Summary
The regulation of metastatic organotropism in pancreatic ductal adenocarcinoma (PDAC) remains poorly understood. We demonstrate, using multiple mouse models, that liver and lung metastatic organotropism is dependent upon p120catenin (p120ctn)-mediated epithelial identity. Mono-allelic p120ctn loss accelerates KrasG12D-driven pancreatic cancer formation and liver metastasis. Importantly, one p120ctn allele is sufficient for E-CADHERIN-mediated cell adhesion. By contrast, cells with bi-allelic p120ctn loss demonstrate marked lung organotropism; however, rescue with p120ctn isoform 1A restores liver metastasis. In a p120ctn-independent PDAC model, mosaic loss of E-CADHERIN expression reveals selective pressure for E-CADHERIN-positive liver metastasis and E-CADHERIN-negative lung metastasis. Furthermore, human PDAC and liver metastases support the premise that liver metastases exhibit predominantly epithelial characteristics. RNA-seq demonstrates differential induction of pathways associated with metastasis and epithelial-to-mesenchymal transition in p120ctn-deficient versus p120ctn-wild-type cells. Taken together, P120CTN and E-CADHERIN mediated epithelial plasticity is an addition to the conceptual framework underlying metastatic organotropism in pancreatic cancer.
Keywords: p120catenin, E-cadherin, pancreatic cancer, epithelial plasticity, metastasis, organotropism
eTOC Blurb
The functional basis of metastatic organotropism sheds light on the properties required for successful colonization of distant organs. Reichert et al demonstrate that epithelial plasticity is a determinant of metastatic organotropism in pancreatic cancer with differing properties required for liver and lung colonization.
Introduction
Pancreatic cancer, chiefly pancreatic ductal adenocarcinoma (PDAC), is responsible for a large disease burden (American Cancer Society 2016) and is projected to be the second leading cause of cancer mortality by 2020 (Rahib et al., 2014). The overwhelming majority of patients are diagnosed after metastasis and nearly all will succumb to disease within 6–12 months of clinical presentation (Hidalgo, 2010).
One of the primary bottlenecks to progress is in understanding the metastatic cascade, especially how tumor cells colonize distant organs and features underlying metastatic organotropism. To that end, we have utilized the main component of the adherens junction, Cdh1 (E-CADHERIN), and its binding partner responsible for stabilizing it, Ctnnd1 (P120CATENIN, P120CTN), to study the role of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions (EMT and MET, respectively) in mediating metastatic colonization and metastatic organotropism. P120CTN is known to stabilize E-CADHERIN at the adherens junctions (Ireton et al., 2002; Ishiyama et al., 2010; Thoreson et al., 2000), and our group has demonstrated that conditional p120ctn loss in the esophagus leads to invasive squamous cell carcinoma (Stairs et al., 2011). Strikingly, p120ctn was identified as a “cancer candidate gene” by analyzing the transposon insertion sites of sleeping beauty mutagenesis-induced pancreatic cancers (Mann et al., 2012). Interestingly, this study also demonstrated that P120CTN loss or mislocalization is associated with worse outcomes (Mann et al., 2012).
Genetic studies in mice have demonstrated that the absence of p120ctn has variable adverse consequences in other tissues, such as skin, salivary gland, and colon by modulating signals that promote tissue proliferation, migration, and inflammation, which may predispose the tissue to dysplasia or cancer (Davis and Reynolds, 2006; Perez-Moreno et al., 2006, 2008; Short et al., 2017; Smalley-Freed et al., 2010). The salivary gland is particularly interesting since its morphology is very similar to that of the exocrine pancreas where acinar cells secrete digestive enzymes into ductal conduits (Reichert and Rustgi, 2011). p120ctn loss in the salivary gland displays a phenotype reminiscent of acinar-to-ductal metaplasia (ADM), a transient phenomenon during pancreatitis and a precursor to pancreatic intraepithelial neoplasia (PanIN) and PDAC (Davis and Reynolds, 2006; Kopp et al., 2012).
