Abstract
As an indispensable factor in DNA damage recognition step of nucleotide excision repair, XPA interacts with a series of proteins to initiate repair process. The expression characteristics of XPA in colorectal cancer (CRC) and its influence on CRC prognosis remain elusive. Tissue specimens of CRC and nontumor adjacent tissues from 283 patients were collected. XPA protein expressions were detected by immunohistochemistry staining. Nonparametric test was used to investigate the difference of XPA expression between CRC and nontumor adjacent tissues, as well as the correlation between XPA expression and clinicopathological parameters of CRC. Univariate and multivariate Cox proportional hazards models were applied to estimate the relationship between XPA expression and CRC prognosis. Meanwhile, we analyzed TCGA data to investigate the relation between XPA mRNA expression and survival of CRC. XPA protein expression was significantly decreased in CRC tissues compared with nontumor adjacent tissues (P = 0.001). Subgroup analysis indicated consistently significant down‐regulation of XPA in CRC tissues in age > 60 (P = 0.026), age ≤ 60 (P = 0.008), colon cancer (P = 0.009), and rectal cancer (P = 0.015) patients and males (P = 0.004). For clinicopathological parameters, CRC patients with drinking habits revealed XPA overexpression than nondrinkers (P = 0.032). For prognosis, CRC patients with high XPA protein expression had longer overall survival (OS) (HR = 0.62, 95%CI: 0.39–0.97, P = 0.037). Stratified analysis suggested a better prognosis in relation to high XPA protein expression in patients over 60 years (adjusted HR = 0.48, P = 0.021), with rectal cancer (HR = 0.56, P = 0.037), without distant metastasis (HR = 0.58, P = 0.033), without tumor deposits (HR = 0.40, P = 0.006; adjusted HR = 0.44, P = 0.028), and with tumor diameter over 4 cm (HR = 0.49, P = 0.023). DNA repair protein XPA is significantly decreased in colorectal cancer tissues than in adjacent nontumor tissues. High expression of XPA protein showed significant relationship with better survival of CRC, especially rectal cancer. XPA might be a novel biomarker but might not be an independent factor to predict prognosis of CRC patients.
Keywords: Colorectal cancer, prognosis, XPA
Introduction
Various human diseases, particularly cancer, mainly derive from an imbalance between DNA damage and repair 1, 2. DNA damage is induced by endogenous or exogenous stimuli 3, 4, while DNA repair is accomplished by systems including nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), and double‐strand break repair (DSBR) 5. NER system, which is versatile and crucial, monitors and restores multiple DNA damage of ultraviolet‐induced cyclobutane pyrimidine dimers, bulky adducts as well as DNA cross‐links 6, 7. And four key procedures participated in the NER pathway are as follows: damage recognition, damage demarcation and unwinding, damage incision, and new strand ligation 8, 9.
Xeroderma pigmentosum group A (XPA) gene, mapped to chromosome 9q22.3, includes six exons and encodes a zinc finger protein of 273 amino acids 10. As an indispensable factor in DNA damage recognition, XPA interacts with a series of NER proteins to initiate repair process 6, 11, 12. It has been revealed that cells or animals lacking XPA cannot accomplish NER 9, 13, 14, 15. Considering the critical role of XPA in NER, a number of studies have been conducted to investigate the effect of XPA on cancer 16, 17, 18. Xiang Fu et al. 19 found that high expression of XPA correlated with poor prognosis in 129 nasopharyngeal carcinoma patients treated with platinum‐based chemoradiotherapy using immunohistochemistry. In metastatic ovarian carcinoma, the results of 67 malignant effusion specimens showed that the overexpression of XPA was associated with better (progression‐free survival) PFS and (overall survival) OS 20. So far, however, the expression characteristics of XPA in CRC, which is the fourth most common cause of cancer mortality and third most frequently diagnosed cancers in both males and females in China 21, and its influence on CRC prognosis remain elusive.
In this study, we detected XPA protein expression levels in the colorectal mucosa tissues and their adjacent nontumor tissues from 283 CRC patients by immunohistochemical staining. Meanwhile, the association between XPA expression with clinicopathological parameters and prognosis in CRC patients was analyzed to clarify the latent effect of XPA on the progression and prognosis of CRC.
Materials and Methods
Patients and tissue specimens
The study was approved by the Institute Research Medical Ethics Committee of the First Affiliated Hospital of China Medical University, and written informed consents were obtained from all individuals. Patients were enrolled from the First Affiliated Hospital of China Medical University who experienced surgical operation between October 2012 and July 2015. Tissue specimens including 283 CRC tissues and the corresponding nontumor adjacent tissues were collected in our study.
On the basis of the World Health Organization criteria, the tissue samples of CRC diagnosed on the account of histological results. International Union Against Cancer (UICC)/American Joint Committee on Cancer (AJCC) (seventh edition, 2010) was used to confirm TNM staging of CRC in the following of postoperative pathological diagnosis. Three criteria were made to exclude CRC patients (1) having XP disease, (2) accepting preoperative chemotherapy or radiation, and (3) having hereditary nonpolyposis colorectal cancer (HNPCC). The follow‐up was performed until August 2017. A total of 266 cases were included to analyze the prognosis (mean survival time was 37.9 months; the time of follow‐up ranged from 1 month to 56 months; 79 of them died), while the rest 17 cases were not included for the OS analysis because loss of follow‐up. The study defines overall survival (OS) as the period from the date of operation to death. The patients who smoke at least one cigarette daily for at least 1 year were regarded as the cases with history of smoking. Meanwhile, the study defines history of drinking as the mean alcohol ingest per day for at least 50 g and lasting for at least 1 year.
