Abstract
Cellulolytic bacteria that produce cellulases, which are active over a range of pH and temperatures, can be used to catalyze hydrolysis of pretreated lignocellulosic material. This is important in the production of second generation biofuels among other biotechnological applications. In this investigation, bacteria isolated from sugarcane bagasse were identified as strains of Enterobacter xiangfangensis, Serratia rubidaea, Klebsiella pneumoniae and a novel species of Citrobacter designated Citrobacter sp. UWIBGS10. The glucose production potential of these strains was studied on thermally and solvent pretreated sugarcane bagasse. This was performed at 24-hour intervals up to 168 hours in the range of pH 5–9 and temperature range 25–40 °C. Maximal concentrations of glucose for Citrobacter sp. UWIBGS10 occurred at pH 6 and 25 °C. For E. xiangfangensis, S. rubidaea, K. pneumoniae glucose concentrations were consistent across the pH and temperature ranges examined. From these results it could be concluded that the bacteria demonstrated ability for lignocellulolytic hydrolysis for the production of glucose and could be further explored for the characterization of commercial cellulolytic enzymes.
Keywords: Microbiology, Biotechnology
1. Introduction
Cellulases derived from microorganisms mainly fungi and bacteria have been applied to various biotechnological applications mainly related to the degradation of cellulose fibers (Béguin and Aubert, 1994; Kuhad et al., 2011; Sadhu and Tushark, 2013). The cellulase enzyme system is composed of exoglucanases, which hydrolyze the reducing and non-reducing ends of cellulose to release glucose monomers; endoglucanases, which hydrolyze internal glycosidic bonds producing cellobiose and short cellulose fragments and β-glucosidases, which mainly hydrolyze cellobiose to release glucose monomers (Sadhu and Tushark, 2013).
In particular, bacteria and their cellulases have demonstrated higher growth rates, and better tolerance to changes in temperature and pH compared to fungi. Therefore their cellulases have been recognized as more suitable for industrial application (Kuhad et al., 2011). Several genera of bacteria produce efficient extracellular cellulases and these include: Pseudomonas, Enterobacter, Bacillus, Klebsiella, Paenibacillus, Rhodococcus, Cellulomonas, Streptomyces and Citrobacter among others (Liang et al., 2014; McCarthy, 1987; Sadhu and Tushark, 2013; Ventorino et al., 2015). Many of these bacteria produce cellulases that tolerate a wide range of pH and temperatures. These include B. brevis, B. subtillis, Paenibacillus sp. and Cellulomonas sp. which are active in the ranges pH 5.5–7.5 and 37–60 °C (Meng et al., 2014). To this effect, bacteria that produce extracellular cellulases that are active across various pH and temperature ranges are important, specifically to the application of biotechnological and industrial processes. These include stone washing and improvement of fabric appearance of clothing and textiles as well as improving pulp yield and reducing environmental problems in the pulp and paper industries and the conversion of cellulose in lignocellulosic material to glucose for fermentation to second generation biofuel to provide an alternative source of liquid fuel (Nigam and Singh, 2011; Sadhu and Tushark, 2013). Therefore, the aim of this investigation was to examine cellulolytic ability of four bacterial species to produce glucose from pretreated sugarcane bagasse over a range of pH and temperatures.
2. Material & methods
2.1. General materials
All chemicals inclusive of broths and reagents used in this investigation were obtained from Sigma Aldrich, St. Louis MO, USA except where otherwise indicated.
2.2. Substrate collection and isolation of bacterial strains
Sugarcane bagasse was used as the lignocellulose substrate for bacterial hydrolysis. This was collected from stores of sugarcane bagasse from Portvale Sugar Factory located in the parish of St. James, Barbados. The total culturable bacterial population of the bagasse was determined by an enrichment culture that was prepared by incubating 2 g of bagasse in tryptic soy broth, at 37 °C and 150 rpm for 48 hours. The culture was serially diluted after 48 h and 40 uL portions of the 10−6 and 10−7 dilutions were spread plated on tryptic soy agar (TSA) plates supplemented with amphotericin B (0.25 μg/mL) and incubated at 30 °C for 48 hours (Anita et al., 2013; Dashtban et al., 2010). Colonies that varied by morphological appearance inclusive of color, size and shape were selected and purified by serial subculturing on TSA plates.
2.3. Screening culture collection for cellulose degrading bacteria
Isolated bacteria were screened for exoglucananse and endoglucanase activity using cellulose and carboxymethyl cellulose (CMC) supplemented agar plates. Agar plates contained (per L): K2HPO4 (2 g), Na2HPO4 (2 g), (NH4)2SO4 (1.25 g), peptone (1 g), bacto agar (7.5 g), 1% cellulose or CMC (Anita et al., 2013; Gupta et al., 2012). Twenty-four hour cultures of bacteria were streaked on to each type of plate. A rapid detection method consisting of staining the plates with Grams iodine for 5 minutes, followed by washing with distilled water, was employed. Cleared zones of hydrolysis on the respective plate indicated cellulase enzyme production and activity (Kasana et al., 2008).
