Abstract
Rising ocean temperatures associated with global climate change induce breakdown of the symbiosis between coelenterates and photosynthetic microalgae of the genus Symbiodinium. Association with more thermotolerant partners could contribute to resilience, but the genetic mechanisms controlling specificity of hosts for particular Symbiodinium types are poorly known. Here, we characterize wild populations of a sea anemone laboratory model system for anthozoan symbiosis, from contrasting environments in Caribbean Panama. Patterns of anemone abundance and symbiont diversity were consistent with specialization of holobionts for particular habitats, with Exaiptasia pallida/S. minutum (ITS2 type B1) abundant on vertical substrate in thermally stable, shaded environments but E. brasiliensis/Symbiodinium sp. (ITS2 clade A) more common in shallow areas subject to high temperature and irradiance. Population genomic sequencing revealed a novel E. pallida population from the Bocas del Toro Archipelago that only harbors S. minutum. Loci most strongly associated with divergence of the Bocas‐specific population were enriched in genes with putative roles in cnidarian symbiosis, including activators of the complement pathway of the innate immune system, thrombospondin‐type‐1 repeat domain proteins, and coordinators of endocytic recycling. Our findings underscore the importance of unmasking cryptic diversity in natural populations and the role of long‐term evolutionary history in mediating interactions with Symbiodinium.
Keywords: 2bRAD, Aiptasia, Cnidaria, co‐evolution, population genomics, symbiosis
1. INTRODUCTION
Rising ocean temperatures threaten the persistence of coral reef ecosystems, invaluable hotspots of marine biodiversity that support the health and economy of human communities (Ferrario et al., 2014; Moberg & Folke, 1999). The fate of coral ecosystems is inextricably tied to the mutualistic symbiosis between corals and photosynthetic microalgae of the genus Symbiodinium, a hyperdiverse taxon classified into nine major clades designated A‐I (Pochon & Gates, 2010; Pochon, Montoya‐Burgos, Stadelmann, & Pawlowski, 2006). The coral‐Symbiodinium symbiosis funnels energy from sunlight through algal photosynthesis and into cnidarian metabolism to generate energy and the calcium carbonate framework of coral reef ecosystems (Gattuso, Allemand, & Frankignouelle, 1999). Symbiodinium living inside host endodermal cells receive access to carbon dioxide, inorganic nutrients, protection from herbivory, and a stable position in the water column to access light (Davy, Allemand, & Weis, 2012). In return, the coral host receives fixed carbon compounds that enhance calcification and support a large portion of host metabolic demands (Muscatine, Falkowski, Porter, & Dubinsky, 1984; Yellowlees, Rees, & Leggat, 2008).
Nutritional symbioses with Symbiodinium have facilitated the success of reef‐building corals in oligotrophic tropical waters, but may also render reef ecosystems particularly susceptible to global climate change (Kiers, Palmer, Ives, Bruno, & Bronstein, 2010; Six, 2009). Elevated temperatures just 1–2°C above summer maxima can trigger bleaching or breakdown of the symbiosis resulting in loss of Symbiodinium pigments or cells from coral host tissues through a variety of mechanisms (Weis, 2008). Widespread bleaching in natural coral populations is most commonly attributed to elevated temperature, through other stressors such as excessively cold temperature (Lirman et al., 2011), changes in salinity (Van Woesik, De Vantier, & Glazebrook, 1995), high solar irradiation (Gleason & Wellington, 1993), and nutrient enrichment (Vega Thurber et al., 2014) can induce or exacerbate bleaching. The frequency and severity of mass coral bleaching events are precipitously increasing (Heron, Maynard, van Hooidonk, & Eakin, 2016; Hughes et al., 2017).
A major obstacle in understanding these crucial symbiosis systems is the challenge of complementing ecological studies with basic insights from laboratory‐based approaches (Weis, Davy, Hoegh‐Guldberg, Rodriguez‐Lanetty, & Pringle, 2008). For many marine species, it is impossible to maintain large laboratory populations, propagate genetically controlled lines, or create different host–symbiont combinations. Because of these challenges, the sea anemone Exaiptasia pallida (Grajales & Rodríguez, 2014), commonly called “Aiptasia,” is emerging as a laboratory model system for studying coral symbiosis, particularly at the cellular level (Goldstein & King, 2016; Weis et al., 2008). These fast‐growing anemones reproduce sexually through broadcast‐spawning and asexually through pedal laceration, allowing large populations of genetically identical individuals to be propagated, experimentally bleached, and reinfected with cultured symbionts. Gene knockdown methods have been developed (Dunn, Phillips, Green, & Weis, 2007), and the lack of calcium carbonate skeleton facilitates in situ imaging impossible in adult reef‐building corals (Weis et al., 2008). Application of high‐throughput sequencing approaches to an experimentally tractable model system has offered a new window into the complex molecular dynamics underlying symbiosis onset and maintenance (Baumgarten et al., 2018; Matthews et al., 2017).
Despite the importance of Exaiptasia for molecular studies of cnidarian symbiosis, natural populations have rarely been studied. The first population genetic study, based on traditional markers, suggested two genetically differentiated populations: a “global” population that forms specific symbioses with Symbiodinium minutum (ITS2 type B1) and a South US Atlantic population that engages in diverse symbioses with S. minutum, S. psygmophilum (ITS2 type B2), and representatives of Symbiodinium clades A and C (Thornhill, Xiang, Pettay, Zhong, & Santos, 2013). A cryptic sister species, E. brasiliensis, has also been described from the southwestern Caribbean Sea and southwestern Atlantic Ocean but was only found in association with Symbiodinium clade A (Grajales, Rodríguez, & Thornhill, 2016).
At finer spatial scales in the Western Atlantic, patterns of symbiont association may be related to local variation in temperature and light environment. Symbiodinium clade A from E. pallida exhibited markedly increased photosynthetic performance at elevated temperatures compared to S. minutum (Goulet, Cook, & Goulet, 2005; Perez, Cook, & Brooks, 2001), a phenomenon that also depends on host genotype (Goulet et al., 2005). Correspondingly, anemone hosts experimentally infected (Perez et al., 2001) or naturally associated (Bellis & Denver, 2017) with Symbiodinium clade A tend to be less susceptible to heat‐induced bleaching. Higher rates of glucose transfer to hosts harboring S. minutum compared to Symbiodinium clade A, however, may indicate potential trade‐offs associated with increased thermal tolerance (see fig. 5a in Burriesci, Raab, & Pringle, 2012).
In this study, we investigate ecological and evolutionary dynamics of Exaiptasia anemones from four sites in Caribbean Panama where extensive long‐term environmental monitoring datasets are available. These locations are characterized by a variety of habitats and thermal regimes ranging from high daily variation often >3°C at a shoaling reef flat near the entrance to the Panama Canal (Cubit, Caffey, Thompson, & Windsor, 1989; http://biogeodb.stri.si.edu/physical_monitoring/research/galeta), to stable daily temperatures with means varying up to 1°C during the warmest months of the year among three mangrove lagoon sites in the Bocas del Toro Archipelago (Kaufmann & Thompson, 2005; http://biogeodb.stri.si.edu/physical_monitoring/research/bocas). To evaluate the hypothesis that environmental variation influences holobiont evolutionary dynamics at regional scales, we compare density and diversity of Symbiodinium from anemone populations at each of these sites. Potential ecological and evolutionary trade‐offs are assessed through estimation of anemone abundance and analyses of host genetic diversity based on type IIb restriction site‐associated DNA sequencing (2bRAD‐Seq; Wang, Meyer, McKay, & Matz, 2012). We further analyze 2bRAD‐Seq data to investigate locus‐specific patterns of host differentiation that could suggest potential roles in adaptation to local environments.
2. MATERIALS AND METHODS
2.1. Sample collection
We surveyed populations of Exaiptasia sea anemones (Figure 1) across a range of habitats in Caribbean Panama. Surveys were conducted between 15 and 19 July 2015 (Galeta) or 30 August and 11 September 2015 (Bocas del Toro) at four sites: Isla Colón, Cayo Roldán, Cayo Agua, and Galeta (Figure 2). Survey areas were chosen based on proximity to long‐term water temperature monitoring sites operated by the Smithsonian Tropical Research Institute's Physical Monitoring Program. Several surveys were also conducted in Portobelo National Park, but as few anemones were found, this site was excluded for the majority of analyses. At each site, 30 m × 1 m belt transects were established parallel to shore. Transects were initiated upon observation of at least one anemone, and surveys were conducted continuously by an observer swimming along the shoreline. Abundance was estimated by counting all anemones per transect, with a maximum of 100 anemones counted per mangrove root. In Galeta, anemones were only found attached to rocks and coral rubble in shallow intertidal areas, so surveys were conducted parallel to shore at <0.5 m depth, with a maximum of 100 anemones counted per 30 m × 1 m belt transect. The median number of anemones per colonized root, the number of colonized roots in a transect, and the number of meters traveled before initiating the next transect were analyzed using generalized linear models (GLMs) and a quasi‐Poisson distribution to account for overdispersion with the “glm” function of R version 3.3.0.
Figure 1.

Mangrove root epibiota including Exaiptasia pallida sea anemones in Bocas del Toro, Panama
Figure 2.