Immunohistochemistry
Immunohistochemistry was performed mainly as previously described 22. Tissues, which were fixed with formalin and embedded with paraffin, were cut into 4‐μm‐thick sections and mounted in a poly‐l‐lysine‐coated glass slides. After routine deparaffinization, rehydration in a graded alcohol series and washing in tap water, the tissue sections were exposed to 10 mmol/L citrate buffer (PH 6.0) for 90 sec in a steam pressure cooker for antigen retrieval. Endogenous peroxidase was blocked using 3% hydrogen peroxide for 10 min, and then, the tissue sections were washed with phosphate‐buffered saline (PBS, PH 7.4). To lessen the nonspecific binding, 10% normal goat serum was subsequently used to block tissue collagen for 10 min. The mouse monoclonal antibody anti‐XPA (ab‐2352, 1:200 dilution; Abcam, Cambridge, U.K.) was used as the primary antibody to detect XPA protein expression and incubated for 60 min at room temperature (24–27°C). After that, the sections were rinsed by PBS for 10 min each and then incubated with biotinylated secondary antibody (goat anti‐rabbit antibody, Maixin Inc., Fujian, China) and streptavidin–biotin–peroxidase for 10 min each at temperature (24–27°C). Slides were stained with DAB (DAB‐0031, Maixin Inc., Fujian, China) chromogenic reagent for 80 sec. At last, the slides were dehydrated and fixed by resin. Meanwhile, we used three ways to control the quality of IHC. First, we used negative (PBS was used to substitute primary and secondary antibodies, respectively) and positive controls in the IHC staining to avoid false‐negative or false‐positive results. Second, the DAB staining was observed by microscope in case that the staining was overestimated or underestimated. Third, two pathologists independently scored the XPA expression level in a double‐blind manner.
Evaluation of immunohistochemistry
XPA protein expressions in the different tissues were read and scored independently by two pathologists, in accordance with the double‐blind principle. On the basis of immunohistochemistry semiquantification method, the pathologists evaluated the area and intensity of the staining results. And if the differences between the results of the pathologists were more than one grade, more scopes would be selected and the final scores would be discussed and concluded by the two pathologists. Semiquantitative scoring criterion was used to evaluate the expression of XPA in nucleus. The staining intensity of cancer cells was graded on a scale of 0–3(I0–I3): I0 (no staining), I1 (light brown), I2 (brown staining), and I3 (heavy brown staining) (intermediary intensity between two levels was defined as I0.5, I1.5, and I2.5); the proportion of stained cells were recorded as (P0–P3): 0–5% (P0), 6–25% (P1), 26–50% (P2), 51–75% (P3), and 76–100% (4). The final IS scores were accumulated by the formula: IS score = In × Pm. At last, the XPA protein expression was graded as follows: negative (–), score = 0; weak expression (+), score = 0.5–4; moderate expression (++), score = 4.5–8; and strong expression (+++), score = 9–12. As the median for immunohistochemistry score, score 4.5 was selected as the cutoff value to distinguish high or low expression for XPA protein.
Obtainment of data from TCGA database
The Cancer Genome Atlas (TCGA) is a publicly available database that has generated comprehensive, multidimensional maps of the important genomic changes in 33 types of cancer. In this study, data of 478 colon adenocarcinoma cases (TCGA‐COAD, provisional) with expression and clinicopathological information were downloaded. Additionally, data of 166 rectum adenocarcinoma cases (TCGA‐READ, provisional) were obtained to analyze the relationship of XPA mRNA expression with CRC prognosis.
Statistical analysis
All statistical analyses were performed using SPSS software, Chicago, IL (version 18.0). The comparison of XPA expression between CRC and nontumor adjacent tissues was assessed by nonparametric test. The correlation between XPA expression and clinicopathological parameters of CRC was also conducted by nonparametric test. The study applied Kaplan–Meier method to visualize the patient survival time and employed log‐rank tests to analyze the difference between groups. Univariate Cox proportional hazards model was applied to estimate the relationship between the expression of XPA and CRC prognosis, and multivariate Cox proportional hazards model was used to evaluate the association adjusted by age, gender, TNM stage, and differentiation degree. P values <0.05 were considered statistically significant.
Results
Baseline characteristics
The baseline characteristics of the 283 CRC patients included are shown in Table 1. Altogether, 165 males and 118 females were enrolled with a median survival time (MST) of 44.55 months and 47.14 months, respectively. Totally, 152 CRC patients were over 60 years of age, while 131 cases were younger than 60. The location of colorectal cancer included colon (80 cases) and rectum (202 cases). TNM staging was as follows: stage I, 73; stage II, 69; stage III, 121; and stage IV, 20.
Table 1.
Clinicopathological parameters and survival in CRC
| Characteristics | CRC | Cases of events | MST | P |
|---|---|---|---|---|
| Gender | ||||
| Male | 165 | 52 | 44.55 | 0.252 |
| Female | 118 | 30 | 47.14 | |
| Age (years) | ||||
| >60 | 152 | 50 | 43.64 | 0.171 |
| ≤60 | 131 | 32 | 47.25 | |
| Smoking | ||||
| Yes | 72 | 19 | 46.66 | 0.578 |
| No | 209a | 63 | 45.19 | |
| Drinking | ||||
| Yes | 31a | 8 | 46.03 | 0.615 |
| No | 237 | 72 | 45.18 | |
| Tumor location | ||||
| Colon | 80 | 24 | 44.41 | 0.889 |
| Rectum | 202a | 57 | 46.29 | |
| TNM stage | ||||
| I | 73 | 14 | 26.62 | <0.001 |
| II | 69 | 50 | 41.32 | |
| III | 121 | 12 | 49.72 | |
| IV | 20 | 6 | 53.35 | |
| Invasive extent | ||||
| T1–2 | 86 | 11 | 52.04 | <0.001 |
| T3–4 | 197 | 71 | 42.77 | |
| Lymph node metastasis | ||||
| Positive | 135 | 62 | 39.29 | <0.001 |
| Negative | 148 | 20 | 51.26 | |
| Distant metastasis | ||||
| Positive | 20 | 14 | 26.62 | <0.001 |
| Negative | 263 | 68 | 46.98 | |
| Tumor deposit | ||||
| Positive | 31 | 17 | 29.16 | <0.001 |
| Negative | 184a | 44 | 44.49 | |
| Perineural invasion | ||||
| Positive | 148 | 49 | 39.75 | 0.004 |
| Negative | 71a | 13 | 49.13 | |
| Vessel carcinoma embolus | ||||
| Positive | 65 | 27 | 38.92 | 0.006 |
| Negative | 218 | 55 | 47.20 | |
| Growth pattern | ||||
| Infiltrative | 163a | 61 | 42.07 | <0.001 |
| Nested/cloddy | 119 | 21 | 50.28 | |
| Differentiation degree | ||||
| Poor/mucinous | 79 | 39 | 36.41 | <0.001 |
| Well/moderate | 191a | 37 | 49.98 | |
| Maximum diameter(cm) | ||||
| >4 | 133 | 45 | 42.92 | 0.036 |
| ≤4 | 149a | 36 | 48.10 | |
| Family history | ||||
| Positive | 57 | 15 | 45.65 | 0.478 |
| Negative | 226 | 67 | 45.43 | |
| Chemotherapy | ||||
| Yes | 107a | 27 | 48.14 | 0.409 |
| No | 111 | 30 | 44.53 | |
CRC, colorectal cancer; MST, median survival time.