2.4. Molecular identification of bacterial strains
Bacterial strains which demonstrated hydrolytic production of glucose from pretreated bagasse were identified by analysis of 16S rRNA gene sequences. DNA was isolated by suspending individual colonies from agar plates in 300 μL of 100 mM Tris-EDTA buffer (pH 8) in 1.5 mL microcentrifuge tubes. Lysozyme (0.1 mg) and RNase A (0.002 mg) were added to the cell suspension and incubated at 30 °C for 30 min.
Cell lysates were extracted with phenol/chloroform/isoamyl alcohol (25:24:1) and the DNA precipitated from the aqueous phase with 70% ethanol after the addition of 1/10 volume of 3 M sodium acetate (pH 5.5). 16S rRNA genes were amplified using the following primers: pA (5′-AGAGTTTGATCCTGGCTCAG) and pH (5′-AAGGAGGTGATCCAGCCGCA). The PCR reaction mixture (50 μL) consisted of: 1X EconoTag Plus Green Master Mix (Lucigen, Middleton, WI, USA), 5% molecular biology grade DMSO (Sigma, Oakville, ON, USA), 1 μM each primer and 25 ng template DNA. 16S rRNA gene amplicons were sequenced using the following primers 530R 5′-GTATTACCGCGGCTGCTGG, 936R (5′-GGGGTTATGCCTGAGCAGTTTG), 1527R (5′-AAGGAGGTGATCCAGCC), 514F (5′-GTGCCAGCASCCGCGG) and 1114F (5′-GCAACGAGCGCAACCC), (Gontang et al., 2007). Sequences were assembled and analyzed using Vector NTI version 11.5 (Life Technologies, Carlsbad, CA). After assembly, nearly complete 16S rRNA gene sequences were analyzed using the EzBioCloud identification tool (Janssen, 2006). Phyologenetic analyses were conducted using MEGA 7 (Kumar et al., 2016). Representative 16S rRNA gene sequences of selected type strains within the Enterobacteriaceae were obtained from GenBank. Aeromonas hydrophilla ATCC7966 (NR_07484) was used as an out group. Sequences were aligned in MEGA7 using ClustalW and alignments were manually edited. A maximum likelihood test to determine the optimal evolutionary model for the dataset identified the Kimura 2 parameter with a gamma distribution with invariant sites was identified as the best model for the dataset (Kimura, 1980). Evolutionary histories were inferred using the Neighbor-Joining, maximum likelihood, maximum parsimony, minimum evolution and unweighted pair group method with arithmetic mean (UPGMA) methods (Saitou and Nei, 1987). To evaluate reproducibility of clades, bootstrap analysis was conducted using 1000 replicates (Felsenstein, 1985).
2.5. Pretreatment of bagasse lignocellulose
Bagasse lignocellulose was subjected to two pretreatment methods, which consisted of a thermal (HT) and a solvent fractionation (ST) process. Bagasse was initially ground using a Waring blender (Shrestha et al., 2011). For the HT pretreatment process, bagasse was transferred to a 1 L flask, mixed with distilled water (1:5, w/v) and autoclaved at 121 °C/60 minutes. The residue was separated and washed with tap water followed by distilled water and air dried for 5 days (Shrestha et al., 2011). In the ST pretreatment process, organic solvent fractionation using ethanol (50%, v/v) was performed. Ground bagasse (100 g) was mixed with 1L (50%, v/v) ethanol at room temperature for 24 hours without stirring. After this time the residue was separated, washed with tap water followed by distilled water and air dried for 5 days (Park et al., 2018; Zhao et al., 2009).
2.6. Glucose production from pretreated sugarcane bagasse by the bacterial strains
The hydrolytic activity of the bacteria on pretreated bagasse was evaluated by the accumulation of hydrolytic product glucose. Glucose concentrations were determined by colorimetric analysis using 3,5-dinitrosalicyclic acid reagent (DNS), (Miller, 1959). The DNS assay was miniaturized to fit a 96-well plate by mixing 125 μL of hydrolysate with 125 μL of DNS reagent in microcentrifuge tubes. The tubes were boiled at 100 °C for 10 minutes. The contents of each tube were then transferred to the wells of a 96 well plate and the absorbance read at 540 nm using a microplate reader (BGM labtech FLUOstar OPTIMA). The concentration of glucose was determined by interpolation of a glucose standard curve (Alvira et al., 2010).
To examine the effect of pH on hydrolysis and glucose concentration, various buffering systems were utilized as follows: citric acid buffer (pH 5–6); phosphate-buffered saline (PBS) (pH 7); phosphate buffer (pH 8) and carbonate buffer (pH 9). For the effects of temperature on hydrolytic production of glucose from pretreated bagasse and glucose concentration, at constant pH the temperature was varied between 25–40 °C.