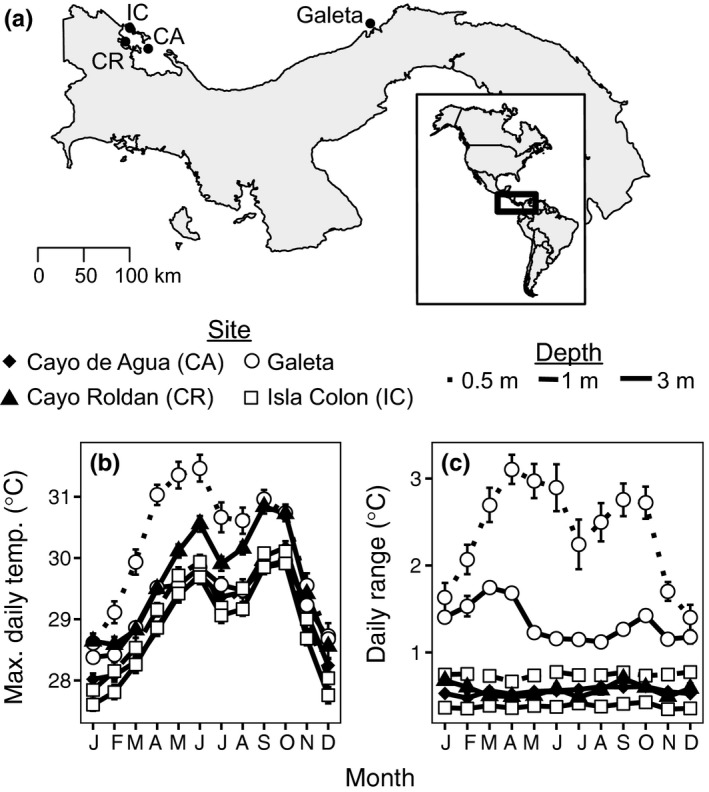
Primary sampling locations in Caribbean Panama. (a) Temperature data displayed in (b) and (c) are averages from 2000 to 2015 (3‐m depth datasets) or 2002 to 2015 (0.5‐m and 1‐m depth datasets) and are derived from data provided by the Physical Monitoring Program of the Smithsonian Tropical Research Institute
2.2. Sample processing
Two anemones from each transect were sampled for genetic analysis and for determination of Symbiodinium density and mitotic index. Samples were homogenized in 200 uL buffer EB (Qiagen, Valencia, CA, USA), and a 100‐μl aliquot of the tissue homogenate was reserved for DNA extraction with the DNeasy Blood and Tissue Kit (Qiagen). The remainder of the homogenate was centrifuged at 5,000 g for 3 min, the pellet suspended in 500 uL filtered seawater (FSW), and the resuspension centrifuged again. The combined supernatants and the resulting algal pellet were frozen for determination of Symbiodinium cell density via hemocytometer counts and normalization to total protein determined using the Pierce BCA Protein Assay (see Appendix S1 for additional detail). Mitotic indices (MI) were estimated as the number of dividing cells per total number of counted cells for each replicate count. MI was analyzed using a generalized linear mixed model with the binomial distribution and logit link function implemented with the “glmer” function of the “lme4” package in R, which enables modeling of both fixed and random effects (Bates, Mächler, Bolker, & Walker, 2015). Site was included as the explanatory variable with a random effect of sample ID to take into account replicate counts (n = 6) from the same anemone. Post hoc comparisons were performed using the “glht” function of the “multcomp” package (Hothorn, Bretz, & Westfall, 2009).
2.3. Anemone genotyping
We created reduced representation sequencing libraries using DNA from 150 anemones from Caribbean Panama. These included anemones from Isla Colón (n = 42), Cayo Roldán (n = 38), Cayo Agua (n = 33), Galeta (n = 32), and Portobelo National Park (n = 5). To facilitate comparison with laboratory strains, we created additional libraries for four previously studied strains CC7, H2, EM5, and MMB (Bellis & Denver, 2017) and a single anemone, N2, from the outdoor aquarium at Naos Island Marine Laboratory in the Bay of Panama. In silico digested reads from low‐coverage whole genome sequencing of 10 laboratory E. pallida strains (Bellis, Howe, & Denver, 2016) were also included.
Genomic libraries were prepared and analyzed following the 2b‐RAD method (Wang et al., 2012). Genomic DNA was digested with the enzyme BcgI (New England BioLabs, Ipswich, MA, USA), which cleaves on either side of the recognition sequence ([N10]CGA[N6]TGC[N12]), to generate 36‐bp fragments. Dual‐indexed libraries were gel‐purified after adaptor ligation and amplification, pooled, and sequenced on the NextSeq 500 at the University of Oregon Genomics and Cell Characterization Core Facility (Eugene, OR, USA). After sequencing, reads were quality‐filtered and aligned to the set of all 44,428 BcgI recognition sites present in the v1.0 reference genome (Baumgarten et al., 2015) using SHRiMP v2.2.2 (Rumble et al., 2009). Genotypes with minor allele frequencies (MAF) below 5% were called as homozygous; those with frequencies above 30% were called as heterozygous. Genotypes were not called if MAF was between 5 and 30% or if coverage was <5×. Variants were further filtered to exclude sites not genotyped in at least 80% of individuals (see Appendix S1 for full methodological details).
Because E. brasiliensis and E. pallida are morphologically indistinguishable (Grajales & Rodríguez, 2016), species assignments were based on phylogenetic analysis (Figures S1‐S2). Phylogenetic trees were inferred using the maximum‐likelihood method based on the Kimura 2‐parameter model in MEGA7, with the discrete Gamma distribution (five categories, estimated α = 0.43) used to model evolutionary rate differences among sites and allowing for invariable sites. Individuals identified as E. brasiliensis were confirmed by sequencing a region of mitochondrial DNA that spanned the 16S ribosomal RNA (16S rRNA), cytochrome c oxidase subunit III (COIII), and cytochrome c oxidase subunit I (COI) genes from several individuals. This region exhibited an interspecific divergence of 0.32% over 3,164 bp, no intraspecific polymorphisms in a trimmed 719‐bp region sequenced in eight individuals, and included a region previously found to distinguish E. brasiliensis and E. pallida (Grajales & Rodríguez, 2016). Mitochondrial DNA was amplified using oligonucleotides MslrRNA‐F (5′‐CTGGAAACTGAAACATCTAAGTACC‐3′) and ApCOI_5‐R (5′‐TCATTCCAGAACCTATTCCAAA‐3′) each at a final concentration of 0.4 μmol/l in 50‐μl reaction volumes containing 1 μl DMSO, 10 μl 5 × MyTaq Reaction Buffer (5 mmol/l dNTPs, 15 mmol/l MgCl2), and 0.25 μl MyTaq DNA Polymerase (Bioline, London, UK). A PCR cycling protocol optimized for amplification of long DNA products was used as follows: 35 cycles of 92°C for 10 s, 55°C for 20 s, and extension at 60°C for 10 m with an additional 10 s per cycle added to the extension time after the first 10 cycles. PCR products were sequenced with BigDye Terminators on the ABI 3730 capillary sequencer at the Center for Genome Research and Biocomputing at Oregon State University (Corvallis, OR, USA) after cycle synthesis with oligonucleotides MslrRNA‐F, ApCOI_5‐R, and internal sequencing primers ApCOIII‐R (5′‐AGACCTTGGAAAGTTGCCTCT‐3′) and AplRNA‐R (5′‐AAGCTCACCTTCGTTACCTTT‐3′).
2.4. Anemone population structure
To investigate population subdivision, we performed hierarchical population structure analyses in STRUCTURE version 2.3.4 (Falush, Stephens, & Pritchard, 2003; Pritchard, Stephens, & Donnelly, 2000). To identify clones, we used “poppr” version 2.3.0, with genotypes contracted based on Prevosti's absolute genetic distance and clusters merged according to the maximum distance in each cluster (Kamvar, Tabima, & Grünwald, 2014). The threshold for clustering clonal genotypes (0.006) was chosen based on genetic distances observed for known clones in the dataset (HI1/HI2 and BM1/BM2 in silico digested libraries from Bellis et al. (2016); CC7 in silico and in vitro digested libraries). STRUCTURE analyses on the clone‐corrected dataset used the admixture model with correlated allele frequencies, which provides greater power to distinguish closely related populations compared with the independent allele frequency model (Falush et al., 2003). Five replicates were run for values of K ranging between 1 and 6, each with 500,000 MCMC steps after a burn‐in period of 100,000. The optimum number of population clusters, K, was determined using the Evanno method implemented in STRUCTURE HARVESTER (Evanno, Regnaut, & Goudet, 2005; Earl & vonHoldt, 2012; Figures S3–S5). Output from replicate STRUCTURE runs was combined with CLUMPP version 1.1.2 (Jakobsson & Rosenberg, 2007) and visualized using DISTRUCT version 1.1 (Rosenberg, 2004). To complement analyses of STRUCTURE, principal components were visualized with “adegenet” version 2.0.1, with allele frequencies scaled and centered to the mean and missing genotype calls replaced by the mean allele frequency (Jombart, 2008).
2.5. Locus‐specific patterns of genetic differentiation
To identify outlier loci in E. pallida most strongly correlated with divergence between local populations, we used the R package “pcadapt” version 3.0.4 (Luu, Bazin, & Blum, 2017). Component‐wise p‐values were computed retaining the first two principal components, chosen based on visual inspection of the scree plot, and false discovery rate correction with a threshold of 0.05. Gene set enrichment analyses were performed with the gene score resampling method on component‐wise p‐values describing the strength of SNP associations with PC1 in ErmineJ version 3.0.3 (Gillis, Mistry, & Pavlidis, 2010). Analyses were based on the median score for each gene set, with full resampling and 200,000 iterations, incorporating testing for multifunctionality. Weir and Cockerham's F ST was calculated using VCFtools for each biallelic locus after removing alleles with frequency below 5% (Danecek et al., 2011).
2.6. Symbiont genotyping
The dominant Symbiodinium in each sample was genotyped as S. sp. clade A, S. minutum type B1, or S. psygmophilum type B2 using a two‐stage PCR banding assay. The first PCR reaction was specific for the chloroplast 23S ribosomal RNA (cp23s) from Symbiodinium clade B. Primers 5′‐TGC TGC TGA CAC TAA AGG ACA‐3′ and 5′‐ATC GCC CCA ATT AAA CAG TG‐3′ were designed to flank an insertion polymorphism between voucher specimens of Symbiodinium type B1 [GenBank accession: JN557991] and type B2 [GenBank accession: JN557995], yielding a 165‐bp band for type B1 or a 205‐bp band for type B2. The second PCR reaction was specific for the chloroplast photosystem II subunit D1 (psbA) from Symbiodinium clade A and did not amplify DNA from types B1 or B2 (primers 5′‐CTT TCT CAG CTC CAG TAG TT‐3′ and 5′‐AGA TTG AGT GAA ATG TCT CCT G‐3′). All PCRs were performed with EconoTaq DNA Polymerase (Lucigen, Middleton, WI, USA) in 25‐μl reaction volumes with an annealing temperature of 56°C.
