Abstract
The inflammasome serves as a mechanism by which the body senses damage or danger. These multiprotein complexes form in the cytosol of myeloid, epithelial and potentially other cell types to drive caspase-1 cleavage and the secretion of the pro-inflammatory cytokines IL-1β and IL-18. Different types of inflammasomes, centered on (and named after) their cytosolic NLRs, respond to signals from bacteria, fungi, and viruses, as well as “sterile inflammatory” triggers. Despite the large body of research accumulated on rodent and human inflammasomes over the past 15 years, only recently have studies expanded to consider the role of inflammasomes in veterinary and wildlife species. Due to the key role of inflammasomes in mediating inflammatory responses observed in humans and rodents, characterization of the similarities and differences between humans/rodents and veterinary species is required to identify genetic and evolutionary influences on disease responses and to develop therapeutic candidates for use in veterinary inflammatory syndromes. Here, we summarize recent findings on inflammasomes in swine, cattle, dogs, bats, small ruminants, and birds. We describe current gaps in our knowledge and highlight promising areas for future research.
Keywords: Inflammasome, veterinary, NLR, IL-1β, caspase, pyroptosis, inflammation
Overview of Inflammasome Research in Rodents and Humans
Inflammasome sensor proteins detect invading pathogens or damage in the body, leading to cell death and/or cytokine release that spur innate immune responses (reviewed by many, including Guo et al., 2015; Man and Kanneganti, 2015; Broz and Dixit, 2016). Inflammasome components, originally thought to be restricted to myeloid cells, are now known to be expressed in a variety of cell types. In response to a pathogen or danger signal, inflammasome sensor proteins recruit adaptor proteins and effector caspases. In canonical inflammasome pathways, caspase-1 activation leads to maturation and release of the pro-inflammatory cytokines interleukin (IL)-1β and IL-18. A wide range of pathogens—bacterial, fungal, and viral—activate the inflammasome, often via specific effectors that are “sensed” by the cell (Franchi et al., 2012; Chen and Ichinohe, 2015). Inflammasome-driven cytokine release also drives sterile inflammation, such as in the response of myeloid cells to uric acid crystals in gout (Martinon et al., 2006), to asbestos crystals (Dostert et al., 2008), and to cholesterol crystals (Duewell et al., 2010). A growing body of literature links inflammasome pathways to chronic diseases such as diabetes, heart disease, and Crohn’s disease, and genetic mutations resulting in over-activation of the inflammasome, as are present in genetic cryopyrinopathies, result in deleterious effects from inappropriate inflammatory responses.
Although some inflammasomes require Toll-like receptor (TLR)-mediated activation (“signal 1”) and downstream NF-κB-driven transcription of pro-IL-1β and pro-IL-18 substrates, other inflammasome sensors do not require a transcriptional activation step. Cell types which constitutively express sufficient levels of cytokine substrates can also respond directly to inflammasome activators (Puren et al., 1999). The activating signal that initiates inflammasome assembly involves a protein sensor present in the cytosol, such as from the NLR family (NLR = nucleotide-binding domain and leucine-rich repeat containing), that can sense pathogen-associated molecular patterns (PAMPs) and/or danger-associated molecular patterns (DAMPs). Different types of inflammasomes involve different NLRs in response to distinct types of stimuli (for reviews of mechanisms described below, see Wen et al., 2013; Chavarria-Smith et al., 2015; Vance, 2015; He et al., 2016). For example, anthrax lethal toxin cleaves the N-terminus of the NLRP1 protein, activating the NLRP1 inflammasome. The NLRP3 inflammasome can be activated by triggers including pore-forming toxins, ATP, and crystalline matter; although the exact mechanism remains unknown, K+ efflux, Ca2+ signaling, and mitochondrial dysfunction have been implicated in NLRP3 activation. The NAIP/NLRC4 inflammasome is activated by binding of bacterial flagellin and/or type III secretion system components to NAIP proteins, and the AIM2 inflammasome binds double-stranded DNA. Note that some inflammasomes (e.g., AIM2) involve sensor proteins outside of the NLR family. Following this initial signal, the sensor proteins drive formation of cytosolic complexes. For the AIM2 and NLRP3 inflammasomes, sensor activation allows for recruitment of the adaptor protein ASC via interaction of pyrin domains on each protein and recruitment and polymerization of caspase-1 via its CARD domain (Cai et al., 2014; Lu et al., 2014). This results in autocatalytic cleavage of caspase-1 into two subunits, activating it as a protease (Fig. 1). Protein components and activators of a sample of major inflammasomes are summarized in Table 1.
Figure 1. Schematic of selected inflammasome mechanisms in the cell.
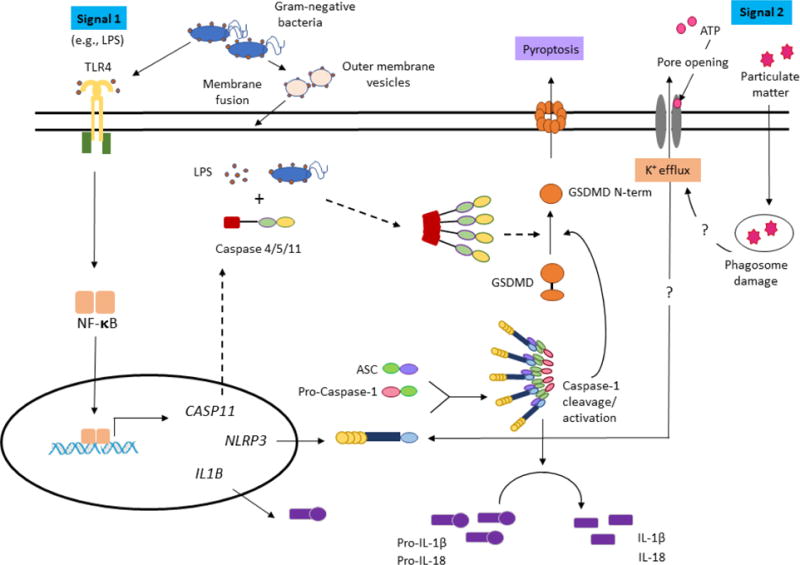
In the canonical NLRP3 inflammasome, two signals are required. While there are multiple potential triggers of the NLRP3 inflammasome, in the activation mechanism depicted in this schematic, the first signal is LPS activation of TLR4; this stimulates NF-κB-driven transcription of inflammasome components like pro-IL-1β (IL1B) and NLRP3 (NLRP3). In the presence of a second signal, the resulting NLRP3 activation triggers formation of complexes with the ASC adaptor protein and caspase-1. The resulting complex induces caspase-1 cleavage, allowing for activation of the caspase to process IL-1β for secretion and to cleave GSDMD to induce pyroptosis. Other canonical inflammasomes act through different pattern-recognition receptors. Elements specific to the non-canonical pathway of caspase-4/5/11 activation, assembly, and cleavage are highlighted with dotted arrows. In the non-canonical inflammasome, the entry of LPS (or Gram-negative bacteria) into the cytoplasm activates the caspase-4/5/11 pathway, with pore formation by cleavage of GSDMD. Here, a mechanism of LPS entry via outer membrane vesicles is depicted, as described by Vanaja et al. (2016). This diagram is based on the schematics of Guo et al. (2015), Ding and Shao (2017), and He et al. (2016). Note that the figure is not designed to be all-inclusive of activation pathways. For graphical depictions of NLRP1 and NLRC4 inflammasome pathways, and the AIM2 inflammasome pathway, see Fig. 1 and Fig. 2 in Broz and Dixit (2016).