The optimal conditions of pH and temperature for bacterial hydrolysis of HT and ST pretreated bagasse were determined by monitoring the concentrations of glucose at 24-hour intervals for 168 hours (Bibi et al., 2014). Three Individual replicate experiments were conducted for each bacterial strain as follows: HT and ST pretreated bagasse (2.5 g, substrate concentration) were each placed in 125 mL conical flasks, to which 45 mL of appropriate buffer was added to the flask and the mixture sterilized at 121 °C for 15 min. A 5 mL aliquot of bacteria cultured in TSB for 24 hours, was washed and resuspended in buffer by centrifugation at 10 000 rpm bringing the total volume to 50 mL (Anita et al., 2013). The flasks were then incubated for 168 h at the appropriate pH and temperature.
2.7. Statistical analysis
Statistical analysis was performed with Statistical Package for Social Sciences (SPSS), version 19 and GraphPad Prism version 7.03 for windows, GraphPad Software, La Jolla California USA. The data were evaluated using two-way repeated measures ANOVA. Where significant differences were found, Tukey HSD tests were performed on pairwise comparisons. All statistical analyses were performed at an alpha of 0.05 and the data were expressed as mean ± SEM. All graphs were constructed using GraphPad Prism version 7.03. Additionally, in cases where the error bars are shorter than the height of the symbol, GraphPad Prism does not draw the error bars.
3. Results
3.1. Bacterial culture collection and screening for cellulolytic bacteria
Twenty-six bacterial strains were isolated from sugarcane bagasse. These strains were examined for production of cellulases using cellulose and CMC plate screens. From these bacteria, 16 strains were selected to test their cellulolytic ability using pretreated sugarcane bagasse. A further 4 strains were identified by molecular analysis for ability to produce glucose over a range of pH (pH 5–9) and temperatures (25–40 °C).
3.2. Molecular identification of bacterial strains
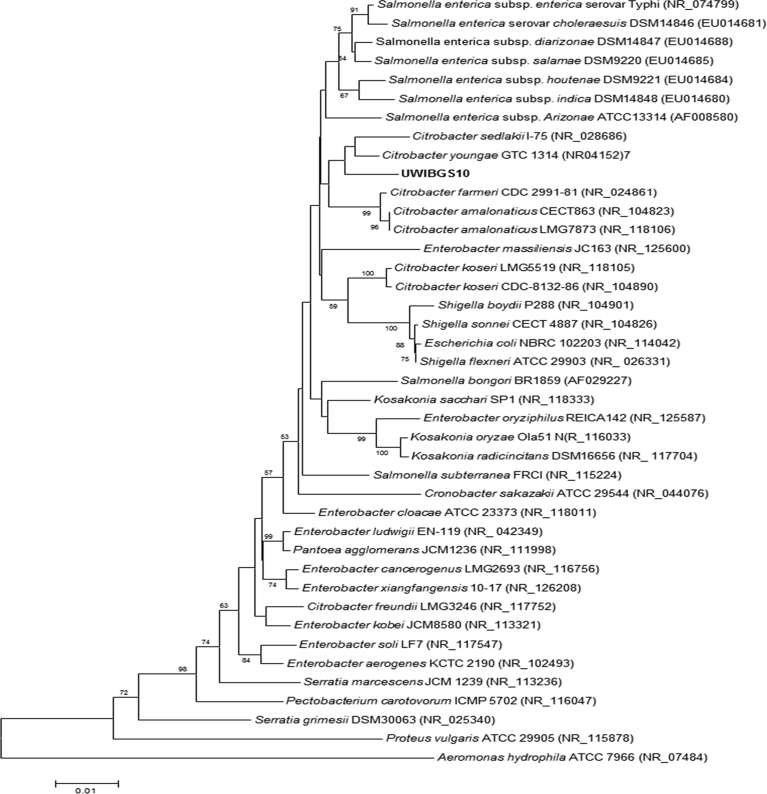
Four bacterial strains designated UWIBGS3, UWIBGS6, UWIBGS10 and UWIBGS12 (accession numbers: MG923793, MG923794, MG923792, MG923795 respectively) isolated from sugarcane bagasse were identified by analysis of nearly complete 16S rRNA gene sequences using the EZBioCloud identifier. The 16S rRNA gene sequence of UWIBGS3 was most similar (99.60%) to that of Enterobacter xiangfangensis LMG 27195T, while those of UWIBGS6 and UWIBGS12 were most similar to Serratia rubidaea JCM 1240T (99.71% similarity) and Klebsiella pneumoniae ATCC 11296T (99.93% similarity), respectively. The huge degree of 16S rRNA gene sequence similarity between these three strains and those of previously described type strains indicated these bacteria are strains of these species. UWIBGS10 was identified as belonging to the genus Citrobacter, however, it exhibited the closest sequence similarity to Cedecea lepagia GTC 346T (98.42%) and Citrobacter sedlakii NBRC 105722T (98.29%). The 16S rRNA sequence of this strain also exhibited >98% identity to several other strains of Citrobacter and