3. RESULTS
3.1. Anemone distribution and abundance
We investigated the distribution and abundance of E. pallida and E. brasiliensis at four sites in Caribbean Panama (Figure 2). E. pallida was found only on vertical substrate and was most abundant on the roots of island‐fringing mangroves (~0.5–2 m depth) at the three studied sites within the Bocas del Toro Archipelago (Table 1). In contrast, E. brasiliensis was abundant in Galeta attached to rocks or coral rubble on a shoaling reef flat (≤0.5 m depth), although four E. brasiliensis specimens were found colonizing mangrove roots in the Bocas del Toro Archipelago (Table 1). No anemones were found attached to mangrove roots in Galeta or in Portobelo National Park.
Table 1.
Summary of samples genotyped. Anemones are classified according to host species and the dominant Symbiodinium type
| Site | Host species | Substrate | Number of anemones | ||
|---|---|---|---|---|---|
| Symbiodinium sp. (clade A) | S. minutum (type B1) | S. psygmophilum (type B2) | |||
| Bocas del Toro | |||||
| Cayo Agua | E. pallida | Mangrove | 0 | 33 | 0 |
| Cayo Roldán | E. brasiliensis | Mangrove | 0 | 1 | 0 |
| E. pallida | Mangrove | 0 | 37 | 0 | |
| Isla Colón | E. brasiliensis | Mangrove | 0 | 3 | 0 |
| E. pallida | Mangrove | 0 | 38 | 1 | |
| Coastal populations | |||||
| Galeta | E. brasiliensis | Reef flat | 28 | 0 | 0 |
| E. pallida | Dock piling | 0 | 4 | 0 | |
| Portobelo | E. brasiliensis | Reef flat | 5 | 0 | 0 |
Within the Bocas del Toro Archipelago, anemones were most abundant at the warmest site, Cayo Roldán (Table 2). A median of 18 anemones was observed per colonized mangrove root in Cayo Roldán, a higher density compared to other sites (p < .01, GLM; Figure 3a). In addition, a greater number of mangrove roots were colonized per transect (p < .001, GLM; Figure 3b), and transects, with placement dependent on anemone distance, were initiated more closely together at Cayo Roldán than the other sites (p < .01, GLM; Figure 3c). The distance between initiated transects in Galeta was similar to Isla Colón and Cayo Agua (data not shown).
Table 2.
Statistical analyses of anemone abundance in Bocas del Toro. Count data for all response variables were modeled using generalized linear models assuming a quasi‐Poisson distribution to account for overdispersion
| Estimate | Std. Error | t‐Value | p‐Value | |
|---|---|---|---|---|
| Anemones per root | ||||
| Intercept | 2.89 | 0.14 | 20.85 | <.001 |
| Site | ||||
| Cayo Agua | −0.65 | 0.24 | −2.68 | .009 |
| Isla Colón | −0.68 | 0.23 | −2.91 | .005 |
| Roots with ≥1 anemone | ||||
| Intercept | 2.47 | 0.10 | 23.76 | <.001 |
| Site | ||||
| Cayo Agua | −1.24 | 0.23 | −5.51 | <.001 |
| Isla Colón | −1.21 | 0.21 | −5.73 | <.001 |
| Distance between clusters (meters) | ||||
| Intercept | 3.40 | 0.22 | 15.534 | <.001 |
| Site | ||||
| Cayo Agua | 0.78 | 0.27 | 2.90 | .005 |
| Isla Colón | 0.80 | 0.26 | 3.05 | .003 |
Figure 3.
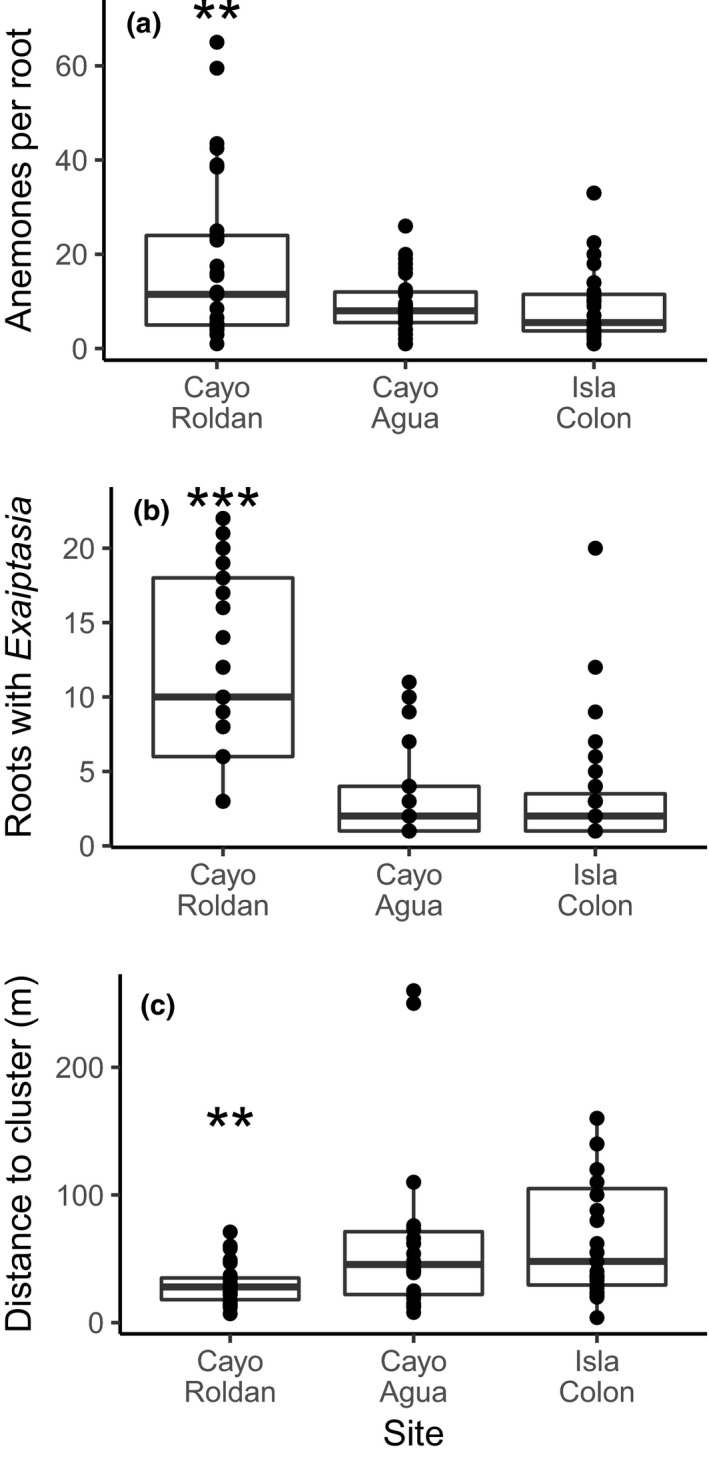
Anemone abundance in Bocas del Toro. Boxplots delimit the 25th and 75th percentiles, and the horizontal bar indicates the median. Data shown in a–c are based on continuous surveys, with 30‐m transects initiated upon observation of at least one anemone. (a) Median number of anemones per colonized mangrove root in each 30‐m transect. (b) Number of colonized roots per transect. (c) Distance between transects. Galeta is not shown, as no anemones were found colonizing mangrove roots, and the majority of individuals genotyped were E. brasiliensis rather than E. pallida. Asterisks indicate significant differences between Cayo Roldán and other sites (p < .001 [***] or p < .01 [**], generalized linear model)
3.2. Symbiodinium density and diversity
We analyzed the occurrence of three Symbiodinium species that associate with E. pallida (Grajales et al., 2016) using a PCR banding approach that targeted cp23s and psbA, two chloroplast genes encoded by separate minicircles (Wisecaver & Hackett, 2011). All sampled E. pallida hosted S. minutum (ITS2 type B1) as the dominant symbiont, with the exception of a single individual from the global E. pallida clade collected in Isla Colón that harbored S. psygmophilum (Table 1) . E. brasiliensis was dominated by S. minutum at sites within the Bocas del Toro Archipelago but Symbiodinium sp. clade A in Galeta (Table 1). Because Exaiptasia was previously reported to associate with multiple genotypes of Symbiodinium clade A (Grajales et al., 2016), we sequenced the psbA region for a subset of samples (n = 6). Four sequences were identical to a Symbiodinium clade A isolated from the jellyfish Cassiopeia xamachana [GenBank: AJ884905], whereas two sequences were identical to sequence for Symbiodinium isolated from a representative of the laboratory E. pallida strain used to construct the reference genome (Baumgarten et al., 2015). Although our PCR analysis does not preclude the presence of additional genotypes or finer scale genetic differences, in all samples, a single dominant Symbiodinium type was discovered, and PCR with alternative primer pairs failed to amplify a discernable band.
Within‐anemone Symbiodinium cell densities per unit protein were similar across sites, but lower division rates were observed in Symbiodinium clade A in anemones from Galeta compared to Symbiodinium type B1 at other sites (Figure 4). Median density ranged between 3.1 and 3.7 × 106 cells per mg protein across the four sites, with a median of 3.3 × 106 cells per mg protein in the pooled sample set. There were no significant differences in within‐anemone Symbiodinium cell density among sites (p = .08, likelihood ratio test for full and intercept‐only model). However, site was a significant predictor of mitotic index (p < .001, likelihood ratio test for comparison of full and intercept‐only model). Average mitotic index of Symbiodinium clade A was lower in anemones from Galeta compared to Symbiodinium type B1 in Cayo Roldán (p < .001, Tukey HSD) or Cayo Agua (p = .014, Tukey HSD) (Figure 4).
Figure 4.
