,435 | nd | nd | nd | |
| RIR | −1.000 | 0.725 | −1.000 | 662 | nd | nd | nd | |
| RKR | −1.000 | −0.991 | −1.000 | 2,451 | nd | nd | nd | |
| 2 | FPF | 0.716 | 0.059 | 0.716 | 177 | 247.0 ± 32.7 | Competitive | YES |
| FPI | 0.716 | 0.059 | 0.725 | 524 | 45.2 ± 5.3 | Competitive | NO | |
| FPW | 0.716 | 0.059 | 1.000 | 67 | 54.9 ± 2.9 | Competitive | YES | |
| IAI | 0.725 | −0.194 | 0.725 | 97 | nd | nd | nd | |
| IPF | 0.725 | 0.059 | 0.716 | 80 | 47.3 ± 12.3 | Competitive | YES | |
| IPW | 0.725 | 0.059 | 1.000 | 31 | 175.3 ± 5.5 | Competitive | YES | |
| WPF | 1.000 | 0.059 | 0.716 | 214 | 159.8 ± 12.8 | Mixed-type | YES | |
| WPI | 1.000 | 0.059 | 0.725 | 633 | 133.0 ± 14.7 | Mixed-type | NO | |
| WPW | 1.000 | 0.059 | 1.000 | 81 | 120.1 ± 13.1 | Mixed-type | YES | |
aPeptide sequences abbreviated with the one letter amino acid code. The first set corresponds to Ile-Pro-Ile and its 15 associated peptide analogs. The second set corresponds to Ile-Ala-Ile and the eight additional peptides tested, which possess the Xaa1-Pro-Xaa3 structure, with Xaa1 and Xaa3 being Ile, Phe, or Trp.
nd, not determined; IC50, DPP-IV half maximal inhibitory concentration; NO, peptide not cleaved during incubation with DPP-IV; YES, peptide cleaved following incubation with DPP-IV.
The experimental values obtained with the 15 peptides were used to develop a predictive multilinear regression (MLR) model linking the % DPP-IV inhibition obtained at 500 µM with the v3 values of the amino acids contained in the peptides. The MLR is described in Eq. 2
| (2) |
with β0 to β9: the coefficients of the model; x, y, and z, the z-centered parameters for the v3 values of Xaa1, Xaa2, and Xaa3, respectively; ε: the residual of the model.
Equation 2 was used to predict the structures of nine tripeptides potentially displaying high DPP-IV inhibition at 500 µM.
DPP-IV Inhibition Assay and Mode of Inhibition
Peptides were dispersed in HPLC grade water at 50 and 500 µM (final concentration). The DPP-IV inhibition assay was carried out in triplicate as outlined by Nongonierma and FitzGerald (18). Briefly, samples (25 μL) were mixed with Gly-Pro-pNA (final concentration 0.200 mM) and DPP-IV (final concentration 0.0025 U mL−1) in a 96-well microplate (Sarstedt, Dublin, Ireland). The microplate was incubated at 37°C for 60 min in a microplate reader (Biotek Synergy HT, Winooski, VT, USA) and absorbance was monitored at 405 nm.
Dose–response curves were determined for peptides displaying a percentage of inhibition > 50% at 500 µM. Peptides were diluted in HPLC water at concentrations ranging from 0.37 to 500 µM (final concentration). The DPP-IV IC50 values were determined by plotting the percentage inhibition as a function of the test compound concentration. Each analysis was carried out in triplicate (n = 3).
The mode of DPP-IV inhibition was determined using the Lineweaver and Burke double reciprocal representation as previously described (18). The most potent DPP-IV inhibitory peptides (i.e., Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ile-Pro-Ala, Lys-Pro-Ala, Phe-Pro-Phe, Phe-Pro-Ile, Phe-Pro-Trp, Ile-Pro-Phe, Ile-Pro-Trp, Trp-Pro-Phe, Trp-Pro-Ile, Trp-Pro-Trp) were assayed at three different final concentrations corresponding to their IC50 value divided by 4, 8, and 16. The peptide solution was substituted with 100 mM Tris–HCl buffer pH 8.0 in the negative control. Gly-Pro-pNA was added at concentrations ranging from 0.1 to 0.6 mM (final concentration). The absorbance at 405 nm was recorded for 30 min at 37°C using a microplate reader (Biotek Synergy HT). Each sample was evaluated four times.
Stability of Peptides to DPP-IV
Stability of the peptides to hydrolysis by DPP-IV was evaluated by liquid chromatography tandem mass spectrometry (LC-MS/MS) using the samples generated during the Lineweaver and Burk analysis containing DPP-IV inhibitory peptides at their IC50 values divided by 4 as previously described (24). Following the 30 min reaction, DPP-IV was heat inactivated by immersing samples in a water bath set at 90°C for 20 min. Test samples were subsequently diluted 40 times in mobile phase A (0.1% FA in MS water). Intact peptides were analyzed at the same concentration as the test samples. The peptide composition of the samples was determined as described previously (25). Briefly, an ultra-high performance liquid chromatography (UHPLC) UltiMate 3000 (Dionex, Camberley, Surrey, UK) fitted with a security guard UHPLC C18 PEPTIDE (2.1 mm, Phenomenex, Cheshire, UK) and an Aeris Peptide XB-C18 column (150 mm × 2.1 mm, 1.7 µm, Phenomenex) was used for peptide separation at 25°C. A sample volume of 2 µL was injected with a solvent delay of 4 min. The UHPLC was coupled to a quadrupole time-of-flight mass spectrometer (Q-TOF, Impact HD™, Bruker Daltonics GmbH, Bremen, Germany) using a 50–600 m/z acquisition range. The MS was fitted with an electrospray ionization source used in positive ion mode. Data acquisition was performed with Hystar software (Bruker Daltonics). Data analysis was performed with Compass Data Analysis 4.2 (Bruker Daltonics).
Molecular Docking of Peptides to the Active Site of DPP-IV
Interactions between the most potent DPP-IV inhibitory peptides and the active site of DPP-IV were investigated using molecular modeling. This analysis was based on docking simulations coupled with rescoring procedures to predict protein–peptide interactions and pharmacophoric analyses to investigate the hydrophobic/hydrophilic properties of the binding site.
The molecular model for porcine DPP-IV was derived from the crystallographic structure deposited in the RCSB Protein Data Bank1 having the PDB code 1ORW (26). The protein and peptides analyzed were processed using the Sybyl software, version 8.12 checking the consistency of atom and bond type assignment. The co-crystallized ligands were removed before proceeding with the analysis.
Specifically, the coupling of GOLD, as docking software, and Hydropathic INTeraction (HINT) (27) as rescoring function, was chosen. This was done on the basis of previous studies (i) demonstrating the high reliability of HINT in predicting ligand interactions with several protein targets (28), (ii) estimating the free energy of protein–ligand complex formation (29), and (iii) estimating enzyme inhibitory activity of small molecules and peptides (30, 31). In particular, the HINT score provides an empirical and quantitative evaluation of protein–ligand interaction as a sum of all single atomic contributions. The HINT score correlates with the free energy of binding. Therefore, the higher the HINT score, the more favorable the protein–ligand interaction. The software settings and the docking protocol reported by Dellafiora et al. (31) were used. Briefly, the protein was kept semi-flexible with the polar hydrogens set free to rotate, while ligands were set to be fully flexible. The maximum number of poses generated for each ligand was set at 25, and all the poses underwent a rescoring procedure with the HINT scoring function. Only the pose with the highest HINT score was kept for each ligand as it was considered to be the most favorable.
The pharmacophoric analysis of the ligand binding site was carried out using the Flapsite tool of the FLAP (Fingerprint for Ligand and Protein) software developed by Molecular Discovery Ltd.3 (32). The GRID algorithm was used to investigate the corresponding pharmacophoric space (33). The DRY probe was used to describe potential hydrophobic interactions, while the sp2 carbonyl oxygen (O) and the neutral flat amino (N1) probes were used to describe the hydrogen bond acceptor and donor capacity of the target, respectively. All images were obtained using the PyMol software version 2.0.4
Statistical Analysis
The significance of the PLSR used in the QSAR modeling (Eq. 1) was assessed using p model, p lack of fit, R2 and R2 cross validation as previously described (17). All statistical analyses were carried out with Matlab.