Salmonella. This level of 16S rRNA sequence similarity suggests UWIBGS10 may represent a novel species in the Enterobacteriaceae. As Cedecea sp. have been shown to belong to the Cedecea clade of the Enterobacteriaceae while Citrobacter and Salmonella belong to the Escherichia clade we further investigated the Cedecea lepagei GTC 346T entry in EzBioCloud (Alnajar and Gupta, 2017). This strain is not the type strain of the species which is Ce. lepagei JCM 1684T. BlastN analysis of the Ce. lepagei GTC 346T (AB273742) sequence revealed it was most similar (98.79%) to Citrobacter youngae GTC 1314. This indicated UWIBGS10 was most closely related to Citrobacter spp. Due to similar levels of 16S rRNA gene sequence similarity to Citrobacter and Salmonella spp., we conducted a phylogenetic analysis to determine the taxonomic affiliation of this strain (Fig. 1). In the Neighbor-Joining tree shown in Fig. 1, UWIBGS10 formed a sister clade to C. youngae and C. sedlakii, albeit with low bootstrap support. The low bootstrap support is consistent with reported poor resolution of 16S rRNA gene sequence based phylogenetic analysis with in the Enterobacteriaceae (Alnajar and Gupta, 2017). Despite the low bootstrap support, UWIBGS10 clustered with C. youngae and C. sedlakii in phylogenetic analyses conducted using the maximum likelihood, maximum parsimony, minimum evolution and UPGMA methods (data not shown). This suggests UWIBGS10 likely represents a novel Citrobacter species. Further polyphasic taxonomic analyses would be required to definitively determine the taxonomic placement of this strain within the Enterobacteriaceae.
Fig. 1.
Neighbor-Joining phylogenetic analysis of 16S rRNA gene sequences of selected bacteria in the Enterobacteriaceae (Saitou and Nei, 1987). The optimal tree with the sum of branch length = 0.44327546 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and are in the units of the number of base substitutions per site (Kimura, 1980). The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 41 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1314 positions in the final dataset.
3.3. Effects of pH and temperature on hydrolysis of pretreated bagasse
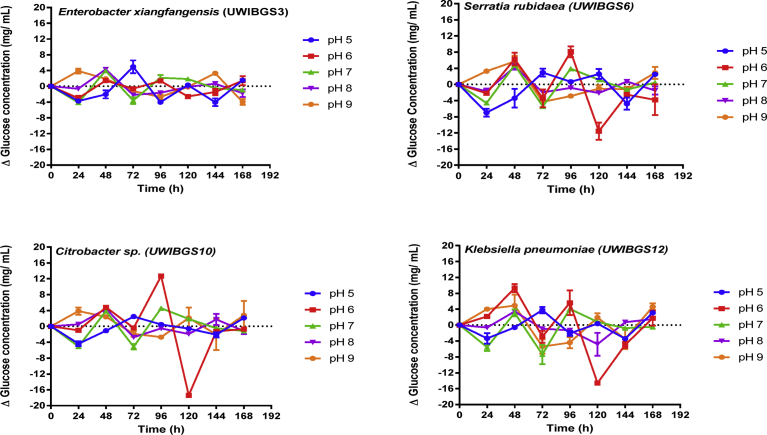
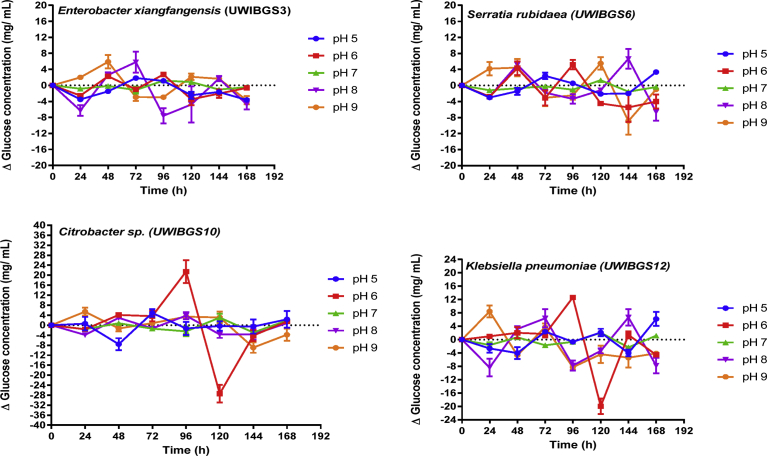
Hydrolysis of pretreated bagasse by the four strains was examined across a range of pH (5–9) and temperatures (25–40 °C). The hydrolysis was monitored by measuring net glucose concentration accumulated during hydrolysis of HT and ST pretreated bagasse every 24 h for 168 h. This was performed by subtracting the starting concentration of glucose available in the media (Ti), from the concentration of glucose at a given time point (Ti+1). Therefore, Δglucose = Ti+1 –Ti is the glucose accumulated as a result of hydrolysis which occurred during a specific time period. The changes in glucose concentrations at different pH levels over 168 h are demonstrated in Figs. 2 and 3 for HT and ST pretreated bagasse respectively. These data indicate the accumulation of glucose which demonstrates hydrolytic ability of the strains in varying conditions of pH and temperatures.