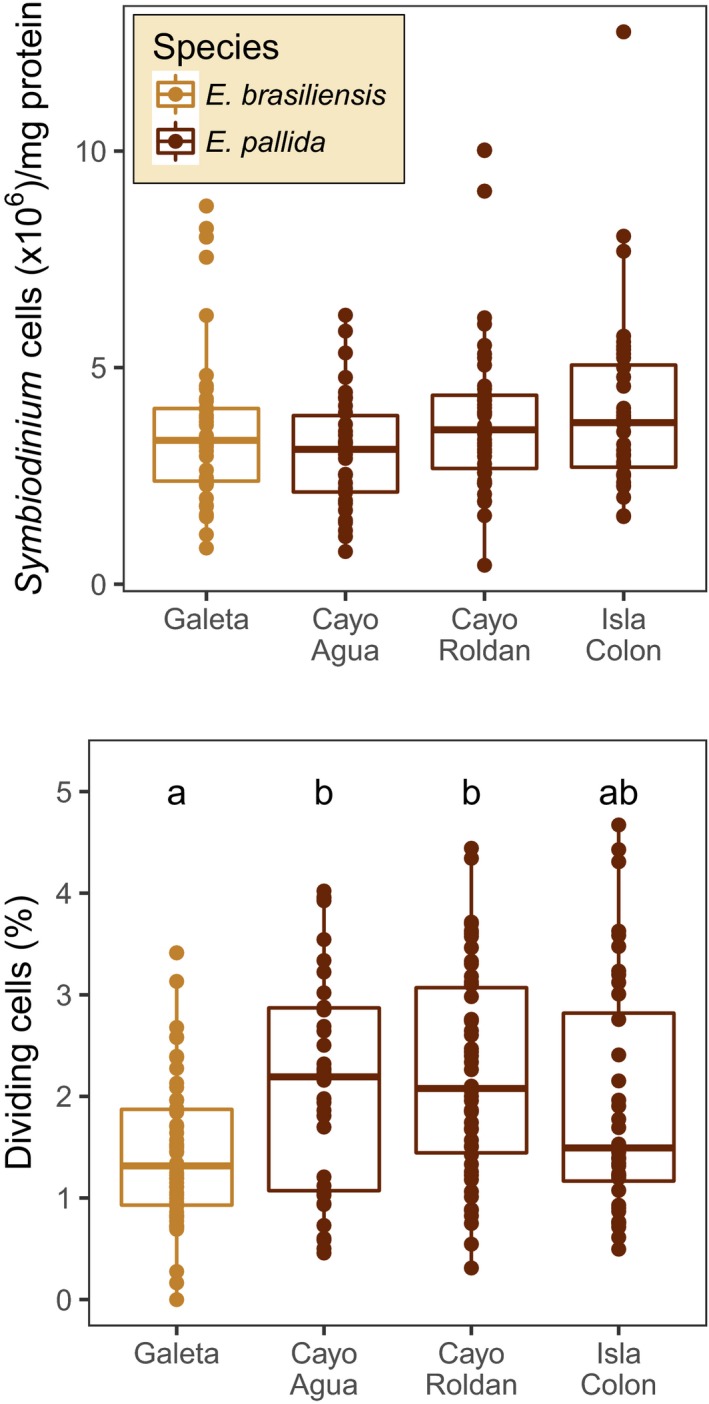
Variation in Symbiodinium cell density (top panel) and mitotic index (lower panel) among sites. Boxplots delimit the 25th and 75th percentiles, and the horizontal bar indicates the median. Strong evidence for differences in Symbiodinium density among sites was not observed (p = .08, generalized linear model). In contrast, site was a significant predictor of mitotic index (p < .001, GLMM), with evidence for lower Symbiodinium division rates in anemones from Galeta compared to Cayo Roldán (p < .001, Tukey HSD) and Cayo Agua (p = .014, Tukey HSD). Anemones from Galeta were primarily E. brasiliensis associated with Symbiodinium sp. clade A, whereas the majority of anemones from the other sites were E. pallida associated with S. minutum type B1. Because not all samples used for density analyses were genotyped, datapoints are colored according to the dominant anemone species at each site
3.3. Host population structure
We investigated population genetic structure of anemones from the Caribbean coast of Panama using type IIb restriction site‐associated DNA sequencing (2bRAD‐Seq). To facilitate comparison with previous studies, sequences from 15 E. pallida isolates collected from geographically widespread locations were also included. On average, 1.2 million reads were generated from each sample. An average of 51.5% (E. brasiliensis) or 64.2% (E. pallida) of reads mapped to the reference genome; significantly fewer reads from E. brasiliensis mapped uniquely compared to E. pallida (32.8% vs. 43.2%, p < .001, Welch two sample t test). Mapping was most successful for the 2bRAD library created from CC7, the strain used to sequence the reference genome (82%), but only 59% of CC7 reads mapped uniquely, suggesting that microbial DNA in the sequencing libraries, divergence from the reference genome, and the short length of the 2bRAD tags all contributed to lower mapping percentages. After filtering, 4640 SNPs were identified in the E. pallida callset and 5391 SNPs in the combined species dataset; of these, 2577 remained for E. pallida after removing sites with an allele frequency below 5%.
Patterns of genetic variation at 2,577 SNPs supported the presence of two distinct E. pallida populations: a local population specific to the Bocas del Toro Archipelago (which we name the “Bocas‐specific” population) and a separate population including individuals we collected in Panama that clustered more closely with geographically widespread E. pallida (which we call the “global population”) (Figure 5). The “global population” cluster in our study included anemones from Florida, Bermuda, and Hawaii as well as four clones from a dock piling in Galeta, ten individuals from sites in the Bocas del Toro Archipelago, and the anemone that was collected from the outdoor aquarium at Naos Island Marine Laboratory in the Bay of Panama (red vertical bars in Figure 5a). Our definition of a global clade, therefore, differs from that of a previous study (Thornhill et al., 2013), which included individuals from Bermuda, Hawaii, Japan, the Red Sea, and Australia but not Florida as part of the global E. pallida group.
Figure 5.
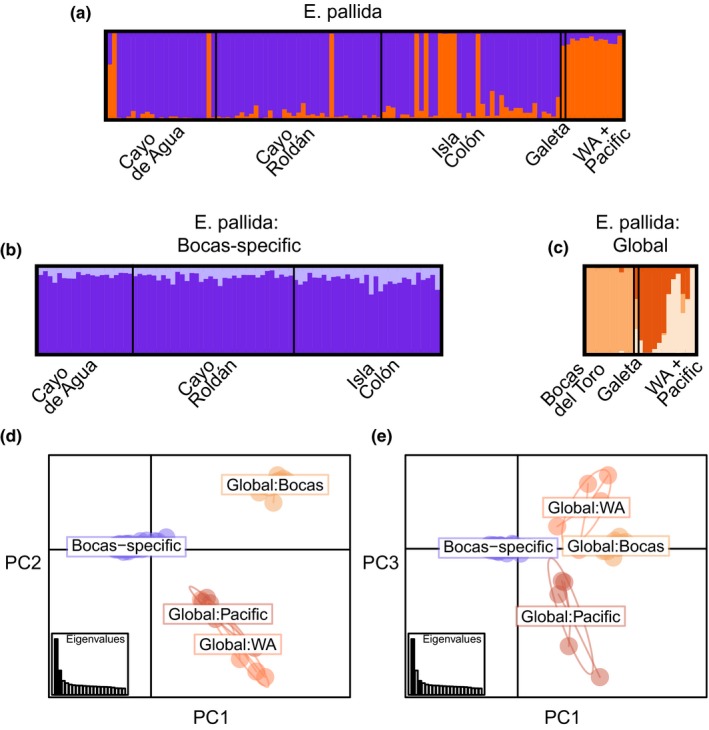
Structure of E. pallida Bocas‐specific and globally widespread populations. STRUCTURE analyses were first performed for all E. pallida samples (a). The two identified populations in (a) were then analyzed individually (b–c). The optimal number of clusters (K = 2 in a and b; K = 3 in c) was chosen based on the Evanno method (Evanno et al., 2005). Representatives of the previously described South U.S. Atlantic and global clades (Grawunder et al., 2015; Thornhill et al., 2013) are included in the Western Atlantic (WA) + Pacific grouping. Results of the principal component analysis (d–e) are shown after excluding three individuals with significant proportions of admixture from multiple populations. Analyses were based on 2,577 SNPs after removal of clones and anemones determined as E. brasiliensis
A median F ST value of 0.10 between the Bocas‐specific and the global clades was in agreement with the distinction of the Bocas‐specific and globally widespread E. pallida populations. The distribution of F ST values among loci was positively skewed, with a similar proportion of SNPs in genic regions compared to the proportion of the reference genome covered by gene models (73% vs. 76%, respectively; Figure 6). Within the Bocas‐specific population, strong genetic differentiation among the three sampled sites in the Bocas del Toro Archipelago was not observed (Figure 5b). In contrast, subpopulation structure was indicated in the global population (Figure 5c), with a median F ST of 0.06 between representatives of the global population collected in Bocas del Toro and individuals from globally widespread locations.
Figure 6.
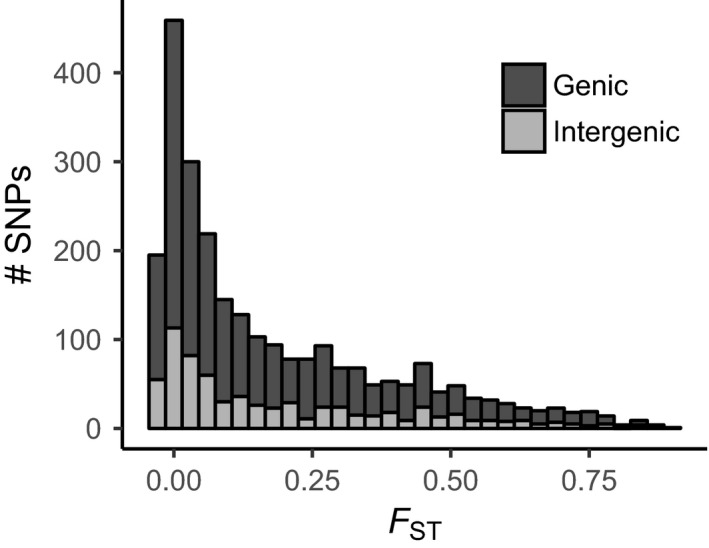
Distribution of pairwise FST values for comparisons between the Bocas‐specific and global E. pallida populations. Single nucleotide polymorphisms in genic regions (n = 1,881) include those present in introns, exons, and 5′‐ and 3′‐untranslated regions
Because Exaiptasia can reproduce clonally through pedal laceration, we removed individuals genotypically identical to other samples in the dataset prior to analyses of population structure. The spatial extent of clonal reproduction in the populations studied appeared to be relatively limited. Among the 112 sampled E. pallida individuals, 87 multilocus genotypes (MLGs) were unique, seven MLGs were shared by two anemones, one MLG was shared by three anemones, and two MLGs were shared by four anemones. All anemone pairs genotyped from the same mangrove root or dock piling were clones (n = 6 pairs). Shared MLGs for E. pallida collected from different transects were observed between the members of two clone pairs from Cayo Roldán, a maximum distance of 50 m apart. Among individuals genotyped from different mangrove roots within the same 30‐m transect, 0%–29% of genotyped pairs were clones, including 0 of 11 transects in Isla Colón, one of 13 transects in Cayo Roldán, and two of seven transects in Cayo Agua. In E. brasiliensis, six clone pairs were identified and a total of 29 unique MLGs among 35 genotyped individuals. Members of four clone pairs were collected <30 m apart, whereas two pairs of clones were collected with their members approximately 100 m apart.