Results
QSAR Model for DPP-IV Inhibitory Peptides
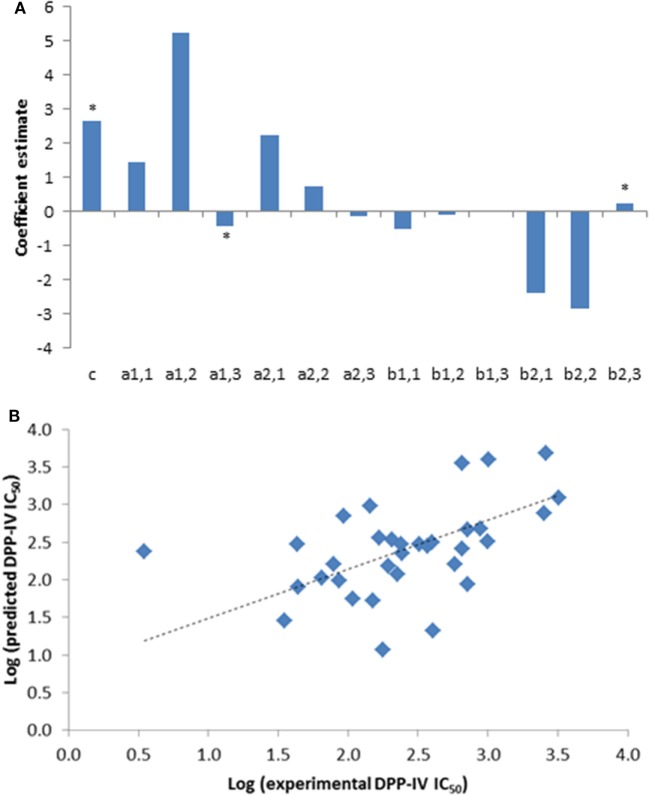
A total of 33 competitive DPP-IV inhibitory peptides were employed to build the PLSR model (Eq. 1; Table 1). The experimentally determined DPP-IV IC50 values of these peptides varied between 3.5 and 3,216.8 µM for Ile-Pro-Ile and Glu-Lys, respectively. The PLSR analysis linking the DPP-IV IC50 value to the amino acid descriptors of the peptides was statistically significant with a p model and p lack of fit value of 0.01 and 0.15, respectively (Table 3). The coefficients of the PLSR model, which were significant (p < 0.05) were the constant (c), a1,3 and b2,3 (Figure 1A). This indicated that the hydrophobicity of the amino acids located at the N-terminal and next to the C-terminal positions of peptides were important for the DPP-IV inhibitory potency of peptides. The relationship (R2 and R2 cross validation 0.66 and 0.57, respectively, Table 3) between the predicted and experimental DPP-IV IC50 value of the peptides used to build the QSAR model (Table 1) is illustrated on Figure 1B.
Table 3.
Statistical significance of the partial least squares regression linking the dipeptidyl peptidase IV (DPP-IV) half maximal inhibitory concentration (IC50) of the peptides used to generate the quantitative structure activity relationship model.
| Parameter | value |
|---|---|
| n | 33 |
| R2 model | 0.66 |
| p model | 0.01 |
| Root mean square error | 0.45 |
| F | 3.16 |
| R2 cross validation | 0.57 |
| p lack of fit | 0.15 |
Figure 1.
(A) Coefficients of the partial least squares regression (PLSR) linking the dipeptidyl peptidase IV (DPP-IV) half maximal inhibitory concentration (IC50) of the peptides used to generate the quantitative structure activity relationship (QSAR) model (Eq. 1). *Coefficients with a p < 0.05 are significantly different from 0. (B) Predicted DPP-IV IC50 value of the peptides used to build the QSAR model (Table 1) as a function of the experimental DPP-IV IC50 value, the line represents the QSAR model (Eq. 1).
DPP-IV Inhibitory Properties of Peptide Analogs of Ile-Pro-Ile
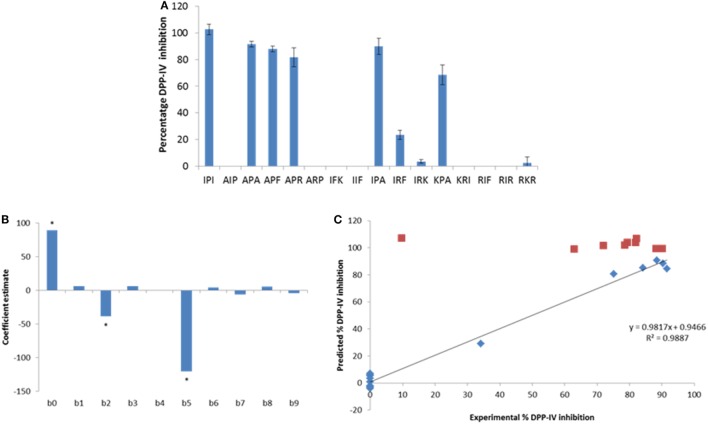
Peptide analogs of Ile-Pro-Ile were designed by varying the hydrophobicity of the constitutive amino acids of the tripeptide following a design of experiments (DOE, Table 2). The % DPP-IV inhibition of the 15 tripeptides when evaluated at 500 µM varied depending on the peptide sequence (Figure 2A). Some peptides (Ala-Ile-Pro, Ala-Arg-Pro, Ile-Phe-Lys, Ile-Ile-Phe, Lys-Arg-Ile, Arg-Ile-Phe, and Arg-Ile-Arg) were not able to inhibit DPP-IV at 500 µM. Arg-Lys-Arg, Ile-Arg-Lys, and Ile-Arg-Phe inhibited DPP-IV by less than 25%. The percentage of DPP-IV inhibition observed with the other peptides (Ile-Pro-Ile, Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ile-Pro-Ala, and Lys-Pro-Ala) when evaluated at 500 µM was > 65% (Figure 2A).
Figure 2.
(A) Percentage of dipeptidyl peptidase IV (DPP-IV) inhibition observed with the peptides of the minimum analog peptide set evaluated at 500 µM. Values are mean ± SD (n = 3). (B) Coefficients of the model (β0–β9; Eq. 2) linking the % DPP-IV inhibition of the peptides at 500 µM to their hydrophobicity (v3). (C) Predicted vs. experimental % DPP-IV inhibition of peptides from set #1 (diamonds) and set #2 (squares) evaluated at 500 µM. The line represents the multilinear regression model determined with data from peptide set #1 (Eq. 2).
Equation 2 was used to predict the % DPP-IV inhibition at 500 µM of the 15 tripeptides from set #1. Two coefficients, β2 and β5, which were associated with the hydrophobicity of the second amino acid of the tripeptides, were significant (p < 0.05, Figure 2B). Peptides with an Ala/Thr/His/Gly/Pro at position 2 were predicted to yield high DPP-IV inhibition when evaluated at 500 µM. Pro at position 2 of peptides has been identified as a feature of potent tripeptides (5). Equation 2 predicted that eight tripeptides (Phe-Pro-Phe, Phe-Pro-Ile, Phe-Pro-Trp, Ile-Pro-Phe, Ile-Pro-Trp, Trp-Pro-Phe, Trp-Pro-Ile, and Trp-Pro-Trp), which had the Xaa1-Pro-Xaa3 structure and similar hydrophobicities as Ile-Pro-Ile would yield high DPP-IV inhibition at 500 µM. Ile-Ala-Ile was also evaluated as Ala has been described as being a preferred amino acid residue at position 2 in DPP-IV peptide substrates (Table 2). The % DPP-IV inhibition at 500 µM of these nine sequences (peptide set #2) is illustrated in Figure 2C. The % inhibition of DPP-IV for these nine sequences was predicted to be >99% using the MLR model (Figure 2C). However, Ile-Ala-Ile could only inhibit 9.6 ± 1.9% of DPP-IV activity when evaluated at 500 µM (Figure 2C). As this peptide was not potent, it was not further evaluated. The % inhibition of DPP-IV by the eight other tripeptides from set #2 varied between 62.9 ± 10.1 and 90.1 ± 1.8% for Phe-Pro-Phe and Phe-Pro-Ile, respectively (Figure 2C).
