Abstract
Hepatocellular carcinoma (HCC) is the sixth most common cancer worldwide, but the overall prognosis remains disappointing especially in the advanced-stage patients. Aberration expression of Aurora kinases is tumorigenic and thus it has attracted interests as therapeutic targets in cancer treatment. Here, we investigated the proteomic response of HCC Hep3B cells to danusertib (Danu), a pan-Aurora kinase inhibitor, and then validated the proteomic results based on stable-isotope labeling by amino acids in cell culture (SILAC). The proteomic data identified that Danu modulated the expression of 542 protein molecules (279 up-regulated; 260 down-regulated; 3 stable). Ingenuity pathway analysis (IPA) and KEGG pathway analysis identified 107 and 24 signaling pathways were regulated by Danu, respectively. IPA analysis showed cellular growth and proliferation, and cell death and survival were among the top five molecular and cellular functions regulated by Danu. The verification experiments showed that Danu inhibited the proliferation of Hep3B cells with a 24-hr IC50 value of 22.03 µM. Danu treatment also arrested Hep3B cells in G2/M phase via regulating the expression of key cell cycle regulators and induced apoptosis via mitochondria-dependent pathway in a dose-dependent manner. Besides, Danu induced a marked autophagy, and inhibition of autophagy enhanced the anticancer effects of Danu, indicating a cyto-protective role of Danu-induced autophagy. Our proteomic data and Western blotting assays showed the PI3K/Akt/mTOR signaling pathway was involved in the inducing effect of Danu on apoptosis and autophagy. Collectively, our findings have demonstrated that the Aurora kinases inhibition with danusertib results in global proteomic response and exerts anticancer effects in Hep3B cells involving regulation of cell cycle, apoptosis and autophagy and associated signaling pathways.
Keywords: Hepatocellular carcinoma; Aurora kinases; danusertib; cell cycle; apoptosis; autophagy, quantitative proteomics.
Introduction
Liver cancer is a major malignancy burden and global public health problem 1, 2. According to the GLOBOCAN 2012 by World Health Organization, there were 782,500 new cases of liver cancer (accounting for 5.6% of all cancers) and 745,500 liver cancer-caused deaths (9.1% of total) globally in 2012 3. This malignancy is the 5th most common cancer in men (554,000 cases) and the 9th in women (228,000 cases). According to the National Cancer Institute, 40,710 patients would be diagnosed with liver cancer in the United States (US) in 2017 and 28,920 would die of this disease. Liver cancer is the 10th most common cancer and the 5th most common cause of cancer death among men; incidence of liver cancer has tripled in the US since 1980 4. In China, liver cancer is the 4th most common cancer after lung, stomach, and esophagus cancer; it is the 3rd leading cause of cancer-related death after lung and stomach cancer 5. The estimated new liver cancer cases were 466,100 (343,700 men and 122,300 women) and estimated deaths due to liver cancer were 422,100 (310,600 men and 111,500 women) in China in 2015 5. The overall 5-year survival rate for liver cancer patients is ~18%; for liver cancer patients who are diagnosed at an early stage, the 5-year survival rate is 31%; but the 5-year survival rate drops down to only 3% if the cancer has metastasized in liver cancer patients in the US 6. The main risks of liver cancer in the US are liver cirrhosis and nonalcoholic fatty liver disease; whereas hepatitis B and C infection and aflatoxin exposure are the major risk factors for liver cancer in China 7, 8. Hepatocellular carcinoma (HCC) accounts for 85-90% of all liver cancers, which can be divided into two subclasses: proliferative and non-proliferative, each depending on distinct pathways 9, 10. Mutations in the TERT promoter, p53, ARID1A, CDKN2A, CTNNB1, AXIN1, and CCND1 are very common in HCC 7, 8, 10. Critical molecular events in HCC pathogenesis include activated WNT signaling pathway due to mutations in CTNNB1 and AXIN1, alterations in p53 and the PI3K/Akt/mTOR pathways, aberrant cell cycle regulation and angiogenesis, and epigenetic changes 10. The main treatment modalities for HCC include surgery, radiofrequency ablation, percutaneous ethanol injection, radiation therapy, chemoembolization and radioembolization, targeted therapy, and immunotherapy 7, 11-15. To date, only sorafenib (Nexavar, co-developed by Bayer and Onyx Pharmaceuticals Inc.) and regorafenib (Stivarga, developed by Bayer HealthCare Pharmaceuticals Inc.), two targeted multi-kinase inhibitors, have been approved by the Food and Drug Administration (FDA) to treat HCC patients 16. Sorafenib has been used as a first-line therapy in patients with advanced stage HCC for a decade and has set the stage for personalized targeted therapy 15, 17. Its role has ranged from monotherapy to neoadjuvant and adjuvant treatment with surgical resection, liver transplantation and chemoembolization. Regorafenib inhibits multiple kinases involved in tumor proliferation and angiogenesis, causing a survival benefit as a second-line therapy in HCC when sorafenib fails 16, 18. However, tumor resistance and adverse effects are two major limiting factors for the targeted therapy of liver cancer 19; the overall prognosis remains dismal in patients with late-stage liver cancer. Therefore, seeking new therapeutic agents for advanced HCC is still mandatory.
Aurora kinases, consisting of three family members, Aurora kinase A, B and C, are one of the serine-threonine kinases and involved in multiple mitotic events 20. In recent years, aberrant expression of these kinases has been proved to be tumorigenic in many types of cancer, including HCC 21, 22. Targeting Aurora kinase has become an attractive target for new anticancer treatments and many Aurora kinase inhibitors have been developed and evaluated in preclinical to clinical studies 23-26. Recently, we have found that the Aurora kinase A inhibitor alisertib induces autophagy and cell cycle arrest and enhances chemosensitivity in HepG2 cells 27. Furthermore, we have reported that danusertib (Danu) 28, a small molecule pan-Aurora kinase inhibitor, kills ovarian, gastric and breast cancer and leukemia cells via induction of apoptosis and autophagy involving AURKB/p70S6K/RPL15 and PI3K/Akt/mTOR signaling pathways 29-32. Several Phase I and II studies have shown that Danu has limited efficacy but is well tolerated by cancer patients 33-37. However, the evidence on its effects on HCC, especially the effect on the global proteomic response, is still limited.
Mass spectrometry-based proteomics is increasingly employed in a quantitative way to investigate protein expression changes in biological samples, often based on labeling of samples with stable isotopes that are introduced chemically or metabolically. Stable-isotope labeling by amino acids in cell culture (SILAC) is a powerful and increasingly popular approach for quantitative proteomics studies in vitro and in vivo 38-41. In the SILAC study, two cell populations are cultured in the presence of heavy or light amino acids (typically lysine and/or arginine), one of them is subject to a perturbation (e.g. drug exposure), and then both are combined, processed, and analyzed. Incorporation of the "heavy" amino acid occurs through cell growth, protein synthesis, and turnover. SILAC allows "light" and "heavy" proteomes to be distinguished by mass spectrometry while avoiding any chemical derivatization and associated purification. SILAC can be applied to systemically evaluate global protein profile, investigate and identify the target networks of drugs, drug toxicity, and new biomarkers for diseases 40, 42, 43. The purpose of this study was to evaluate the proteomic responses and validated the molecular targets of Danu in Hep3B cells using a combination of proteomic and cell-based approaches, with a focus on cell cycle progression, apoptosis, and autophagy.