EMT appears to be important for tumor initiation (Craene and Berx, 2013). At the same time, some have advocated that it is dispensable in PDAC (Zheng et al., 2015), lung cancer (Fischer et al., 2015), breast cancer (Ocaña et al., 2012), and skin cancer (Tsai et al., 2012), although this premise has garnered divergent viewpoints (Aiello et al., 2017; Ye et al., 2017). The reverse process, MET, may foster metastatic colonization and outgrowth (Aiello et al., 2016a; Takano et al., 2016). The role of P120CTN in mediating epithelial identity makes it a powerful tool to study the role of EMT-MET in pancreatic cancer dissemination, and, in particular, whether EMT and MET may play roles in mediating metastatic organotropism.
Herein, we demonstrate that monoallelic p120ctn loss accelerates KrasG12D-driven PDAC formation and liver metastasis. By contrast, bi-allelic p120ctn loss prevents pancreatic cancer cells from acquiring a MET-phenotype required for liver metastases; however, bi-allelic p120ctn loss is permissive for lung metastases. An independent genetic approach utilizing mosaic Cdh1/E-cadherin loss in a KrasG12D background results in predominantly E-CADHERIN-negative, mesenchymal lung metastasis but E-CADHERIN-positive, epithelial liver metastasis. Furthermore, restoration of the p120ctn1A isoform in a p120ctn null background restores liver metastatic tropism. Finally, human liver metastases show increased membranous E-CADHERIN relative to paired primary PDAC, providing a new prism through which to view metastatic organotropism in PDAC.
Results
Monoallelic p120ctn loss accelerates KrasG12D-driven PDAC formation and metastatic dissemination
Given the fact that chronic pancreatitis is a major risk factor of PDAC (Guerra et al., 2007), we first investigated the effects of p120ctn under homeostatic conditions and injury using the Pdx1-cre; p120ctnfl/fl; Rosa26YFP mouse (Figure S1A). We confirmed mosaic recombination in ducts as well as loss in the exocrine compartment (Figure S1B) and observed a significant impairment of exocrine compartment maintenance over a period of one year (Figure S1C). However, in a cohort of 31 Pdx1-cre;p120ctnwt/fl;Rosa26YFP and 20 Pdx1-cre;p120ctnfl/fl;Rosa26YFP mice that were aged to a maximum of 74 weeks and 65 weeks of age, respectively, we detected no cancer. In addition, while the exocrine compartment was compromised by p120ctn loss, the endocrine compartment was not affected (Figure S1D and S1E). In a cerulein model of acute pancreatitis, p120ctn loss led to impaired regeneration and a prolonged ADM phenotype (Figures S1F, S1G and S1H).