The IC50 values of the most potent peptides, which inhibited DPP-IV by >50% at 500 µM, were determined (Table 2). The QSAR model predicted and the experimentally determined DPP-IV IC50 values of the peptides are shown in Table 2. The DPP-IV IC50 values of the peptides varied between 3.9 ± 1.0 and 247.0 ± 32.7 µM for Ile-Pro-Ile and Phe-Pro-Phe, respectively (Table 2). The QSAR analysis predicted DPP-IV IC50 values for the peptides ranging from 15 to 67,699 µM for Ala-Pro-Arg and Lys-Arg-Ile, respectively (Table 2).
Mode of DPP-IV Inhibition and Stability of Peptides to DPP-IV
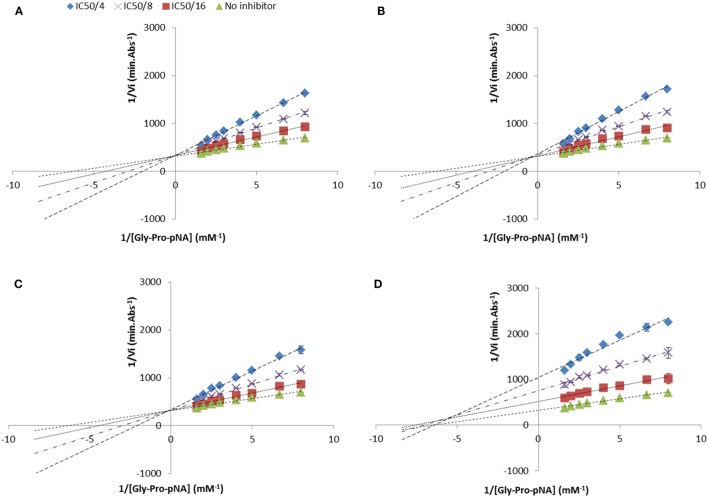
The mode of inhibition of DPP-IV was determined using Lineweaver and Burke double reciprocal plots (Figure 3). Most of the peptides studied in relation to their mode of inhibition (Ile-Pro-Ile, Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ile-Pro-Ala, Lys-Pro-Ala, Phe-Pro-Phe, Phe-Pro-Ile, Phe-Pro-Trp, Ile-Pro-Phe, and Ile-Pro-Trp) were competitive inhibitors of DPP-IV. However, three peptides (Trp-Pro-Phe, Trp-Pro-Ile, and Trp-Pro-Trp) were mixed-type inhibitors of DPP-IV (Table 2).
Figure 3.
Lineweaver and Burke double reciprocal representation of (A) Ile-Pro-Ile (competitive), (B) Ala-Pro-Phe (competitive), (C) Ile-Pro-Trp (competitive), and (D) Trp-Pro-Trp (mixed-type) evaluated at their half maximal inhibitory concentration divided by 4, 8, and 16. Values are the mean reciprocal initial rate ± SD (n = 4).
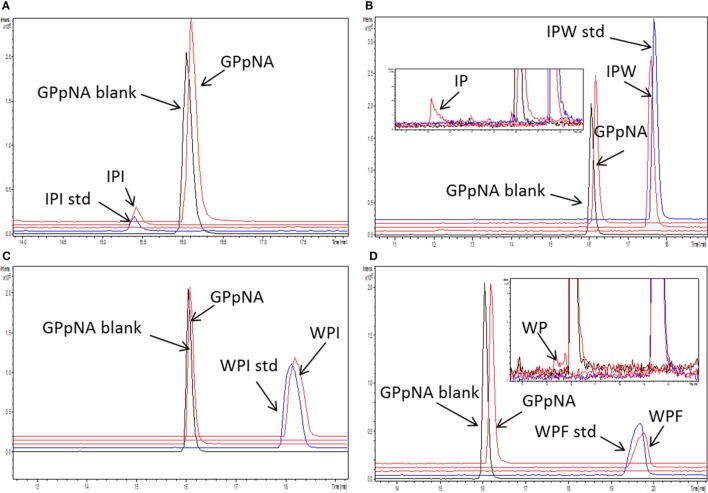
Peptide stability to DPP-IV was evaluated with the samples generated during the analysis of the mode of inhibition. Several peptides (Ile-Pro-Ile, Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ile-Pro-Ala, Lys-Pro-Ala, Phe-Pro-Ile, and Trp-Pro-Ile) were not hydrolyzed by DPP-IV (Table 2; Figures 4A,C). Six peptides (Phe-Pro-Phe, Phe-Pro-Trp, Ile-Pro-Phe, Ile-Pro-Trp, Trp-Pro-Phe, and Trp-Pro-Trp) were partly hydrolyzed by DPP-IV following cleavage at the C-terminal side of the Pro residue (Table 2; Figures 4B,D).
Figure 4.
Stability of the peptides [concentration 1/4 of their half maximal inhibitory concentration (IC50)] during incubation with dipeptidyl peptidase IV (DPP-IV) and Gly-Pro-pNA for 30 min at 37°C in Tris–HCl buffer pH 8.0. Extracted ion chromatogram of the ultra-high performance liquid chromatography tandem mass spectrometry (MS/MS) of (A) Ile-Pro-Ile, (B) Ile-Pro-Trp, (C) Trp-Pro-Ile, and (D) Trp-Pro-Phe. peptide_std: peptide, which was not incubated with DPP-IV; GPpNA_blank: Gly-Pro-pNA incubated with DPP-IV; peptides are abbreviated with the one letter amino acid code.
Molecular Modeling of DPP-IV Binding Interactions With Peptides
The geometry of binding of the most potent peptides within the DPP-IV binding site was investigated using molecular modeling based on pharmacophoric analysis and docking simulations coupled to rescoring procedures. The workflow used has already proved successful in computing the binding architecture of low-molecular mass ligands (34, 35). However, a fit-for-purpose validation was carried out to check the case-specific reliability. To this end, the 3D structures of porcine DPP-IV bound with non-covalent competitive inhibitors, which are available in the RCSB PDB database (36) (see text footnote 1), were taken as validation reference. Specifically, four structures were found (database last accessed on January 25th 2018). The chemical structures of the inhibitors are reported in Figure S1 in Supplementary Material. The 3D structures of the inhibitors (1-methylamine-1-benzyl-cyclopentane, (S)-2-[(R)-3-amino-4-(2-fluoro-phenyl)-butyryl]-1,2,3,4-tetrahydro-isoquinoline-3-carboxylic acid amide, 7-benzyl-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-purine-2,6-dione and 8-[(3~{R})-3-azanylpiperidin-1-yl]-7-[(2-bromophenyl)methyl]-1,3-dimethyl-purine-2,6-dione) were retrieved from the PDB chemical component repository and underwent the docking simulation study and the rescoring procedures. As shown in Figure S2 in Supplementary Material, all the computed poses were in strong agreement with the crystallographic data supporting the case-specific reliability of the procedure in computing the binding architectures of the ligands.