Table 1.
Summary of key inflammasome components. Note that representative activators are listed, and an exhaustive list is not provided. Abbreviations are based on those in the review by Jha and Ting (2009).
| Abbreviation | Full name | Description | Examples of Activators |
|---|---|---|---|
| Caspase-1 | Protease | ||
| Caspase-11 | Protease | ||
| ASC | Apoptosis associated speck-like protein containing a caspase recruitment domain | Adaptor/Scaffold | |
| NLRC4 | NLR family, CARD-containing (inflammasome)-4 | Adaptor/Scaffold | |
| NLRP1 | NLR family, pyrin domain-containing (inflammasome)-1 | Sensor | Rodent: Anthrax lethal toxin, Toxoplasma gondii, depletion of cytosolic ATP |
| NLRP3 | NLR family, pyrin domain-containing (inflammasome)-3 | Sensor | ATP, pore-forming toxins, crystalline matter |
| AIM2 | Absent in melanoma-2 | Sensor | Double-stranded DNA, from host, viruses (e.g., vaccinia) or intracellular bacteria (e.g., Francisella tularensis novicida) |
| NAIPs (various) | NLR apoptosis inhibitory protein | Sensor | Bacterial flagellin, and Type III secretion system components |
Caspase-1 cleavage of pro-IL-1β and pro-IL-18 leads to secretion of the active form of these cytokines from macrophages. In some inflammasomes, caspase-1 activation also leads to the induction of pyroptosis, a controlled inflammatory cell death process that results in osmotic lysis of the cell due to pore formation in the cell membrane (coined by Cookson and Brennan, 2001; Fink and Cookson, 2006). Pyroptosis provides the opportunity for release of intracellular bacterial pathogens, exposing them to neutrophil-mediated killing (Miao et al., 2010), but can also result in systemic tissue and organ damage (Aziz et al., 2014). Therefore, careful regulation of induction of pyroptosis in response to inducing signals is critical. Cell lysis in pyroptosis is tied to activation of inflammatory caspases by their action on the gasdermin D (GSDMD) protein; caspase-1 and -11 cleavage of GSDMD liberates the N-terminal domain of GSDMD to induce cell lysis (Shi et al., 2015; Kayagaki et al., 2015) by pore formation (Aglietti et al., 2016; Ding et al., 2016; Liu et al., 2016; Sborgi et al., 2016). Interestingly, activation of gasdermins by caspases classically associated with apoptosis can also lead to rapid cell lysis in a variety of cell types, indicating that gasdermin expression may be the primary required component for pyroptotic death (Shi et al., 2015; Wang et al., 2017).
While much literature has focused on the canonical inflammasome pathway of activation, recent work has discovered and elucidated the role of non-canonical inflammasomes, involving inflammatory caspases-4, -5, and -11, in responses of rodents and humans to invasion by pathogens (reviewed by Crowley et al., 2017). Humans express caspases-4 and -5, while mice express the orthologue caspase-11; these caspases have similar roles, but exhibit differences in expression patterns (Crowley et al., 2017). Caspase-11 becomes activated via direct binding of lipopolysaccharide (LPS) in the host cell cytoplasm (Shi et al., 2014), inducing pyroptosis via cleavage of GSDMD, as described above (Fig. 1); it can also drive secretion of IL-1β and IL-18 via non-canonical activation of the NLRP3 inflammasome (Kayagaki et al., 2011). Therefore, the caspase-11 pathway reflects a response to cytoplasmic LPS that is independent of signaling through TLRs and canonical pathways. Caspase-11-dependent pathways are important in responses to invasion by Gram-negative bacteria, including infection with Burkholderia sp. (Aachoui et al., 2013) and Salmonella enterica Typhimurium (Knodler et al., 2010; Knodler et al. 2014).
A large number of pathogens that activate inflammasome pathways are of veterinary importance, including Bacillus anthracis (anthrax), Francisella tularensis (tularemia), and Salmonella enterica Typhimurium (salmonellosis). Veterinary species, both domestic and wildlife, play important roles in transmission of zoonotic diseases; therefore, explorations of how current knowledge of inflammasomes applies to veterinary species can inform both human and animal health. For example, differences in inflammasome components between genomes could aid in explaining species differences in responses to pathogen invasion or in immune responses to vaccination. Additionally, genetic differences in inflammasome components or regulators could help explain breed-based differences in production traits (e.g., response to stress, gut function). In the context of its potentially broad implications in vivo, this review summarizes the current state of research on inflammasomes in multiple animal species, including cattle, swine, small ruminants, bats, birds, and dogs. We have focused on the NLRs and associated inflammasome complexes, as previous reviews have summarized functions of TLRs in diverse species (although we note that many examples of NLRP3 upregulation discussed below are the result of TLR signaling as a transcriptional “priming” signal, prior to inflammasome complex formation). New avenues of veterinary research are proposed to apply knowledge gained in rodents and humans to production livestock and wildlife species.
Inflammasome Research in Swine
Of livestock species, the most extensive inflammasome characterization has been conducted in pigs, presumably due to their potential use as models of the human immune system (Dawson et al., 2017). Extracellular ATP, nigericin, and crystals (both alum and calcium pyrophosphate dihydrate (CPPD)), known activators of the inflammasome in mice and humans, activate inflammasomes in LPS-primed porcine cells (PBMCs; Kim et al., 2014). High levels of NLRP3 expression were detected in porcine Peyer’s patches and mesenteric lymph nodes, with upregulation in gut-associated lymphoid tissues in the presence of probiotic Lactobacillus (Tohno et al., 2011). However, the AIM2 and NAIP/NLRC4 inflammasome pathways are absent in pigs (Dawson et al., 2017; Sakuma et al., 2017).