Materials and methods
Chemicals and Reagents
Danu was purchased from Selleckchem Inc (Houston, TX, USA). Dulbecco's phosphate buffered saline (PBS), fetal bovine serum (FBS), thiazolyl blue tetrazolium bromide (MTT), RNase A, propidium iodide, 13C6 L-lysine, 13C6 15N4 L-arginine and L-arginine, and chloroquine (CQ) were purchased from Sigma-Aldrich Inc. (St Louis, MO, USA). Dulbecco's Modified Eagle's Medium (DMEM) was obtained from Corning Cellgro Inc (Herndon, VA, USA). Phenol red-free culture medium and 4,6-diamidino-2-phenylindole were bought from Invitrogen Inc. (Carlsbad, CA, USA). The Cyto-ID® autophagy detection kit was obtained from Enzo Life Sciences Inc. (Farmingdale, NY, USA). The Pierce bicinchoninic acid (BCA) protein assay kit, skim milk, and Western blot substrate were purchased from Thermo Scientific Inc. (Waltham, MA, USA). Polyvinylidene difluoride (PVDF) membrane was purchased from EMD Millipore Inc. (Bedford, MA, USA). Primary antibodies against human cyclin B1, p-cyclin B1 at Ser 133, cell division cycle protein 2 homologue (CDC2), p-CDC2 at Tyr15, p-CDC25C at Ser216, cytochrome c, Bcl-2-like protein 4/Bcl-2-associated X protein (Bax), B-cell lymphoma-extra large (Bcl-xl), B-cell lymphoma 2 (Bcl-2), cleaved caspase 9, cleaved caspase 3, p53 up-regulated modulator of apoptosis (PUMA), cleaved poly-ADP-ribose polymerase (cleaved-PARP), Akt, p-Akt at Ser473, mammalian target of rapamycin (mTOR), p-mTOR at Ser2448, PI3K, p-PI3K/p85 at Tyr458, beclin1, SQSTM1/p62, microtubule-associated protein 1A/1B-light chain 3 (LC3)-I, and LC3-II were all purchased from Cell Signaling Technology Inc. (Beverly, MA, USA). The antibody against human β-actin was obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA).
Cell line, cell culture and Danu treatment
Hep3B cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM medium supplemented with 10% heat-inactivated FBS and 1% penicillin/streptomycin. The cells were maintained in a 5% CO2/95% air-humidified incubator at 37°C. Danu was dissolved in DMSO with a stock concentration of 100 mM and the stock solution was stored at -20°C. Danu was freshly diluted to the predetermined concentrations with culture medium. The control cells received the vehicle only. Hep3B cells were treated with Danu at 0.01, 0.1, and 0.5 µM for 24 hr, and then subject to flow cytometric analysis and confocal microscopic examination. The protein samples were subject to Western blotting assay.
Quantitative proteomic study using SILAC
Quantitative proteomic experiments were performed using a SILAC-based approach as described previously 29, 44. Briefly, Hep3B cells were cultured in DMEM-F12 medium (for SILAC) with (heavy) or without (light) stable isotope labeled amino acids (13C6 L-lysine and 13C6 15N4 L-arginine) and 10% dialyzed FBS. Hep3B cells cultured in heavy medium were treated with 0.5 µM Danu for 24 hr after six cell doubling times. After treatment with Danu, Hep3B cells were harvested and lysed with hot lysis buffer (100 mM Tris base, 4% sodium dodecyl sulfate [SDS], and 100 mM dithiothreitol), and protein concentration was determined using ionic detergent compatibility reagent. Subsequently, equal amounts of heavy and light protein samples were combined to reach a total volume of 30-60 µL containing 300-600 µg protein. The combined protein sample was digested using a filter-aided sample prep (FASP™) protein digestion kit and desalted using a C18 solid-phase extraction column. The peptide mixtures (5 µL) were subject to the hybrid linear ion trap (LTQ Orbitrap XL™, Thermo Fisher Scientific Inc.). Liquid chromatography-tandem mass spectrometry was performed using a 10 cm long, 75 µm (inner diameter) reversed-phase column packed with 5 µm diameter C18 material having a pore size of 300 Å (New Objective Inc., Woburn, MA, USA) with a gradient mobile phase of 2-40% acetonitrile in 0.1% formic acid at 200 µL per min for 125 min. The Orbitrap full mass spectrometry scanning was performed at a mass (m/z) resolving power of 60,000, with positive polarity in profile mode (M + H+). Peptide SILAC ratio was calculated using MaxQuant version 1.2.0.13. The SILAC ratio was determined by averaging all peptide SILAC ratios from peptides identified of the same protein. The protein IDs were identified using Scaffold 4.3.2 from Proteome Software Inc. (Portland, OR, USA).
Pathway analysis using IPA and DAVID
The pathway was analyzed using ingenuity pathway analysis (IPA) from QIAGEN (Redwood City, CA, USA) and the Database for Annotation, Visualization and Integrated Discovery (DAVID) 45. DAVID was also used to provide biological functional interpretation of the potential targets of Danu. Enrichment scores and Fisher's exact test P-values (and corresponding false discovery rate [FDR]) were calculated to identify which functional related gene groups were significantly enriched in the target list. These significant enriched gene groups could explain the mechanism of action of Danu systematically.
Cell viability assay
The MTT assay was performed to examine Hep3B cells viability. Hep3B cells were seeded in 96-well culture plates at a density of 8,000 cells per well. After incubation for 24 hr, the cells were treated with Danu at different concentrations ranging from 0.01 to 50 μM for 24 hr. In the autophagy inhibition experiments, the cells were coincubated with Danu and CQ for 24 hr. Absorbance at the 450 nm wavelength was measured with a Synergy H4 Hybrid microplate reader (BioTek Inc., Winooski, VT, USA). IC50 values were determined using the relative viability over Danu concentration curve by GraphPad Prism 6.0 (GraphPad Software Inc., La Jolla, CA, USA).
Cell cycle distribution analysis
The effect of Danu on cell cycle distribution of Hep3B cells was examined using flow cytometry as previously described 46. Propidium iodide is used as a DNA stain in cell cycle analysis. A total number of 1×104 cells were subject to cell cycle analysis using a flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA).
Quantification of cellular apoptosis
The effect of Danu on the apoptosis of Hep3B cells was quantitated using the annexin V: PE apoptosis detection kit (BD Biosciences Inc.) according to the manufacturer's instruction. In brief, the cells were collected after Danu treatment at different concentrations over 24 hr, or evaluated for different time intervals, and resuspended and incubated in 100 μL 1 × binding buffer containing 5 μL annexin V:PE and 5 μL 7-amino-actinomycin D (7-AAD) in the dark at room temperature for 15 min. The number of apoptotic cells was analyzed by flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA) within 1 hr.
Quantification of cellular autophagy
To determine the effect of Danu on autophagy in Hep3B cells, the intracellular autophagy level was examined using the Cyto-ID® autophagy detection kit (No. ENZ-51031-K200) according to the manufacturer's instructions. In brief, cells were collected after Danu treatment at different conditions and resuspended in 250 μL of assay buffer containing 5% FBS. Following with the addition of 250 μL of the diluted Cyto-ID® Green stain solution, cells were incubated at room temperature in the dark for 20 min, then cells were collected and washed with 1 × assay buffer. The percentage of autophagy cells was analyzed using the green (FL1) channel of a flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA).
