Abstract
Toll-like receptors comprise a family of cell surface receptors that play a crucial role in the innate immune recognition of both Drosophila and mammals. Previous studies have shown that Drosophila Toll-1 mediates the induction of antifungal peptides during fungal infection of adult flies. Through genetic studies, Tube, Pelle, Cactus, and Dif have been identified as downstream components of the Toll-1 signaling pathway. Here we report characterization of a Drosophila homologue of human MyD88, dMyD88. We show that dMyD88 is an adapter in the Toll signaling pathway that associates with both the Toll receptor and the downstream kinase Pelle. Expression of dMyD88 in S2 cells strongly induced activity of a Drosomycin reporter gene, whereas a dominant-negative version of dMyD88 potently inhibited Toll-mediated signaling. We also show that dMyD88 associates with the death domain-containing adapter Drosophila Fas-associated death domain-containing protein (dFADD), which in turn interacts with the apical caspase Dredd. This pathway links a cell surface receptor to an apical caspase in invertebrate cells and therefore suggests that the Toll-mediated pathway of caspase activation may be the evolutionary ancestor of the death receptor-mediated pathway for apoptosis induction in mammals.
Pathogen detection by the innate immune system is mediated by a set of germ-line encoded receptors that, on recognition of invariant microbial structures, signal activation of NF-κB and other proinflammatory signaling pathways (1). The best characterized of these receptors are the Toll receptors, a family of transmembrane proteins found in Drosophila and mammals. Through genetic analyses of mutant flies and more recent studies in mammalian systems, Toll receptors have been shown to be essential components of both Drosophila and mammalian immunity (2, 3). Human and Drosophila Tolls contain extracellular leucine-rich repeat domains. Their cytoplasmic domains, which are homologous to the corresponding domain of the IL-1 receptor (IL-1R), are referred to as the Toll/IL-1R (TIR) domains. Mammalian Toll-like receptors (TLRs) signal through the adapter protein MyD88 (4–6). Human MyD88 contains an N-terminal death domain, followed by an intermediate domain and a C-terminal TIR domain. MyD88 is recruited to activated Toll and IL-1Rs through homophilic interactions between the TIR domains of the receptor and MyD88 (4, 5). The death domain of MyD88, in turn, associates with the death domain of the serine threonine protein kinase IL-1R-associated kinase (IRAK) (7–9). Recruitment of IRAK leads to its activation and association with another adapter, tumor necrosis factor receptor-associated factor (TRAF6) (10). Oligomerization of TRAF6 results in activation of IκB kinases, followed by phosphorylation and degradation of IκB. On IκB degradation, NF-κB factors translocate into the nucleus, where they induce transcription of target genes (11).
The mammalian and Drosophila Toll signaling pathways are remarkably conserved. On ligand binding, Drosophila Toll-1 activates the pathway consisting of the adapter protein Tube, the protein kinase Pelle (a homologue of human IRAK), the IκB homologue Cactus, and the NF-κB protein Dif. One notable difference between the Toll pathway of mammals and flies is the adaptor used by the two receptors. Although mammalian Toll signals through MyD88, in flies, Tube functions downstream of Toll-1 and upstream of Pelle (12, 13). Tube is therefore thought to be the functional equivalent of MyD88 in the mammalian Toll pathway.