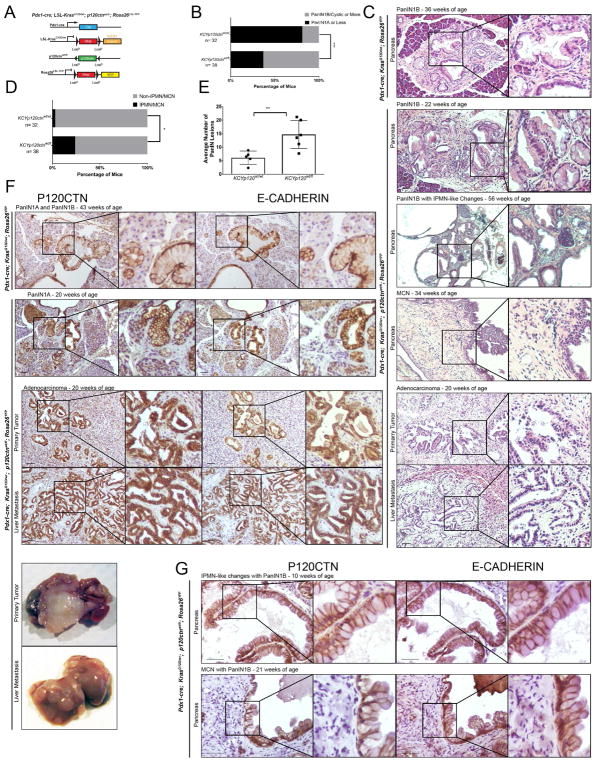
We next introduced the mutant LSL-KrasG12D allele in to Pdx1-cre; p120ctnfl/fl; R26YFP mice (Figure 1A). Pdx1cre;LSL-KrasG12D/wt;p120ctnfl/fl;Rosa26YFP mice were not viable. Timed matings indicated that embryos died between E8.5 and E14.5 (data not shown). Pdx1-cre; LSL-KrasG12D/wt; p120ctnwt/fl; Rosa26YFP mice (from here on referred to as KCYp120ctnwt/fl) were born according to Mendelian ratios. Across an age-matched cohort ranging from 3 to 58 weeks of age (Tables S1 and S2), KCYp120ctnwt/fl mice (n = 38) show a significant acceleration of KrasG12D-driven pathology compared to control KCYp120ctnwt/wt animals (n = 32) (Figures 1B, 1C, and S1A). Remarkably, KCYp120ctnwt/fl mice harbor the entire spectrum of PDAC-precursor lesions, including PanINs 1–3, mucinous cystic neoplasm (MCNs)-like lesions, and intraductal papillary mucinous neoplasm (IPMN)-like lesions. MCN-like and IPMN-like lesions are significantly more frequent in KCYp120ctnwt/fl (9/38) compared to controls (1/32) (Figure 1D) and, in 4/9 cases, mice exhibited simultaneous PanIN1A or 1B lesions. Mono-allelic p120ctn loss results in twice as many PanIN lesions compared to controls (14.67 ± 4.71 vs 6.1 ± 2.15, mean ± SD, respectively) (Figure 1E). Interestingly, it has been demonstrated that PDAC arising from cystic neoplasms might harbor a distinct cell-of-origin than that in the PanIN–PDAC sequence (von Figura et al., 2014; Kopp et al., 2012). Specifically, the ductal compartment may serve as the cellular origin of IPMN. Moreover, 2/38 KCYp120ctnwt/fl mice presented with frank invasive PDAC with evidence of metastasis in one mouse (the liver of the other mouse was not available for analysis). The phenotype in control KCYp120ctnwt/wt mice was restricted almost entirely to ductalization, acinar-to-ductal metaplasia (ADM) and PanIN 1A/B lesions with a single mouse exhibiting MCN-like lesions.
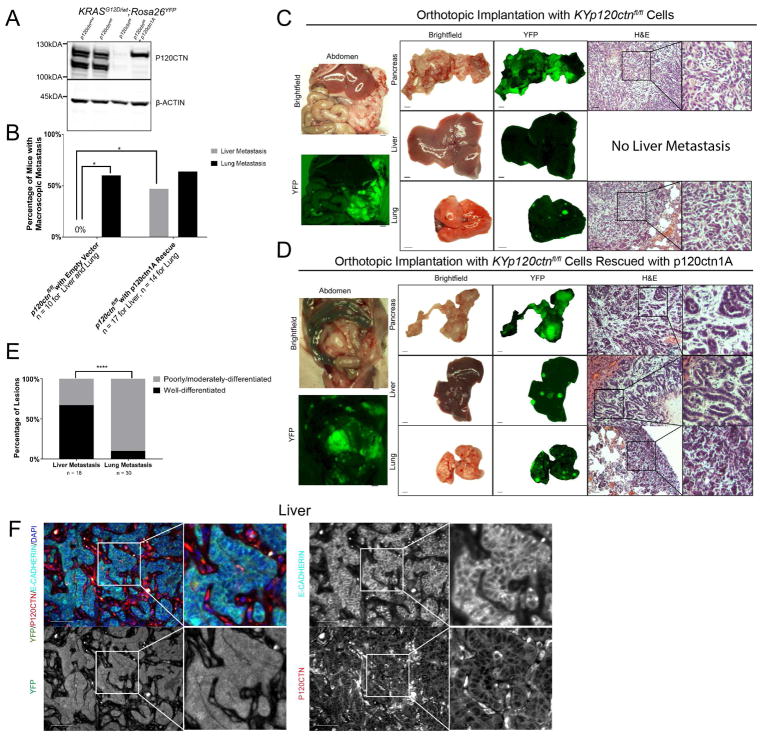
Figure 1. Monoallelic p120ctn loss in a KrasG12D background leads to accelerated pancreatic pathology.