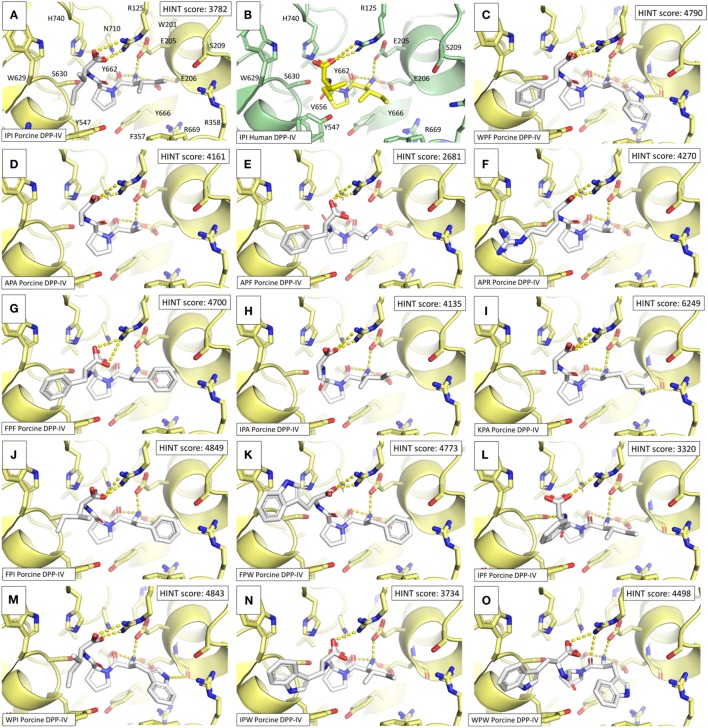
In addition, a pharmacophoric analysis of the porcine DPP-IV-binding site was carried out to map the space available for ligands in terms of hydrophobic and hydrophilic environment distributions. As shown in Figure 5, the ligand-binding sites showed an environment mainly able to receive hydrophobic and H-bond donor groups, with a limited space suitable for receiving H-bond acceptor groups. The binding architecture of the well-known DPP-IV inhibitory peptide, Ile-Pro-Ile, was calculated along with that of the other DPP-IV inhibitory peptide analogs. As shown in Figures 6A,B, the computed architecture of Ile-Pro-Ile was found to be in strong agreement with the crystallographic pose observed within human DPP-IV (37). In particular, both the ligand arrangement and the polar interactions network computed by the procedure were found to be consistent with those reported in the crystallographic data currently available (Figures 6A,B). This result further confirmed the reliability of the procedure. In addition, the binding pose of Ile-Pro-Ile complied with the pharmacophoric requirements of the binding pocket as the two hydrophobic side chains of Ile were found to be arranged into the hydrophobic space available for ligands, while the C- and N-terminal groups were found to be arranged into regions suitable to receive H-bond acceptor and donor groups, respectively (Figure 7). All the other inhibitory peptides were found to adopt an architecture of binding comparable to Ile-Pro-Ile. In addition, with other peptides, polar interactions between Arg125, Glu205, Glu206, and Tyr662 and Ile-Pro-Ile were also observed (Figure 6).
Figure 5.
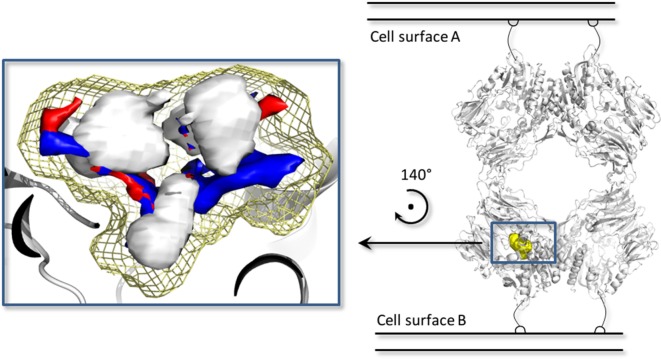
The biological assembly of the porcine dipeptidyl peptidase IV (DPP-IV) (PDB ID 1ORW) (26) is represented in cartoon on the right side, while a close up of pharmacophoric environment of the binding site is represented on the left. The yellow mesh retraces the shape of the binding site, while the white, blue, and red contours identify regions sterically and energetically favorable for hydrophobic, hydrogen bond donor and hydrogen bond acceptor groups, respectively.
Figure 6.
Binding architectures of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. The protein is represented in cartoon while peptides and amino acid side chains are represented in sticks. Yellow dotted lines indicate the protein–peptide polar interactions. (A) Ile-Pro-Ile. (B) Crystallographic pose of Ile-Pro-Ile within human DPP-IV [PDB structure 1WCY (37)]. (C) Trp-Pro-Phe. (D) Ala-Pro-Ala. (E) Ale-Pro-Phe. (F) Ala-Pro-Arg. (G) Phe-Pro-Phe. (H) Ile-Pro-Ala. (I) Lys-Pro-Ala. (J) Phe-Pro-Ile. (K) Phe-Pro-Trp. (L) Ile-Pro-Phe. (M) Trp-Pro-Ile. (N) Ile-Pro-Trp. (O) Trp-Pro-Trp.
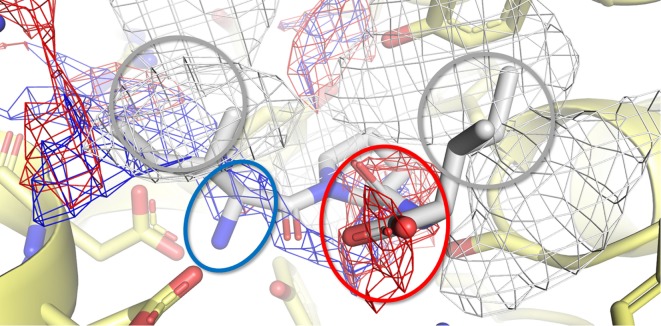
Figure 7.
Calculated binding pose of Ile-Pro-Ile within the active site of porcine dipeptidyl peptidase IV (DPP-IV). The protein is represented in cartoon, while the amino acids constitutive of the binding site (yellow) and Ile-Pro-Ile (white) are shown in sticks. The white, blue, and red meshes identify regions sterically and energetically favorable for hydrophobic, hydrogen bond donor and hydrogen bond acceptor groups, respectively. The proper placement of the Ile-Pro-Ile groups in respect to the pharmacophoric requirements of the pocket is highlighted with rings. Specifically, gray indicates hydrophobic regions, while red and blue indicate regions suitable to receive hydrogen bond acceptor and donor groups, respectively.
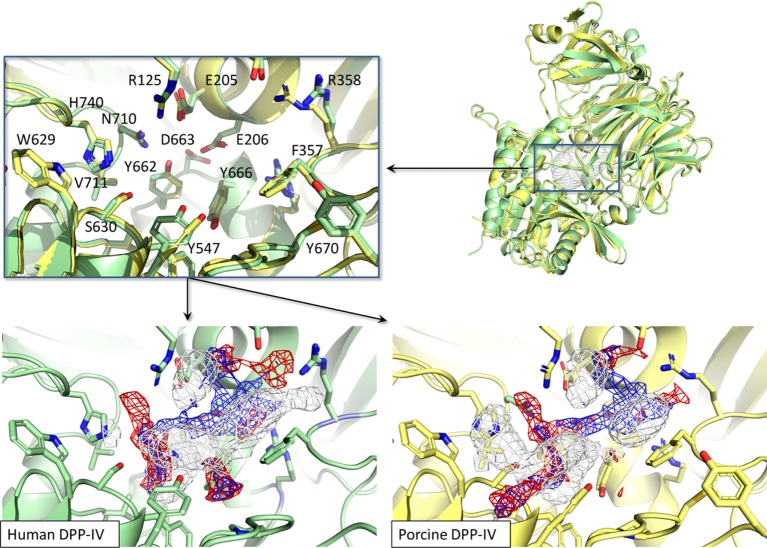
The binding site of porcine DPP-IV was compared to that of the human orthologous. The porcine and human enzymes have an identical length of 766 residues and the overall sequence identity is 88% (26) with a strongly conserved 3D organization (Figure 8). In relation to the catalytic site, those residues forming the space available for ligands showed 100% conservation in terms of residue composition and organization (Figure 8). Consequently, the pharmacophoric fingerprints of the two binding pockets were found to be highly comparable (Figure 8).
Figure 8.
Structural and pharmacophorical comparison between porcine [PDB ID 1ORW (26)] and human [PDB ID 1WCY (37)] dipeptidyl peptidase IV (DPP-IV). Proteins are represented in cartoon while amino acids constitutive of the binding site are represented in sticks. The white, blue, and red meshes identify regions sterically and energetically favorable for hydrophobic, hydrogen bond donor and hydrogen bond acceptor groups, respectively.
Discussion
The ability of Pro-containing tripeptides to inhibit DPP-IV in vitro was investigated. It has been suggested that Pro-containing peptides are relatively stable to gastrointestinal enzymes (38). Specific Pro-containing tripeptides may also be bioavailable in humans. For instance, Ile-Pro-Pro and Val-Pro-Pro have been identified in human sera following the ingestion of a milk beverage enriched in lactotripeptides (39). Ile-Pro-Ile is the most potent DPP-IV inhibitory peptide identified to date in the literature (5). Therefore, a MAPS-based approach was employed herein to design peptide analogs of Ile-Pro-Ile with the view to (i) study the role of tripeptide structures on DPP-IV inhibition and (ii) identify novel potent inhibitors of DPP-IV. A range of in silico methodologies including DOE, QSAR modeling, and molecular docking were used to design and study peptide analogs of Ile-Pro-Ile.