The best characterized trigger of inflammasome responses in pigs is the porcine reproductive and respiratory syndrome virus (PRRSV), which affects swine herds worldwide and is the largest economic concern for pork producers. Inoculation of pigs with a PRRSV vaccine strain upregulates genes in the NLRP3 inflammasome pathway (Islam et al., 2016). PRRSV infection causes IL-1β secretion in the pig (Lunney et al., 2010), with IL-1β secretion from porcine alveolar macrophages (PAMs) upregulated shortly after infection (Qiao et al., 2011; Bi et al., 2014). The virus’ small envelope protein E activates the inflammasome by acting as an ion channel; Zhang et al. (2013b) hypothesize that drugs that block the PRRSV protein E ion channel may reduce inflammasome-mediated tissue damage in PRRSV infection. The PRRSV-encoded nsp11 protein can in turn inhibit IL-1β expression and secretion in PAMs (Wang et al., 2015), suggesting that PRRSV components can modulate inflammasome function. Additionally, variation in the porcine gene sequence for guanylate binding protein 5 (GBP5), an activator of NLRP3 inflammasome assembly (Shenoy et al., 2012), is associated with differences in responses (viremia, body weight) of pigs to PRRSV (Boddicker et al., 2012; Koltes et al., 2015). Porcine caspase-1 and IL-1β secretion can also be activated by the p7 protein of classical swine fever virus (Lin et al., 2014). The p7 protein is part of a class of hydrophobic proteins expressed by many animal viruses known as viroporins, which form oligomeric pores in host cell membranes (Nieva et al., 2012); this finding is consistent with other work demonstrating a role for viroporins in inflammasome induction, similar to other pore-forming toxins.
In humans, over-activation of inflammasomes can lead to inflammatory disease and tissue damage. Recent findings suggest potential for application of inflammasome research to porcine production stress and other chronic conditions. In lactating sows, feed reduction was demonstrated to reduce levels of expression of NLRP3, CASP1, and IL1B in liver approximately two-fold, indicating potential for influences of environment on porcine inflammasomes (Gessner et al., 2015). Hypoxia in piglets leads to brain injury and is associated with caspase-1 activation and small increases (<2-fold) in IL-1β release in the cerebral cortex (Angelis et al., 2014). Additionally, activation of NLRP3, ASC (PYCARD), and IL1B transcription in aortal tissue was observed in a porcine atherosclerosis model (Li et al., 2013); the authors indicate that this may be due to cholesterol crystals. In human cells, effects of cholesterol are post-transcriptional, as pre-priming (i.e., with LPS) is required for upregulation of cytokine substrates and NLRP3 prior to inflammasome activation by cholesterol crystals (Duewell et al., 2010; Rajamaki et al., 2010). Overall, characterization of porcine inflammasome responses to physiological and metabolic factors may be useful for studies aiming to improve swine production and health, but our level of understanding is at a nascent stage.
Inflammasome Research in Cattle
Despite characterized roles of inflammasomes in rodent and/or cell culture models of important bovine pathogens like Mycobacterium bovis, Bacillus anthracis, and Salmonella, very few studies have attempted to characterize inflammasome responses in cattle. The NLRP3 inflammasome has been demonstrated to be activated by ATP in cultured cattle monocytes; distinct from mice and humans (Di Virgilio et al., 2017), this process in bovine cells does not require formation of membrane pores by the P2X7 receptor (Hussen et al., 2012). Studies have also examined activation of inflammasome components by bovine viruses; Wang et al. (2014) demonstrated caspase-1 activation upon in vitro infection of bovine kidney cells with bovine herpesvirus 1, and Schaut et al. (2016) demonstrated IL-1β secretion from bovine monocyte-derived macrophages in the presence of bovine viral diarrhea virus type 2 and LPS. Recently, alum was demonstrated to activate IL-1β secretion in bovine peripheral blood mononuclear cell culture via NLRP3 activation in the absence of TLR stimulation (Harte et al., 2017).
Cattle are a natural host for anthrax, an activator of the NLRP1 inflammasome. Consistent with the closer evolutionary relationship of bovine and bison NLRP1 sequences to the human NLRP1 protein, bovine and bison macrophages were resistant to the cleavage event that activates rodent NLRP1 (Levinsohn et al., 2012), failing to undergo pyroptotic responses observed in sensitive rodent macrophages and associated with protection from anthrax spore infection in mice (Vrentas et al., in preparation). This result suggests that the NLRP1 activation response to anthrax has not evolved in cattle, which, like in the human host, could be the basis for their sensitivity to anthrax. Differences in evolution of the NLRP1 sequence may have resulted from long-term population exposures to other activators of the NLRP1 inflammasome, such as the parasite Toxoplasma gondii or a general activation condition such as ATP depletion (as described by Liao and Mogridge, 2013; Neiman-Zenevich et al., 2014).
Other recent studies in cattle have identified breed-related differences in inflammasome-related responses that may correlate with differences in responses to infection. Macrophages from Brown Swiss as compared to Holstein cattle differed in their responses to bacterial and fungal infections, including IL-1β secretion, production of reactive nitrogen species, and ability to kill intracellular Salmonella enterica Typhimurium (Gibson et al., 2016). In Angus cattle, a copy number variation in the genome adjacent to (and in the same haplotype block with) the bovine NLRP3 gene correlates with the level of resistance to infection with gastrointestinal nematodes, suggesting that genetic variation in NLRP3 could contribute to responses to parasite invasion (Xu et al., 2014).
Finally, studies have also investigated the relationship between probiotic bacteria and the bovine innate immune system. The probiotic strain Lactobacillus rhamnosus GR-1 reduces E. coli adhesion to bovine mammary epithelial cells, suggesting a potential therapeutic approach to prevent mastitis (Wu et al., 2015), an inflammatory condition with huge economic impact to the dairy industry. One effect of the probiotic was attenuation of NLRP3 activation associated with E. coli infection, although downstream effects were not assessed.