3.4. Locus‐specific patterns of host differentiation
Under a model of divergence among three populations, in which two populations share more recent evolutionary history compared to a third, outlier loci strongly correlated with principal components that separate these populations may be promising targets of differential selection pressure (Duforet‐Frebourg, Luu, Laval, Bazin, & Blum, 2016). To identify loci in E. pallida most strongly correlated with divergence between the Bocas‐specific and global populations, we performed outlier detection using principal component analysis (Luu et al., 2017). This analysis was based on 2577 SNPs that remained after removing biallelic sites with minor allele frequency below 5%. To further reduce false positives, we report only those loci that were also significantly associated with divergence between the Bocas‐specific and global E. pallida populations in comparisons using the Mahalanobis distance, which may provide higher power for hierarchically structured populations compared to other methods when there is admixture (Luu et al., 2017).
Five loci showed strong signatures of association with the first principal component (PC1) at an FDR threshold of 0.05 (Table 3). These included two sites in intergenic regions and three loci located in genic regions. One locus was identified in the coding region of cytoplasmic FMR1‐interacting protein 2, but was not predicted to cause a change to the protein sequence (Table 3). The nearest predicted gene to one of the identified intergenic sites was an uncharacterized protein approximately 1 kb away from this locus. The other intergenic site was in the midst of a ~4.7 kb intergenic region between genes identified as Frizzled‐5 and General transcription factor IIF subunit 2. While none of the identified polymorphisms were expected to result in changes to protein sequence, these sites could be linked to nonsynonymous polymorphisms, influence gene expression, or impact protein structure through alternative splicing (Table 3).
Table 3.
Individual outlier loci associated with divergence of the Bocas‐specific population in the principal component analysis. F ST values for each locus are also given and reflect comparisons between the global E. pallida population (including individuals from Bocas del Toro, Bermuda, Florida, and Hawaii) and the Bocas‐specific population
| Scaffold | Position | Region | Identifier | F ST | Annotation |
|---|---|---|---|---|---|
| 10 | 1258482 | 5′UTR | AIPGENE19408 | 0.85 | Myoferlin |
| 143 | 149323 | Intergenic | N/A | 0.90 | N/A |
| 227 | 50641 | Intron | AIPGENE5586 | 0.83 | N‐glycanase 1 |
| 47 | 283015 | Intergenic | N/A | 0.83 | N/A |
| 60 | 564146 | CDS | AIPGENE172 | 0.86 | Cytoplasmic FMR1‐interacting protein 2 |
Because individual outlier loci were present in genes with possible but unverified roles in cnidarian symbiosis (see Discussion), we further tested for enrichment of polymorphisms strongly associated with divergence of the Bocas‐specific population in previously identified symbiosis‐related genes. After excluding SNPs in intergenic regions, this analysis was based on 1,432 distinct genes in our dataset that contained one or more high‐quality SNPs with a minor allele frequency of at least 5%. Based on previous studies of symbiosis in Exaiptasia, we tested four gene sets for enrichment that had SNP markers in at least three genes in our dataset. These gene sets included a list of scavenger receptors (three genes present; Neubauer, Poole, Weis, & Davy, 2016), genes involved in the complement pathway (three genes present; Baumgarten et al., 2015), genes encoding proteins with thrombospondin‐type‐1 repeat motifs (14 genes present; Neubauer et al., 2017), and genes differentially regulated in larvae during symbiosis establishment (38 genes present; Wolfowicz et al., 2016).
Both thrombospondin‐type‐1 repeat domain‐containing proteins (Neubauer et al., 2017) and genes involved in activating the complement system (Poole, Kitchen, & Weis, 2016) were more likely than expected by chance to exhibit strong associations with PC1 after correcting for multiple comparisons (p = .01 and p = .04, respectively; Table 4). We observed no evidence in our dataset indicating overall enrichment of SNPs strongly associated with PC1 in scavenger receptors (p = .31; Neubauer et al., 2016) or genes differentially regulated during symbiosis establishment in larvae (p = .52; Wolfowicz et al., 2016).
Table 4.
Genes in symbiosis‐related gene sets enriched for SNP associations with PC1. F ST values for each locus are also given. Only loci with F ST values in comparisons between the Bocas‐specific and the global E. pallida populations above the genomewide median of 0.1 are listed
| Classification | Scaffold | Position | Gene ID | Annotation | F ST |
|---|---|---|---|---|---|
| VWA‐domain TSR | 46 | 59254 | AIPGENE29239 | EGF‐like repeat and discoidin I‐like domain‐containing protein 3 | 0.88 |
| ADAMTS‐like | 105 | 237954 | AIPGENE15743 | ADAM‐9 | 0.77 |
| Complement | 90 | 100854 | AIPGENE2377 | Complement component 3 (C3) | 0.72 |
| VWA‐domain TSR | 280 | 116748 | AIPGENE20111 | Lactadherin | 0.70 |
| ADAMTS‐like | 32 | 161937 | AIPGENE3437 | ADAMTS‐6 | 0.66 |
| ADAMTS‐like | 207 | 241923 | AIPGENE19810 | ADAMTS‐6 | 0.66 |
| ADAMTS‐like | 32 | 123333 | AIPGENE3355 | ADAMTS‐6 | 0.54 |
| ADAMTS‐like | 32 | 79397 | AIPGENE3413 | ADAMTS‐7 | 0.51 |
| VWA‐domain TSR | 280 | 122504 | AIPGENE20111 | Lactadherin | 0.38 |
| ADAMTS‐like | 78 | 399268 | AIPGENE26625 | ADAMTS‐19 | 0.36 |
| TSR domain only | 139 | 186746 | AIPGENE10970 | Hemicentin‐1 | 0.33 |
| Complement | 343 | 122144 | AIPGENE1009 | Venom factor | 0.32 |
| Complement | 343 | 133790 | AIPGENE1009 | Venom factor | 0.14 |
4. DISCUSSION
This study offers a fine‐scale view into ecological and evolutionary dynamics of wild Exaiptasia sea anemones. Our findings were consistent with specialization of closely related host species and their symbiont communities for particular habitats. Furthermore, we discovered a distinct, locally abundant population of E. pallida that harbored a single symbiont species. Markers present in candidate symbiosis‐related genes were among the most highly differentiated in our dataset, pointing to intraspecific variation in host specificity associated with divergence from the globally distributed population. These findings support an important role of environmental variation in shaping co‐evolutionary dynamics of Exaiptasia holobionts at regional scales.
Our results contribute to the broader perspective of genetic diversity emerging for natural Exaiptasia populations, crucial for interpreting the rapidly growing body of scientific literature surrounding this important laboratory symbiosis system (Goldstein & King, 2016; Hoang, Morran, & Gerardo, 2016). Until recently, evolutionary perspectives on phylogenetic relationships and genetic diversity of the anemone host have been few (but see Thornhill et al., 2013; Bellis et al., 2016; Grajales & Rodríguez, 2016). Focusing on a previously identified geographic area of high diversity, we discovered a unique population of E. pallida that may be endemic to the Bocas del Toro Archipelago in Caribbean Panama (Figure 5). This population is genetically distinct from the previously described “global” and “South US Atlantic” networks (Grawunder et al., 2015; Thornhill et al., 2013). In general, the patterns of genetic diversity described here and elsewhere (Bellis et al., 2016; Thornhill et al., 2013) are consistent with a large, genetically diverse population located in the Western Atlantic that has occasionally given rise to new populations throughout the evolutionary history of Exaiptasia.
We describe the Bocas‐specific population as genetically distinct; it may represent an incipient species. Some evidence was observed for admixture from the global population (Figure 5a), and despite a large proportion of highly structured polymorphisms (Figure 6), the genomewide median F ST between the two populations was moderate (~0.10). There was no indication of variation in mitochondrial sequence, although mitochondrial DNA divergence is not very useful for resolving closely related anthozoan species, because it evolves very slowly (Shearer, Van Oppen, Romano, & Wörheide, 2002). The Bocas‐specific population may have a strong competitive advantage in its home environment, judging by the paucity of individuals from the global population particularly at the inshore site Cayo Roldán (Figure 5a). Anemone population density was higher than any other site sampled (Figures 3), and there was no evidence of reductions in symbiont density when populations were sampled (Figure 4), although Cayo Roldán typically reaches the highest temperatures of the study sites in the Bocas del Toro Archipelago (Figure 2).
We also observed associations between abiotic environment and variation in symbiont community assemblages in natural populations. Clade B1 Symbiodinium dominated both E. pallida and E. brasiliensis individuals throughout the thermally stable Bocas del Toro Archipelago in areas shaded by mangrove canopy and at a shaded location in Galeta (Table 1). In contrast, E. brasiliensis harbored at least two genotypes of Symbiodinium clade A but not S. minutum in a high light, variable temperature site on a shoaling reef flat at Galeta (Table 1). S. minutum may therefore be favored in thermally stable, intermediate light environments, with Symbiodinium clade A predominating if exposure to high light and temperature is likely. This pattern of distribution is consistent with previously reported photophysiological characteristics of representatives of these Symbiodinium lineages (Goulet et al., 2005; Suggett et al., 2015). Similar variation in symbiont type according to abiotic environment is well documented in natural populations of corals (Keshavmurthy et al., 2012; Rowan, Knowlton, Baker, & Jara, 1997).