A. Genetic schematic of Pdx1-cre; KrasG12D/wt; p120ctnwt/fl; Rosa26YFP (KCYp120wt/fl) mice. B. A comparison of PanIN, IPMN, and MCN lesion development in KCY and KCYp120wt/fl mice. From 32 KCY mice aged between 3 weeks and one year, 26 exhibited lesions equal to or less severe than PanIN1As, while 6 exhibited more severe lesions. In an age-matched cohort of 38 KCYp120wt/fl mice, 14 exhibited lesions less or equal to PanIN1As, while 24 harbored PanIN1B, PDAC, IPMN, or MCN lesions. Fisher’s Exact Test. C. Representative pathology from KCY and KCYp120wt/fl mice. D. When only IPMN or MCN lesions (with or without concomitant PanIN lesion presence) are considered, 1 out of 32 KCY mice harbored an IPMN/MCN-like lesion, while 9 KCYp120wt/fl mice harbored such lesions. Fisher’s Exact Test. E. Three high-powered fields (10X) across a cohort of age-matched mice (n = 5 KCY, n = 6 KCYp120wt/fl) were quantified by two independent observers for the number of PanIN lesions. KCYp120wt/fl mice harbored more than two times as many PanIN lesions than KCY mice (14.67 ± 4.71 vs 6.1 ± 2.15, mean ± SD). Unpaired t-test. F. Membranous P120CTN and E-CADHERIN expression are present at all stages of disease: PanINs in KCY mice and PanINs, PDAC, and liver metastasis in KCYp120wt/fl mice. G. IPMN and MCN lesions also display membranous P120CTN and E-CADHERIN. 50 μm scale bar. *, p < .05, ** < .01, *** < .001. See Supplemental Figures 1 and 2 and Supplemental Tables 1 and 2.
Given the remarkably accelerated phenotype of KCYp120ctnwt/fl mice (Figure S2A), we next asked whether the second allele of p120ctn might be lost during the progression from preneoplastic lesions to primary PDAC formation to metastasis. Surprisingly, we were able to demonstrate that the remaining wild-type p120ctn allele is retained in all stages of PanIN to PDAC metastases by immunohistochemistry (Figure 1F) and immunofluorescence (co-staining of YFP/E-CADHERIN/P120CTN) (Figure S2B), indicating that p120ctn loss-of-heterozygosity (LOH) did not occur. This was also true of the epithelial lining of MCN/IPMN lesions (Figure 1G). In all all pathological states, including liver metastasis, P120CTN co-localizes with E-CADHERIN at the cell membrane, thereby indicating that a single p120ctn allele is sufficient to stabilize E-CADHERIN.
p120ctn impacts metastatic organotropism in a mouse model of accelerated PDAC
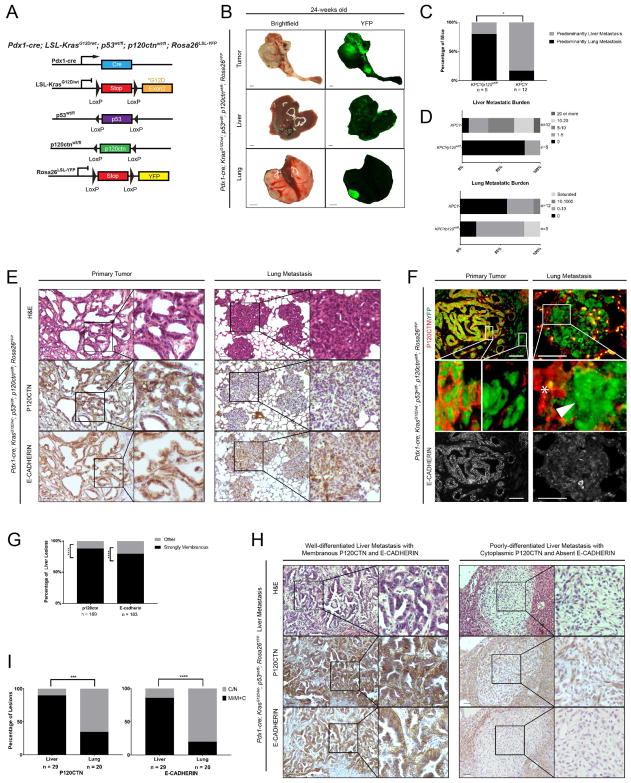
In order to investigate the effect of p120ctn loss on metastatic colonization, we utilized a mouse model of accelerated PDAC by additionally deleting one allele of Trp53 (Pdx1-cre; KrasG12D/wt; p120ctnwt/fl; p53wt/fl; Rosa26YFP, KPCYp120ctnwt/fl) (Figure 2A). The KPCYp120ctnwt/fl mice exhibit a marked propensity lung metastasis relative to liver metastasis (Figure 2B). Mono-allelic p120ctn loss significantly shifted metastatic burden from the liver to the lung (4/5 lung in the KPCYp120ctnwt/fl group versus 9/11 liver in the PCYp120ctnwt/wt group) (Figure 2C). Additionally, the absolute number of metastases in liver and lungs shows a much higher burden in liver for KPCYp120ctnwt/wt and lung for KPCYp120ctnwt/fl mice (Figure 2D). The primary tumors in KPCYp120ctnwt/fl mice express P120CTN to a varying degree. Differentiated (ductal/tubular) regions express membranous P120CTN that co-localizes with E-CADHERIN. By contrast, delaminated and invading tumor cells undergoing EMT are devoid of both P120CTN and E-CADHERIN (Figure 2E). Lung metastases in KPCYp120ctnwt/fl mice do not express P120CTN or E-CADHERIN, thereby suggesting that pancreatic cancer cells maintain their mesenchymal phenotype and fail to undergo MET in the lung (Figure 2E). This was confirmed by immunofluorescence, which revealed that YFP+ lung metastases do not express P120CTN or E-CADHERIN (Figure 2F). A total of 169 liver metastatic lesions were analyzed for P120CTN and 183 liver metastatic lesions for E-CADHERIN, with both showing robust membranous staining in almost all lesions (148/169 for P120CTN and 146/183 for E-CADHERIN) (Figure 2G). P120CTN and E-CADHERIN staining frequently tracked with each other, with non-membranous P120CTN staining correlating with absent or low E-CADHERIN staining (Figure 2H), which underscores the crucial role for P120CTN in fostering the MET phenotype in liver metastasis.
Figure 2. Monoallelic p120ctn loss in a KrasG12D; p53wt/fl background leads to a metastatic shift characterized by P120CTN-deficient lung lesions.
A. Genetic schematic of Pdx1-cre; KrasG12D/wt; p53wt/fl; p120ctnwt/fl; Rosa26YFP (KPCYp120ctnwt/fl) mice. B. Gross brightfield and YFP images of primary PDAC, liver, and lung from a KPCYp120ctnwt/fl mice. C. One slide of liver and lung each from KPCYp120wt/fl (n = 5) and KPCY (n = 12) mice was quantified at 10X magnification. Each mouse was assigned to either lung or liver dominant category. One mouse of each genotype, which had a single lesion in both liver and lung, was excluded. Mice with no tumor burden in either organ were excluded. Fisher’s Exact Test. D. Stratification of mice by the burden of tumor metastasis shows an increase in lung metastasis and diminution of liver metastasis in KPCYp120ctnwt/fl versus KPCY mice. E. Primary tumors and lung metastasis from KPCYp120ctnwt/fl mice show retention of membranous P120CTN and E-CADHERIN staining at the primary site but loss in the lungs. F. Immunofluorescence of primary and lung metastasis in KPCYp120ctnwt/fl mice reveals absent P120CTN (red) in YFP-positive (green) lung metastasis with retained E-CADHERIN (lower panel) in P120CTN-positive primary tumor areas and absent in lung metastasis. P120CTN-deficient lung metastasis (arrowhead) is surrounded by P120CTN-positive lung tissue (star). G. Quantification of P120CTN (n = 169) and E-CADHERIN (n = 183) localization in KPCY liver metastasis (n = 14 mice), which were analyzed by two independent observers. Binomial test. H. Examples from KPCY mice of a well-differentiated liver lesion with strongly membranous P120CTN/E-CADHERIN and a poorly-differentiated liver lesion with cytoplasmic P120CTN and absent E-CADHERIN. 1 mm scale bar for gross pathology. I. Quantification of P120CTN and E-CADHERIN localization in liver and lung metastases (n = 29 for liver, n = 20 for lung) (n = 4 mice) in a tet-on, Elastase KRASG12V PDAC model was done by two independent observers. Fisher’s Exact Test. *p < .05, **p < .01, ***p < .001, ****p < .0001. 50 μm scale bar for histology. See Supplemental Figure 3 and Supplemental Tables 3 and 4.