Fig. 2.
Net glucose concentration at 24-hour intervals during hydrolysis thermally pretreated (HT) bagasse in the range pH 5–9 for four bacterial strains.
Fig. 3.
Net glucose concentration at 24-hour intervals during hydrolysis of solvent pretreated (ST) bagasse in the range pH 5–9 for four bacterial strains.
Statistical analysis consisting of two-way repeated measures ANOVA was conducted to determine effects of pretreatment, strain and pH on hydrolytic production of glucose from pretreated bagasse and glucose concentration. These results are displayed in Table 1, indicating a significant interaction effect (F (4, 27) = 7.48, p = 0.00) of pretreatment and pH on glucose concentration during cellulose hydrolysis. Further post hoc analysis consisting of Tukey HSD tests were conducted on pairwise comparisons of mean glucose concentrations. This was for the range pH 5–9 for each bacterial strain for HT and ST pretreated bagasse. It was determined that for Citrobacter sp.UWIBGS10, the concentration of glucose at pH 6 (21.46 ± 2.50 mg/mL, mean ± SEM) between 72–96 hours was significantly higher (Tukey, p < 0.05) than glucose concentrations at pH 5, 7, 8 and 9 for HT and ST bagasse. Additionally, glucose concentration for Citrobacter sp.UWIBGS10 between 72–96 hours for HT and ST bagasse was significantly higher (Tukey, p < 0.05) than glucose concentrations accumulated for E. xiangfangensis, S. rubidaea, and K. pneumoniae (Figs. 2 and 3).
Table 1.
Repeated measures ANOVA for effects of pretreatment, strain and pH on glucose concentration for four bacterial strains.
| Source | Type III sum of | df | Mean square | F | p |
|---|---|---|---|---|---|
| Pretreatment | 4.54 | 1 | 4.54 | 7.83 | 0.01 |
| Strain | 3.03 | 3 | 1.01 | 1.74 | 0.18 |
| pH | 25.63 | 4 | 6.41 | 11.04 | 0.00 |
| Pretreatment*Strain | 0.62 | 3 | 0.21 | 0.36 | 0.79 |
| Pretreatment*pH | 17.35 | 4 | 4.34 | 7.48 | 0.00 |
| Strain*pH | 11.05 | 12 | 0.92 | 1.59 | 0.16 |
| Pretreatment*strain*pH | 4.76 | 6 | 0.79 | 1.37 | 0.26 |
| Error | 15.67 | 27 | 0.58 |
The bold highlights reflect the highest level of statistically significant interaction effect of the independent variables on the dependent variable.
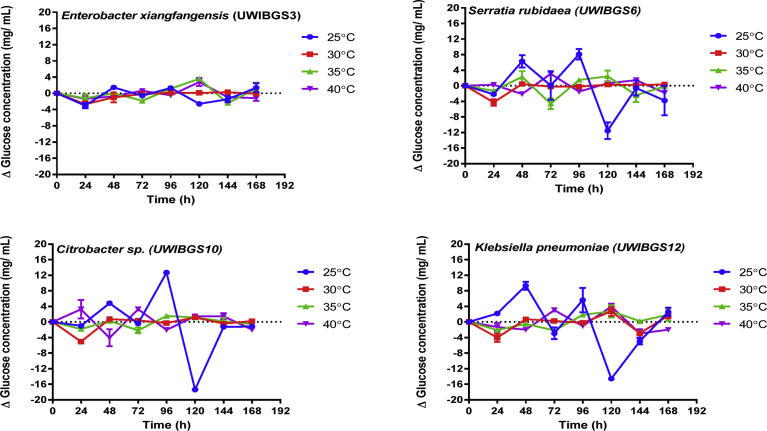
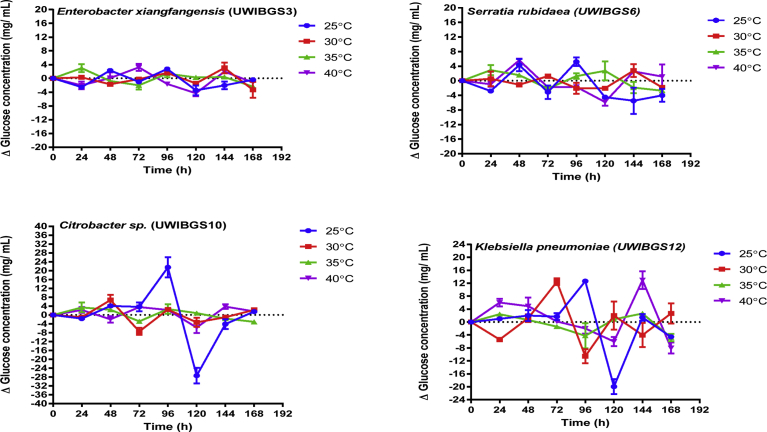
The effects of temperature on hydrolytic ability of thermally and solvent pretreated bagasse by four bacterial strains were displayed in Figs. 4 and 5. These data were subjected to two-way repeated measures ANOVA analysis to determine the effects of pretreatment, strain and temperature on hydrolytic production of glucose from pretreated bagasse. This analysis indicated a significant interaction effect (F (5, 19) = 3.16, p = 0.03) of pretreatment, strain and temperature on glucose concentration as highlighted in Table 2. Tukey HSD tests were performed on pairwise comparisons of glucose concentrations for HT and ST pretreated bagasse for the strains in the temperature range 25–40 °C. These comparisons indicated that glucose concentration for Citrobacter sp. at 25 °C (21.46 ± 2.50 mg/mL; 72–96 hours; Tukey, p < 0.05) was also significantly higher for HT and ST bagasse compared to glucose concentrations at 30, 35 and 40 °C (Fig. 5). Also, glucose concentrations for Citrobacter sp.UWIBGS10 between 72–96 hours were significantly higher (Tukey, p < 0.05) compared to glucose concentrations for E. xiangfangensis, S. rubidaea and K. pneumoniae (Figs. 4 and 5). These results indicate that for Citrobacter sp., optimal conditions are pH 6 and a temperature of 25 °C for release of maximal concentrations of glucose from the pretreated bagasse.