Confocal fluorescence microscopic examination
The cellular autophagy level was further detected using confocal fluorescence microscopy. The cells were also dyed with Cyto-ID® green detection and Hoechst 33342 nuclear stain reagent contained in the Cyto-ID® autophagy detection kit (No. ENZ-51031-K200) according to the manufacturer's instructions. The cells were examined using a Leica TCS SP2 laser scanning confocal microscopy (Leica Microsystems, Wetzlar, Germany) using a standard FITC filter set for imaging the autophagic signal at wavelengths of 405/488 nm.
Western blotting analysis
The expression levels of various cellular proteins were determined using Western blotting assays. Protein samples were collected in the RIPA buffer (50 mmol HEPES at pH 7.5, 150 mmol NaCl, 10% glycerol, 1.5 mmol MgCl2, 1% Triton-X 100, 1 mmol EDTA at pH 8.0, 10 mmol sodium pyrophosphate, 10 mmol sodium fluoride) containing the protease inhibitor and phosphatase inhibitor cocktails, and centrifuged at 3,000 × g for 10 min at 4oC. Protein concentrations were determined using the BCA assay and 20 μg samples were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer and electrophoresed on 7-12% SDS-PAGE mini-gel after thermal denaturation at 95oC for 5 min. Proteins were transferred onto PVDF membrane at 400 mA for 2 h at 4oC. Membranes were probed with indicated primary antibody overnight at 4oC and then blotted with respective secondary anti-mouse or anti-rabbit antibody. Visualization was performed using Bio-Rad ChemiDocTM XRS system (Hercules, CA, USA) with enhanced chemiluminescence substrate. The blots were analyzed using ImageLab 3.0 (Hercules) and protein level was normalized to the matching densitometric value of β-actin as internal control.
Statistical analysis
Data are presented as the mean ± standard deviation (SD). Comparisons of multiple groups were evaluated by one-way analysis of variance followed (ANOVA) by Tukey's multiple comparison procedure. A value of P<0.05 was considered statistically different. Assays were performed at least three times independently.
Results
Proteomic response to Danu treatment in Hep3B cells
First, we performed a SILAC-based proteomic study to quantitatively determine the general proteomic response to 0.5µM Danu treatment in Hep3B cells. Danu treatment increased the expression level of 279 protein molecules, but decreased the expression level of 260 protein molecules (Table S1). Subsequently, these proteins were subject to IPA and DAVID analysis. The IPA results showed that 109 signaling pathways were regulated by Danu in Hep3B cells (Table S2, Figure S1). KEGG pathway analysis showed that 24 signaling pathways were modulated by Danu (Table 2). As shown in Table S3, 18 functional clusters were identified to be significantly enriched (enrichment score > 5). The top five molecular and cellular functions regulated by Danu in Hep3B cells (IPA analysis) were cellular growth and proliferation, protein synthesis, cell death and survival, RNA post-transcriptional modification, and gene expression (Table 1). Then we focused on analyzing the effect of Danu on cell cycle distribution, apoptosis, and autophagy, which are related to cellular growth and proliferation, cell death, and survival.
Table 2.
The KEGG pathways by the DAVID database for the target list of Danu in Hep3B cells
| Pathway | Gene count | P-Value | Fold Enrichment | |
|---|---|---|---|---|
| 1 | Spliceosome | 26 | 5.24×10-10 | 4.34 |
| 2 | Aminoacyl-tRNA biosynthesis | 12 | 1.93×10-6 | 6.15 |
| 3 | Ribosome | 17 | 2.28×10-6 | 4.11 |
| 4 | Pathogenic Escherichia coli infection | 11 | 2.86×10-4 | 4.06 |
| 5 | Glycolysis / Gluconeogenesis | 11 | 4.40×10-4 | 3.85 |
| 6 | Antigen processing and presentation | 12 | 1.68×10-3 | 3.04 |
| 7 | Methane metabolism | 4 | 1.89×10-3 | 14.01 |
| 8 | Fatty acid metabolism | 8 | 2.39×10-3 | 4.20 |
| 9 | Cysteine and methionine metabolism | 7 | 4.67×10-3 | 4.33 |
| 10 | Pentose phosphate pathway | 6 | 5.58×10-3 | 5.04 |
| 11 | Proteasome | 8 | 6.10×10-3 | 3.58 |
| 12 | Citrate cycle (TCA cycle) | 6 | 1.41×10-2 | 4.07 |
| 13 | Oocyte meiosis | 12 | 1.44×10-2 | 2.29 |
| 14 | Prion diseases | 6 | 2.32×10-2 | 3.60 |
| 15 | Tight junction | 13 | 2.42×10-2 | 2.04 |
| 16 | Glutathione metabolism | 7 | 2.94×10-2 | 2.94 |
| 17 | Pyruvate metabolism | 6 | 3.89×10-2 | 3.15 |
| 18 | Tryptophan metabolism | 6 | 3.89×10-2 | 3.15 |
| 19 | Systemic lupus erythematosus | 10 | 4.39×10-2 | 2.12 |
| 20 | Regulation of actin cytoskeleton | 17 | 4.77×10-2 | 1.66 |
| 21 | Fatty acid elongation in mitochondria | 3 | 5.18×10-2 | 7.88 |
| 22 | Focal adhesion | 16 | 5.2×10-2 | 1.67 |
| 23 | Cell cycle | 11 | 7.13×10-2 | 1.85 |
| 24 | Base excision repair | 5 | 8.16×10-2 | 3.00 |
Abbreviations: KEGG, Kyoto encyclopedia of genes and genomes; Danu, danusertib
Table 1.
Top five molecular and cellular functions regulated by Danu in Hep3B cells.
| Names | P-value range | Molecules |
|---|---|---|
| Cellular growth and proliferation | 4.19×10-4 - 7.19×10-33 | 263 |
| Protein synthesis | 1.69×10-4 - 3.29×10-30 | 138 |
| Cell death and survival | 4.60×10-4 - 8.60×10-30 | 258 |
| RNA post-transcriptional modification | 3.34×10-4 - 8.30×10-22 | 54 |
| Gene expression | 5.47×10-4 - 4.95×10-20 | 153 |
Danu inhibits the proliferation of Hep3B cells
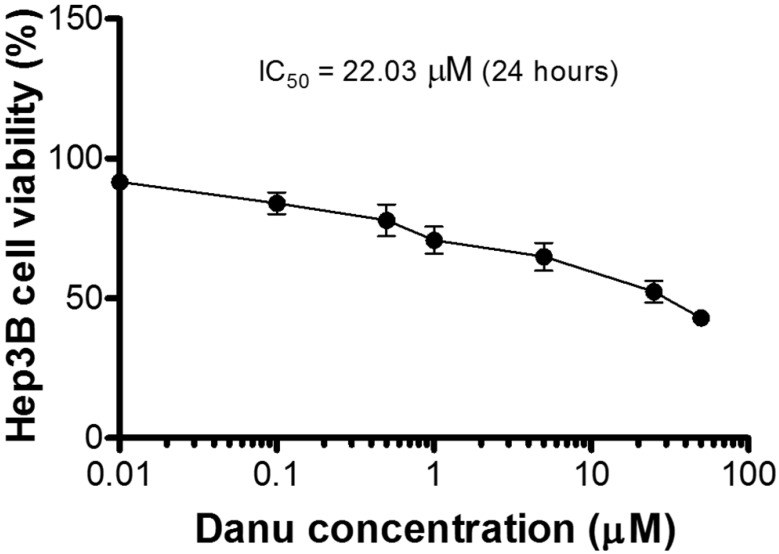
We first verified the effect of Danu on the proliferation of Hep3B cells using MTT assay. A concentration-dependent inhibitory effect of Danu on the growth of Hep3B cells was observed (Figure 1).