Abiotic environment and symbiont type were also associated with cryptic host diversity. Both species of Exaiptasia were identified in both regions, but E. pallida was only found attached to vertical substrate (e.g., mangrove roots or dock pilings), whereas E. brasiliensis was more common attached to rocks and coral rubble in Galeta (Table 1). Because no anemones were found on rocks or coral rubble in Bocas del Toro in our study, sampling site was confounded with substrate type. It is therefore unclear to what extent the observed patterns of distribution could result from variation in host preference for different symbiont types, abiotic environments, and/or substrates. However, this study adds to a growing body of research in cnidarians that has uncovered cryptic host diversity associated with both habitat and symbiont type in natural populations of symbiotic cnidarians (Prada et al., 2014; Warner, Van Oppen, & Willis, 2015).
The potential variation in habitat preference described here has important practical implications given the inability to induce larval settlement in the laboratory, a major challenge for further development of Exaiptasia as a genetic model system. One possibility is that early uptake of different Symbiodinium types could influence the type of substrate upon which anemone larvae prefer to settle. Larval settlement of Fungia scutaria, a scleractinian coral that also acquires symbionts horizontally, occurred earlier when larvae were colonized by Symbiodinium (Schwarz, Krupp, & Weis, 1999). Exaiptasia larvae similarly take up Symbiodinium prior to settlement and metamorphosis (Wolfowicz et al., 2016). Additionally, both temperature and the identity of ex‐hospite symbiont types can influence settlement behavior and substrate choice in larvae of broadcast‐spawning corals (Winkler, Pandolfi, & Sampayo, 2015). In the lagoon sites we studied, most propagation beyond individual mangrove roots was attributable to sexual reproduction, implying that E. pallida larvae actively seek out shaded, vertical, and/or fibrous substrates for settlement or that they differentially survive in such habitats. Variation in Exaiptasia settlement success may also be influenced by the strength of ocean currents at particular sites. Clonal propagation in the Bocas del Toro Archipelago was highest in the exposed offshore site Cayo Agua, suggesting an important role of wave exposure in mediating rates of asexual reproduction and eco‐evolutionary dynamics. Future investigations of variation in habitat preference among Exaiptasia populations and species could provide useful guidance for identifying the cues necessary to induce larval settlement under controlled conditions.
Despite a relatively limited genomic sequencing effort, patterns of genetic variation suggested strong differentiation of polymorphisms in putative symbiosis‐related genes. While providing high resolution for population genetic studies, RAD‐based approaches survey only a small proportion of the genome (Lowry et al., 2017). We are therefore likely to have missed the vast majority of loci that may be under selection between the global and Bocas‐specific populations. We expected to sequence at most ~1% of the E. pallida reference genome based on a priori predicted locations of all BcgI tags. Considering blocks of linkage disequilibrium extending approximately 5 kb on either side of genotyped loci as documented for another broadcast‐spawning anthozoan (Shinzato, Mungpakdee, Arakaki, & Satoh, 2015), our callset of 2.6k SNPs might reflect differentiation across up to 10% of the 260 Mb Exaiptasia genome. Assuming strong selection and a complex genetic architecture of symbiosis involving hundreds of genes, identification of a handful of symbiosis‐related outlier loci in our study may not be surprising.
Concordantly, outlier loci in previously identified candidate symbiosis genes were among the most highly differentiated SNPs in our dataset. Gene set enrichment analyses revealed overrepresentation of loci in the complement pathway and thrombospondin‐type‐1 repeat (TSR) domain genes (Table 4). A locus in complement C3, a protein that opsonizes foreign microbes and then enhances their phagocytosis by binding to receptors on the surface of host cells (Davy et al., 2012; Poole et al., 2016), was highly differentiated in our dataset. Loci in several TSR domain genes were also highly differentiated. In a previous study, blocking TSR domains decreased colonization of Exaiptasia anemones by symbionts, whereas addition of exogenous TSRs resulted in a “supercolonized” phenotype (Neubauer et al., 2017). Most of the differentiated TSR loci in our study were in ADAMTS‐like genes, a family of proteases involved in remodeling of the extracellular matrix (Kelwick, Desanlis, Wheeler, & Edwards, 2015). However, the most differentiated TSR locus was present in EGF‐like repeat and discoidin I‐like domain‐containing protein 3 (a.k.a. DEL‐1), a secreted calcium‐binding molecule that mediates cell adhesion through binding to integrins (Hidai et al., 1998). A role in blocking complement‐dependent phagocytosis in vertebrate immune cells has been proposed for DEL‐1 (Mitroulis et al., 2014).
Our analyses further revealed variation in genes that have not been previously implicated in cnidarian symbiosis. The individual locus most strongly associated with differentiation of the Bocas‐specific population was present in myoferlin (Table 3). Myoferlin is a calcium‐activated protein that regulates lysosomal exocytosis. Myoferlin‐deficient mice accumulate phagolysosomes (Song & Hanayama, 2016), mirroring the early arrest of phagosomal maturation that leads to persistence of symbiosomes within cnidarian cells (Chen, Cheng, Sung, Kuo, & Fang, 2003). Loci in N‐glycanase 1 and cytoplasmic FMR1‐interacting protein 2 (CYFIP2) were also strongly associated with divergence of the Bocas‐specific population (Table 3). N‐glycanase 1 is an enzyme that cleaves N‐linked glycans preferentially from high‐mannose glycoproteins (Zhou et al., 2006) and may be of interest given evidence for an important role of glycan/lectin interactions in recognition of cnidarian symbionts (Davy et al., 2012; Wood‐Charlson, Hollingsworth, Krupp, & Weis, 2006). CYFIP2 is a pro‐apoptotic gene involved in adhesion of human immune cells (Mayne et al., 2004) and is part of WAVE1, a protein complex that activates actin filament reorganization and regulates formation of cytoplasmic projections prior to phagocytosis. These genes may represent interesting targets for further study at cellular levels given their predicted roles in innate immunity, phagocytosis, and regulation of postphagocytic symbiont cell removal.
Although we did not quantify infection rates in this study, differentiation of genes involved in mediating symbiosis could suggest local adaptation of host populations through evolutionary fine‐tuning of interactions with regionally abundant Symbiodinium. Theory predicts the existence of co‐evolutionary hotspots where the intensity of reciprocal selection on symbiotic interactions is strong, drift is weak, and patterns of gene flow are conducive to local adaptation (Thompson, 1999). In cnidarian–dinoflagellate symbioses, reciprocal selection is likely to be strong in the tropics, where exogenous nutrients are generally low and irradiance and temperature are high year‐round compared to more temperate latitudes (Muller‐Parker & Davy, 2001). Anemones in the tropics may be more reliant on autotrophy, and Symbiodinium densities exhibit greater sensitivity to variation in temperature and irradiance compared to temperate anemone symbioses (Muller‐Parker & Davy, 2001). The extent to which particular regions such as the Bocas del Toro Archipelago may serve as co‐evolutionary hotspots more broadly among symbiotic broadcast‐spawning invertebrates remains to be seen.
In conclusion, this study underscores the importance of characterizing cryptic species diversity in cnidarians and natural variation in laboratory model systems. As global temperatures rise, improved understanding of coral resilience mechanisms such as adaptation or partner shifts is vital for informing management efforts (van Oppen, Oliver, Putnam, & Gates, 2015). Our results emphasize the potential for genetic variation in host specificity not only between cnidarian species but also among populations, and the importance of long‐term evolutionary history in shaping coral–Symbiodinium interactions. Integrating studies of natural variation with the extensive molecular resources available for Exaiptasia promises to continue providing a wealth of future insights on the cellular, ecological, and evolutionary dynamics of marine symbioses.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
ESB contributed to research design, data acquisition, and analysis, and drafted the initial manuscript. RBE and HKB acquired and analyzed data. HAL and DRD contributed to research design, data interpretation, and critical review of the manuscript. All authors read and approved the final manuscript.
DATA ACCESSIBILITY
DNA sequences are available from GenBank for 2bRAD sequencing reads [NCBI SRA: SRP111971] and Sanger sequences [NCBI GenBank MF942419‐MF942434]. Sampling locations, anemone abundance, Symbiodinium cell count, and genotype data files are available through the Dryad Digital Repository (https://doi.org/10.5061/dryad.g8t9240). Custom scripts are available in Appendix S1.
Supporting information
ACKNOWLEDGMENTS
This work was funded by an NSF Graduate Research Fellowship [0946928], a STRI Short‐Term Fellowship, and an OSU Zoology Research Fund award to ESB. The authors wish to thank the Smithsonian Tropical Research Institute particularly A. Calderón, R. Collin, P. Gondola, and G. Thomas for logistical support during sample collections, E. Meyer and H. Elder for assistance with 2bRAD, A. Liston, J. Spatafora, and the MCB525 class of 2016 for helpful discussions, D. Turnbull and the University of Oregon Genomics and Cell Characterization Core Facility for sequencing, and V. Weis for conversations about cnidarian innate immune pathways. Samples were collected and exported with permission from the Ministerio de Ambiente of the Republic of Panama under permit numbers SC/A‐25‐15 and SEX/A‐73‐15.