Inflammasome Research in Other Mammals
Currently, data on inflammasomes in sheep and goats is limited to NLRP3 expression data. NLRP3 mRNA expression in goat tissues was recently profiled by Zhang et al. (2017). In Merino sheep, vaccination with the TLR ligand poly(I:C) plus diphtheria toxoid induced upregulation of NLRP3 inflammasome pathway genes in cells in afferent lymph vessels (Burke et al., 2014), which likely simply reflected a signal 1 upregulation event.
Multiple studies have demonstrated activation of NLRP3 in dogs in association with infectious and non-communicable diseases. Activation of the P2X7 receptor in canine monocytes induces IL-1β release via the NLRP3 inflammasome (Jalilian et al., 2012). NLRP3 and CASP1 are downregulated in dogs with chronic enteropathy (an inflammatory bowel disease/IBD), while IL1B expression is unchanged, distinct from human IBD (Schmitz et al., 2015). In dogs demonstrating glomerulonephritis from chronic infection with the parasite Leishmania infantum, both NLRP3 granular staining and histologic changes associated with inflammation were observed in the glomeruli, suggesting a possible role for the inflammasome in renal damage during L. infantum infection (Esch et al., 2015). Finally, activation of the NLRP3 inflammasome, including IL-1β and IL-18 secretion, is associated with myocardial ischemia in canine heart tissue (Hu et al., 2015), proposed to be via oxidative stress and TXNIP binding (Zhou et al., 2010).
Inflammasome research has also expanded to bats, which are important vectors for zoonotic viruses. Sequence analysis of bat genomes has revealed the absence of the PYHIN gene family, including the AIM2 inflammasome (Zhang et al., 2013a; Ahn et al., 2016). The PYHIN gene family is involved in responses to foreign DNA (Cridland et al., 2012), which are important in immune responses to viral infection (Rathinam et al., 2010). Due to the presence of an AIM2 gene in a common bat ancestor, the authors suggest the PYHIN gene family was lost from bats during evolution, possibly as an adaptation to metabolic changes and reactive oxygen species production during flight. Intriguingly, the authors suggest that the loss of the family may be related to the “asymptomaticity” of bats to most viruses.
Inflammasome Research in Birds
Some inflammasome genes are also expressed in birds; however, the PYHIN family is only found in mammals, and an ortholog of ASC is absent. Due to the role of birds in transmission of a range of zoonotic viruses, avian responses to viral nucleic acid are of particular interest. Despite the absence of the DNA-activated PYHIN proteins in birds, introduction of foreign DNA into chicken macrophages rapidly induces lytic cell death, a response that is also observed in Drosophila cells (Vitak et al., 2015); the authors propose that this reflects a distinct pathway from mammalian responses (Vitak et al., 2016). The RIG-I foreign DNA sensor (Yoneyama et al., 2004; Poeck et al., 2010) is present in ducks but not chickens; Barber et al. (2010) suggest that the presence of RIG-I may explain the differential resistance of ducks to influenza.
Additionally, Ye et al. (2015) identified expression of NLRP3 in chicken tissues and found 54% sequence homology to mammalian NLRP3, and the NLR family member NLRC5 is associated with inflammatory pathways in chicken (reviewed by Chen et al., 2013). However, little else is known about the role of NLRs in avian responses.
Implications and Avenues for Further Research
The majority of inflammasome findings in veterinary species report on NLRP3 in myeloid cells, with the most detailed elucidation for the impact of PRRSV on the porcine NLRP3 inflammasome. It is important to note that many studies defined inflammasome activation by increases in NLRP3, IL1B, or CASP1 expression; however, this only provides an initial priming step, and activation of NLRP3 by a stimulus like pore-forming toxins or crystals is needed for downstream inflammatory responses like pyroptosis and cytokine release. Therefore, additional research focusing on events subsequent to NLR activation is needed. One common limitation to this and other work in veterinary systems is reagent availability. Table 2 summarizes reagents used in published veterinary studies, to facilitate future work.
Table 2.
Selected examples of antibodies against inflammasome components used in published manuscripts to characterize veterinary inflammasome responses.
| Reference | Species | Protein Target | Reagent | Company | Product Number | Applications |
|---|---|---|---|---|---|---|
| Tohno et al., 2016 | Porcine | Secreted IL-1β | IL-1β Quantikine ELISA kit | R & D Systems (Minneapolis) | PLB00B | ELISA detection of porcine IL-1β in cell supernatants |
| Hussen et al., 2012 | Bovine | Secreted IL-1β | Anti-ovine IL-1β (capture Ab) and anti-bovine IL-1β (detection antibody) | Antibodies from AbD Serotec | Specific antibody not specified—company purchased by Bio-Rad | ELISA detection of bovine IL-1β in supernatants |
| Hussen et al., 2012 | Bovine | – | Purified bovine recombinant IL-1β | AbD Serotec | Specific product number not specified—company purchased by Bio-Rad | Control for ELISAs |
| Gibson et al., 2016 | Bovine | Secreted IL-1β | Bovine IL-1β ELISA kit | ThermoFisher | ESS0027 | ELISA detection of bovine IL-1β in cell supernatants |
| Schaut et al., 2015; Schaut et al., 2016 | Bovine | Secreted IL-1β | Bovine IL-1β ELISA Array | Aushon Biosystems (Billerica, MA) | Specific product number not specified | Chemiluminescent ELISA detection of bovine IL-1β in cell supernatants |
| Jalilian et al., 2012 | Canine | Secreted IL-1β | Canine IL-1β VetSet ELISA kit | Kingfisher Biotech (St. Paul, MN) | VS0130D-002B | ELISA detection of canine IL-1β in cell supernatants |
| Esch et al., 2015 | Canine | Secreted IL-1β | Anti-IL-1β antibody | Abbiotec (San Diego, CA) | 250716 | Western blot detection of IL 1β-from cell homogenates |
| Esch et al., 2015 | Canine | NLRP3 | Anti-NLRP3 antibody (Mouse IgG2; designed for mouse and human NLRP3) | AdipoGen Life Sciences (San Diego, CA) | AG-20B-0014-C100 | Western blot detection of canine NLRP3 |
| Esch et al., 2015 | Canine | ASC | Anti-ASC antibody (Rabbit; designed for mouse and human ASC) | AdipoGen Life Sciences | AG-25B-0006-C100 | Western blot detection of canine ASC |
| Esch et al., 2015 | Canine | NLRP3 | Anti-NLRP3 antibody (Goat; Designed for mouse and human NLRP3) | Abcam (Cambridge, MA) | ab4207 | Immunofluorescence detection of NLRP3 |
| Hu et al., 2015 | Canine | Secreted IL-1β, 18-IL | IL-1β and IL-18 ELISA kits | IBL (Minneapolis, MN) | No product numbers/description specified | ELISA detection of canine IL-1β and IL-18 for heart tissue-derived samples |
Despite limitations, currently available data indicate similarities between the inflammasomes of humans, rodents, and other animal species, suggesting the potential for development of veterinary therapeutics similar to those being developed to modulate human inflammasomes. Animal inflammasome responses that might be targeted by new therapeutics include: (1) insufficient activation, compromising clearance of the pathogen; (2) and over-activation, leading to tissue damage and possibly chronic disease (Guo et al., 2015). In regards to insufficient activation of innate immunity, molecules that trigger inflammasome activation could serve as novel adjuvants in veterinary vaccines. Immune over-activation can be in response to DAMPs in the absence of pathogens, or “sterile inflammation” (Chen and Nunez, 2010). In humans, blockade of IL-1β has proven to be an effective treatment for inflammasome-based autoinflammatory diseases like CAPS (cryopyrin-associated autoinflammatory syndrome) (Jesus and Goldbach-Mansky, 2014); anakinra, a IL-1 receptor antagonist, has been successfully used for treatment of gout and rheumatoid arthritis (Furst, 2004; Schlesinger, 2014), and the anti-IL-1β monoclonal antibody canakinumab has been used for treatment of atherosclerotic disease (Ridker et al., 2017). Table 3 identifies examples of veterinary autoinflammatory diseases that may be of interest for further inflammasome-related study.