Several structural and physicochemical parameters of molecules have been shown to affect their ability to inhibit DPP-IV (40). The hydrophobicity of amino acids located within specific positions of peptides is thought to be of importance for the inhibition of DPP-IV (9, 41, 42). The QSAR model (Eq. 1) demonstrated that the hydrophobicity (v3) of the amino acids at the N-terminal and next to the C-terminal positions of peptides played a role in their DPP-IV inhibitory properties (Figure 2A). For tripeptides, the hydrophobicity (v3) of the second amino acid was shown to be linked with DPP-IV inhibition by the peptides (Eq. 2; Figure 2B). The models (Eqs 1 and 2) developed here were all significant (p < 0.05). However, they had limitations in terms of their predictive ability for DPP-IV inhibitory properties of peptides (Table 2; Figure 2C). Similar results have been obtained during the development of a QSAR model, which showed differences between predicted and experimental DPP-IV IC50 values of peptides (17). Similarly, with other metabolic enzymes such as ACE, major differences between QSAR predicted and experimental IC50 values have been reported for peptides on a number of occasions (43, 44). These differences are likely due to the fact that physicochemical parameters, other than those that are used to build the models, may play an important role in the inhibitory properties studied.
Other than hydrophobicity, aromaticity and steric hindrance of amino acid residues may play a role in the ability of peptides to inhibit DPP-IV. In a recent review paper on DPP-IV inhibitory properties of molecules, Ojeda-Montes et al. (40) suggested that potent DPP-IV inhibitors included molecules possessing aromatic rings, which can establish hydrophobic interactions with the S1 subsite of the DPP-IV active site. Earlier studies have reported on the existence of exclusion volumes in the S1 pocket of DPP-IV (45). These exclusion volumes may be responsible for the limited access of bulky amino acids residues to the active site of DPP-IV. This may prevent interactions between certain peptides and the active site of DPP-IV and/or the entry path to the catalytic site. Nevertheless, based on the molecular docking outcomes, all tripeptides studied herein were predicted to interact in a very similar manner with the active site of DPP-IV (Figure 6), and no evident steric interferences due to the aromatic or aliphatic residues in positions Xaa1 and Xaa3 were found. This finding is consistent with the availability of space to arrange side chains at positions #1 and #3 as both subsites are exposed to the bulk solvent with a large capability to arrange sidechains reducing steric interferences. On the other side, the Xaa2 position is likely to be more affected by steric restraints being buried into the protein core with a limited capability to accept bulky side chains (Figure S3 in Supplementary Material). On this basis, the discrepancies between the calculated and experimental inhibitory activity of the peptides considered herein might be due to additional factors beyond the inherent capability to interact with the binding site. This may relate to the capability to go through the entry path of the binding site and/or the capability of peptide binding to induce molecular rearrangement of DPP-IV, which in turn may affect enzymatic activity.
The % DPP-IV inhibition at 500 µM measured with tripeptides from set #1 confirmed that the presence of a Pro at position 2 was important to observe substantial inhibition of DPP-IV (Figures 2A,C). In fact, all tripeptides from sets #1 and 2, which possessed a Pro in position 2 inhibited DPP-IV while peptides possessing a different amino acid in position 2 were not able to inhibit DPP-IV or were not potent DPP-IV inhibitors (Figures 2A,C; Table 2). These results are consistent with earlier findings reporting on the features of potent (IC50 < 100 µM) DPP-IV inhibitory tripeptides, which also revealed the occurrence of a Pro residue at position 2 (5). Other analogs of Ile-Pro-Ile have been shown earlier to be relatively potent inhibitors of DPP-IV (Table S1 in Supplementary Material). For instance, Ile-Pro-Ala, also known as β-lactosin A (an antihypertensive peptide), had previously been reported for its ability to inhibit DPP-IV (46). The DPP-IV IC50 value of Ile-Pro-Ala reported herein was slightly lower than that reported by Tulipano et al. (46), i.e., 28.3 (Table 2) vs. 49.0 µM. However, this was consistent with the DPP-IV IC50 value of Ile-Pro-Ile, which was 3.9 vs. 7 µM (46). Several novel peptides displaying a DPP-IV IC50 value < 100 µM (Ala-Pro-Ala, Ala-Pro-Phe, Lys-Pro-Ala, Phe-Pro-Ile, Phe-Pro-Trp, and Ile-Pro-Trp, Table 2) were reported herein for the first time. The presence of a Pro residue at position 2 has often been associated with DPP-IV inhibition. However, it is not a guarantee for high DPP-IV inhibitory potency (Table 2; Table S1 in Supplementary Material). The amino acids located at the N- and C-terminal side of tripeptides also appear to play an important role in their DPP-IV inhibitory properties. This can be seen with the variation in the DPP-IV IC50 value of peptides differing in either their N- or C-terminal amino acid (Table 2).
Ile-Pro-Ile possesses the structure of preferred DPP-IV inhibitory substrates. Incubation of Ile-Pro-Ile with DPP-IV was reported to induce its breakdown over time. For instance, after 36 min incubation, 37% of the initial amount of Ile-Pro-Ile was cleaved by DPP-IV (47). These results are in contrast with our findings, showing no hydrolysis of Ile-Pro-Ile following 30 min incubation with DPP-IV, which is consistent with earlier reports (21, 24). Other tripeptides (Ala-Pro-Ala, Ala-Pro-Phe, Ala-Pro-Arg, Ile-Pro-Ala, Lys-Pro-Ala, Phe-Pro-Ile, and Trp-Pro-Ile) studied herein, which possess the structure of preferred DPP-IV inhibitory substrates, were also not hydrolyzed by DPP-IV after 30 min incubation (Table 2; Figure 4). In contrast, Phe-Pro-Phe, Phe-Pro-Trp, Ile-Pro-Phe, Ile-Pro-Trp, Trp-Pro-Phe, and Trp-Pro-Trp were partly hydrolyzed by DPP-IV during 30 min incubation. Substrate-type inhibitors are generally susceptible to hydrolysis by the enzyme (48). Several milk protein-derived peptides with a penultimate Pro have been shown to be degraded during incubation with DPP-IV (21, 49, 50). An earlier study has demonstrated that the extent of cleavage of peptides possessing the structure of DPP-IV substrates depended on the enzyme to substrate ratio (E:S) and the peptide sequence (21). A higher E:S generally resulted in higher extents of peptide cleavage. This may explain the differences seen in the extent of cleavage of Ile-Pro-Ile between the study from Rahfeld et al. (47) and our results. Surprisingly, Ile-Ala-Ile, which also possesses the structure of preferred DPP-IV substrates, could not significantly inhibit DPP-IV (<10% DPP-IV inhibition at 500 µM). Further investigation is warranted to explain this result.
As expected, most peptides with a Pro at position 2 were able to bind the active site of DPP-IV as demonstrated by their competitive mode of inhibition (Table 2). However, peptides possessing a Trp at their N-terminal position, i.e., Trp-Pro-Phe, Trp-Pro-Ile, and Trp-Pro-Trp were mixed-type inhibitors of DPP-IV. Several peptides having a Trp residue at their N-terminal position have been shown to be non-competitive inhibitors of DPP-IV (18, 22, 51). It has been suggested in earlier studies that alternative binding sites, differing from the active site of DPP-IV exist. The outcomes of molecular docking studies suggested that a number of peptides with a Trp N-terminus can bind secondary sites of DPP-IV. The location of this secondary binding site is nearby the active site of DPP-IV (51, 52). Pro-Phe, Trp-Pro-Ile, and Trp-Pro-Trp possess the structure of preferred substrates of DPP-IV (i.e., Pro residue at position 2). This is further confirmed by the fact that Trp-Pro-Phe and Trp-Pro-Trp were hydrolyzed by DPP-IV (Table 2). The mixed mode of inhibition of the peptides studied herein may arise from the fact that Trp-Pro-Phe, Trp-Pro-Ile, and Trp-Pro-Trp are also able to bind both to the active and secondary sites of DPP-IV.