Bellis ES, Edlund RB, Berrios HK, Lessios HA, Denver DR. Molecular signatures of host specificity linked to habitat specialization in Exaiptasia sea anemones. Ecol Evol. 2018;8:5413–5426. https://doi.org/10.1002/ece3.4058
REFERENCES
- Bates, D. , Mächler, M. , Bolker, B. , & Walker, S. (2015). Fitting linear mixed‐effects models using lme4. Journal of Statistical Software, 67, 1–48. [Google Scholar]
- Baumgarten, S. , Cziesielski, M. J. , Thomas, L. , Michell, C. T. , Esherick, L. Y. , Pringle, J. R. , … Voolstra, C. R. (2018). Evidence for miRNA‐mediated modulation of the host transcriptome in cnidarian‐dinoflagellate symbiosis. Molecular Ecology, 2, 403–418. [DOI] [PubMed] [Google Scholar]
- Baumgarten, S. , Simakov, O. , Esherick, L. Y. , Liew, Y. J. , Lehnert, E. M. , Michell, C. T. , … Voolstra, C. R. (2015). The genome of Aiptasia, a sea anemone model for coral biology. Proceedings of the National Academy of Sciences, 112, 11893–11898. https://doi.org/10.1073/pnas.1513318112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis, E. S. , & Denver, D. R. (2017). Natural variation in responses to acute heat and cold stress in a sea anemone model system for coral bleaching. The Biological Bulletin, 233, 168–181. https://doi.org/10.1086/694890 [DOI] [PubMed] [Google Scholar]
- Bellis, E. S. , Howe, D. K. , & Denver, D. R. (2016). Genome‐wide polymorphism and signatures of selection in the symbiotic sea anemone Aiptasia . BMC Genomics, 17, 160 https://doi.org/10.1186/s12864-016-2488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burriesci, M. S. , Raab, T. K. , & Pringle, J. R. (2012). Evidence that glucose is the major transferred metabolite in dinoflagellate‐cnidarian symbiosis. The Journal of Experimental Biology, 215, 3467–3477. https://doi.org/10.1242/jeb.070946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M.‐C. , Cheng, Y.‐M. , Sung, P.‐J. , Kuo, C.‐E. , & Fang, L.‐S. (2003). Molecular identification of Rab7 (ApRab7) in Aiptasia pulchella and its exclusion from phagosomes harboring zooxanthellae. Biochemical and Biophysical Research Communications, 308, 586–595. https://doi.org/10.1016/S0006-291X(03)01428-1 [DOI] [PubMed] [Google Scholar]
- Cubit, J. D. , Caffey, H. M. , Thompson, R. C. , & Windsor, D. M. (1989). Meteorology and hydrography of a shoaling reef flat on the Caribbean coast of Panama. Coral Reefs, 8, 59–66. https://doi.org/10.1007/BF00301804 [Google Scholar]
- Danecek, P , Auton, A , Abecasis, G , Albers, CA , Banks, E , DePristo, MA , … 1000 Genomes Project Analysis Group (2011). The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. https://doi.org/10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, S. K. , Allemand, D. , & Weis, V. M. (2012). Cell biology of cnidarian‐dinoflagellate symbiosis. Microbiology and Molecular Biology Reviews, 76, 229–261. https://doi.org/10.1128/MMBR.05014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duforet‐Frebourg, N. , Luu, K. , Laval, G. , Bazin, E. , & Blum, M. G. B. (2016). Detecting genomic signatures of natural selection with principal component analysis: Application to the 1000 genomes data. Molecular Biology and Evolution, 33, 1082–1093. https://doi.org/10.1093/molbev/msv334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, S. R. , Phillips, W. S. , Green, D. R. , & Weis, V. M. (2007). Knockdown of actin and caspase gene expression by RNA interference in the symbiotic anemone Aiptasia pallida . Biological Bulletin, 212, 250–258. https://doi.org/10.2307/25066607 [DOI] [PubMed] [Google Scholar]
- Earl, D. A. , & vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. https://doi.org/10.1007/s12686-011-9548-7 [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. , & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario, F. , Beck, M. W. , Storlazzi, C. D. , Micheli, F. , Shepard, C. C. , & Airoldi, L. (2014). The effectiveness of coral reefs for coastal hazard risk reduction and adaptation. Nature Communications, 5, 3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattuso, J. , Allemand, D. , & Frankignouelle, M. (1999). Photosynthesis and calcification at cellular, organismal and community levels in coral reefs: A review on interactions and control by carbonate chemistry. American Zoologist, 39, 160–183. https://doi.org/10.1093/icb/39.1.160 [Google Scholar]
- Gillis, J. , Mistry, M. , & Pavlidis, P. (2010). Gene function analysis in complex data sets using ErmineJ. Nature Protocols, 5, 1148–1159. https://doi.org/10.1038/nprot.2010.78 [DOI] [PubMed] [Google Scholar]
- Gleason, D. F. , & Wellington, G. M. (1993). Ultraviolet radiation and coral bleaching. Nature, 365, 836–838. https://doi.org/10.1038/365836a0 [Google Scholar]
- Goldstein, B. , & King, N. (2016). The future of cell biology: Emerging model organisms. Trends in Cell Biology, 26, 818–824. https://doi.org/10.1016/j.tcb.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet, T. L. , Cook, C. B. , & Goulet, D. (2005). Effect of short‐term exposure to elevated temperatures and light levels on photosynthesis of different host‐symbiont combinations in the Aiptasia pallida‐Symbiodinium symbiosis. Limnology and Oceanography, 50, 1490–1498. https://doi.org/10.4319/lo.2005.50.5.1490 [Google Scholar]
- Grajales, A. , & Rodríguez, E. (2014). Morphological revision of the genus Aiptasia and the family Aiptasiidae (Cnidaria, Actiniaria, Metridioidea). Zootaxa, 3826, 55–100. https://doi.org/10.11646/zootaxa.3826.1.2 [DOI] [PubMed] [Google Scholar]
- Grajales, A. , & Rodríguez, E. (2016). Elucidating the evolutionary relationships of the Aiptasiidae, a widespread cnidarian–dinoflagellate model system (Cnidaria: Anthozoa: Actiniaria: Metridioidea). Molecular Phylogenetics and Evolution, 94, 252–263. https://doi.org/10.1016/j.ympev.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Grajales, A. , Rodríguez, E. , & Thornhill, D. J. (2016). Patterns of Symbiodinium spp. associations within the family Aiptasiidae, a monophyletic lineage of symbiotic of sea anemones (Cnidaria, Actiniaria). Coral Reefs, 35, 345–355. https://doi.org/10.1007/s00338-015-1352-5 [Google Scholar]
- Grawunder, D. , Hambleton, E. A. , Bucher, M. , Wolfowicz, I. , Bechtoldt, N. , & Guse, A. (2015). Induction of Gametogenesis in the Cnidarian Endosymbiosis Model Aiptasia sp. Scientific Reports, 5, 15677 https://doi.org/10.1038/srep15677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron, S. F. , Maynard, J. A. , van Hooidonk, R. , & Eakin, C. M. (2016). Warming trends and bleaching stress of the world's coral reefs 1985–2012. Scientific Reports, 6, 38402 https://doi.org/10.1038/srep38402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidai, C. , Zupancic, T. , Penta, K. , Mikhail, A. , Kawana, M. , Quertermous, E. E. , … Quertermous, T. (1998). Cloning and characterization of developmental endothelial locus‐ 1: An embryonic endothelial cell protein that binds the alphavbeta3 integrin receptor. Genes & Development, 12, 21–33. https://doi.org/10.1101/gad.12.1.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, K. L. , Morran, L. T. , & Gerardo, N. M. (2016). Experimental evolution as an underutilized tool for studying beneficial animal‐microbe interactions. Frontiers in Microbiology, 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn, T. , Bretz, F. , & Westfall, P. (2008). Simultaneous inference in general parametric models. Biometrical Journal, 50, 346–363. [DOI] [PubMed] [Google Scholar]
- Hughes, T. P. , Kerry, J. , Álvarez‐Noriega, M. , Álvarez‐Romero, J. G. , Anderson, K. D. , Baird, A. H. , … Wilson, S. K. (2017). Global warming and recurrent mass bleaching of corals. Nature, 543, 373–378. https://doi.org/10.1038/nature21707 [DOI] [PubMed] [Google Scholar]
- Jakobsson, M. , & Rosenberg, N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. https://doi.org/10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jombart, T. (2008). Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405. https://doi.org/10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Kamvar, Z. N. , Tabima, J. F. , & Grünwald, N. J. (2014). Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ, 2, e281 https://doi.org/10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, K. W. , & Thompson, R. C. (2005). Water temperature variation and the meteorological and hydrographic environment of Bocas del Toro, Panama. Caribbean Journal of Science, 41, 392–413. [Google Scholar]
- Kelwick, R. , Desanlis, I. , Wheeler, G. N. , & Edwards, D. R. (2015). The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biology, 16, 113 https://doi.org/10.1186/s13059-015-0676-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavmurthy, S. , Hsu, C.‐M. , Kuo, C.‐Y. , Meng, P. J. , Wang, J. T. , & Chen, C. A. (2012). Symbiont communities and host genetic structure of the brain coral Platygyra verweyi, at the outlet of a nuclear power plant and adjacent areas. Molecular Ecology, 21, 4393–4407. https://doi.org/10.1111/j.1365-294X.2012.05704.x [DOI] [PubMed] [Google Scholar]
- Kiers, T. E. , Palmer, T. M. , Ives, A. R. , Bruno, J. F. , & Bronstein, J. L. (2010). Mutualisms in a changing world: An evolutionary perspective. Ecology Letters, 13, 1459–1474. https://doi.org/10.1111/j.1461-0248.2010.01538.x [DOI] [PubMed] [Google Scholar]
- Lirman, D. , Schopmeyer, S. , Manzello, D. , Gramer, L. J. , Precht, W. F. , Muller‐Karger, F. , … Thanner, S. (2011). Severe 2010 cold‐water event caused unprecedented mortality to corals of the Florida reef tract and reversed previous survivorship patterns. PLoS ONE, 6, e23047 https://doi.org/10.1371/journal.pone.0023047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, D. B. , Hoban, S. , Kelley, J. L. , Lotterhos, K. E. , Reed, L. K. , Antolin, M. F. , & Storfer, A. (2017). Breaking RAD: An evaluation of the utility of restriction site associated DNA sequencing for genome scans of adaptation. Molecular Ecology Resources, 17, 142–152. https://doi.org/10.1111/1755-0998.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, K. , Bazin, E. , & Blum, M. G. B. (2017). pcadapt: An R package to perform genome scans for selection based on principal component analysis. Molecular Ecology Resources, 17, 67–77. https://doi.org/10.1111/1755-0998.12592 [DOI] [PubMed] [Google Scholar]