Figure 1.
Cytotoxicity of Danu towards Hep3B cells determined by the MTT assay.
Danu regulates the cell cycle distribution of Hep3B cells
Compared to the control cells (100%), the percentage of the viability of Hep3B cells decreased to 91.7, 84.1, 77.9, 70.9, 64.8, 52.4, and 43.1%, respectively, when cells were treated with Danu at 0.01, 0.1, 0.5, 1, 5, 25, and 50 µM for 24 hr. The IC50 value was 22.03 µM. The results show that Danu inhibits the proliferation of Hep3B cells.
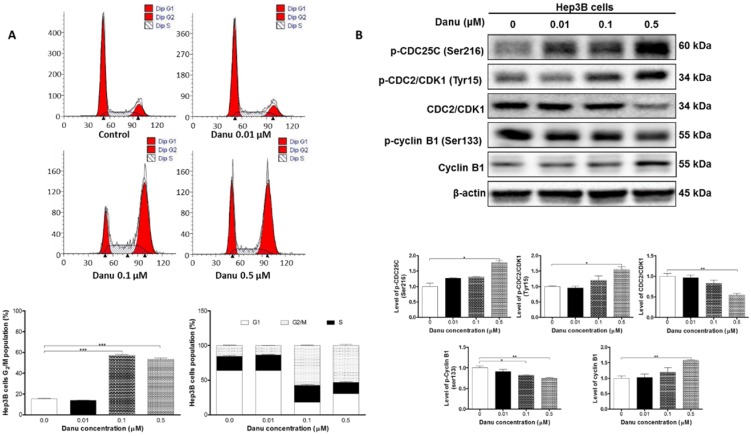
As shown in Table S2 and Table 2, both IPA (Pathway No. 30) and DAVID (Pathway No. 23) analysis have identified the effect of Danu on the cell cycle distribution. We next used flow cytometry to validate this effect. The data showed that Danu treatment induced cell cycle arrest at G2/M phase in a concentration-dependent manner. Compared to the basal level (15.4%), the percentage of Hep3B cells arrested at G2/M phase ascended to 57.3% (P<0.001, Figure 2A) and 53.3% (P<0.001, Figure 2A), when cells were treated with Danu at 0.1 and 0.5 μM for 24 hr, respectively. On the contrary, there was a marked reduction in the number of cells at G1 phase when treated with 0.1 and 0.5 μM Danu for 24 hr.
Figure 2.
Danu treatment arrested Hep3B cells in G2/M phase via regulating the expression of key cell cycle regulators. Hep3B cells were treated with Danu at 0.01, 0.1, and 0.5 µM for 24 hr, and then subject to flow cytometric analysis. The protein samples were subject to Western blotting assay. Representative flow cytometric plots of cell cycle distribution (A) and blots of p-CDC25C (Ser216), p-CDC2/CDK1 (Tyr15), CDC2/CDK1, p-cyclin B1 (Ser133), and cyclin B1 (B). Bar graphs show the percentage of Hep3B cells in G1, S, and G2/M phases and the relative level of the above proteins. β-Actin was used as the internal control. Data are the mean ± SD of three independent experiments. *P<0.05, **P<0.001 and ***P<0.001 by one-way ANOVA.
Danu alters the expression of key cell cycle regulators in Hep3B cells
We next used Western blotting assay to further validate the effect of Danu on the cell cycle distribution. Our results demonstrated treatment with Danu at 0.5 μM resulted in a 45.8% (P<0.01, Figure 2B) decrease in the total level of CDC2 and a 55.3% (P<0.05, Figure 2B) increase in the phosphorylation (at Tyr15) level. There was a remarkable decrease (57%, P<0.01, Figure 2B) in the level of p-cyclin B1 at Ser133, while there was a marked increase (30.2%, P<0.01, Figure 2B) in the total level of cyclin B1 when cells were treated with 0.5 μM Danu. We further detected the level of p-CDC25C (Ser216), a kinase responsible for the de-phosphorylation of CDC2 (Tyr15). Treatment with Danu at 0.5 μM up-regulated the expression level of p-CDC25C (Ser216) by 77.4% (P<0.05, Figure 2B). These findings indicate Danu treatment results in an inactivation of CDC2/CDK1-cyclin B1 complex, which leads to cell arrest at G2/M phase.
Danu induces apoptosis in Hep3B cells
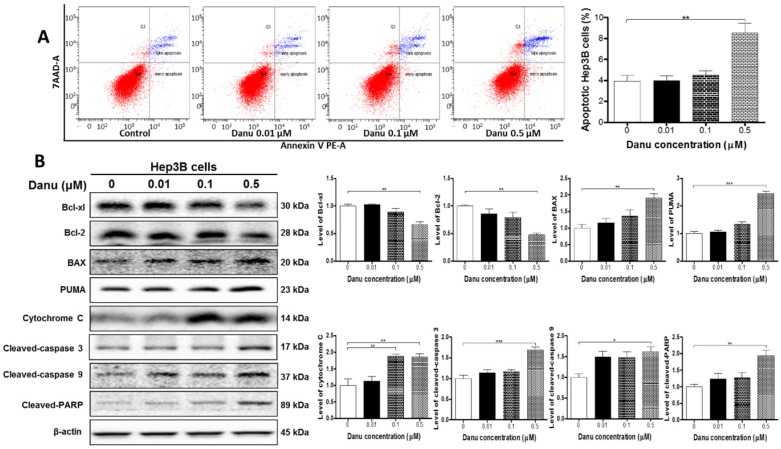
Both IPA (Table S2, Pathway No. 63 and No. 66) and DAVID (Table S3, Cluster No. 18) analysis have revealed the effect of Danu on the apoptosis of Hep3B cells. We next used flow cytometry and Western blotting assay to validate the pro-apoptotic effect of Danu. As shown in Figure 3A, compared to the control cells, treatment with Danu at 0.5 μM resulted in a 2.16-fold increase in the overall apoptotic rate (P<0.01, Figure 3A). The Western blotting assays showed that Danu treatment at 0.5 μM significantly up-regulated the level of cleaved PARP 1.9-fold, a marker of cellular apoptosis.
Figure 3.
Danu treatment induced apoptosis via mitochondria-dependent pathway. Hep3B cells were treated with Danu at 0.01, 0.1, and 0.5 µM for 24 hr, and then subject to flow cytometric analysis. The protein samples were subject to Western blotting assay. Representative flow cytometric plots of cell cycle distribution (A) and blots of cytochrome c, Bax, Bcl-xl, Bcl-2, cleaved caspase 9, cleaved caspase 3, PUMA, and cleaved PARP (B). Bar graphs show the percentage of autophagic Hep3B cells and the relative level of the above proteins. β-Actin was used as the internal control. Data are the mean ± SD of three independent experiments. *P<0.05, **P<0.001 and ***P<0.001 by one-way ANOVA.