Incomplete information.
Down‐regulation of XPA in CRC tissues than nontumor adjacent tissues
The representative immunohistochemistry staining of CRC tissue and nontumor adjacent tissue is shown in Figure 1 (Figure 1A and B), respectively. Figure 2 demonstrated four different staining grades as negative (−), light positive (+), positive (++), and strong positive (+++). The detailed results of the expression profile of XPA in CRC and nontumor adjacent tissues are summarized in Table 2. According to the Mann–Whitney U‐test, XPA protein expression was significantly decreased in CRC tissues compared with nontumor adjacent tissues (P = 0.001), which is visualized by scatter plots in Figure 3.
Figure 1.
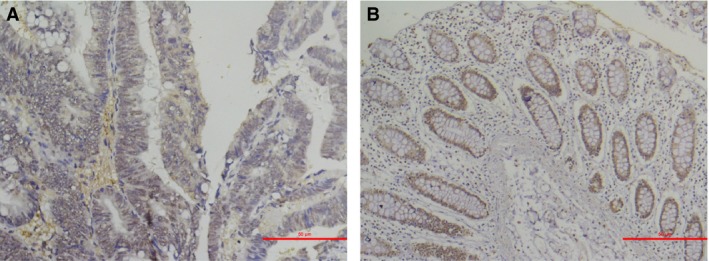
Representative photomicrographs of immunohistochemical staining of XPA in CRC specimens and adjacent nontumor specimens. (A) Colorectal cancer tissues and (B) adjacent nontumor tissues of CRC. Original magnification, ×200.
Figure 2.
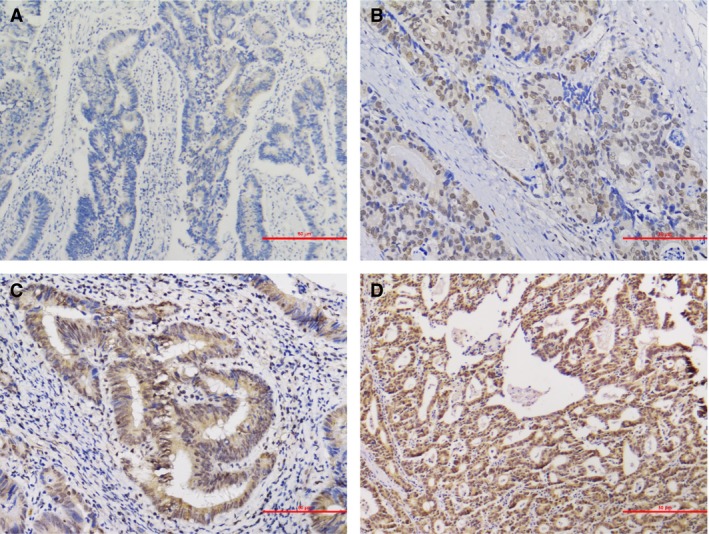
Different XPA expression levels in CRC tissues. (A) negative (−), (B) weakly positive (+), (C) moderately positive (++), and (D) strongly positive (+++). Magnification, ×200.
Table 2.
XPA expression in CRC and nontumor adjacent tissues
| Category | Group | Cases | (−) | (+) | (++) | (+++) | PR (%) | P |
|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | |||||
| Overall | CRC | 275 | 11 (4.0) | 120 (43.6) | 108 (39.3) | 36 (13.1) | 96.0 | 0.001 |
| Adjacent | 275 | 27 (9.8) | 64 (23.3) | 123 (44.7) | 61 (22.2) | 90.2 | ||
| Male | CRC | 161 | 7 (4.3) | 72 (44.7) | 59 (36.6) | 23 (14.3) | 95.7 | 0.004 |
| Adjacent | 161 | 17 (10.6) | 36 (22.4) | 70 (43.5) | 38 (23.6) | 89.4 | ||
| Female | CRC | 114 | 4 (3.5) | 48 (42.1) | 49 (43.0) | 13 (11.4) | 96.5 | 0.067 |
| Adjacent | 114 | 10 (8.8) | 28 (24.6) | 53 (46.5) | 23 (20.2) | 91.2 | ||
| ≤60 | CRC | 130 | 5 (3.8) | 65 (50.0) | 43 (33.1) | 17 (13.1) | 96.2 | 0.008 |
| Adjacent | 130 | 14 (10.8) | 29 (22.3) | 63 (48.5 | 24 (18.5) | 89.2 | ||
| >60 | CRC | 145 | 6 (4.1) | 55 (37.9) | 65 (44.8) | 19 (13.1) | 95.9 | 0.026 |
| Adjacent | 145 | 13 (9.0) | 35 (24.1) | 60 (41.4) | 37 (25.5) | 91.0 | ||
| Colon | CRC | 78 | 2 (2.6) | 38 (48.7) | 29 (37.2) | 9 (11.5) | 97.4 | 0.009 |
| Adjacent | 78 | 9 (11.5) | 15 (19.2) | 38 (48.7) | 16 (20.5) | 88.5 | ||
| Rectum | CRC | 196 | 9 (4.6) | 82 (41.8) | 78 (39.8) | 27 (13.8) | 95.4 | 0.015 |
| Adjacent | 196 | 18 (9.2) | 48 (24.5) | 85 (43.4) | 45 (23.0) | 90.8 |
PR, positive rate. Negative (−), light positive (+), positive (++), strong positive (+++) staining. Mann–Whitney U‐test of nonparametric test to compare the XPA protein expression between CRC and adjacent tissues.