In Drosophila, the main effectors of the humoral immune responses are the antimicrobial peptides induced in the fat body and hemocytes (14). These antimicrobial peptides fall into distinct categories depending on their activity. Drosomycin, for example, has potent antifungal properties, whereas diptericin and attacin preferentially target bacterial pathogens (14). Interestingly, infection of Drosophila with a specific pathogen leads to a preferential induction of those peptides that are active against the infecting agent (15, 16). Moreover, in the fat body of adult flies, the Toll-Tube-Pelle-Cactus pathway is critically important for the expression of Drosomycin in response to fungal infection (16–18). However, these flies are comparable to wild-type flies in their ability to resist infection by Gram-negative bacterial pathogens (16, 18). Induction of antibacterial peptides is mediated by a distinct pathway, which involves a product of the imd gene (19) and another Drosophila NF-κB protein, Relish (20), as well as the caspase Dredd (21, 22). Interestingly, however, induction of Drosomycin in epithelial cells is independent of Toll and appears to be controlled instead by the imd/Relish pathway (23). Available information suggests, therefore, that the Toll/Dif and imd/Relish pathways control antifungal and antibacterial peptide gene expression, respectively, in the fat bodies and hemocytes of adult flies, although this distinction may not apply to all cell types and developmental stages.
Here we present characterization of the Drosophila homologue of MyD88, dMyD88. dMyD88 potently activates a Drosomycin reporter gene and associates with the Toll receptor via TIR domains and with Pelle through homophilic death domain interactions. dMyD88, like its mammalian homologue, is therefore an adapter in the Toll signaling pathway. In addition, dMyD88 can interact with the Drosophila Fas-associated death-domain containing protein (dFADD) homologue, dFADD, which in turn associates with the apical caspase Dredd. We thus provide evidence suggestive of a receptor-mediated pathway of caspase activation in Drosophila. Moreover, the intriguing similarity of this pathway with that of the mammalian death receptors may be indicative of a possible evolutionary relationship between these two pathways.
Materials and Methods
Plasmids.
Toll 10b, the pJL1 vector, and the luciferase reporter constructs were kindly provided by J. Hoffmann (Institute of Molecular and Cellular Biology, France) and were previously described (24). All other constructs were isolated from S2 cDNA by reverse transcription–PCR. All constructs were subcloned into pAc5.1/HisA V5 (Invitrogen) and or/pJL1-Flag vectors by using standard methods. The sequence encoding the Flag epitope tag of pJL1-Flag was obtained from pFlagCMV2 vector.
Transfections and Luciferase Reporter Assays.
Cells used for all transfection assays were Invitrogen S2 cells that were maintained at 25°C in Schneider's Drosophila medium supplemented with 10% FCS and 10% Penn Strep. Cells were plated in 6-well plates 1 day before transfection. Cells were transfected by using either Fugene 6 or Lipofectamine 2000 Reagent (GIBCO/BRL) according to the manufacturer's protocol and by using 2–2.5 μg of expression vector and 0.1 μg of reporter plasmid per well. All transfections were done in duplicates. Cells were harvested 24–36 h later, washed in PBS, and then lysed in TNT lysis buffer (20 mm Tris·HCl, pH 8.0/150 mm NaCl/1% Triton X-100) or RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS). Luciferase activity was measured in a luminometer after addition of luciferase substrate.
Coimmunoprecipitations and Western Blot Analyses.
For immunoprecipitations, lysates prepared from cells transfected as above were incubated 5–16 h with either Flag beads (Sigma) or Protein G Sepharose and α-Toll serum, washed 5 times with TNT lysis buffer or RIPA buffer, and then denatured and boiled and resolved on 8–12% polyacrylamide gels. For the dToll/dMyD88 immunoprecipitation, lysates were split in two, one-half for incubation with Protein G Sepharose and rabbit a-Toll serum and one-half with Protein G Sepharose and rabbit serum of irrelevant specificity. The α-Toll serum was a kind gift from Carl Hashimoto (Cell Biology Department, Yale University). Proteins were transferred onto poly(vinylidene difluoride) membranes by electroblotting and then probed by using V5 antibody (Invitrogen), Flag M5 antibody (Sigma) and/or rabbit α-Toll serum, as appropriate, and visualized by enhanced chemiluminescence.
Results
dMyD88 Is a Homologue of Mammalian MyD88.