Table 3.
Examples of veterinary autoinflammatory diseases that may be of interest for future study of the role of the inflammasome in these conditions. List is based on Gershwin, 2007, and the Merck Veterinary Manual. In each of these cases, research on human and/or mouse systems has examined potential links between the corresponding disease and inflammasome pathways. Note that inflammasome mechanisms are not necessarily the primary cause of disease, but rather may be linked to the diseases in mice or humans in a variety of possible ways. For a review of autoinflammatory diseases linked to human and rodent inflammasomes, see Shaw et al. (2011).
| Disease Name |
|---|
| SLE (Systemic lupus erythematosus) |
| Immune-mediated thrombocytopenia |
| Myasthenia gravis |
| Diabetes mellitus |
| Inflammatory bowel disease (chronic enteropathies) |
| Contact allergic dermatitis |
Many current human therapeutics are recombinant proteins and may not be economically practical for veterinary applications. However, small-molecule inflammasome inhibitors offer greater promise for veterinary medicine. The diabetes drug glyburide was the first small-molecule drug demonstrated to inhibit the NLRP3 inflammasome (Lamkanfi et al., 2009), and this therapeutic was followed by identification of numerous other small-molecule inflammasome inhibitors (reviewed by Guo et al., 2015), including NLRP3 inhibitors CY-09 (Jiang et al., 2017) and MCC950 (Coll et al., 2015), and caspase-1 inhibitors (e.g., Wannamaker et al., 2007; Juliana et al., 2010; MacKenzie et al., 2010). Probiotic bacteria serve as another potential veterinary intervention with promise for future research; a new study in pigs examined the impact of an Enterococcus faecium probiotic on transcription of NLRP3 inflammasome components and IL-1β secretion in jejunum sections after Gram-negative pathogen challenge, with a reduction of IL-1β secretion observed with probiotic pre-incubation (Kern et al., 2017). In the swine industry, minimizing gut inflammation through feed supplementation may be of particular interest for growth promotion. Feed modification has been extensively used previously and could be easily implemented.
As there are notable differences in inflammasome responses across species, there is potential that studies in other species, such as our analysis of NLRP1 inflammasomes in cattle or the characterization of the bat inflammasome, may enhance understanding related to the pathogenesis of zoonotic infections. In the case of NLRP1, other unique B. anthracis or T. gondii hosts may offer clues to the role of this inflammasome. Preliminary work on breed-dependent differences in inflammasome responses also suggests potential use of genetic selection in animals to enhance preferred immune responses. For example, in humans, polymorphisms in NLRP3 have been linked to different disease responses (for example, Roberts et al., 2011; Ji et al., 2012), and a study in rabbits associated NLRP3 single nucleotide polymorphisms (SNPs) with susceptibility to digestive disorders (Yang et al., 2013). A recent study in pigs described NLRP3 SNPs across six breeds, identifying an NLRP3-Q969R mutation associated with increased IL-1β expression in a reconstituted porcine inflammasome system that may be linked to phenotypic differences across breeds, such as E. coli susceptibility (Tohno et al., 2016).
Finally, we did not identify any assessment in the literature of the role of noncanonical inflammasomes in livestock, despite the prevalence of Gram-negative caspase-11 activators like E. coli and Salmonella in these species. Based on BLAST alignment, the bovine genome carries a caspase-4, but not -5, ortholog that is closer in sequence to human caspase-4 than to murine caspase-11. Intriguingly, the closest porcine sequence to caspase-4, -5, and -11 is annotated as caspase-13. Along with studies of roles of inflammasomes in non-myeloid cell types, this avenue of research is wide open for exploration by veterinary immunologists.
Acknowledgments
Funding Sources
This work was supported by intramural funding from the U.S. Department of Agriculture, Agricultural Research Service (C.E.V., R.G.S., P.M.B, S.C.O); National Institutes of Health grant R01 AI118719 (F.S.S.), and the intramural program at the National Institute of Allergy and Infectious Diseases, National Institutes of Health (M.M.). Inclusion of product names does not constitute endorsement by the U.S. federal government.