Since DAVID (Table S3, Cluster No. 18) analysis have demonstrated a number of protein molecules involved in the regulation of apoptosis, we further examined the expression levels of pro-apoptosis and anti-apoptosis proteins. As shown in Figure 3B, compared to the control cells, there were a 33.5% and 52.1% reduction, respectively, in the level of Bcl-xl and Bcl-2 (P<0.01, Figure 3B), while there was a 1.9-fold elevation in the level of Bax (P<0.01, Figure 3B), when Hep3B cells were treated with 0.5 μM Danu. Mitochondrial dysfunction (Table 1, Pathway No. 42) following remarkable increase in the ratio of Bax/Bcl-2 leads to the release of cytochrome c. Indeed, we observed that Danu treatment at 0.1 and 0.5 μM increased the level of cytosolic cytochrome c by 88.2% and 85.6%, respectively (P<0.01, Figure 3B). Subsequently, we observed a remarkable increase in the level of cleaved caspases 9 (1.62-fold) and 3 (1.69-fold). Additionally, Danu also up-regulated the negative regulator of Bcl-2 family, PUMA, in Hep3B cells. Treatment of cells with Danu at 0.5 μM resulted in 2.4-fold increase in the level of PUMA compared to the control cells (P<0.001, Figure 3B). Taken together, the results indicate that Danu exhibits via activation of mitochondria-dependent pathway.
Danu induces autophagy in Hep3B cells
As shown in Table S2 and Table 2, a number of pathways were regulated by Danu treatment based on the IPA and DAVID analysis. Of note, near one third (IPA analysis) or half (DAVID analysis) of them were involved in the nutrition and energy metabolism. Functional cluster No. 18 is also in regard to nutrition and energy metabolic process (Table S3). These findings indicate the regulatory role of Danu in intracellular hemostasis.
We speculated that Danu treatment would exert a significant effect on the autophagy in Hep3B cells. In order to validate the pro- or anti-autophagic effect of Danu, flow cytometry, confocal microscopic examination, and Western blotting assay were performed.
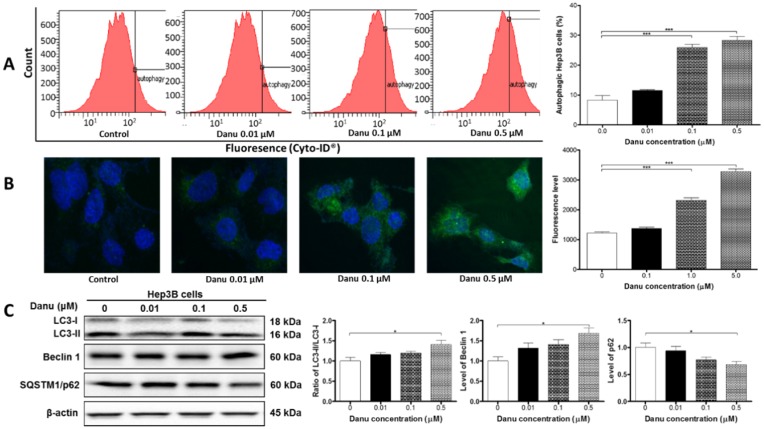
As shown in Figure 4, treatment with Danu markedly increased the level of autophagy in a concentration-dependent manner. Our flow cytometric data showed treatment with Danu at 0.1 and 0.5 µM for 24 hr resulted in a 3.1- and 3.4-fold increase in the percentage of autophagic cells compared to the control cells (basal level = 8.3%) (P<0.001, Figure 4A). The confocal microscopic examination showed that autophagic level was increased 1.9- and 2.7-fold when Hep3B cells were treated with Danu at 0.1 and 0.5 µM for 24 hr, respectively (P<0.001, Figure 4B).
Figure 4.
Danu treatment induced autophagy. Hep3B cells were treated with Danu at 0.01, 0.1, and 0.5 µM for 24 hr, and then subject to flow cytometric analysis and confocal fluorescence microscopic examination. The protein samples were subject to Western blotting assay. (A) Histograms show autophagy of HepG2 cells and bar graphs show the percentage of autophagic Hep3B cells. (B) Representative confocal microscopic images show autophagy of Hep3B cells and bar graph shows the fluorescence level. (C) Representative blots of LC3-I, LC3-II, beclin 1, and p62 determined by Western blotting assay. β-Actin was used as the internal control. Bar graphs show the relative level of the above proteins. Data are the mean ± SD of three independent experiments. *P<0.01, **P<0.001, and ***P<0.001 by one-way ANOVA.
As shown in Figure 4C, the result of LC3 turnover assay was positive, since the ratio of LC3-II/LC3-I was elevated by 41% when cells were treated with 0.5 µM Danu (P<0.05, Figure 4C). The expression level of another autophagic marker, beclin 1, was up-regulated (P<0.05, Figure 4C). SQSTM1/p62 accumulates when autophagy is inhibited, and decreased level can be observed when autophagy is induced. Treatment of Hep3B cells with Danu at 0.5 μM resulted in a 32% decrease in the level of SQSTM1/p62 (P<0.05, Figure 4C). In aggregate, these results demonstrate the inducing effect of Danu on autophagy in Hep3B cells.
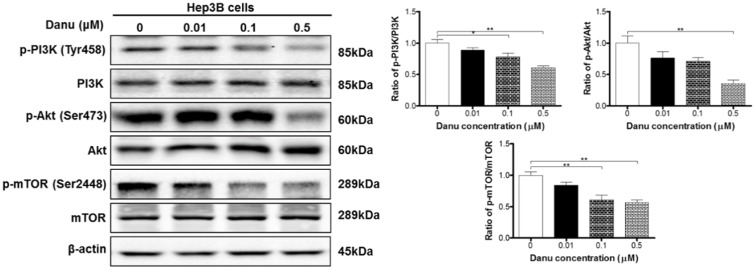
Danu regulates the PI3K/Akt/mTOR signaling pathway in Hep3B cells
Accumulating evidence suggest that the PI3K/Akt/mTOR signaling pathway is involved in the regulation of both apoptosis and autophagy 47. Our data have demonstrated that Danu induced apoptosis and autophagy. As shown in Table 1, mTOR signaling, one of the top five IPA canonical pathways, along with PI3K/Akt signaling (Pathway No. 29), have been regulated by Danu treatment. So we next used Western blotting assay to validate the regulatory effect of Danu on this well-known pathway. Danu treatment at 0.1 and 0.5 µM markedly inhibited the phosphorylation of PI3K at Tyr199 in Hep3B cells, but did not impact the expression of total PI3K, leading to a 22.1% and 39.3% decrease in the ratio of p-PI3K/PI3K, respectively (P<0.05 or 0.01, Figure 5). Danu inactivated PI3K resulted in an inhibition of the phosphorylation of Akt at Ser473. Consequently, the ratio of p-Akt/Akt decreased 64.6% when cells were exposed to 0.5 µM Danu for 24 hr (P<0.01, Figure 5). Inhibition of mTOR was detected following the inactivation of PI3K/Akt pathway in these HCC cells. Danu inhibited the phosphorylation of mTOR and the ratio of p-mTOR/mTOR was decreased 39.2% and 43.6% when treated 0.1 and 0.5 µM Danu, respectively (P<0.01, Figure 5). Collectively, these findings indicate that Danu induces apoptosis and autophagy via the PI3K/Akt/mTOR signaling pathway in Hep3B cells.