The bold values: P<0.05
Figure 3.
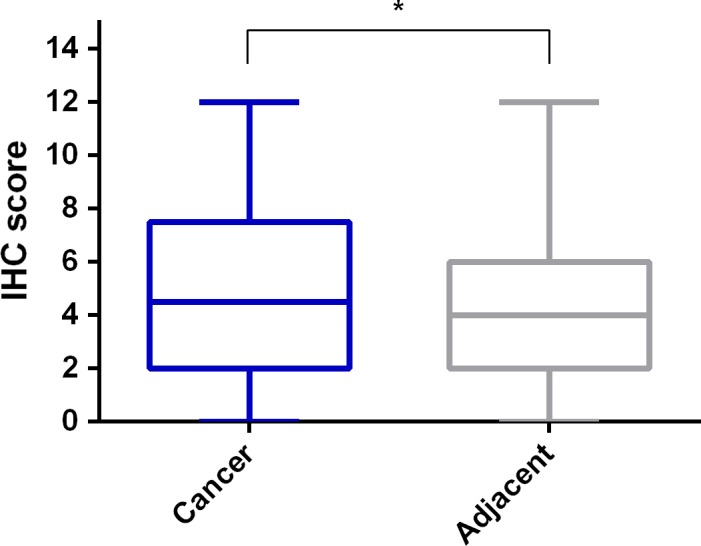
XPA protein expression was significantly decreased in CRC tissues compared with nontumor adjacent tissues. According to the Mann–Whitney U‐test, the differential expression of XPA between CRC specimens and nontumor adjacent specimens was visualized by scatter plots. *: P<0.05
Subgroup analysis based on age and tumor location suggested consistently significant down‐regulation of XPA in CRC tissues than in their adjacent tissues in age > 60 (P = 0.026), age ≤ 60 (P = 0.008), colon cancer (P = 0.009), and rectal cancer (P = 0.015). In addition, male patients showed low XPA expression in CRC tissues compared with adjacent tissues (P = 0.004), but no significant difference was observed in female individuals (P = 0.067).
Association between XPA protein expression and clinicopathological parameters of CRC patients
CRC patients were stratified according to variables including gender, age, smoking, drinking, TNM stage, and tumor invasion depth, and Mann–Whitney U‐test was performed to explore the differential expression of XPA between groups (Table 3). The results indicated that XPA protein expression correlated with drinking status: CRC patients with drinking habits revealed XPA overexpression than nondrinkers (P = 0.032). However, most comparisons of other clinicopathological parameters of CRC did not demonstrate significant difference (P > 0.05).
Table 3.
Association between XPA expression and clinicopathological parameters in CRC
| Variables | Cases | (−) | (+) | (++) | (+++) | PR (%) | P |
|---|---|---|---|---|---|---|---|
| n | n | n | n | ||||
| Gender | |||||||
| Male | 165 | 8 (4.8) | 73 (44.2) | 61 (37.0) | 23 (13.9) | 95.2 | 0.734 |
| Female | 118 | 5 (4.2) | 50 (42.4) | 50 (42.4) | 13 (11.0) | 95.8 | |
| Age (years) | |||||||
| >60 | 152 | 8 (5.3) | 57 (37.5) | 68 (44.7) | 19 (12.5) | 94.7 | 0.218 |
| ≤60 | 131 | 5 (3.8) | 66 (43.4) | 43 (28.3) | 17 (13.0) | 96.2 | |
| Smoking | |||||||
| Yes | 72 | 2 (2.8) | 7 (9.7) | 31 (20.4) | 12 (16.7) | 97.2 | 0.102 |
| No | 209a | 11 (2.1) | 95 (45.5) | 80 (38.3) | 23 (11.0) | 94.7 | |
| Drinking | |||||||
| Yes | 31a | 1 (3.2) | 9 (29.0) | 14 (45.2) | 7 (22.6) | 96.8 | 0.032 |
| No | 237 | 12 (5.1) | 108 (45.6) | 92 (38.8) | 25 (10.5) | 94.9 | |
| Tumor location | |||||||
| Colon | 80 | 2 (2.5) | 39 (48.8) | 30 (37.5) | 9 (11.3) | 97.5 | 0.372 |
| Rectum | 202a | 11 (5.4) | 84 (41.6) | 80 (39.6) | 27 (13.4) | 94.6 | |
| TNM stage | |||||||
| I | 73 | 2 (2.7) | 33 (45.2) | 30 (31.1) | 8 (11.0) | 97.3 | 0.863 |
| II | 69 | 5 (7.2) | 25 (36.2) | 31 (44.9) | 8 (11.6) | 92.8 | |
| III | 121 | 4 (43.3) | 58 (47.9) | 44 (36.4) | 15 (12.4) | 96.7 | |
| IV | 20 | 2 (10.0) | 7 (35.0) | 6 (30.0) | 5 (25.0) | 90.0 | |
| Invasive depth | |||||||
| T1–2 | 86 | 2 (2.3) | 38 (44.2) | 37 (43.0) | 9 (10.5) | 97.7 | 0.653 |
| T3–4 | 197 | 11 (5.6) | 85 (43.1) | 74 (37.6) | 27 (13.7) | 94.4 | |
| Lymph node metastasis | |||||||
| Positive | 135 | 6 (4.4) | 64 (47.4) | 48 (35.6) | 17 (12.6) | 95.6 | 0.552 |
| Negative | 148 | 7 (4.7) | 59 (39.9) | 63 (42.6) | 19 (12.8) | 95.3 | |
| Distant metastasis | |||||||
| Positive | 20 | 2 (10.0) | 7 (35.0) | 6 (30.0) | 5 (25.0) | 90.0 | 0.997 |
| Negative | 263 | 11 (4.2) | 116 (44.1) | 105 (39.9) | 31 (11.8) | 95.8 | |
| Tumor deposits | |||||||
| Positive | 31 | 1 (3.2) | 11 (35.5) | 13 (41.9) | 6 (19.4) | 96.8 | 0.098 |
| Negative | 184a | 11 (6.0) | 81 (44.0) | 73 (39.7) | 19 (10.3) | 94.0 | |
| Perineural invasion | |||||||
| Positive | 148 | 10 (6.8) | 67 (45.3) | 53 (35.8) | 18 (12.2) | 93.2 | 0.146 |
| Negative | 71a | 2 (2.8) | 28 (39.4) | 33 (46.5) | 8 (11.3) | 97.2 | |
| Lymphatic/venous invasion | |||||||
| Positive | 65 | 3 (4.6) | 27 (41.5) | 25 (38.5) | 10 (15.4) | 95.4 | 0.729 |
| Negative | 218 | 10 (4.6) | 96 (44.0) | 86 (39.4) | 26 (11.9) | 95.4 | |