Molecular docking showed that Ile-Pro-Ile and its associated peptide analogs established similar interactions with the active site of porcine DPP-IV (Figure 6). These interactions are consistent with those (i.e., with Glu205, Glu206, Tyr662) that have been demonstrated earlier with the S1 pocket of the active site of DPP-IV (40). Nevertheless, there was no correlation between the computed scores and the experimentally determined DPP-IV IC50 values (Figure 6; Table 2). Bearing in mind that the HINT score correlates with the free energy of binding (29), this result might suggest that the energy of interaction within the catalytic site of DPP-IV may have a secondary role in determining inhibition of the enzyme. This may particularly be the case here since the peptides also possess the structure of preferred DPP-IV substrates and, therefore, the mechanisms of inhibition may be more complex than a simple binding of peptides to the active site. This hypothesis is further supported by the unexpected stability of the peptides during incubation with DPP-IV (Table 2; Figure 4). It appears that the manner in which peptides bind to the active site of DPP-IV may protect them from extensive degradation. It is interesting to note that all peptides cleaved by DPP-IV possess bulky amino acid residues (i.e., Trp and Phe) at their C-terminal side.
The results herein are relevant to glycemic regulation and potentially food intake in humans. This is particularly the case as the study herein reports on potent DPP-IV inhibitory peptides (Ile-Pro-Ile and Ile-Pro-Ala) having the characteristics of peptides, which have previously been shown to be relatively stable to gastrointestinal enzymes (53, 54). Several of these peptides were also shown to be relatively stable to DPP-IV (Table 2; Figure 4). Therefore, they may be resistant to the action of brush border DPP-IV in the intestine and protect incretins from early degradation by DPP-IV. There was 100% conservation for the composition and organization of residues forming the space available for ligands within the crystal structures of porcine and human DPP-IV (Figure 8). Therefore, it may be assumed that peptides able to interact with the porcine DPP-IV active site may also interact with that of the human ortholog. As a consequence, inhibition of human DPP-IV activity may be expected for the most potent peptides studied herein. Nonetheless, due to the non-conserved residues at other regions in DPP-IV, effects in reaching the binding site and/or in regulating the protein dynamics, and the reaction kinetics cannot be excluded. Accordingly, differences in the magnitude of the inhibition between the two orthologous may occur. In this regard, it is worth mentioning, however, that short peptides (<5 residues) have been shown to inhibit human and porcine DPP-IV in a similar manner (55). The ability of peptide analogs of Ile-Pro-Ile to play a role as DPP-IV inhibitors in humans needs to be evaluated during human intervention studies.
In recent studies, the utilization of Pro-rich protein substrates has been employed as a strategy to develop relatively potent DPP-IV inhibitory hydrolyzates. The 1 kDa permeate of an ultrafiltration fraction from a porcine skin gelatin hydrolyzate had a DPP-IV IC50 value of 1.50 mg mL−1 (56). Salmon gelatin hydrolyzates having DPP-IV IC50 values between 0.08 ± 0.01 and 0.71 ± 0.09 mg mL−1 have been reported (57). Wheat gluten hydrolyzates, which are also rich in Pro-containing peptides, had DPP-IV IC50 values of 0.136 (58) and 0.24 ± 0.02 to 0.66 ± 0.06 mg mL−1 (59). Food (milk, porcine skin gelatin, and wheat gluten) hydrolyzates were shown to contain numerous tripeptides with a Pro at position 2, some of which had DPP-IV IC50 values < 100 µM (53, 54, 56, 58–61). While the effects of these hydrolyzates in humans have not been evaluated, they may represent natural alternatives to synthetic drugs for glycemic regulation.
Conclusion
Within this work, the utilization of MAPS has been described for the study of analogs of Ile-Pro-Ile, the most potent DPP-IV inhibitory peptide identified to date. While relatively potent DPP-IV inhibitory peptides were identified, none were as potent as Ile-Pro-Ile. This study has confirmed the importance of a Pro residue in position 2 of tripeptides in displaying potent DPP-IV inhibitory properties. Molecular docking showed very similar interactions between Ile-Pro-Ile and its associated peptide analogs. However, the HINT scores did not correlate with the DPP-IV IC50 values obtained experimentally. This result, together with the surprisingly high stability of Ile-Pro-Ile and its associated peptide analogs to the hydrolytic action of DPP-IV suggests that complex mechanisms are involved in the inhibition of DPP-IV by these peptides. The peptides investigated in this study may have potential applications for glycemic management and energy homeostasis in humans.
Author Contributions
AN, LD, SP, RF, PC, and GG contributed to the conception and design of the study. AN, LD, and SP carried out the experimental work, data analysis, and interpretation. AN and LD wrote the first draft of the manuscript. SP, PC, GG, and RF critically revised the manuscript. All authors listed have directly contributed to the manuscript, revised, read, and approved the submitted version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The work described herein was partly supported by Enterprise Ireland under Grant Number TC2013-0001.
1http://www.rcsb.org (Accessed: January 27, 2018).
2www.certara.com (Accessed: February 26, 2017).
3http://www.moldiscovery.com (Accessed: January 8, 2017).
4http://www.pymol.org (Accessed: March 6, 2017).
Supplementary Material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fendo.2018.00329/full#supplementary-material.
References
- 1.Cahn A, Cernea S, Raz I. An update on DPP-4 inhibitors in the management of type 2 diabetes. Expert Opin Emerg Drugs (2016) 21(4):409–19. 10.1080/14728214.2016.1257608 [DOI] [PubMed] [Google Scholar]
- 2.Juillerat-Jeanneret L. Dipeptidyl peptidase IV and its inhibitors: therapeutics for type 2 diabetes and what else? J Med Chem (2014) 57(6):2197–212. 10.1021/jm400658e [DOI] [PubMed] [Google Scholar]
- 3.Sánchez-Garrido MA, Brandt SJ, Clemmensen C, Müller TD, DiMarchi RD, Tschöp MH. GLP-1/glucagon receptor co-agonism for treatment of obesity. Diabetologia (2017) 60(10):1851–61. 10.1007/s00125-017-4354-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacroix IME, Li-Chan ECY. Food-derived dipeptidyl-peptidase IV inhibitors as a potential approach for glycemic regulation – current knowledge and future research considerations. Trends Food Sci Technol (2016) 54:1–16. 10.1016/j.tifs.2016.05.008 [DOI] [Google Scholar]
- 5.Nongonierma AB, FitzGerald RJ. Features of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from dietary proteins. J Food Biochem (2018). 10.1111/jfbc.12451 [DOI] [PubMed] [Google Scholar]
- 6.Nongonierma AB, FitzGerald RJ. Dipeptidyl peptidase IV inhibitory properties of a whey protein hydrolysate: influence of fractionation, stability to simulated gastrointestinal digestion and food-drug interaction. Int Dairy J (2013) 32(1):33–9. 10.1016/j.idairyj.2013.03.005 [DOI] [Google Scholar]