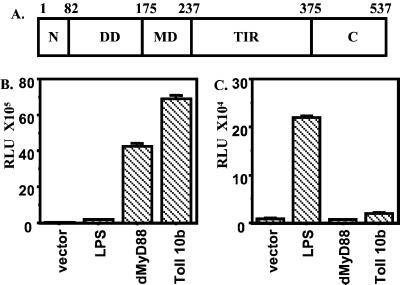
A blast search of the Drosophila genome identified the sequence encoding dMyD88 (accession no. AE003834), a Drosophila homologue of human MyD88. Similar to its human homologue, dMyD88 contains an N-terminal death domain, an intermediate domain, and a TIR domain. However, unlike human MyD88, dMyD88 contains an additional 81 amino acids preceding the death domain and a 162-aa-long C-terminal region following the TIR domain (Fig. 1A).
Figure 1.
DMyD88 domain structure and signaling. (A) dMyD88 consists of a death domain (DD, amino acids 82–175), a middle domain (MD, amino acids 176–237), and a TIR domain (amino acids 238–375), as well as an N-terminal region (N, amino acids 1–81) and a C-terminal region (C, amino acids 375–537) of unknown functions. (B and C) dMyD88 induces a Drosomycin, but not an Attacin, reporter gene. S2 cells were transiently transfected with 2.0 μg of empty plasmid, dMyD88, or Toll 10b, a constitutively active form of Toll-1, and 0.1 μg of either a Drosomycin (B) or an Attacin (C) luciferase reporter gene.
dMyD88 Preferentially Induces a Drosomycin Reporter.
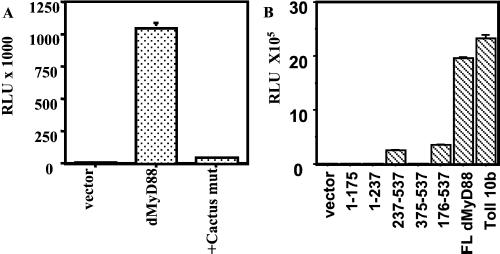
Transfection of dMyD88 into Drosophila S2 cells potently induces a Drosomycin reporter gene but not an Attacin reporter gene (Fig. 1 B and C). This preferential ability to induce an antifungal gene is similar to that of Toll 10b, a constitutively active form of Toll (25), as shown here and as previously described (24, 25), and suggests that dMyD88 may be a component of the Toll-Tube-Pelle-Cactus-Dif signaling pathway. Previous studies have demonstrated that Toll-mediated Drosomycin induction requires the nuclear translocation of Dif (17, 18). Dif is normally retained in the cytoplasm by the IκB inhibitor Cactus and is released only in response to signal-dependent degradation of Cactus. To test whether dMyD88-mediated Drosomycin induction would also depend on Cactus degradation, we made a Cactus mutant that contains mutations of the conserved serine residues that, in mammalian IκB, are the targets of signal-dependent phosphorylation. As shown in Fig. 2A, a Cactus mutant inhibited Drosomycin induction by dMyD88 and, as expected, by Toll (data not shown). This result indicates that, similar to Toll, dMyD88 regulates Drosomycin induction through the Cactus-dependent pathway.
Figure 2.
Signaling by dMyD88. (A) A Cactus mutant that cannot be phosphorylated inhibits induction of a Drosomycin reporter by dMyD88. S2 cells were transfected with either 2.0 μg of empty plasmid or 0.2 μg of dMyD88 with or without 0.9 μg of the Cactus mutant and appropriate amounts of filler DNA. (B) Activation of a Drosomycin reporter gene by dMyD88 deletion constructs. dMyD88 (1), (1), and (375) did not induce expression of a Drosomycin reporter gene, whereas TIR domain-containing constructs dMyD88 (237) and (176) retained some residual activity.