| Growth pattern | |||||||
| Infiltrative | 163a | 11 (6.7) | 74 (45.4) | 59 (36.2) | 19 (11.7) | 93.3 | 0.085 |
| Cloddy/nested | 119 | 2 (1.7) | 49 (41.2) | 51 (42.9) | 17 (14.3) | 98.3 | |
| Differentiation degree | |||||||
| Poor/mucinous | 79 | 7 (8.9) | 35 (44.3) | 30 (38.0) | 7 (8.9) | 91.1 | 0.332 |
| Well/moderate | 191a | 6 (3.1) | 83 (43.5) | 76 (39.8) | 26 (13.6) | 96.9 | |
| Maximum diameter (cm) | |||||||
| >4 | 133 | 8 (6.0) | 53 (39.8) | 54 (40.6) | 18 (13.5) | 94.0 | 0.521 |
| ≤4 | 149a | 5 (3.4) | 69 (46.3) | 57 (38.3) | 18 (12.1) | 96.6 | |
| Family history | |||||||
| Positive | 57 | 2 (3.5) | 26 (45.6) | 21 (36.8) | 8 (14.0) | 96.5 | 0.911 |
| Negative | 226 | 11 (4.9) | 97 (42.9) | 90 (39.8) | 28 (12.4) | 95.1 | |
PR, positive rate. Negative (−), light positive (+), positive (++), strong positive (+++) staining.
The association of XPA expression with TNM stage was analyzed by Kruskal–Wallis H‐test of nonparametric test. For other clinicopathological parameters, Mann–Whitney U‐test of nonparametric test was used.
Incomplete information.
The bold values: P<0.05
Relationship between XPA expression and CRC prognosis
The cutoff value of IS was 4.5 in this study as it was the median score for immunohistochemistry staining of XPA in CRC (IS ≥ 4.5 means high expression, and IS < 4.5 means low expression). To investigate whether XPA protein expression could indicate CRC prognosis, Cox proportional hazards model was applied to estimate the relationship between the expression of XPA and CRC survival (Table 4). Univariate Cox proportional hazards model revealed that CRC patients with high XPA protein expression had longer overall survival (OS) (HR = 0.62, 95%CI: 0.39–0.97, P = 0.037, Fig. 4A). Multivariate Cox proportional hazards model adjusting for age, gender, TNM stage, and differentiation degree did not show significant relation with CRC survival (adjusted HR = 0.68, 95% CI: 0.42–1.09, P = 0.107).
Table 4.
Correlation between XPA expression and survival in CRC
| Cases | Cases of events | MST | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||||
| XPA expression | |||||||||
| Low (IS < 4.5) | 129 | 46 | 42.25 | ||||||
| High (IS ≥ 4.5) | 134 | 32 | 47.52 | 0.62 | 0.39–0.97 | 0.037 | 0.68 | 0.42–1.09 | 0.107 |
| Stratification | |||||||||
| Age | |||||||||
| >60 | |||||||||
| Low | 63 | 27 | 39.76 | ||||||
| High | 82 | 22 | 45.74 | 0.58 | 0.33–1.01 | 0.055 | 0.48 | 0.26–0.89 | 0.021 |
| ≤60 | |||||||||
| Low | 66 | 19 | 44.63 | ||||||
| High | 52 | 10 | 49.23 | 0.59 | 0.28–1.28 | 0.183 | 0.81 | 0.38–1.77 | 0.601 |
| Location | |||||||||
| Rectum | |||||||||
| Low | 90 | 32 | 43.06 | ||||||
| High | 96 | 21 | 48.54 | 0.56 | 0.32–0.97 | 0.037 | 0.59 | 0.33–1.05 | 0.072 |
| Colon | |||||||||
| Low | 39 | 14 | 40.24 | ||||||
| High | 37 | 10 | 45.20 | 0.70 | 0.31–1.57 | 0.390 | 0.85 | 0.36–2.02 | 0.710 |
| Distant metastasis | |||||||||
| Positive | |||||||||
| Low | 9 | 7 | 20.44 | ||||||
| High | 11 | 7 | 27.18 | 0.58 | 0.19–1.74 | 0.330 | 0.71 | 0.19–2.72 | 0.618 |
| Negative | |||||||||
| Low | 120 | 25 | 43.66 | ||||||
| High | 123 | 39 | 49.02 | 0.58 | 0.35–0.96 | 0.033 | 0.61 | 0.36–1.04 | 0.072 |
| Tumor deposits | |||||||||
| Positive | |||||||||
| Low | 10 | 6 | 27.90 | ||||||
| High | 19 | 10 | 28.47 | 0.89 | 0.32–2.45 | 0.823 | 1.22 | 0.43–3.47 | 0.711 |
| Negative | |||||||||
| Low | 88 | 29 | 41.34 | ||||||
| High | 85 | 13 | 46.72 | 0.40 | 0.21–0.77 | 0.006 | 0.44 | 0.21–0.92 | 0.028 |
| Max diameter (cm) | |||||||||
| >4 | |||||||||
| Low | 58 | 27 | 37.37 | ||||||
| High | 61 | 16 | 45.91 | 0.49 | 0.26–0.91 | 0.023 | 0.62 | 0.32–1.18 | 0.143 |
| ≤4 | |||||||||
| Low | 70 | 18 | 46.67 | ||||||
| High | 73 | 16 | 48.29 | 0.83 | 0.42–1.63 | 0.589 | 0.82 | 0.39–1.70 | 0.586 |
CI, confidence interval; HR, hazard radio; MST, median survival time. IS, the immunohistochemistry score.