Abbreviations
- TLR
Toll-like Receptor
- NLR
Nucleotide-binding domain and leucine-rich repeat containing (protein)
- GSDMD
Gasdermin D
- LPS
Lipopolysaccharide
- CPPD
Calcium pyrophosphate dihydrate
- PRRSV
Porcine reproductive and respiratory syndrome virus
- PAMs
Porcine alveolar macrophages
- SNPs
Single nucleotide polymorphisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
None
References
- Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, Tan MH, Cotter PA, Vance RE, Aderem A, Miao EA. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Duber EC. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A. 2016;113:7858–7863. doi: 10.1073/pnas.1607769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn M, Cui J, Irving AT, Wang LF. Unique loss of the PYHIN gene family in bats amongst mammals: Implications for inflammasome sensing. Sci Rep. 2016;6:21722. doi: 10.1038/srep21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelis D, Fontanez-Nieves TD, Delivoria-Papadopoulos M. The role of SRC kinase in the caspase-1 pathway after hypoxia in the brain of newborn piglets. Neurochem Res. 2014;39:2118–2126. doi: 10.1007/s11064-014-1404-1. [DOI] [PubMed] [Google Scholar]
- Aziz M, Jacob A, Wang P. Revisiting caspases in sepsis. Cell Death Dis. 2014;5:e152658. doi: 10.1038/cddis.2014.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber MRW, Aldridge JR, Webster RG, Magor KE. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci. 2010;107:5913–5917. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Song S, Fang L, Wang D, Jing H, Gao L, Cai Y, Luo R, Chen H, Xiao S. Porcine reproductive and respiratory syndrome virus induces IL-1β production depending on TLR4/MyD88 pathway and NLRP3 inflammasome in primary porcine alveolar macrophages. Mediators Inflamm. 2014;2014:403515. doi: 10.1155/2014/403515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddicker N, Waide EH, Rowland RR, Lunney JK, Garrick DJ, Reecy JM, Dekkers JC. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J Anim Sci. 2012;90:1733–1746. doi: 10.2527/jas.2011-4464. [DOI] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signaling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Burke ML, de Veer M, Pleasance J, Neeland M, Elhay M, Harrison P, Meeusen E. Innate immune pathways in afferent lymph following vaccination with poly(I:C)-containing liposomes. Innate Immun. 2014;20:501–510. doi: 10.1177/1753425913501213. [DOI] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156:1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Smith J, Vance RE. The NLRP1 inflammasomes. Immunol Rev. 2015;265:22–34. doi: 10.1111/imr.12283. [DOI] [PubMed] [Google Scholar]
- Chen S, Cheng A, Wang M. Innate sensing of viruses by pattern recognition receptors in birds. Vet Res. 2013;44:82. doi: 10.1186/1297-9716-44-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IY, Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Nunez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA, O’Neill LA. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9:113–114. doi: 10.1016/s0966-842x(00)01936-3. [DOI] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley SM, Vallance BA, Knodler LA. Noncanonical inflammasomes: Antimicrobial defense that does not play by the rules. Cell Microbiol. 2017;19:e12730. doi: 10.1111/cmi.12730. [DOI] [PubMed] [Google Scholar]
- Dawson HD, Smith AD, Chen C, Urban JF., Jr An in-depth comparison of the porcine, murine and human inflammasomes; lessons from the porcine genome and transcriptome. Vet Microbiol. 2017;202:2–15. doi: 10.1016/j.vetmic.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang DC, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- Ding J, Shao F. SnapShot: The noncanonical inflammasome. Cell. 2017;168:544. doi: 10.1016/j.cell.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity. 2017;47:15–31. doi: 10.1016/j.immuni.2017.06.020. [DOI] [PubMed] [Google Scholar]
- Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, Latz E. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch KJ, Schaut RG, Lamb IM, Clay G, Morais Lima AL, do Nascimento PRP, Whitley EM, Jeronimo SMB, Sutterwala FS, Haynes JS, Petersen CA. Activation of autophagy and nucleotide-binding domain leucine-rich repeat-containing-like receptor family, pyrin domain-containing 3 inflammasome during Leishmania infantum-associated glomerulonephritis. Am J Pathol. 2015;185:2105–2117. doi: 10.1016/j.ajpath.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8:1812–1815. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- Franchi L, Munoz-Planillo R, Nunez G. Sensing and reacting to microbes via the inflammasomes. Nat Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DE. Anakinra: review of recombinant human interleukin-I receptor antagonist in the treatment of rheumatoid arthritis. Clin Ther. 2004;26:1960–1975. doi: 10.1016/j.clinthera.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Gershwin LJ. Autoimmune diseases in domestic animals. Ann N Y Acad Sci. 2007;1109:109–116. doi: 10.1196/annals.1398.013. [DOI] [PubMed] [Google Scholar]
- Gessner DK, Grone B, Rosenbaum S, Most E, Hillen S, Becker S, Erhardt G, Reiner G, Ringseis R, Eder K. Effect of a negative energy balance induced by feed restriction on pro-inflammatory and endoplasmic reticulum stress signaling pathways in the liver and skeletal muscle of lactating sows. Arch Anim Nutr. 2015;69:411–423. doi: 10.1080/1745039X.2015.1075670. [DOI] [PubMed] [Google Scholar]
- Gibson AJ, Woodman S, Pennelegion C, Patterson R, Stuart E, Hosker N, Siviter P, Douglas C, Whitehouse J, Wilkinson W, Pegg SA, Villareal-Ramos B, Werling D. Differential macrophage function in Brown Swiss and Holstein Friesian cattle. Vet Immunol Immunopathol. 2016;15:15–23. doi: 10.1016/j.vetimm.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Callaway JB, Ting JPY. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte C, Gorman AL, McCluskey S, Carty M, Bowie AG, Scott CJ, Meade KG, Lavelle EC. Alum activates the bovine NLRP3 inflammasome. Front Immunol. 2017;9:1494. doi: 10.3389/fimmu.2017.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Hara H, Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Wei B, Wei L, Hua K, Yu X, Li H, Ji H. Sodium tanshinone IIA sulfonate ameliorates ischemia-induced lipid accumulation in Beagle dogs through NLRP3 inflammasome. Int J Cardiol. 2015;196:183–192. doi: 10.1016/j.ijcard.2015.05.152. [DOI] [PubMed] [Google Scholar]