Figure 5.
Danu regulates the PI3K/Akt/mTOR signaling pathway. Hep3B cells were treated with Danu at 0.01, 0.1, and 0.5 µM for 24 hr, and the protein samples were subject to Western blotting assay. Representative blots of phosphorylation level of PI3K, Akt, and mTOR and the total levels of PI3K, Akt, and mTOR, determined by Western blotting assay. Bar graphs show the ratio of p-PI3K/PI3K, p-Akt/Akt, and p-mTOR/mTOR in Hep3B cells. β-Actin was used as the internal control. Data are the mean ± SD of three independent experiments. *P<0.05, **P<0.001, and ***P<0.001 by one-way ANOVA.
Inhibition of autophagy enhances the anticancer effect of Danu in Hep3B cells
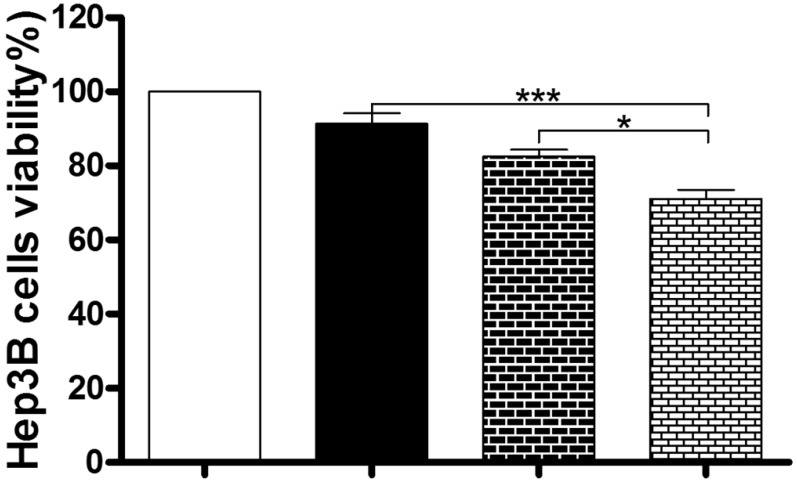
Under most circumstances, autophagy represents a pro-survival process against apoptosis; but in other special settings, it culminates in alternative cell death 47. So we next used the MTT assay to examine the role of autophagy in the anticancer effect of Danu in Hep3B cells. Coincubation with 40 μM CQ and 0.5 µM Danu markedly decreased the viability of Hep3B cells (71.0%), compared to the group treated with 0.5 µM Danu (91.4%) or 40 μM CQ (82.5%) alone (P<0.001 or 0.05, Figure 6). These preliminary data suggest that Danu might trigger a cyto-protective autophagy in Hep3B cells.
Figure 6.
Inhibition of autophagy enhanced the anticancer effect of Danu. Hep3B cells were incubated with Danu or CQ alone, or coincubated for 24 hr, and then subject to the MTT assay. Data are the mean ± SD of three independent experiments. *P<0.05, **P<0.001, and ***P<0.001 by one-way ANOVA.
Discussion
Studies have showed that Aurora kinases A and B are overexpressed in human HCCs which is associated with aggressive tumor characteristics and poor prognosis 22, 48, 49. These findings indicate the importance of Aurora kinases as potential targets for the treatment of HCC. A series of Aurora kinase inhibitors have been tested for HCC treatment in preclinical and clinical studies 28, 33-35, 50-52. In the present study, we evaluated the global proteomic responses to Danu, including the related molecular targets and signaling pathways, in Hep3B cells using a SILAC-based quantitative proteomic approach. The proteomic study and subsequent validating assays revealed that Danu inhibited the proliferation and induced cell cycle arrest, apoptosis, and autophagy in Hep3B cells, with the involvement of a number of function proteins and pathways.
SILAC-based proteomics can quantitatively and globally evaluate the proteomic response to a given compound and identify its potential targets and related signaling pathways in vitro and in vivo 38-41. Our proteomic data demonstrated the molecular targets regulated by Danu were involved in a number of important cellular functions, with cellular growth and proliferation, cell death, and survival being listed in the top five. Besides, several signaling pathways relevant to these functions have been revealed, such as cell cycle regulation, apoptosis signaling, and Myc-mediated apoptosis signaling. The functional cluster analysis based on the DAVID also depicted these regulatory effects of Danu in Hep3B cells. The proteomic results suggest that Danu may target these signaling molecules to elicit its anticancer effects in the treatment of HCC. Notably, we further validated the proteomic responses to Danu treatment in Hep3B cells.
Danu inhibits all three members of the Aurora kinase family, Aurora A, B, and C 28. Inhibition of Aurora kinases lead to improper mitotic progression, including G2/M phase arrest 28. We examined the effect of Danu on the cell cycle distribution and found that Danu treatment induced cell cycle arrest at G2/M phase in Hep3B cells, which also verified the proteomic data. In general, a cell with suppressed CDC2/CDK1-cyclin B1 complex activity would tend to be arrested in the G2/M phase, whereas a cell with elevated CDC2/CDK1-cyclin B1 complex activity would be favored to enter mitosis. We have detected inhibition of the dephosphorylation of CDC2/CDK1 at Tyr15, inhibition of the phosphorylation of cyclin B1 at Ser133 and increased expression level of total CDC2/CDK1, resulting in inactivation of CDC2/CDK1-cyclin B1 complex and consequently the inhibition of the cell cycle progression. Our data also showed that, the upstream regulator of CDC2/CDK1 and CDC25C (Ser216) were involved. In aggregate, the proteomics and verification data both indicate that Danu exerts a marked cell cycle arresting effect via regulation of key functional proteins.
Disruption of mitochondrial function and the resultant cytochrome c release initiate apoptosis process, with the latter being activated caspase cascade, including activation of caspases 9 and 3 53, 54. Our proteomic data demonstrated that Danu regulated mitochondrial function and apoptotic cell death. In the present study, the flow cytometric and Western blotting results have verified the pro-apoptotic effect of Danu. The finding also showed cytosolic level of cytochrome c was significantly increased and caspase cascade was markedly activated in response to Danu treatment, which contributes to Danu-induced apoptosis of Hep3B cells. Pro- and anti-apoptotic members of the Bcl-2 family are highly involved in apoptosis 53. Anti-apoptotic members of Bcl-2 are suppressed by post-translational modification and/or by increased expression of PUMA 55. The proteomic study also showed a number of proteins were involved in the regulation of apoptosis. The Western blotting assay showed that Danu disrupted the balance between Bax and Bcl-2 with involvement of PUMA.
Autophagy is the major intracellular degradation system and plays a critical role in maintaining intracellular hemostasis 56-58. Under most circumstances, autophagy emerges as a biological process that promotes cell survival 56, 57. In recent years, the distinct role of autophagy in cell growth, survival, and death has been demonstrated to be context-dependent in cancer cells 59. Under certain circumstances, the pro-survival functions of autophagy may be deleterious and lead to cell death 59. Our proteomics and verification data both demonstrated that Danu induced autophagy in Hep3B cells. The preliminary study showed inhibition of autophagy with chloroquine markedly decreased the viability of Hep3B cells. These data suggest that Danu might trigger a cyto-protective autophagy in Hep3B cells and supports potential application of autophagic inhibitors for HCC therapy.