We next utilized another independent genetic model, which uses an inducible KRASG12V under the control of the elastase promoter (Guerra et al., 2007). These mice had either mono-allelic or bi-allelic Trp53 loss under the control of either a FLP-FRT or CRE-LOXP Elastase-tTA/tetO (Elas-tTA/tetO-cre; KrasG12V/wt; p53wt/fl or fl/fl or Elas-tTA/tetO-flp; KrasG12V/wt; p53wt/frt or frt/frt). A total of 29 liver lesions (n = 4 mice) were analyzed for both P120CTN and E-CADHERIN, with metastases demonstrating overwhelming membranous staining (26/29 for P120CTN and 25/29 for E-CADHERIN) (Figure 2I). Analysis of 20 lung lesions across the same 4 mice for P120CTN and E-CADHERIN demonstrated a shift from membranous expression (7/20 for P120CTN and 4/20 for E-CADHERIN). These data support the notion that the lung organotropism observed in the KPCYp120ctnwt/fl mouse is not dependent upon MET.
Single circulating tumor cells exhibit differing epithelial properties compared to clusters of cells
We next asked whether the profile of circulating tumor cells mimics that of metastases, insofar as they exhibit a spectrum of epithelial and mesenchymal properties. Both single cells and clusters of circulating tumor cells were isolated from KPCY mice and stained for E-CADHERIN and additional markers, including P120CTN (Figure S3A) and β-CATENIN (Figure S3B). Additionally, clusters were stained for CYTOKERATIN-19 (Figure S3C). Clusters of tumor cells exhibited membranous localization of the markers, whereas individual tumor cells did not.
Sequential genetic manipulation of E-cadherin in pancreas cancer reveals the requirement of epithelial integrity to establish liver metastases in vivo
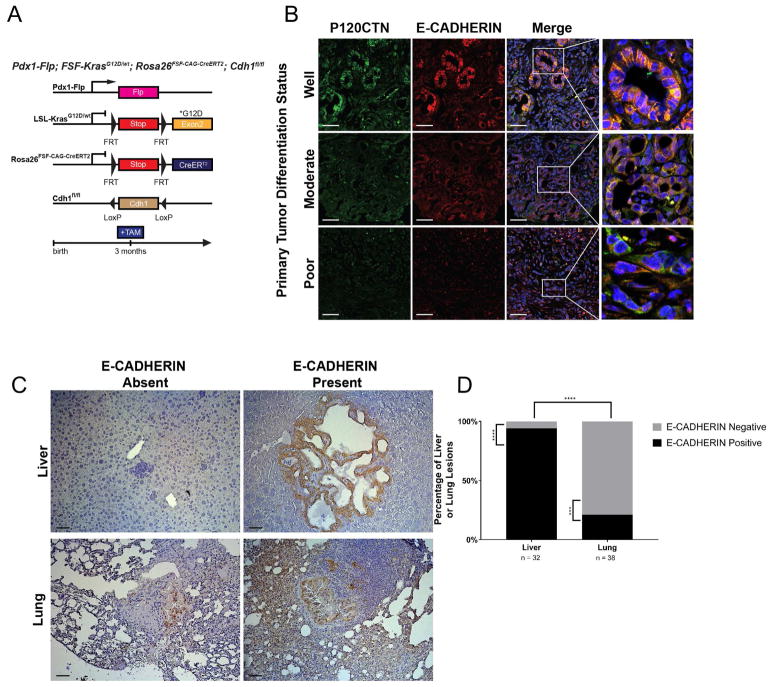
In order to verify that the metastatic shift caused by p120ctn loss is due to the functional role of P120CTN in stabilizing E-CADHERIN, we next utilized an inducible, conditional dual-recombination system in which E-cadherin (Cdh1) can be manipulated independently from the PDAC-initiating KrasG12D mutation (Pdx1-Flp; FSF-KrasG12D/wt; FSF-R26GAG-CreERT2;CDH1fl/fl) (Figure 3A) (Schönhuber et al., 2014). Mice were induced with tamoxifen at 3 months of age and maintained until moribund. We did not observe a survival difference between mice with wild-type CDH1 and heterozygous CDH1 knockout (data not shown). Tumors show heterogeneous E-CADHERIN expression by immunofluorescence, thus indicating a variable degree of E-CADHERIN loss (Figure 3B). Well-differentiated tumor regions display high E-CADHERIN expression levels and membranous E-CADHERIN and P120CTN. Poorly-differentiated areas within the same tumor lack E-CADHERIN and P120CTN.