The bold values: P<0.05
Figure 4.
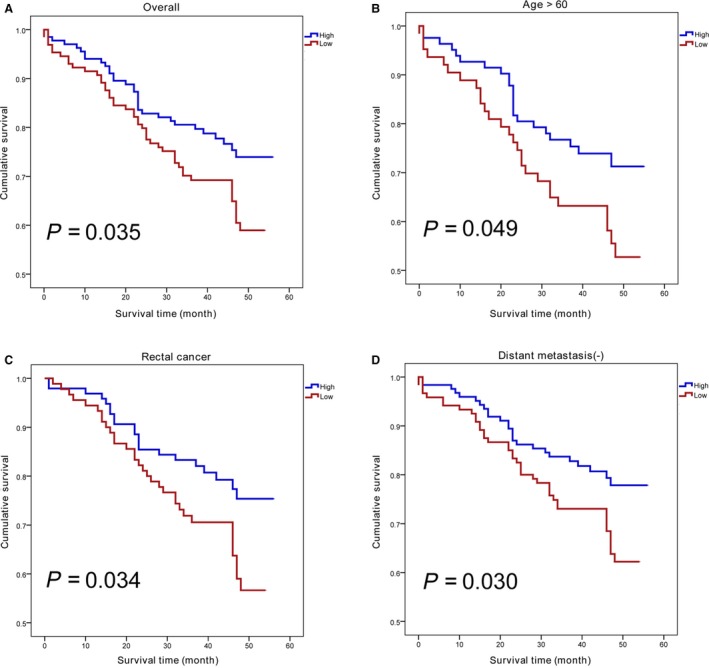
High expression of XPA correlates with the prognosis in CRC patients. (A) Kaplan–Meier analysis and log‐rank test for overall survival according to XPA expression level; (B) patients over 60 years of age with high XPA expression exhibited longer survival time than those with low XPA expression; (C) rectal cancer individuals who expressed higher XPA protein demonstrated favorable prognosis; (D) subgroup without distant metastasis also identified XPA expression as a good indicator for CRC prognosis.
Stratified analysis based on age and tumor location suggested that patients over 60 years of age with high XPA expression exhibited longer survival time than those with low XPA expression (adjusted HR = 0.48, 95% CI: 0.26–0.89, P = 0.021, Fig. 4B); rectal cancer individuals who expressed higher XPA protein demonstrated favorable prognosis (HR = 0.56, 95% CI: 0.32–0.97, P = 0.037, Fig. 4C). In the subgroup without distant metastasis, high XPA expression showed significant association with better OS (HR = 0.58, 95% CI: 0.35–0.96, P = 0.033, Fig. 4D). Both univariate and multivariate analyses indicated significant correlation between high XPA expression and decreased hazards of death in the CRC cases without tumor deposits (HR = 0.40, 95% CI: 0.21–0.77, P = 0.006, adjusted HR = 0.44, 95% CI: 0.21–0.92, P = 0.028). Besides, the subgroup with tumor diameter over 4 cm also identified XPA expression as a good indicator for CRC prognosis (HR = 0.49, 95% CI: 0.26–0.91, P = 0.023). However, no significant relation was observed according to the subgroup analysis of TNM stage, invasion depth, lymph node metastasis, growth pattern, differentiation degree, and chemotherapy after initial surgical operation (Table S1). According to the TCGA results, the association between XPA mRNA expression and survival of CRC was not statistically significant (Table S2).
Discussion
XPA, containing a zinc‐finger domain, displays a damaged DNA‐binding activity, which is essential for assembly of the preincision complex during nucleotide excision repair 11, 23. It has been reported that XPA exerts regulatory role not only by recognizing the existence of DNA damage, but, along with its interaction partner RPA, also in monitoring proper three‐dimensional arrangement of NER complex ahead of activation of endonuclease subunits 3, 24, 25, 26. Considering its important function in NER pathway, XPA is probably implicated in diseases related to imbalance between DNA damage and repair. However, the specific role of XPA in the progression and prognosis in CRC was still ambiguous. In this study including 283 CRC patients in China, we, for the first time, elucidated that DNA repair protein XPA is significantly decreased in colorectal cancer tissues than adjacent nontumor tissues. Univariate Cox proportional hazards model revealed that CRC patients with high XPA protein expression had longer overall survival (OS), but the association was not statistically significant in multivariate analysis. Besides, no significant relation was observed between XPA mRNA expression and survival of CRC according to TCGA results. According to the results of multivariate analysis and TCGA data, we suggested that XPA might be a promising biomarker but might not be an independent factor to predict prognosis of CRC patients.