Fig. 4.
Net glucose concentration at 24-hour intervals during hydrolysis thermally pretreated (HT) bagasse in the temperature range 25–40 °C for four bacterial strains.
Fig. 5.
Net glucose concentration at 24-hour intervals during hydrolysis of solvent pretreated (ST) bagasse in the temperature range 25–40 °C for four bacterial strains.
Table 2.
Repeated measures ANOVA for effects of pretreatment, strain and temperature on glucose concentration for four bacterial strains.
| Source | Type III sum of | df | Mean square | F | p |
|---|---|---|---|---|---|
| Pretreatment | 0.02 | 1 | 0.02 | 0.08 | 0.78 |
| Strain | 1.77 | 3 | 0.59 | 2.71 | 0.07 |
| Temperature | 19.65 | 3 | 6.55 | 30.10 | 0.00 |
| Pretreatment*Strain | 2.98 | 3 | 0.99 | 4.56 | 0.14 |
| Pretreatment*Temperature | 5.57 | 3 | 1.86 | 8.53 | 0.00 |
| Strain*Temperature | 3.05 | 8 | 0.38 | 1.75 | 0.15 |
| Pretreatment*strain*Temperature | 3.44 | 5 | 0.69 | 3.16 | 0.03 |
| Error | 4.13 | 19 | 0.22 |
The bold highlights reflect the highest level of statistically significant interaction effect of the independent variables on the dependent variable.
Further analyses consisting of paired t-tests were conducted to compare hydrolysis between the HT and ST pretreated bagasse by the bacterial strains. These results indicated that ST pretreated bagasse in the pH range 5–9 generally produced higher concentrations of glucose however, these were not statistically significantly different (t = 1.44, p > 0.05) from glucose produced from thermally pretreated bagasse. Alternatively, higher concentrations (t = 7.23, p < 0.05) of glucose were produced from hydrolysis of ST pretreated bagasse in the temperature range 25–40 °C by the bacterial strains examined.
4. Discussion
In this study, bacterial strains isolated from sugarcane bagasse were examined for cellulolytic ability. Four cellulolytic bacteria were identified of the Enterobacteriaceae family, which are Gram negative, facultatively anaerobic, commonly found in water and soil, and can cause opportunistic infection (Liang et al., 2014; Sadhu and Tushark, 2013). E. xiangfangensis, originally isolated from sourdough in Heilongjiang Province, China, has not been previously investigated for hydrolysis of pretreated bagasse lignocellulose for glucose production as was observed in this examination (Gu et al., 2014). S. rubidaea, which has a distinctive characteristic of production of red pigment prodigiosin, has been associated with production of cellulolytic enzymes as well as, K. pneumoniae (Berlemont and Martiny, 2013; Chen et al., 2007). This work has also resulted in the identification of a novel species of Citrobacter which has demonstrated ability to hydrolyze pretreated sugarcane bagasse to produce glucose. Previous studies have indicated several species of Citrobacter as possessing cellulolytic ability (Fontes-Perez et al., 2015; Sethi et al., 2013). However, in this work we examine this ability using sugarcane bagasse as the lignocellulose substrate.