- Hussen J, Duvel A, Koy M, Schuberth HJ. Inflammasome activation in bovine monocytes by extracellular ATP does not require the purinergic receptor P2X7. Dev Comp Immunol. 2012;38:312–320. doi: 10.1016/j.dci.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Islam MA, Grobe-Brinkhaus C, Proll MJ, Uddin MJ, Rony SA, Tesfaye D, Tholen E, Holker M, Schellander K, Neuhoff C. Deciphering transcriptome profiles of peripheral blood mononuclear cells in response to PRRSV vaccination in pigs. BMC Genomics. 2016;17:641. doi: 10.1186/s12864-016-2849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalilian I, Peranec M, Curtis BL, Seavers A, Spildrejorde M, Sluyter V, Sluyter R. Activation of the damage-associated molecular pattern receptor P2X7 induces interleukin-1β release from canine monocytes. Vet Immunol Immunopathol. 2012;149:86–91. doi: 10.1016/j.vetimm.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Jesus AA, Goldbach-Mansky R. IL-1 blockade in autoinflammatory syndromes. Annu Rev Med. 2014;65:223–244. doi: 10.1146/annurev-med-061512-150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Ting JPY. Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J Immunol. 2009;183:7623–7629. doi: 10.4049/jimmunol.0902425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Hou Z, Wang T, Jin K, Fan J, Luo C, Chen M, Han R, Ni C. Polymorphisms in inflammasome genes and risk of coal workers’ pneumoconiosis in a Chinese population. PLoS One. 2012;7:e47949. doi: 10.1371/journal.pone.0047949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, He H, Chen Y, Huang W, Cheng J, Ye J, Wang A, Tao J, Wang C, Liu Q, Jin T, Jiang W, Deng X, Zhou R. Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J Exp Med. 2017;214:3219. doi: 10.1084/jem.20171419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliana C, Fernandes-Alnemri T, Wu J, Datta P, Solorzano L, Yu JW, Meng R, Quong AA, Latz E, Scott CP, Alnemri ES. Anti-inflammatory compounds parthenolide and Bay 11-7082 are direct inhibitors of the inflammasome. J Biol Chem. 2010;285:9792–9802. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CM, Line S, Merck & Co . The Merck Veterinary Manual. Merck & Co; 2010. p. 2945. [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, Zhang J, Lee WP, Roose-Girma M, Dixit VM. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, Dixit VM. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kern M, Aschenbach JR, Tedin K, Pieper R, Loss H, Lodemann U. Characterization of inflammasome components in pig intestine and analysis of the influence of probiotic Enterococcus faecium during an Escherichia coli challenge. Immunol Invest. 2017;46:742–757. doi: 10.1080/08820139.2017.1360341. [DOI] [PubMed] [Google Scholar]
- Kim J, Ahn H, Woo HM, Lee E, Lee GS. Characterization of porcine NLRP3 inflammasome activation and its upstream mechanism. Vet Res Comm. 2014;38:193–200. doi: 10.1007/s11259-014-9602-5. [DOI] [PubMed] [Google Scholar]
- Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. PNAS. 2010;107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler LA, Crowley SM, Sham HP, Yang H, Wrande M, Ma C, Ernst RK, Steele-Mortimer O, Celli J, Vallance BA. Noncanonical inflammasome activation of caspase-4/caspase-11 mediated epithelial defenses against enteric bacterial pathogens. Cell Host Microbe. 2014;16:249–256. doi: 10.1016/j.chom.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltes JE, Fritz-Waters E, Eisley CJ, Choi I, Bao H, Kommadath A, Serao NV, Boddicker NJ, Abrams SM, Schroyen M, Loyd H, Tuggle CK, Plastow GS, Guan L, Stothard P, Lunney JK, Liu P, Carpenter S, Rowland RR, Dekkers JC, Reecy JM. Identification of a putative quantitative trait nucleotide in guanylate binding protein 5 for host response to PRRS virus infection. BMC Genomics. 2015;28:412. doi: 10.1186/s12864-015-1635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Mueller JL, Vitari AC, Misaghi S, Federova A, Deshayes K, Lee WP, Hoffman HM, Dixit VM. Glyburide inhibits the cryopyrin/Nalp3 inflammasome. J Cell Biol. 2009;187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu S, Jiang B, Cohen RA, Zang M. Activation of sterol regulatory element binding protein and NLRP3 inflammasome in atherosclerotic lesion development in diabetic pigs. PLoS One. 2013;8:e67532. doi: 10.1371/journal.pone.0067532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao KC, Mogridge J. Activation of the Nlrp1b inflammasome by reduction of cytosolic ATP. Infect Immun. 2013;81:570–579. doi: 10.1128/IAI.01003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Liang W, Kang K, Li H, Cao Z, Zhang Y. Classical swine fever virus and p7 protein induce secretion of IL-1β in macrophages. J Gen Virol. 2014;95:2693–2699. doi: 10.1099/vir.0.068502-0. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney JK, Fritz ER, Reecy JM, Kuhar D, Prucnal E, Molina R, Christopher-Hennings J, Zimmerman J, Rowland RR. Interleukin-8, interleukin-1beta, and interferon-gamma levels are linked to PRRS virus clearance. Viral Immunol. 2010;23:127–134. doi: 10.1089/vim.2009.0087. [DOI] [PubMed] [Google Scholar]
- MacKenzie SH, Schipper JL, Clark AC. The potential for caspases in drug discovery. Curr Opin Drug Discov Devel. 2010;13:568–576. [PMC free article] [PubMed] [Google Scholar]
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman-Zenevich J, Liao KC, Mogridge J. Distinct regions of NLRP1B are required to respond to anthrax lethal toxin and metabolic inhibition. Infect Immun. 2014;82:3697–3703. doi: 10.1128/IAI.02167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Feng L, Bao D, Guo J, Wan B, Xiao Z, Yang S, Zhang G. Porcine reproductive and respiratory syndrome virus and bacterial endotoxin act in synergy to amplify the inflammatory response of infected macrophages. Vet Micro. 2011;149:213–220. doi: 10.1016/j.vetmic.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Puren AJ, Fantuzzi G, Dinarello CA. Gene expression, synthesis, and secretion of interleukin 18 and interleukin 1β are differentially regulated in human blood mononuclear cells and mouse spleen cells. Proc Natl Acad Sci. 1999;96:2256–2261. doi: 10.1073/pnas.96.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT, Eklund KK. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: A novel link between cholesterol metabolism and inflammation. PLoS One. 2010;5:e11765. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolany-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Everett BM, Thuren T, MacFayden JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, Kastelein JJP, Cornel JH, Pais P, Pella D, Genest J, Cifkova R, Lorenzatti A, Forster T, Kobalava Z, Vida-Simiti L, Flather M, Shimokawa H, Ogawa H, Dellborg M, Rossi PRF, Torquay RPT, Libby P, Glynn RJ, CANTOS Trial Group Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Van Rij AM, Phillips LV, Young S, McCormick SP, Merriman TR, Jones GT. Interaction of the inflammasome genes CARD8 and NLRP3 in abdominal aortic aneurysms. Atherosclerosis. 2011;218:123–126. doi: 10.1016/j.atherosclerosis.2011.04.043. [DOI] [PubMed] [Google Scholar]