In the present study, differential expression of XPA between colorectal cancer and nontumor adjacent tissues was explored. We found that XPA protein expression was significantly decreased in CRC tissues compared with nontumor adjacent tissues. Subgroup analysis suggested consistently significant difference in age over 60 years, age less than 60 years, colon cancer, rectal cancer, and males except that female individuals showed borderline significance (P = 0.067). These consistent findings ensure the phenomenon of decreased XPA expression in CRC tissues than in adjacent normal tissues, regardless of other factors. Previous studies on other types of cancer also came out with similar results: One research investigated twenty DNA repair pathway genes in 52 Dukes’ C colorectal cancer in Americans and revealed that only XPA had a lower RNA level in tumor samples than in matched normal ones 27; another study in Italians found significantly lower transcriptional expression of XPA in 50 nonsmall cell lung cancer (NSCLC) specimens compared with normal matched samples 28; bladder cancer also expressed low XPA of both mRNA and protein levels than nontumor bladder tissue, which was closely related to chromosomal aberrations 29. Taken together, the above‐mentioned results from other types of cancer could, at least in part, confirm our findings of XPA down‐regulation in CRC.
The association of XPA protein expression with the overall survival of CRC was also explored in this study. After classifying the CRC patients into high and low XPA expression groups by immunohistochemistry scores, we revealed a significantly increased survival time of individuals with high XPA protein expression. As for tumor location, the relationship was more obvious in rectal cancer rather than in colon cancer. Previously, the predictive role of high XPA expression for better prognosis has also been found in other types of cancers other than CRC: An Italian study investigated 171 ovarian cancer cases and suggested a longer OS and progression‐free survival (PFS) in cases that overexpressed XPA mRNA; similarly, high XPA protein expression in ovarian cancer has been regarded as an indicator for favorable prognosis according to a Norwegian research 20; Hyo Jung Cho et al. 30 found in 50 liver cancer cases in Korean population that low XPA mRNA level confers to worse survival. From this point of view, the correlation of up‐regulation of XPA with increased survival time might be applicable to not only CRC but also other types of cancers, the molecular mechanism of which requires further investigations to elucidate. Additionally, CRC patient subgroups without distant metastasis, without tumor deposits, or with tumor diameter over 4 cm demonstrated a more significant relationship with better overall survival. Thus, the influence of certain clinicopathological parameters on the implication of XPA in CRC progression is an intriguing direction for future researches.
The observations of differential expression of XPA in CRC and its predictive potential for overall survival enlighten our understanding of the complex participation of NER in the development and progression of CRC. Considering the core position of XPA in NER pathway, we assumed that the down‐regulation of XPA in CRC tissues might arise from the impairment of NER capacity upon colorectal carcinogenesis and the low XPA protein expression, which indicates degraded nuclear expression repair in CRC patients, might help create poor prognosis. On the contrary, sufficient NER ability did not benefit cancer patients from the aspect of chemotherapy, because platinum‐based chemotherapeutic regimens destroy cancer cells mainly via DNA damage. As the one of the toughest challenges for cancer treatment, chemotherapeutic resistance for platinum has been detected in XPA‐overexpressed nasopharyngeal cancer 19. Whether XPA contributes to CRC chemotherapeutic resistance remains to be clarified in the future. Biomarkers that could predict survival of cancer patients are urgently in need for clinical doctors to make individualized treatment plans and follow‐up management. In this study, the cutoff value (4.5) we used was based on our group of patients. More reliable cutoff value should be explored by multiple investigations based on different ethnicities. The obvious relation between XPA protein overexpression and favorable CRC prognosis in our study might provide useful clues for elucidating colorectal development, offering novel idea for effective treatment and improving survival.
Conclusion
In summary, DNA repair protein XPA is significantly decreased in colorectal cancer tissues than adjacent nontumor tissues. High expression of XPA protein showed significant relationship with better survival of CRC, especially rectal cancer. XPA might be a novel biomarker but might not be an independent factor to predict prognosis of CRC patients.
Conflict of Interest
All authors declare that there is no conflict of interest.
Supporting information
Table S1. Correlation between XPA expression and survival in CRC.
Table S2. Correlation between XPA mRNA expression and survival in colon cancer and rectal cancer ( based on TCGA).
Cancer Medicine 2018; 7(6):2339–2349
Contributor Information
Yuan Yuan, Email: yuanyuan@cmu.edu.cn.
Chengzhong Xing, Email: xcz1966@126.com.