- 7.Watson LE, Wu T, Horowitz M, Rayner CK. Whey protein and diabetes. In: Watson RR, Collier RJ, Preedy VR, editors. Dairy in Human Health and Disease Across the Lifespan. Oxford, UK: Academic Press; (2017). p. 197–209. [Google Scholar]
- 8.Umezawa H, Aoyagi T, Ogawa K, Naganawa H, Hamada M, Takeuchi T. Diprotins A and B, inhibitors of dipeptidyl aminopeptidase IV, produced by bacteria. J Antibiot (Tokyo) (1984) 37(4):422–5. 10.7164/antibiotics.37.422 [DOI] [PubMed] [Google Scholar]
- 9.Nongonierma AB, FitzGerald RJ. An in silico model to predict the potential of dietary proteins as sources of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem (2014) 165:489–98. 10.1016/j.foodchem.2014.05.090 [DOI] [PubMed] [Google Scholar]
- 10.Nongonierma AB, FitzGerald RJ. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci Technol (2017) 69(B):289–305. 10.1016/j.tifs.2017.03.003 [DOI] [Google Scholar]
- 11.Nongonierma AB, FitzGerald RJ. The scientific evidence for the role of milk protein-derived bioactive peptides in humans: a review. J Funct Foods (2015):640–56. 10.1016/j.jff.2015.06.021 [DOI] [Google Scholar]
- 12.Boutrou R, Henry G, Sanchez-Rivera L. On the trail of milk bioactive peptides in human and animal intestinal tracts during digestion: a review. Dairy Sci Technol (2015) 95(6):815–29. 10.1007/s13594-015-0210-0 [DOI] [Google Scholar]
- 13.Wermuth CG. Analogues as a means of discovering new drugs. In: Fischer J, Ganellin CR, editors. Analogue-Based Drug Discovery. Germany: Wiley-VCH Verlag GmbH & Co. KGaA; (2006). p. 1–23. [Google Scholar]
- 14.Lan VTT, Ito K, Ito S, Kawarasaki Y. Trp-Arg-Xaa tripeptides act as uncompetitive-type inhibitors of human dipeptidyl peptidase IV. Peptides (2014) 54:166–70. 10.1016/j.peptides.2014.01.027 [DOI] [PubMed] [Google Scholar]
- 15.Hellberg S, Eriksson L, Jonsson J, Lindgren F, Sjöström M, Skagerberg B, et al. Minimum analogue peptide sets (MAPS) for quantitative structure-activity relationships. Chem Biol Drug Des (1991) 37(5):414–24. [DOI] [PubMed] [Google Scholar]
- 16.Hellberg S, Sjoestroem M, Skagerberg B, Wold S. Peptide quantitative structure-activity relationships, a multivariate approach. J Med Chem (1987) 30(7):1126–35. 10.1021/jm00390a003 [DOI] [PubMed] [Google Scholar]
- 17.Nongonierma AB, FitzGerald RJ. Structure activity relationship modelling of milk protein-derived peptides with dipeptidyl peptidase IV (DPP-IV) inhibitory activity. Peptides (2016) 79:1–7. 10.1016/j.peptides.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 18.Nongonierma AB, FitzGerald RJ. Dipeptidyl peptidase IV inhibitory and antioxidative properties of milk-derived dipeptides and hydrolysates. Peptides (2013) 39:157–63. 10.1016/j.peptides.2012.11.016 [DOI] [PubMed] [Google Scholar]
- 19.Lin ZH, Long HX, Bo Z, Wang YQ, Wu YZ. New descriptors of amino acids and their application to peptide QSAR study. Peptides (2008) 29(10):1798–805. 10.1016/j.peptides.2008.06.004 [DOI] [PubMed] [Google Scholar]
- 20.Li Y-W, Li B. Characterization of structure–antioxidant activity relationship of peptides in free radical systems using QSAR models: key sequence positions and their amino acid properties. J Theor Biol (2013) 318:29–43. 10.1016/j.jtbi.2012.10.029 [DOI] [PubMed] [Google Scholar]
- 21.Nongonierma AB, FitzGerald RJ. Susceptibility of milk protein-derived peptides to dipeptidyl peptidase IV (DPP-IV) hydrolysis. Food Chem (2014) 145(15):845–52. 10.1016/j.foodchem.2013.08.097 [DOI] [PubMed] [Google Scholar]
- 22.Nongonierma AB, FitzGerald RJ. Inhibition of dipeptidyl peptidase IV (DPP-IV) by tryptophan containing dipeptides. Food Funct (2013) 4(12):1843–9. 10.1039/c3fo60262a [DOI] [PubMed] [Google Scholar]
- 23.Nongonierma AB, FitzGerald RJ. Inhibition of dipeptidyl peptidase IV (DPP-IV) by proline containing peptides. J Funct Foods (2013) 5(4):1909–17. 10.1016/j.jff.2013.09.012 [DOI] [PubMed] [Google Scholar]
- 24.Nongonierma AB, Paolella S, Mudgil P, Maqsood S, FitzGerald RJ. Identification of novel dipeptidyl peptidase IV (DPP-IV) inhibitory peptides in camel milk protein hydrolysates. Food Chem (2018) 244:340–8. 10.1016/j.foodchem.2017.10.033 [DOI] [PubMed] [Google Scholar]
- 25.Nongonierma AB, Paolella S, Mudgil P, Maqsood S, FitzGerald RJ. Dipeptidyl peptidase IV (DPP-IV) inhibitory properties of camel milk protein hydrolysates generated with trypsin. J Funct Foods (2017) 34:49–58. 10.1016/j.jff.2017.04.016 [DOI] [Google Scholar]
- 26.Engel M, Hoffmann T, Wagner L, Wermann M, Heiser U, Kiefersauer R, et al. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc Natl Acad Sci U S A (2003) 100(9):5063–8. 10.1073/pnas.0230620100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellogg GE, Abraham DJ. Hydrophobicity: is LogPo/w more than the sum of its parts? Eur J Med Chem (2000) 35(7–8):651–61. 10.1016/S0223-5234(00)00167-7 [DOI] [PubMed] [Google Scholar]
- 28.Dellafiora L, Warth B, Schmidt V, Del Favero G, Mikula H, Fröhlich J, et al. An integrated in silico/in vitro approach to assess the xenoestrogenic potential of alternaria mycotoxins and metabolites. Food Chem (2018) 248:253–61. 10.1016/j.foodchem.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 29.Cozzini P, Fornabaio M, Marabotti A, Abraham DJ, Kellogg GE, Mozzarelli A. Simple, intuitive calculations of free energy of binding for protein− ligand complexes. 1. Models without explicit constrained water. J Med Chem (2002) 45(12):2469–83. 10.1021/jm0200299 [DOI] [PubMed] [Google Scholar]
- 30.Dellafiora L, Paolella S, Dall’Asta C, Dossena A, Cozzini P, Galaverna G. Hybrid in silico/in vitro approach for the identification of angiotensin I converting enzyme inhibitory peptides from Parma dry-cured ham. J Agric Food Chem (2015) 63(28):6366–75. 10.1021/acs.jafc.5b02303 [DOI] [PubMed] [Google Scholar]
- 31.Dellafiora L, Marchetti M, Spyrakis F, Orlandi V, Campanini B, Cruciani G, et al. Expanding the chemical space of human serine racemase inhibitors. Bioorg Med Chem Lett (2015) 25(19):4297–303. 10.1016/j.bmcl.2015.07.081 [DOI] [PubMed] [Google Scholar]
- 32.Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for ligands and proteins (FLAP): theory and application. J Chem Inf Model (2007) 47(2):279–94. 10.1021/ci600253e [DOI] [PubMed] [Google Scholar]
- 33.Carosati E, Sciabola S, Cruciani G. Hydrogen bonding interactions of covalently bonded fluorine atoms: from crystallographic data to a new angular function in the GRID force field. J Med Chem (2004) 47(21):5114–25. 10.1021/jm0498349 [DOI] [PubMed] [Google Scholar]
- 34.Dellafiora L, Galaverna G, Dall’Asta C, Cozzini P. Hazard identification of cis/trans-zearalenone through the looking-glass. Food Chem Toxicol (2015) 86:65–71. 10.1016/j.fct.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 35.Dellafiora L, Galaverna G, Reverberi M, Dall’Asta C. Degradation of aflatoxins by means of laccases from Trametes versicolor: an in silico insight. Toxins (2017) 9(1):17. 10.3390/toxins9010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berman H, Westbrook J, Feng Z, Gilliland G, Bhat T, Weissig H, et al. The protein data bank. Nucleic Acids Res (2000) 28(1):235–42. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiramatsu H, Yamamoto A, Kyono K, Higashiyama Y, Fukushima C, Shima H, et al. The crystal structure of human dipeptidyl peptidase IV (DPPIV) complex with diprotin A. Biol Chem (2004) 385(6):561–4. 10.1515/BC.2004.068 [DOI] [PubMed] [Google Scholar]