In the examination of hydrolytic production of glucose from pretreated bagasse in the range of pH 5–9 the data indicate that glucose concentrations for Citrobacter sp. UWIBGS10 are highest between 72–96 hours at pH 6. This demonstrates that hydrolysis of pretreated sugarcane bagasse for glucose production at this pH was optimal. Similar studies have previously indicated that hydrolysis of pretreated sugarcane bagasse for glucose production is optimal in the range pH 4–7.5. This has been documented for B. subtilis (pH 5.5), B. licheniformis (pH 6.1), and Cellulomonas sp. ASN2 (pH 7.5) (Sadhu and Tushark, 2013). Glucose accumulated for the strains E. xiangfangensis, S. rubidaea and K. pneumoniae were consistent in the pH range during cellulose hydrolysis. This is useful for industrial applications, which can require cellulolytic enzymes that tolerate changes in pH. Bacteria have also been documented to produce cellulolytic enzymes that tolerate alkaline pH conditions between pH 8–11 such as Bacillus sp. HSH-810 (pH 10) (Sadhu and Tushark, 2013; Ventorino et al., 2015). Alkaline tolerant cellulases have been documented in Pseudomonas fluorescens (pH 10), Serratia marcescens (pH 10), and other Bacillus species, inclusive of Bacillus sp N-4 (pH 9), Bacillus KSM-522 (pH 7–10), and B. subtilis (pH 10) (Ladeira et al., 2015; Sethi et al., 2013).
It was also observed that during hydrolytic production of glucose from pretreated bagasse for Citrobacter sp. at pH 6, glucose concentration decreased after 96 hours. Factors that affect hydrolytic production of glucose from pretreated bagasse causing decreases in glucose concentrations include glucose consumption for metabolism and the presence of metabolic by-products that inhibit enzyme production and activity (Yue et al., 2004). Hydrolysis is also affected by the high concentrations of glucose, which cause end-product inhibition. Furthermore, if sufficient glucose resources are available in the media, bacteria can decrease production of cellulases during that time period, which results in lower concentrations of glucose (Lynd et al., 2002).
For hydrolytic production of glucose from pretreated bagasse in the temperature range of 25–40 °C Citrobacter sp. UWIBGS10 demonstrated optimal hydrolysis of pretreated sugarcane bagasse for glucose production by accumulating higher concentrations of glucose at 25 °C between 72–96 hours. Glucose concentrations accumulated for E. xiangfangensis, S. rubidaea and K. pneumoniae however, were consistent during the experimental period. These results are consistent with previous studies where it was found that cellulolytic enzymes from bacterial sources exhibit optimal hydrolytic ability between 25–50 °C (Kafle et al., 2015; Sadhu and Tushark, 2013; Tamaki et al., 2016). Other studies have, however, demonstrated that at temperatures approaching 50 °C, cellulolytic enzymes tend to lose 60% of their activity and lose all activity at temperatures approaching 80 °C as the enzymes become denatured (Sethi et al., 2013).
Overall, between the HT and the ST pretreatment, results indicated that higher concentrations of glucose were produced from hydrolysis of ST pretreated bagasse for the bacterial strains. This was significant for the temperature range 25–40 °C and indicated that the ST pretreatment made the bagasse lignocellulose more amenable for hydrolysis. This pretreatment process favored the disruption and removal of lignin, to increase pore volume, which facilitates improved hydrolysis indicated by higher concentrations of glucose (Zhao et al., 2009).
5. Conclusion
This work has identified a novel species of Citrobacter that could hydrolyze sugarcane bagasse at optimal conditions of pH 6 and 25 °C. This along with E. xiangfangensis, S. rubidaea and K. pneumoniae that could act over a broad range of pH and temperature, has significant potential for commercial exploitation for the production of cellulolytic enzymes. This is important in decreasing environmental impact associated with lignocellulose conversion to fermentable sugars among other biotechnological processes, in a low energy and environmentally friendly manner.
Declarations
Author contribution statement
Jamila A.D. Jones: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
R.G. Kerr: Performed the experiments; Contributed reagents, materials, analysis tools or data.
B.A. Haltli: Performed the experiments; Analyzed and interpreted the data.
Winston F. Tinto: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Alnajar S., Gupta R.S. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect. Genet. Evol. 2017;54:108–127. doi: 10.1016/j.meegid.2017.06.024. [DOI] [PubMed] [Google Scholar]
- Alvira P., Negro M.J., Sáez F., Ballesteros M. Application of a microassay method to study enzymatic hydrolysis of pretreated wheat straw. J. Chem. Technol. Biotechnol. 2010;85(9):1291–1297. [Google Scholar]
- Anita B.B., Thatheyus A.J., Ramya D. Biogradation of carboxymethly cellulose using Aspergillus flavus. Sci. Int. 2013;1(4):85–91. [Google Scholar]
- Béguin P., Aubert J.-P. The biological degradation of cellulose. FEMS Microbiol. Rev. 1994;13(1):25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- Berlemont R., Martiny A.C. Phylogenetic distribution of potential cellulases in bacteria. Appl. Environ. Microbiol. 2013;79(5):1545–1554. doi: 10.1128/AEM.03305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi Z., Ansari A., Zohra R.R., Aman A., Ul Qader S.A. Production of xylan degrading endo-1, 4-β-xylanase from thermophilic Geobacillus stearothermophilus KIBGE-IB29. J. Radiat. Res. Appl. Sci. 2014;7(4):478–485. [Google Scholar]