For further analyses, we generated various deletion mutants of dMyD88 (Fig. 2B). Two of the deletion mutants, one containing the TIR domain and the C-terminal domain (amino acids 237–537) and another containing the intermediate, TIR, and C-terminal domains (amino acids 176–537), activated the Drosomycin reporter weakly (10-fold) in comparison to full length dMyD88, indicating that the intact protein is required for optimal activity (Fig. 2B). However, that these truncation mutants can still induce signaling is surprising, as they lack the death domain that mediates interactions with downstream signaling components (see below). Moreover, similar analyses of human MyD88 showed that a combination of the death domain and the intermediate domain was sufficient to induce signaling activity comparable of that of the wild-type protein (4). An equivalent truncation of dMyD88 (amino acids 1–237) retained no residual activity despite being well expressed, suggesting that there are some differences in domain function between human and Drosophila MyD88 proteins.
dMyD88 Is a Component of the Toll Signaling Pathway.
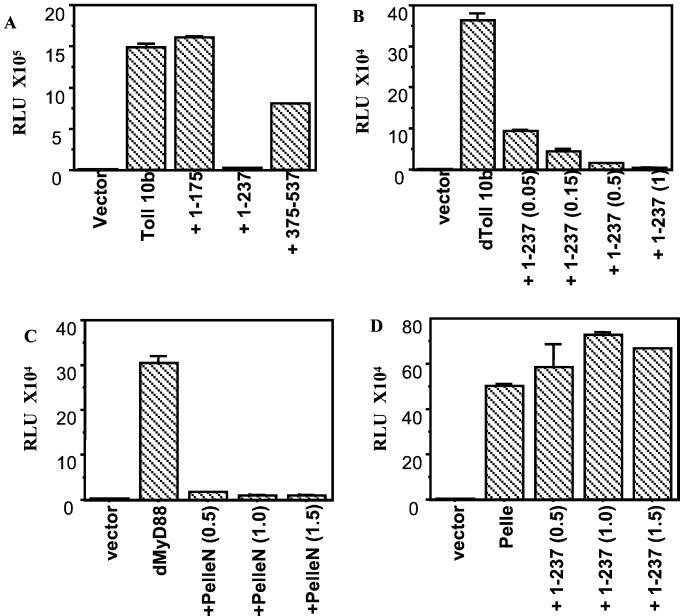
To determine whether dMyD88 is a component of the Toll signaling pathway, we next sought to identify a deletion mutant that would have dominant-negative activity. Therefore, the three dMyD88 deletion mutants that did not activate the Drosomycin reporter were tested for their ability to inhibit Toll-mediated Drosomycin induction (Fig. 3A). The strongest inhibitor was the death domain- and middle domain-containing construct (amino acids 1–237), which at low concentrations potently inhibited Toll-mediated Drosomycin induction in a dose-dependent manner (Fig. 3B).
Figure 3.
dMyD88 is downstream of Toll but upstream of Pelle. (A) S2 cells were transfected with either 2.0 μg of empty plasmid or 0.25 μg of Toll 10b and 1.75 μg of the dMyD88 deletion constructs. dMyD88 (1) completely abolished Toll-mediated Drosomycin induction. (B) dMyD88 1–237 inhibits Toll-mediated Drosomycin induction in a dose-dependent manner. S2 cells were transfected with 0.25 μg of Toll and increasing concentrations of dMyD88 (1), as indicated. (C) PelleN, a dominant-negative form of Pelle, inhibits activation of a Drosomycin reporter by dMyD88. (D) dMyD88 (1) does not inhibit Pelle-induced Drosomycin activation, indicating that Pelle is downstream of dMyD88. Cells were transfected with 0.5 μg of dMyD88 (C) or Pelle (D) and the indicated concentrations of the inhibitors.
To order dMyD88 in the pathway with respect to Pelle, dMyD88 was tested for its ability to be inhibited by PelleN, a dominant-negative form of Pelle that consists of the N-terminal death domain-containing region of Pelle. dMyD88, like Toll (data not shown), was strongly inhibited by PelleN (Fig. 3C). dMyD88 (1), however, did not inhibit Pelle (Fig. 3D), demonstrating that, similar to the mammalian pathway, dMyD88 functions upstream of Pelle.