- 38.Kim Y, Birtwhistle W, Kim Y. Peptide hydrolases in the brush border and soluble fractions of small intestinal mucosa of rat and man. J Clin Invest (1972) 51(6):14–9. 10.1172/JCI106938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foltz M, Meynen EE, Bianco V, van Platerink C, Koning TMMG, Kloek J. Angiotensin converting enzyme inhibitory peptides from a lactotripeptide-enriched milk beverage are absorbed intact into the circulation. J Nutr (2007) 137(4):953–8. 10.1093/jn/137.4.953 [DOI] [PubMed] [Google Scholar]
- 40.Ojeda-Montes MJ, Gimeno A, Tomas-Hernández S, Cereto-Massagué A, Beltrán-Debón R, Valls C, et al. Activity and selectivity cliffs for DPP-IV inhibitors: lessons we can learn from SAR studies and their application to virtual screening. Med Res Rev (2018). 10.1002/med.21499 [DOI] [PubMed] [Google Scholar]
- 41.Tulipano G, Faggi L, Nardone A, Cocchi D, Caroli AM. Characterisation of the potential of β-lactoglobulin and α-lactalbumin as sources of bioactive peptides affecting incretin function: in silico and in vitro comparative studies. Int Dairy J (2015) 48:66–72. 10.1016/j.idairyj.2015.01.008 [DOI] [Google Scholar]
- 42.Nongonierma AB, Mooney C, Shields DC, FitzGerald RJ. In silico approaches to predict the potential of milk protein-derived peptides as dipeptidyl peptidase IV (DPP-IV) inhibitors. Peptides (2014) 57:43–51. 10.1016/j.peptides.2014.04.018 [DOI] [PubMed] [Google Scholar]
- 43.Sagardia I, Iloro I, Elortza F, Bald C. Quantitative structure–activity relationship based screening of bioactive peptides identified in ripened cheese. Int Dairy J (2013) 33(2):184–90. 10.1016/j.idairyj.2012.12.006 [DOI] [Google Scholar]
- 44.Jing P, Qian B, He Y, Zhao X, Zhang J, Zhao D, et al. Screening milk-derived antihypertensive peptides using quantitative structure activity relationship (QSAR) modelling and in vitro/in vivo studies on their bioactivity. Int Dairy J (2014) 35(1):95–101. 10.1016/j.idairyj.2013.10.009 [DOI] [Google Scholar]
- 45.Lu I, Tsai K-C, Chiang Y-K, Jiaang W-T, Wu S-H, Mahindroo N, et al. A three-dimensional pharmacophore model for dipeptidyl peptidase IV inhibitors. Eur J Med Chem (2008) 43(8):1603–11. 10.1016/j.ejmech.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 46.Tulipano G, Sibilia V, Caroli AM, Cocchi D. Whey proteins as source of dipeptidyl dipeptidase IV (dipeptidyl peptidase-4) inhibitors. Peptides (2011) 32(4):835–8. 10.1016/j.peptides.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 47.Rahfeld J, Schierborn M, Hartrodt B, Neubert K, Heins J. Are diprotin A (Ile-Pro-Ile) and diprotin B (Val-Pro-Leu) inhibitors or substrates of dipeptidyl peptidase IV? Biochim Biophys Acta (1991) 1076(2):314–6. 10.1016/0167-4838(91)90284-7 [DOI] [PubMed] [Google Scholar]
- 48.Fujita H, Yoshikawa M. LKPNM: a prodrug-type ACE-inhibitory peptide derived from fish protein. Immunopharmacology (1999) 44(1):123–7. 10.1016/S0162-3109(99)00118-6 [DOI] [PubMed] [Google Scholar]
- 49.Miyamoto Y, Ganapathy V, Barlas A, Neubert K, Barth A, Leibach FH. Role of dipeptidyl peptidase IV in uptake of peptide nitrogen from beta-casomorphin in rabbit renal BBMV. Am J Physiol Renal Physiol (1987) 252(4):F670–7. 10.1152/ajprenal.1987.252.4.F670 [DOI] [PubMed] [Google Scholar]
- 50.Tiruppathi C, Miyamoto Y, Ganapathy V, Leibach FH. Genetic evidence for role of DPP IV in intestinal hydrolysis and assimilation of prolyl peptides. Am J Physiol (1993) 265(1):G81–9. 10.1152/ajpgi.1993.265.1.G81 [DOI] [PubMed] [Google Scholar]
- 51.Lorey S, Stöckel-Maschek A, Faust J, Brandt W, Stiebitz B, Gorrell MD, et al. Different modes of dipeptidyl peptidase IV (CD26) inhibition by oligopeptides derived from the N-terminus of HIV-1 Tat indicate at least two inhibitor binding sites. Eur J Biochem (2003) 270(10):2147–56. 10.1046/j.1432-1033.2003.03568.x [DOI] [PubMed] [Google Scholar]
- 52.Nongonierma AB, Mooney C, Shields DC, FitzGerald RJ. Inhibition of dipeptidyl peptidase IV and xanthine oxidase by amino acids and dipeptides. Food Chem (2013) 141(1):644–53. 10.1016/j.foodchem.2013.02.115 [DOI] [PubMed] [Google Scholar]
- 53.Le Maux S, Nongonierma AB, Murray B, Kelly PM, FitzGerald RJ. Identification of short peptide sequences in the nanofiltration permeate of a bioactive whey protein hydrolysate. Food Res Int (2015) 77:534–9. 10.1016/j.foodres.2015.09.012 [DOI] [Google Scholar]
- 54.Le Maux S, Nongonierma AB, FitzGerald RJ. Improved short peptide identification using HILIC–MS/MS: retention time prediction model based on the impact of amino acid position in the peptide sequence. Food Chem (2015) 173:847–54. 10.1016/j.foodchem.2014.10.104 [DOI] [PubMed] [Google Scholar]
- 55.Lacroix IME, Li-Chan ECY. Comparison of the susceptibility of porcine and human dipeptidyl-peptidase IV to inhibition by protein-derived peptides. Peptides (2015) 69:19–25. 10.1016/j.peptides.2015.03.016 [DOI] [PubMed] [Google Scholar]
- 56.Hsu K-C, Tung Y-S, Huang S-L, Jao C-L. Dipeptidyl peptidase-IV inhibitory activity of peptides in porcine skin gelatin hydrolysates. In: Hernández-Ledesma B, editor. Bioactive Food Peptides in Health and Disease. In Tech; Available from: http://www.intechopen.com/books/bioactive-food-peptides-in-health-and-disease/dipeptidyl-peptidase-iv-inhibitory-activity-of-peptides-in-porcine-skin-gelatin-hydrolysates (Accessed: December 7, 2017). [Google Scholar]
- 57.Neves AC, Harnedy PA, O’Keeffe MB, Alashi MA, Aluko RE, FitzGerald RJ. Peptide identification in a salmon gelatin hydrolysate with antihypertensive, dipeptidyl peptidase IV inhibitory and antioxidant activities. Food Res Int (2017) 100:112–20. 10.1016/j.foodres.2017.06.065 [DOI] [PubMed] [Google Scholar]
- 58.Taga Y, Hayashida O, Kusubata M, Ogawa-Goto K, Hattori S. Production of a novel wheat gluten hydrolysate containing dipeptidyl peptidase-IV inhibitory tripeptides using ginger protease. Biosci Biotechnol Biochem (2017) 81(9):1823–8. 10.1080/09168451.2017.1345615 [DOI] [PubMed] [Google Scholar]
- 59.Nongonierma AB, Hennemann M, Paolella S, FitzGerald RJ. Generation of wheat gluten hydrolysates with dipeptidyl peptidase IV (DPP-IV) inhibitory properties. Food Funct (2017) 8(6):2249–57. 10.1039/c7fo00165g [DOI] [PubMed] [Google Scholar]
- 60.Nongonierma AB, Lalmahomed M, Paolella S, FitzGerald RJ. Milk protein isolate (MPI) as a source of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides. Food Chem (2017) 231:202–11. 10.1016/j.foodchem.2017.03.123 [DOI] [PubMed] [Google Scholar]
- 61.Nongonierma AB, Mazzocchi C, Paolella S, FitzGerald RJ. Release of dipeptidyl peptidase IV (DPP-IV) inhibitory peptides from milk protein isolate (MPI) during enzymatic hydrolysis. Food Res Int (2017) 94:79–89. 10.1016/j.foodres.2017.02.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.