- Chen Y., Stipanovic A.J., Winter W.T., Wilson D.B., Kim Y.J. Effect of digestion by pure cellulases on crystallinity and average chain length for bacterial and microcrystalline celluloses. Cellulose. 2007;14 [Google Scholar]
- Dashtban M., Maki M., Leung K.T., Mao C., Qin W. Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 2010;30 doi: 10.3109/07388551.2010.490938. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fontes-Perez H., Olvera-García M., Chávez-Martínez A., Rodriguez-Almeida F.A., Arzola-Alvarez C.A., Sanchez-Flores A., Corral-Luna A. Genome sequence of Citrobacter sp. CtB7.12, isolated from the gut of the desert subterranean termite Heterotermes aureus. Genome Announc. 2015;3(6) doi: 10.1128/genomeA.01290-15. e01290–01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontang E., Fenical W., Jensen P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C.T., Li C.Y., Yang L.J., Huo G.C. Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough, and reclassification of Enterobacter sacchari Zhu et al. 2013 as Kosakonia sacchari comb. nov. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 8):2650–2656. doi: 10.1099/ijs.0.064709-0. [DOI] [PubMed] [Google Scholar]
- Gupta P., Samant K., Sahu A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Food Microbiol. 2012;2012:5. doi: 10.1155/2012/578925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafle K., Shin H., Lee C.M., Park S., Kim S.H. Progressive structural changes of Avicel, bleached softwood, and bacterial cellulose during enzymatic hydrolysis. Sci. Rep. 2015;5 doi: 10.1038/srep15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasana R.C., Salwan R., Dhar H., Dutt S., Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram's iodine. Curr. Microbiol. 2008;57(5):503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kuhad R.C., Gupta R., Singh A. Microbial cellulases and their industrial applications. Enzym. Res. 2011;2011:10. doi: 10.4061/2011/280696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeira S.A., Cruz E., Delatorre A.B., Barbosa J.B., Martins M.L.L. Cellulase production by thermophilic Bacillus sp. SMIA-2 and its detergent compatibility. Electron. J. Biotechnol. 2015;18(2):110–115. [Google Scholar]
- Liang Y.-L., Zhang Z., Wu M., Wu Y., Feng J.-X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. BioMed Res. Int. 2014;2014:13. doi: 10.1155/2014/512497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd L., Weimer P., van Zyl W., Pretorius I. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A.J. Lignocellulose-degrading actinomycetes. FEMS Microbiol. Lett. 1987;46(2):145–163. [Google Scholar]
- Meng F., Ma L., Ji S., Yang W., Cao B. Isolation and characterization of Bacillus subtilis strain BY-3, a thermophilic and efficient cellulase-producing bacterium on untreated plant biomass. Lett. Appl. Microbiol. 2014;59(3):306–312. doi: 10.1111/lam.12276. [DOI] [PubMed] [Google Scholar]
- Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31 [Google Scholar]
- Nigam P.S., Singh A. Production of liquid biofuels from renewable resources. Prog. Energy Combust. Sci. 2011;37(1):52–68. [Google Scholar]
- Park S.Y., Kim J.Y., Youn H.J., Choi J.W. Fractionation of lignin macromolecules by sequential organic solvents systems and their characterization for further valuable applications. Int. J. Biol. Macromol. 2018;106:793–802. doi: 10.1016/j.ijbiomac.2017.08.069. [DOI] [PubMed] [Google Scholar]
- Sadhu S., Tushar M.K. Cellulase production by bacteria: a review. Br. Microbiol. Res. J. 2013;3(3):235–258. [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sethi S., Datta A., Gupta B.L., Gupta S. Optimization of cellulase production from bacteria isolated from soil. ISRN Biotechnol. 2013;2013:7. doi: 10.5402/2013/985685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha P., Timothy S.M., Thomas B.D., John T.W. Systematic search for cultivatable fungi that best deconstruct cell walls of Miscanthus and sugarcane in the field. Appl. Environ. Microbiol. 2011;77(15):5490–5504. doi: 10.1128/AEM.02996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki F.K., Araujo É.M., Rozenberg R., Marana S.R. A mutant β-glucosidase increases the rate of the cellulose enzymatic hydrolysis. Biochem. Biophys. Rep. 2016;7:52–55. doi: 10.1016/j.bbrep.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventorino V., Aliberti A., Faraco V., Robertiello A., Giacobbe S., Ercolini D. Exploring the microbiota dynamics related to vegetable biomasses degradation and study of lignocellulose-degrading bacteria for industrial biotechnological application. Sci. Rep. 2015;5 doi: 10.1038/srep08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Bin W., Baixu Y., Peiji G. Mechanism of cellobiose inhibition in cellulose hydrolysis by cellobiohydrolase. Sci. China C Life Sci. 2004;47(1):18–24. doi: 10.1360/02yc0163. [DOI] [PubMed] [Google Scholar]
- Zhao X., Cheng K., Liu D. Organosolv pretreatment of lignocellulosic biomass for enzymatic hydrolysis. Appl. Microbiol. Biotechnol. 2009;82(5):815–827. doi: 10.1007/s00253-009-1883-1. [DOI] [PubMed] [Google Scholar]