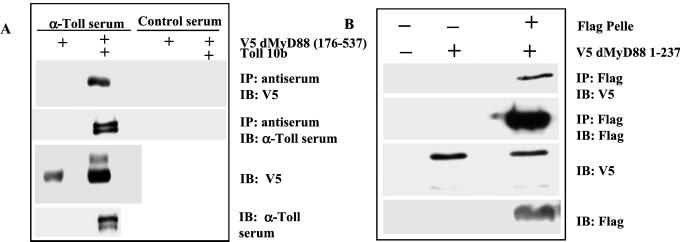
To further establish dMyD88 as a component of the Toll pathway, we tested whether dMyD88 would interact with Toll by coimmunoprecipitation assays. As shown in Fig. 4A, the TIR domain-containing dMyD88 construct (176) was detected in anti-Toll immunoprecipitates. Interestingly, dMyD88 (176), when cotransfected with Toll 10b, reproducibly appeared as two distinct bands, a slower migrating upper band that may correspond to phosphorylated dMyD88 (176) and a faster migrating lower band. The predominant form of dMyD88 detected in immunoprecipitates is the faster migrating species. dMyD88 therefore associates with Toll, presumably through TIR domains, and is a component of the active receptor complex.
Figure 4.
dMyD88 associates with Toll and Pelle. (A) S2 cells were transiently transfected with dMyD88 (176) tagged with a V5 epitope and Toll 10b. Cell lysates were incubated overnight with Protein G Sepharose and either Toll-specific rabbit immune serum or rabbit serum of irrelevant specificity. Immunoprecipitates were analyzed by SDS/PAGE immunoblot with a V5-specific antibody. dMyD88 (176) can be detected in anti-Toll immunoprecipitates (Top, lane 2) but not in control immunoprecipitates from cells transfected with dMyD88 alone (Top, lane 1) or in controls immunoprecipitated with the irrelevant antiserum (Top, lanes 3 and 4). Whole cell lysates (50 μg/lane) immunoblotted with V5-specific antibody (Bottom) and Toll-specific antiserum (Bottom) confirm expression of the indicated proteins. dMyD88 therefore can be detected in active Toll receptor complexes and the middle, TIR, and C-terminal domains are sufficient for association of dMyD88 with Toll. (B) S2 cells were transfected with dMyD88 (1) tagged with a V5 epitope and Flag-tagged Pelle. Lysates were incubated overnight with anti-Flag M2 agarose beads, and immunoprecipitates were analyzed by SDS/PAGE followed by immunoblotting with V5 antibody and Flag antibody. dMyD88 (1) coimmunoprecipitated with Flag Pelle (Top, lane 3). No dMyD88 (1) could be detected in immunoprecipitated lysates of untransfected cells (Top, lane 1) or cells transfected with dMyD88 (1) alone (Top, lane 2). Immunoblotting of whole cell lysates show appropriate expression of proteins (bottom two sections). The death domain and middle domain of dMyD88 are therefore sufficient for interaction with Pelle.
dMyD88 Associates with Pelle.
Because human MyD88 associates with IRAK through death domains, a likely immediate downstream target of dMyD88 is the IRAK homologue Pelle. We therefore looked for interaction between the death domain-containing dMyD88 construct (amino acids 1–237) and Pelle. As shown in Fig. 4B, dMyD88 (1) was detected in Pelle immunoprecipitates, indicating that dMyD88 interacts with Pelle, presumably through their death domains.