- Matthews, J. L. , Crowder, C. M. , Oakley, C. A. , Lutz, A. , Roessner, U. , Meyer, E. , … Davy, S. K. (2017). Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian‐dinoflagellate symbiosis. Proceedings of the National Academy of Sciences, 114, 201710733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne, M. , Moffatt, T. , Kong, H. , Kong, H. , McLaren, P. J. , Fowke, K. R. , … Melanson, M. (2004). CYFIP2 is highly abundant in CD4 + cells from multiple sclerosis patients and is involved in T cell adhesion. European Journal of Immunology, 34, 1217–1227. https://doi.org/10.1002/(ISSN)1521-4141 [DOI] [PubMed] [Google Scholar]
- Mitroulis, I. , Kang, Y. , Gahmberg, C. G. , Siegert, G. , Hajishengallis, G. , Chavakis, T. , & Choi, E.‐Y. (2014). Developmental endothelial locus‐1 attenuates complement‐dependent phagocytosis through inhibition of Mac‐1‐integrin. Thrombosis and Haemostasis, 111, 1004–1006. https://doi.org/10.1160/TH13-09-0794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg, F. , & Folke, C. (1999). Ecological goods and services of coral reef ecosystems. Ecological Economics, 29, 215–233. https://doi.org/10.1016/S0921-8009(99)00009-9 [Google Scholar]
- Muller‐Parker, G. , & Davy, S. K. (2001). Temperate and tropical algal‐sea anemone symbioses. Invertebrate Biology, 120, 104–123. https://doi.org/10.1111/j.1744-7410.2001.tb00115.x [Google Scholar]
- Muscatine, L. , Falkowski, P. , Porter, J. , & Dubinsky, Z. (1984). Fate of photosynthetic fixed carbon in light‐ and shade‐adapted colonies of the symbiotic coral Stylophora pistillata . Proceedings of the Royal Society of London Series B, Biological Sciences, 222, 181–202. https://doi.org/10.1098/rspb.1984.0058 [Google Scholar]
- Neubauer, E. F. , Poole, A. Z. , Neubauer, P. , Detournay, O. , Tan, K. , Davy, S. K. , & Weis, V. M. (2017). A diverse host thrombospondin‐type‐1 repeat protein repertoire promotes symbiont colonization during establishment of cnidarian‐dinoflagellate symbiosis. eLife, 6, e24494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, E. F. , Poole, A. Z. , Weis, V. M. , & Davy, S. K. (2016). The scavenger receptor repertoire in six cnidarian species and its putative role in cnidarian‐dinoflagellate symbiosis. PeerJ, 4, e2692 https://doi.org/10.7717/peerj.2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen, M. J. H. , Oliver, J. K. , Putnam, H. M. , & Gates, R. D. (2015). Building coral reef resilience through assisted evolution. Proceedings of the National Academy of Sciences, 112, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, S. F. , Cook, C. B. , & Brooks, W. R. (2001). The role of symbiotic dinoflagellates in the temperature‐induced bleaching response of the subtropical sea anemone Aiptasia pallida . Journal of Experimental Marine Biology and Ecology, 256, 1–14. https://doi.org/10.1016/S0022-0981(00)00282-3 [DOI] [PubMed] [Google Scholar]
- Pochon, X. , & Gates, R. D. (2010). A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai'i. Molecular Phylogenetics and Evolution, 56, 492–497. https://doi.org/10.1016/j.ympev.2010.03.040 [DOI] [PubMed] [Google Scholar]
- Pochon, X. , Montoya‐Burgos, J. I. , Stadelmann, B. , & Pawlowski, J. (2006). Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium . Molecular Phylogenetics and Evolution, 38, 20–30. https://doi.org/10.1016/j.ympev.2005.04.028 [DOI] [PubMed] [Google Scholar]
- Poole, A. Z. , Kitchen, S. A. , & Weis, V. M. (2016). The role of complement in cnidarian‐dinoflagellate symbiosis and immune challenge in the sea anemone Aiptasia pallida . Frontiers in Microbiology, 7, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada, C. , McIlroy, S. E. , Beltrán, D. M. , Valint, D. J. , Ford, S. A. , Hellberg, M. E. , & Coffroth, M. A. (2014). Cryptic diversity hides host and habitat specialization in a gorgonian‐algal symbiosis. Molecular Ecology, 23, 3330–3340. https://doi.org/10.1111/mec.12808 [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A. (2004). DISTRUCT: A program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138. [Google Scholar]
- Rowan, R. , Knowlton, N. , Baker, A. C. , & Jara, J. (1997). Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature, 388, 265–269. https://doi.org/10.1038/40843 [DOI] [PubMed] [Google Scholar]
- Rumble, S. M. , Lacroute, P. , Dalca, A. V. , Fiume, M. , Sidow, A. , & Brudno, M. (2009). SHRiMP: Accurate mapping of short color‐space reads. PLoS Computational Biology, 5, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, J. A. , Krupp, D. A. , & Weis, V. M. (1999). Late larval development and onset of symbiosis in the Scleractinian coral Fungia scutaria . Biological Bulletin, 196, 70–79. https://doi.org/10.2307/1543169 [DOI] [PubMed] [Google Scholar]
- Shearer, T. L. , Van Oppen, M. J. H. , Romano, S. L. , & Wörheide, G. (2002). Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Molecular Ecology, 11, 2475–2487. https://doi.org/10.1046/j.1365-294X.2002.01652.x [DOI] [PubMed] [Google Scholar]
- Shinzato, C. , Mungpakdee, S. , Arakaki, N. , & Satoh, N. (2015). Genome‐wide SNP analysis explains coral diversity and recovery in the Ryukyu Archipelago. Scientific Reports, 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six, D. L. (2009). Climate change and mutualism. Nature Reviews Microbiology, 7, 686 https://doi.org/10.1038/nrmicro2232 [DOI] [PubMed] [Google Scholar]
- Song, J. , & Hanayama, R. (2016). Mechanisms of lysosomal exocytosis by immune cells In Miyasaka M. & Takatsu K. (Eds.), Chronic inflammation: Mechanisms and regulation (pp. 369–378). Berlin, Germany: Springer; https://doi.org/10.1007/978-4-431-56068-5 [Google Scholar]
- Suggett, D. J. , Goyen, S. , Evenhuis, C. , Szabó, M. , Tye Pettay, D. , Warner, M. E. , & Ralph, P. J. (2015). Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytologist, 208, 370–381. https://doi.org/10.1111/nph.13483 [DOI] [PubMed] [Google Scholar]
- Thompson, J. N. (1999). Specific hypotheses on the Geographic Mosaic of Coevolution. The American Naturalist, 153, S1–S14. https://doi.org/10.1086/303208 [Google Scholar]
- Thornhill, D. J. , Xiang, Y. , Pettay, D. T. , Zhong, M. , & Santos, S. R. (2013). Population genetic data of a model symbiotic cnidarian system reveal remarkable symbiotic specificity and vectored introductions across ocean basins. Molecular Ecology, 22, 4499–4515. https://doi.org/10.1111/mec.12416 [DOI] [PubMed] [Google Scholar]
- Vega Thurber, R. L. , Burkepile, D. E. , Fuchs, C. , Shantz, A. A. , McMinds, R. , & Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Global Change Biology, 20, 544–554. https://doi.org/10.1111/gcb.12450 [DOI] [PubMed] [Google Scholar]
- Wang, S. , Meyer, E. , McKay, J. , & Matz, M. (2012). 2b‐RAD: A simple and flexible method for genome‐wide genotyping. Nature Methods, 9, 808–810. https://doi.org/10.1038/nmeth.2023 [DOI] [PubMed] [Google Scholar]
- Warner, P. A. , Van Oppen, M. J. H. , & Willis, B. L. (2015). Unexpected cryptic species diversity in the widespread coral Seriatopora hystrix masks spatial‐genetic patterns of connectivity. Molecular Ecology, 24, 2993–3008. https://doi.org/10.1111/mec.13225 [DOI] [PubMed] [Google Scholar]
- Weis, V. M. (2008). Cellular mechanisms of Cnidarian bleaching: Stress causes the collapse of symbiosis. The Journal of Experimental Biology, 211, 3059–3066. https://doi.org/10.1242/jeb.009597 [DOI] [PubMed] [Google Scholar]
- Weis, V. M. , Davy, S. K. , Hoegh‐Guldberg, O. , Rodriguez‐Lanetty, M. , & Pringle, J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends in Ecology & Evolution, 23, 369–376. https://doi.org/10.1016/j.tree.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Winkler, N. S. , Pandolfi, J. M. , & Sampayo, E. M. (2015). Symbiodinium identity alters the temperature‐dependent settlement behaviour of Acropora millepora coral larvae before the onset of symbiosis. Proceedings of the Royal Society B, 282, 20142260 https://doi.org/10.1098/rspb.2014.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisecaver, J. H. , & Hackett, J. D. (2011). Dinoflagellate genome evolution. Annual Review of Microbiology, 65, 369–387. https://doi.org/10.1146/annurev-micro-090110-102841 [DOI] [PubMed] [Google Scholar]
- Van Woesik, R. , De Vantier, L. M. , & Glazebrook, J. S. (1995). Effects of Cyclone “Joy” on nearshore coral communities of the Great Barrier Reef. Marine Ecology Progress Series, 128, 261–270. https://doi.org/10.3354/meps128261 [Google Scholar]
- Wolfowicz, I. , Baumgarten, S. , Voss, P. A. , Hambleton, E. A. , Voolstra, C. R. , Hatta, M. , & Guse, A. (2016). Aiptasia sp. larvae as a model to reveal mechanisms of symbiont selection in cnidarians. Scientific Reports, 6, 32366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood‐Charlson, E. M. , Hollingsworth, L. L. , Krupp, D. A. , & Weis, V. M. (2006). Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cellular Microbiology, 8, 1985–1993. https://doi.org/10.1111/j.1462-5822.2006.00765.x [DOI] [PubMed] [Google Scholar]
- Yellowlees, D. , Rees, T. A. V. , & Leggat, W. (2008). Metabolic interactions between algal symbionts and invertebrate hosts. Plant, Cell & Environment, 31, 679–694. https://doi.org/10.1111/j.1365-3040.2008.01802.x [DOI] [PubMed] [Google Scholar]
- Zhou, X. , Zhao, G. , Truglio, J. J. , Wang, L. , Li, G. , Lennarz, W. J. , & Schindelin, H. (2006). Structural and biochemical studies of the C‐terminal domain of mouse peptide‐N‐glycanase identify it as a mannose‐binding module. Proceedings of the National Academy of Sciences, 103, 17214–17219. https://doi.org/10.1073/pnas.0602954103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences are available from GenBank for 2bRAD sequencing reads [NCBI SRA: SRP111971] and Sanger sequences [NCBI GenBank MF942419‐MF942434]. Sampling locations, anemone abundance, Symbiodinium cell count, and genotype data files are available through the Dryad Digital Repository (https://doi.org/10.5061/dryad.g8t9240). Custom scripts are available in Appendix S1.