References
- 1. Sandovici, I. , Buhusi M. C., Stoica O., and Covic M.. 2002. DNA repair pathways and their involvement in human diseases. Rev. Med. Chir. Soc. Med. Nat. Iasi 107:247–257. [PubMed] [Google Scholar]
- 2. Iyama, T. , and Wilson D. M. 3rd. 2013. DNA repair mechanisms in dividing and non‐dividing cells. DNA Repair 12:620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fadda, E. 2016. Role of the XPA protein in the NER pathway: a perspective on the function of structural disorder in macromolecular assembly. Comput. Struct. Biotechnol. J. 14:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Riedl, T. , Hanaoka F., and Egly J. M.. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goode, E. L. , Ulrich C. M., and Potter J. D.. 2002. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol. Biomark. Prev. 11:1513–1530. [PubMed] [Google Scholar]
- 6. Friedberg, E. C. 2001. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1:22–33. [DOI] [PubMed] [Google Scholar]
- 7. Slyskova, J. , Korenkova V., Collins A. R., Prochazka P., Vodickova L., Svec J., et al. 2012. Functional, genetic, and epigenetic aspects of base and nucleotide excision repair in colorectal carcinomas. Clin. Cancer Res. 18:5878–5887. [DOI] [PubMed] [Google Scholar]
- 8. de Laat, W. L. , Jaspers N. G., and Hoeijmakers J. H.. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768–785. [DOI] [PubMed] [Google Scholar]
- 9. Liu, J. , He C., Xing C., and Yuan Y.. 2014. Nucleotide excision repair related gene polymorphisms and genetic susceptibility, chemotherapeutic sensitivity and prognosis of gastric cancer. Mutat. Res. 765:11–21. [DOI] [PubMed] [Google Scholar]
- 10. Sugitani, N. , Sivley R. M., Perry K. E., Capra J. A., and Chazin W. J.. 2016. XPA: a key scaffold for human nucleotide excision repair. DNA Repair 44:123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sugasawa, K. 2008. Xeroderma pigmentosum genes: functions inside and outside DNA repair. Carcinogenesis 29:455–465. [DOI] [PubMed] [Google Scholar]
- 12. Ma, X. J. , Shang L., Zhang W. M., Wang M. R., and Zhan Q. M.. 2016. Mitotic regulator Nlp interacts with XPA/ERCC1 complexes and regulates nucleotide excision repair (NER) in response to UV radiation. Cancer Lett. 373:214–221. [DOI] [PubMed] [Google Scholar]
- 13. Camenisch, U. , and Nageli H.. 2008. XPA gene, its product and biological roles. Adv. Exp. Med. Biol. 637:28–38. [DOI] [PubMed] [Google Scholar]
- 14. Miyauchi‐Hashimoto, H. , Tanaka K., and Horio T.. 1996. Enhanced inflammation and immunosuppression by ultraviolet radiation in xeroderma pigmentosum group A (XPA) model mice. J. Invest. Dermatol. 107:343–348. [DOI] [PubMed] [Google Scholar]
- 15. Nakane, H. , Takeuchi S., Yuba S., Saijo M., Nakatsu Y., Murai H., et al. 1995. High incidence of ultraviolet‐B‐or chemical‐carcinogen‐induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377:165–168. [DOI] [PubMed] [Google Scholar]
- 16. He, L. , Deng T., and Luo H.. 2015. XPA A23G polymorphism and risk of digestive system cancers: a meta‐analysis. OncoTargets Therapy 8:385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding, D. , Zhang Y., Yu H., Guo Y., Jiang L., He X., et al. 2012. Genetic variation of XPA gene and risk of cancer: a systematic review and pooled analysis. Int. J. Cancer 131:488–496. [DOI] [PubMed] [Google Scholar]
- 18. Mellon, I. , Hock T., Reid R., Porter P. C., and States J. C.. 2002. Polymorphisms in the human xeroderma pigmentosum group A gene and their impact on cell survival and nucleotide excision repair. DNA Repair 1:531–546. [DOI] [PubMed] [Google Scholar]
- 19. Fu, X. , Hu J., Han H. Y., Hua Y. J., Zhou L., Shuai W. D., et al. 2015. High expression of XPA confers poor prognosis for nasopharyngeal carcinoma patients treated with platinum‐based chemoradiotherapy. Oncotarget 6:28478–28490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stevens, E. V. , Raffeld M., Espina V., Kristensen G. B., Trope C. G., Kohn E. C., et al. 2005. Expression of xeroderma pigmentosum A protein predicts improved outcome in metastatic ovarian carcinoma. Cancer 103:2313–2319. [DOI] [PubMed] [Google Scholar]
- 21. Chen, W. , Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., et al. 2016. Cancer statistics in China, 2015. CA Cancer J. Clin. 66:115–132. [DOI] [PubMed] [Google Scholar]
- 22. Deng, N. , Liu J. W., Sun L. P., Xu Q., Duan Z. P., Dong N. N., et al. 2014. Expression of XPG protein in the development, progression and prognosis of gastric cancer. PLoS ONE 9:e108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartels, C. L. , and Lambert M. W.. 2007. Domains in the XPA protein important in its role as a processivity factor. Biochem. Biophys. Res. Commun. 356:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feltes, B. C. , and Bonatto D.. 2015. Overview of xeroderma pigmentosum proteins architecture, mutations and post‐translational modifications. Mutat. Res., Rev. Mutat. Res. 763:306–320. [DOI] [PubMed] [Google Scholar]
- 25. Ge, R. , Liu L., Dai W., Zhang W., Yang Y., Wang H., et al. 2016. XPA promotes autophagy to facilitate cisplatin resistance in melanoma cells through the activation of PARP1. J. Invest. Dermatol. 136:S115. [DOI] [PubMed] [Google Scholar]
- 26. Marchetto, M. C. , Muotri A. R., Burns D. K., Friedberg E. C., and Menck C. F.. 2004. Gene transduction in skin cells: preventing cancer in xeroderma pigmentosum mice. Proc. Natl Acad. Sci. USA 101:17759–17764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu, J. , Mallon M. A., Zhang W., Freimuth R. R., Marsh S., Watson M. A., et al. 2006. DNA repair pathway profiling and microsatellite instability in colorectal cancer. Clin. Cancer Res. 12:5104–5111. [DOI] [PubMed] [Google Scholar]
- 28. Saviozzi, S. , Ceppi P., Novello S., Ghio P., Lo Iacono M., Borasio P., et al. 2009. Non‐small cell lung cancer exhibits transcript overexpression of genes associated with homologous recombination and DNA replication pathways. Can. Res. 69:3390–3396. [DOI] [PubMed] [Google Scholar]
- 29. Zhi, Y. , Ji H., Pan J., He P., Zhou X., Zhang H., et al. 2017. Downregulated XPA promotes carcinogenesis of bladder cancer via impairment of DNA repair. Tumour Biol. 39:1010428317691679. [DOI] [PubMed] [Google Scholar]
- 30. Cho, H. J. , Kim S. S., Wang H. J., Kim B. W., Cho H., Jung J., et al. 2016. Detection of Novel genomic markers for predicting prognosis in hepatocellular carcinoma patients by integrative analysis of copy number aberrations and gene expression profiles: results from a long‐term follow‐up. DNA Cell Biol. 35:71–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Correlation between XPA expression and survival in CRC.
Table S2. Correlation between XPA mRNA expression and survival in colon cancer and rectal cancer ( based on TCGA).