Our results therefore demonstrate that dMyD88 is an adaptor in the Toll signaling pathway downstream of the receptor and upstream of Pelle. From genetic analyses, the adaptor protein Tube has also been implicated to be downstream of Toll and upstream of Pelle in the Toll signaling pathway. The death domain of Tube also interacts with Pelle (12, 13). Because Tube and dMyD88 also contain death domains that could potentially mediate their interaction, we also tested for association between these two proteins in immunoprecipitation assays and found that Tube and dMyD88 (1) do indeed interact (data not shown). Therefore, dMyD88 and Tube both function as adaptors downstream of Toll, exist in the same active complex along with Pelle, and are probably both involved in the recruitment and/or activation of Pelle. Understanding functional differences between these two adapters will require further analysis.
dMyD88 Associates with a Death Domain-Containing Protein dFADD.
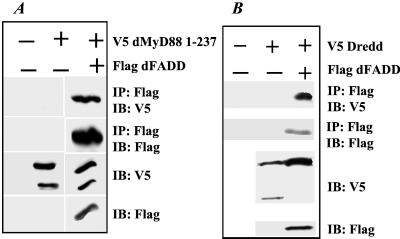
To identify other potential downstream targets of dMyD88, we searched the Drosophila genome for other sequences that encode death domain-containing proteins that may interact with dMyD88. One such sequence encodes a protein with a death domain as well as a death effector domain and appears to be a homologue of mammalian FADD (accession no. AAF55950). While our manuscript was in preparation, this cDNA was independently identified and named dFADD (26). We first tested whether dFADD could interact with dMyD88. Lysates from S2 cells transfected with either dMyD88 (1) and dMyD88 alone were incubated with anti-Flag beads to immunoprecipitate dFADD, and immunoprecipitates were blotted with anti-V5 antibody to look for associated dMyD88 (1). As shown in Fig. 5A, a strong band corresponding to dMyD88 (1) was observed, indicating that dMyD88 can interact with dFADD through death domains. Overexpression of dFADD in S2 cells, however, did not lead to activation of either the Drosomycin or Attacin reporters (data not shown).
Figure 5.
dMyD88 interacts with dFADD and Dredd. (A) dMyD88 (1) associates with dFADD. Cells were transfected with either V5 dMyD88 (1) or V5 dMyD88 (1) and Flag dFADD. Lysates were subjected to immunoprecipitation by using anti-Flag beads as before. dMyD88 (1) strongly associates with dFADD, presumably through their respective death domains (Top, lane 3). Immunoblots of whole cell lysates show appropriate expression of all proteins (bottom two sections). (B) dFADD associates with the caspase Dredd. S2 cells were transfected with the indicated constructs. Full length V5-tagged Dredd is coimmunoprecipitated with Flag dFADD (Top, lane 3).
dFADD Associates with Dredd, a Caspase Involved in Apoptosis and Antibacterial Immune Defense.
Mammalian FADD is recruited to the tumor necrosis factor receptor complex through homophilic death domain interactions with the adapter TNFR-associated death domain-containing protein (TRADD) (27). In turn, FADD recruits procaspase-8 through homophilic death effector domain associations (27). We speculated that dFADD may likewise recruit a Drosophila caspase to the Toll receptor complex. A potential candidate caspase is Dredd, an apical caspase with a long prodomain shown to be essential for induction of antibacterial genes (21, 22, 28). Indeed, analysis of immunoprecipitated lysates from cells cotransfected with dFADD, and either full length Dredd or the death effector domain of Dredd showed strong association of Dredd with dFADD (Fig. 5B). An independent study published while our manuscript was in preparation also showed interaction of dFADD with Dredd (26).