Although apoptosis and autophagy are regulated by distinct signaling pathways, they have substantial interconnections in determining cell fate 60. In recent years, various signaling pathways have been implicated in the up-regulating or down-regulating of apoptosis and autophagy, respectively. The PI3K/Akt/mTOR signaling is a well-known pathway involved in the regulation of both apoptosis and autophagy and may contribute to simultaneous or sequential induction of both processes 47. Our proteomic study revealed that PI3K/Akt/mTOR signaling pathway was regulated by Danu. Subsequent data demonstrated the Danu inhibited the phosphorylation of PI3K, Akt, and mTOR. We deduce that PI3K/Akt/mTOR signaling pathway might be involved in regulating apoptosis and autophagy in Hep3B cells, although further studies are mandatory. Apart from the signaling pathways, the dual regulatory roles of some key interconnected molecules in these two pathways have been identified, such as Bcl-2 family members, beclin 1, caspases, and p53 47, 61, 62. In the present study, the expression of Bcl-2, Bcl-xl, and beclin 1 was regulated by Danu; but their roles in the crosstalk between apoptosis and autophagy remain to be clarified in future studies.
In conclusion, this SILAC proteomic study shows that Danu regulates a number of functional proteins and molecular signaling pathways, Danu inhibits cell proliferation, induces cell cycle arrest, apoptosis, and cyto-protective autophagy. Inhibition of autophagy increases the sensitivity of Hep3B cells to Danu. More functional and mechanistic studies are needed to elucidate the role of Danu in the treatment of HCC.
Supplementary Material
Supplementary figures and tables.
References
- 1.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet; 2018. [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–8. doi: 10.1016/j.canlet.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Beal EW, Tumin D, Kabir A, Moris D, Zhang XF, Chakedis J, Trends in the mortality of hepatocellular carcinoma in the United States. J Gastrointest Surg; 2017. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 6.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–89. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 7.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 8.Ringehan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci; 2017. p. 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Are C, Meyer B, Stack A, Ahmad H, Smith L, Qian B. et al. Global trends in the burden of liver cancer. J Surg Oncol. 2017;115:591–602. doi: 10.1002/jso.24518. [DOI] [PubMed] [Google Scholar]
- 10.Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017;16:149. doi: 10.1186/s12943-017-0712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–9. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Longo V, Gnoni A, Casadei Gardini A, Pisconti S, Licchetta A, Scartozzi M. et al. Immunotherapeutic approaches for hepatocellular carcinoma. Oncotarget. 2017;8:33897–910. doi: 10.18632/oncotarget.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allaire M, Nault JC. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29:288–95. doi: 10.1097/CCO.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 14.Kim DW, Talati C, Kim R. Hepatocellular carcinoma (HCC): beyond sorafenib-chemotherapy. J Gastrointest Oncol. 2017;8:256–65. doi: 10.21037/jgo.2016.09.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai JR, Ochoa S, Prins PA, He AR. Systemic therapy for advanced hepatocellular carcinoma: an update. J Gastrointest Oncol. 2017;8:243–55. doi: 10.21037/jgo.2017.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo HY, Yoo SY, Heo J. New chemical treatment options in second-line hepatocellular carcinoma: what to do when sorafenib fails? Expert Opin Pharmacother. 2017;18:35–44. doi: 10.1080/14656566.2016.1261825. [DOI] [PubMed] [Google Scholar]
- 17.Ziogas IA, Tsoulfas G. Evolving role of sorafenib in the management of hepatocellular carcinoma. World J Clin Oncol. 2017;8:203–13. doi: 10.5306/wjco.v8.i3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trojan J, Waidmann O. Role of regorafenib as second-line therapy and landscape of investigational treatment options in advanced hepatocellular carcinoma. J Hepatocell Carcinoma. 2016;3:31–6. doi: 10.2147/JHC.S112537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galun D, Srdic-Rajic T, Bogdanovic A, Loncar Z, Zuvela M. Targeted therapy and personalized medicine in hepatocellular carcinoma: drug resistance, mechanisms, and treatment strategies. J Hepatocell Carcinoma. 2017;4:93–103. doi: 10.2147/JHC.S106529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 21.Hilton JF, Shapiro GI. Aurora kinase inhibition as an anticancer strategy. J Clin Oncol. 2014;32:57–9. doi: 10.1200/JCO.2013.50.7988. [DOI] [PubMed] [Google Scholar]
- 22.Jeng YM, Peng SY, Lin CY, Hsu HC. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clin Cancer Res. 2004;10:2065–71. doi: 10.1158/1078-0432.ccr-1057-03. [DOI] [PubMed] [Google Scholar]
- 23.Borisa AC, Bhatt HG. A comprehensive review on Aurora kinase: Small molecule inhibitors and clinical trial studies. Eur J Med Chem. 2017;140:1–19. doi: 10.1016/j.ejmech.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 24.Bayliss R, Burgess SG, McIntyre PJ. Switching Aurora-A kinase on and off at an allosteric site. FEBS J. 2017;284:2947–54. doi: 10.1111/febs.14069. [DOI] [PubMed] [Google Scholar]
- 25.Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8:23937–54. doi: 10.18632/oncotarget.14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q, Yu X, Zhou ZW, Zhou C, Chen XW, Zhou SF. Inhibition of Aurora A kinase by alisertib induces autophagy and cell cycle arrest and increases chemosensitivity in human hepatocellular carcinoma HepG2 cells. Curr Cancer Drug Targets. 2017;17:386–401. doi: 10.2174/1568009616666160630182344. [DOI] [PubMed] [Google Scholar]
- 28.Payton M, Bush TL, Chung G, Ziegler B, Eden P, McElroy P. et al. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res. 2010;70:9846–54. doi: 10.1158/0008-5472.CAN-10-3001. [DOI] [PubMed] [Google Scholar]
- 29.He SJ, Shu LP, Zhou ZW, Yang T, Duan W, Zhang X. et al. Inhibition of Aurora kinases induces apoptosis and autophagy via AURKB/p70S6K/RPL15 axis in human leukemia cells. Cancer Lett. 2016;382:215–30. doi: 10.1016/j.canlet.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Zi D, Zhou ZW, Yang YJ, Huang L, Zhou ZL, He SM. et al. Danusertib induces apoptosis, cell cycle arrest, and autophagy but inhibits epithelial to mesenchymal transition involving PI3K/Akt/mTOR signaling pathway in human ovarian cancer cells. Int J Mol Sci. 2015;16:27228–51. doi: 10.3390/ijms161126018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan CX, Zhou ZW, Yang YX, He ZX, Zhang X, Wang D. et al. Danusertib, a potent pan-Aurora kinase and ABL kinase inhibitor, induces cell cycle arrest and programmed cell death and inhibits epithelial to mesenchymal transition involving the PI3K/Akt/mTOR-mediated signaling pathway in human gastric cancer AGS and NCI-N78 cells. Drug Des Devel Ther. 2015;9:1293–318. doi: 10.2147/DDDT.S74964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JP, Yang YX, Liu QL, Zhou ZW, Pan ST, He ZX. et al. The pan-inhibitor of Aurora kinases danusertib induces apoptosis and autophagy and suppresses epithelial-to-mesenchymal transition in human breast cancer cells. Drug Des Devel Ther. 2015;9:1027–62. doi: 10.2147/DDDT.S74412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoffski P, Besse B, Gauler T, de Jonge MJ, Scambia G, Santoro A. et al. Efficacy and safety of biweekly i.v. administrations of the Aurora kinase inhibitor danusertib hydrochloride in independent cohorts of patients with advanced or metastatic breast, ovarian, colorectal, pancreatic, small-cell and non-small-cell lung cancer: a multi-tumour, multi-institutional phase II study. Ann Oncol. 2015;26:598–607. doi: 10.1093/annonc/mdu566. [DOI] [PubMed] [Google Scholar]