- Sakuma C, Toki D, Shinkai H, Sato M, Kitani H, Uenishi H. Pig lacks functional NLRC4 and NAIP genes. Immunogenetics. 2017;69:125–130. doi: 10.1007/s00251-016-0955-5. [DOI] [PubMed] [Google Scholar]
- Sborgi L, Ruhl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Muller DJ, Broz P, Hiller S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016;35:1766–1778. doi: 10.15252/embj.201694696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaut RG, McGill JL, Neill JD, Ridpath JF, Sacco RE. Bovine viral diarrhea virus type 2 in vivo infection modulates TLR4 responsiveness in differentiated myeloid cells which is associated with decreased MyD88 expression. Virus Res. 2015;208:44–55. doi: 10.1016/j.virusres.2015.05.017. [DOI] [PubMed] [Google Scholar]
- Schaut RG, Ridpath JF, Sacco RE. Bovine viral diarrhea virus type 2 impairs macrophage responsiveness to Toll-like receptor ligation with the exception of Toll-like receptor 7. PLoS One. 2016;11:e0159491. doi: 10.1371/journal.pone.0159491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger N. Anti-interleukin-1 therapy in the management of gout. Curr Rheumatol Rep. 2014;16:398. doi: 10.1007/s11926-013-0398-z. [DOI] [PubMed] [Google Scholar]
- Schmitz S, Werling D, Allenspach K. Effects of ex-vivo and in-vivo treatment with probiotics on the inflammasome in dogs with chronic enteropathy. PLoS One. 2015;10:e0120779. doi: 10.1371/journal.pone.0120779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, McDermott MF, Kanneganti TD. Inflammasomes and autoimmunity. Trends Mol Med. 2011;17:57–64. doi: 10.1016/j.molmed.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, MacMicking JD. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science. 2012;336:481–485. doi: 10.1126/science.1217141. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Solle M, Labasi J, Perregaux DG, Stam E, Petrushova N, Koller BH, Griffiths RJ, Gabel CA. Altered cytokine production in mice lacking P2X7 receptors. J Biol Chem. 2001;276:125–132. doi: 10.1074/jbc.M006781200. [DOI] [PubMed] [Google Scholar]
- Tohno M, Shimosato T, Aso H, Kitazawa H. Immunobiotic Lactobacillus strains augment NLRP3 expression in newborn and adult porcine gut-associated lymphoid tissues. Vet Immunol Immunopathol. 2011;144:410–416. doi: 10.1016/j.vetimm.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Tohno M, Shinkai H, Toki D, Okumura N, Tajima K, Uenishi H. Identification of the Q969R gain-of-function polymorphism in the gene encoding porcine NLRP3 and its distribution in pigs of Asian and European origin. Immunogenetics. 2016;68:693–701. doi: 10.1007/s00251-016-0917-y. [DOI] [PubMed] [Google Scholar]
- Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, Rathinam VAK. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. 2016;165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance RE. The NAIP/NLRC4 inflammasomes. Curr Opin Immunol. 2015;32:84–89. doi: 10.1016/j.coi.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak N, Johnson KN, Sester DP, Stacey KJ. A novel pathway of cell death in response to cytosolic DNA in Drosophila cells. J Innate Immun. 2015;7:212–222. doi: 10.1159/000368276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak N, Hume DA, Chappell KJ, Sester DP, Stacey KJ. Induction of interferon and cell death in response to cytosolic DNA in chicken macrophages. Dev Comp Immunol. 2016;59:145–152. doi: 10.1016/j.dci.2016.01.023. [DOI] [PubMed] [Google Scholar]
- Wang J, Alexander J, Wiebe M, Jones C. Bovine herpesvirus 1 productive infection stimulates inflammasome formation and caspase 1 activity. Virus Res. 2014;185:72–76. doi: 10.1016/j.virusres.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shi X, Zhang X, Wang A, Wang L, Chen J, Deng R, Zhang G. The endoribonuclease activity essential for the nonstructural protein 11 of porcine reproductive and respiratory syndrome virus to inhibit NLRP3 inflammasome-mediated IL-1β induction. DNA Cell Biol. 2015;34:728–735. doi: 10.1089/dna.2015.2929. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- Wannamaker W, Davies R, Namchuk M, Pollard J, Ford P, Ku G, Decker C, Charifson P, Weber P, Germann UA, Kuida K, Randle JC. (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}-3,3-dimethyl-butanoyl)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)-converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1beta and IL-18. J Pharmacol Exp Ther. 2007;321:509–516. doi: 10.1124/jpet.106.111344. [DOI] [PubMed] [Google Scholar]
- Wen H, Miao EA, Ting JP. New mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013;39 doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Liu MC, Yang J, Wang JF, Zhu YH. Lactobacillus rhamnosus GR-1 ameliorates Escherichia coli-induced inflammation and cell damage via attenuation of ASC-dependent NLRP3 inflammasome activation. Appl Environ Microbiol. 2015;82:1173–1182. doi: 10.1128/AEM.03044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hou Y, Bickhart DM, Song J, Van Tassell CP, Sonstegard TS, Liu GE. A genome-wide survey reveals a deletion polymorphism associated with resistance to gastrointestinal nematodes in Angus cattle. Funct Integr Genomics. 2014;14:333–339. doi: 10.1007/s10142-014-0371-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang GW, Chen SY, Peng J, Lai SJ. Polymorphism of NLRP3 gene and association with susceptibility to digestive disorders in rabbit. Asian-Australas J Anim Sci. 2013;26:455–462. doi: 10.5713/ajas.2012.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Yu M, Zhang K, Liu J, Wang Q, Tao P, Jia K, Liao M, Ning Z. Tissue-specific expression pattern and histological distribution of NLRP3 in Chinese yellow chicken. Vet Res Commun. 2015;39:171–177. doi: 10.1007/s11259-015-9641-6. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, Tachedjian M, Zhu Y, Zhou P, Jiang X, Ng J, Yang L, Wu L, Xiao J, Feng Y, Chen Y, Sun X, Zhang Y, Marsh GA, Crameri G, Broder CC, Frey KG, Wang LF, Wang J. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013a;339:456–460. doi: 10.1126/science.1230835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Hou Q, Zhong Z, Li X, Chen H, Li W, Wen J, Liu W, Zhong F. Porcine reproductive and respiratory syndrome virus activates inflammasomes of porcine alveolar macrophages via its small envelope protein E. Virology. 2013b;442:156–162. doi: 10.1016/j.virol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang K, Tao P, Liu J, Wang Q, Ge S, Ning Z. Distinct expression profile and histological distribution of NLRP3 inflammasome components in the tissues of Hainan black goat suggest a site-specific role in the inflammatory response. Acta Vet Hung. 2017;65:402–416. doi: 10.1556/004.2017.038. [DOI] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]