Discussion
In this report, we provide evidence that dMyD88 is an adapter in the Toll signaling pathway. dMyD88 associates with both Toll and Pelle and functions upstream of Pelle. Tube is known from genetic studies to be an adapter in the Toll pathway that functions upstream of Pelle (2). Why Toll should signal through dMyD88 and Tube, two receptor-proximal adapters with seemingly similar functions, is not yet clear. We show that dMyD88 associates with the receptor Toll as well as the downstream adapter dFADD that in turn interacts with the apical caspase Dredd. Because caspases are essential executioners of the apoptotic machinery in organisms from nematodes to mammals, and because Dredd has been shown to be involved in apoptosis during Drosophila development (28), it is possible that Toll-1 or some of the other eight Tolls that exist in Drosophila may induce apoptosis (or another Dredd-dependent pathway) through the dMyD88/dFADD/Dredd pathway in a cell-type specific and/or developmental stage-specific manner. The pathway comprised of Toll, dMyD88, dFADD, and Dredd would be the first description of a pathway in invertebrates that links a cell surface receptor to an apical caspase. Such a pathway, if it exists, would enable extracellular stimuli, perhaps ligands secreted by other cells during development or pathogen-derived products during infection, to instruct invertebrate cells to undergo cell death. In addition, the Toll/dMyD88/dFADD/Dredd pathway we propose here is remarkably similar to that activated by the receptors of the tumor necrosis factor receptor (TNFR) superfamily in mammals, in which FADD-mediated recruitment of caspase-8 leads to induction of apoptosis. As the Drosophila genome does not encode any cell surface receptors homologous to TNFRs, it appears that the Toll/dMyD88/dFADD/Dredd pathway is the evolutionary ancestor of the mammalian death receptor pathways. This possibility is further supported by the recent finding that human TLR2 can induce apoptosis through the MyD88/FADD/Caspase-8 pathway (29).
Acknowledgments
We thank Jules Hoffmann and Carl Hashimoto for gifts of reagents, and Liz Kopp and Chaoqun Chen for help with this work. This work was supported by the Howard Hughes Medical Institute and by National Institutes of Health Grant AI46688–01. T.H. is supported by a predoctoral fellowship from the Howard Hughes Medical Institute; R.M. is a Searle Scholar.
Abbreviations
- TIR
Toll/IL-1 receptor
- TLR
Toll-like receptor
References
- 1.Medzhitov R, Janeway C A., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson K V. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- 3.Imler J L, Hoffmann J A. Curr Opin Microbiol. 2000;3:16–22. doi: 10.1016/s1369-5274(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Natoli G, Saccani S, Levrero M, Mantovani A. J Exp Med. 1998;187:2097–2101. doi: 10.1084/jem.187.12.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 7.Cao Z, Henzel W J, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 8.Wesche H, Henzel W J, Shillinglaw W, Li S, Cao Z. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 9.Muzio M, Ni J, Feng P, Dixit V M. Science. 1997;278:1612–1615. [Google Scholar]
- 10.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 12.Grosshans J, Bergmann A, Haffter P, Nusslein-Volhard C. Nature (London) 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- 13.Xiao T, Towb P, Wasserman S A, Sprang S R. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann J A, Kafatos F C, Janeway C A, Ezekowitz R A. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 15.Lemaitre B, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 17.Meng X, Khanuja B S, Ip Y T. Genes Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutschmann S, Jung A C, Hetru C, Reichhart J M, Hoffmann J A, Ferrandon D. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 19.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart J M, Hoffmann J A. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedengren M, Asling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 21.Elrod-Erickson M, Mishra S, Schneider D. Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- 22.Leulier F, Rodriguez A, Khush R S, Abrams J M, Lemaitre B. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrandon D, Jung A C, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann J A. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tauszig S, Jouanguy E, Hoffmann J A, Imler J L. Proc Natl Acad Sci USA. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. . (First Published September 5, 2000; 10.1073/pnas.18013079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider D S, Hudson K L, Lin T Y, Anderson K V. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 26.Hu S, Yang X. J Biol Chem. 2000;275:30761–30764. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- 27.Wallach D, Varfolomeev E E, Malinin N L, Goltsev Y V, Kovalenko A V, Boldin M P. Annu Rev Immunol. 1999;17:331–367. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 28.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 29.Aliprantis A O, Yang R B, Weiss D S, Godowski P, Zychlinsky A. EMBO J. 2000;19:3325–3336. doi: 10.1093/emboj/19.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]