- 34.Borthakur G, Dombret H, Schafhausen P, Brummendorf TH, Boissel N, Jabbour E. et al. A phase I study of danusertib (PHA-739358) in adult patients with accelerated or blastic phase chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia resistant or intolerant to imatinib and/or other second generation c-ABL therapy. Haematologica. 2015;100:898–904. doi: 10.3324/haematol.2014.115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meulenbeld HJ, Bleuse JP, Vinci EM, Raymond E, Vitali G, Santoro A. et al. Randomized phase II study of danusertib in patients with metastatic castration-resistant prostate cancer after docetaxel failure. BJU Int. 2013;111:44–52. doi: 10.1111/j.1464-410X.2012.11404.x. [DOI] [PubMed] [Google Scholar]
- 36.Cohen RB, Jones SF, Aggarwal C, von Mehren M, Cheng J, Spigel DR. et al. A phase I dose-escalation study of danusertib (PHA-739358) administered as a 24-hour infusion with and without granulocyte colony-stimulating factor in a 14-day cycle in patients with advanced solid tumors. Clin Cancer Res. 2009;15:6694–701. doi: 10.1158/1078-0432.CCR-09-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steeghs N, Eskens FA, Gelderblom H, Verweij J, Nortier JW, Ouwerkerk J. et al. Phase I pharmacokinetic and pharmacodynamic study of the aurora kinase inhibitor danusertib in patients with advanced or metastatic solid tumors. J Clin Oncol. 2009;27:5094–101. doi: 10.1200/JCO.2008.21.6655. [DOI] [PubMed] [Google Scholar]
- 38.Tyanova S, Mann M, Cox J. MaxQuant for in-depth analysis of large SILAC datasets. Methods Mol Biol. 2014;1188:351–64. doi: 10.1007/978-1-4939-1142-4_24. [DOI] [PubMed] [Google Scholar]
- 39.Mann M. Fifteen years of stable isotope labeling by amino acids in cell culture (SILAC) Methods Mol Biol. 2014;1188:1–7. doi: 10.1007/978-1-4939-1142-4_1. [DOI] [PubMed] [Google Scholar]
- 40.Geiger T, Wisniewski JR, Cox J, Zanivan S, Kruger M, Ishihama Y. et al. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat Protoc. 2011;6:147–57. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 41.Ong SE, Mann M. Stable isotope labeling by amino acids in cell culture for quantitative proteomics. Methods Mol Biol. 2007;359:37–52. doi: 10.1007/978-1-59745-255-7_3. [DOI] [PubMed] [Google Scholar]
- 42.Zanivan S, Maione F, Hein MY, Hernandez-Fernaud JR, Ostasiewicz P, Giraudo E. et al. SILAC-based proteomics of human primary endothelial cell morphogenesis unveils tumor angiogenic markers. Mol Cell Proteomics. 2013;12:3599–611. doi: 10.1074/mcp.M113.031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiger T, Madden SF, Gallagher WM, Cox J, Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72:2428–39. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- 44.Qiu JX, Zhou ZW, He ZX, Zhao RJ, Zhang X, Yang L. et al. Plumbagin elicits differential proteomic responses mainly involving cell cycle, apoptosis, autophagy, and epithelial-to-mesenchymal transition pathways in human prostate cancer PC-3 and DU145 cells. Drug Des Devel Ther. 2015;9:349–417. doi: 10.2147/DDDT.S71677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J. et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding YH, Zhou ZW, Ha CF, Zhang XY, Pan ST, He ZX. et al. Alisertib, an Aurora kinase A inhibitor, induces apoptosis and autophagy but inhibits epithelial to mesenchymal transition in human epithelial ovarian cancer cells. Drug Des Devel Ther. 2015;9:425–64. doi: 10.2147/DDDT.S74062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao GX, Pan H, Ouyang DY, He XH. The critical molecular interconnections in regulating apoptosis and autophagy. Ann Med. 2015;47:305–15. doi: 10.3109/07853890.2015.1040831. [DOI] [PubMed] [Google Scholar]
- 48.Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, Lai PL. et al. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B Overexpression in HCC. BMC Cancer. 2010;10:461. doi: 10.1186/1471-2407-10-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Chen J, Chen D, Huang J, Feng B, Han S. et al. Aurora-A promotes chemoresistance in hepatocelluar carcinoma by targeting NF-kappaB/microRNA-21/PTEN signaling pathway. Oncotarget. 2014;5:12916–35. doi: 10.18632/oncotarget.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aihara A, Tanaka S, Yasen M, Matsumura S, Mitsunori Y, Murakata A. et al. The selective Aurora B kinase inhibitor AZD1152 as a novel treatment for hepatocellular carcinoma. J Hepatol. 2010;52:63–71. doi: 10.1016/j.jhep.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Lin ZZ, Hsu HC, Hsu CH, Yeh PY, Huang CY, Huang YF. et al. The Aurora kinase inhibitor VE-465 has anticancer effects in pre-clinical studies of human hepatocellular carcinoma. J Hepatol. 2009;50:518–27. doi: 10.1016/j.jhep.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 52.Benten D, Keller G, Quaas A, Schrader J, Gontarewicz A, Balabanov S. et al. Aurora kinase inhibitor PHA-739358 suppresses growth of hepatocellular carcinoma in vitro and in a xenograft mouse model. Neoplasia. 2009;11:934–44. doi: 10.1593/neo.09664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boehning D, Patterson RL, Sedaghat L, Glebova NO, Kurosaki T, Snyder SH. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5:1051–61. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
- 54.Fesik SW, Shi Y. Structural biology. Controlling the caspases. Science. 2001;294:1477–8. doi: 10.1126/science.1062236. [DOI] [PubMed] [Google Scholar]
- 55.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J. et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–8. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 56.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 58.Ravanan P, Srikumar IF, Talwar P. Autophagy: The spotlight for cellular stress responses. Life Sci; 2017. [DOI] [PubMed] [Google Scholar]
- 59.Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87–95. doi: 10.1038/cdd.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song S, Tan J, Miao Y, Li M, Zhang Q. Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress. J Cell Physiol. 2017;232:2977–84. doi: 10.1002/jcp.25785. [DOI] [PubMed] [Google Scholar]
- 61.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pihan P, Carreras-Sureda A, Hetz C. BCL-2 family: integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017;24:1478–87. doi: 10.1038/cdd.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.