Figure 3. Sequential and mosaic Cdh1 deletion in a KrasG12D background demonstrates selective pressure for E-CADHERIN-positive liver metastasis and E-CADHERIN-deficient lung metastasis.
A. Genetic schematic of the Pdx1-Flp;FSF-KrasG12D/wt;FSF-R26GAG-CreERT2;CDH1fl/fl mouse. This dual-recombinase system allows FLP recombinase-mediated recombination of the FRT sites at the KRASG12D/wt and Rosa26YFP loci as well as independent manipulation of CDH1 through tamoxifen-mediated induction of CreERT2. B. Examples of well-, moderately-, and poorly-differentiated tumor regions in Pdx1-Flp;FSF-KrasG12D/wt;FSF-R26GAG-CreERT2;CDH1fl/fl mice. Tumors show strong correlation between membranous co-localization of P120CTN and E-CADHERIN. These areas, in turns, are more highly differentiated. C. Immunohistochemistry of representative metastatic lesions in liver and lung by E-CADHERIN status. D. Liver (n = 32) and lung (n = 38) metastatic lesions, quantified across 5 mice, were classified by E-CADHERIN status. Liver metastases showed a strong predisposition towards E-CAHDERIN-positivity (n = 30/32), whereas lung metastases showed a strong predisposition towards E-CADHERIN-negativity (n = 30/38). Comparison between liver and lung showed a statistically significant switch in E-CADHERIN status. Binomial test for comparisons within each organ. Fisher’s Exact Test for comparison between organs. 50 μm scale bar.
We employed the genetic heterogeneity regarding the recombination of the Cdh1 locus to investigate the role of E-CADHERIN in PDAC metastasis evolution within each individual mouse. A given cancer cell that lacks E-CADHERIN would only be able to establish E-CADHERIN-negative metastases. By contrast, cancer cells with retained E-CADHERIN expression can establish E-CADHHERIN-positive or -negative metastases. We next analyzed the number and distribution of metastases in these mice. There was no statistical difference between Cdh1 heterozygous animals; however, when liver and lung metastases were stained for E-CADHERIN, liver metastases uniformly expressed membranous E-CADHERIN (n = 30/32) (Figures 3C and 3D). Only a negligible number of micro-metastases present within the liver lacked E-CADHERIN expression indicating that E-CADHERIN loss leads to a negative selection pressure restricting the capability of PDAC cells to establish liver metastases. Conversely, lung metastases demonstrate mixed E-CADHERIN expression, thereby suggesting E-CADHERIN-mediated epithelial integrity is dispensable for lung metastasis (Figures 3C and 3D).
Taken together, the results obtained in an entirely independent genetic mouse model of PDAC emphasize that epithelial integrity mediated by P120CTN, and its role regulating E-CADHERIN turnover, determines metastatic outgrowth in the liver.
One p120ctn allele is sufficient to maintain epithelial integrity in vitro and in vivo
Our in vivo data suggest one p120ctn allele is required to re-establish epithelial integrity or MET in the liver. In order to address this mechanistically, we performed 3D organoid culture with primary pancreatic cells isolated from LSL-KrasG12D/wt;p120ctnwt/wt;Rosa26YFP (wild-type p120ctn, KYp120ctnwt/wt) (cell line ID: 611, 793, 801), LSL-KrasG12D/wt; p120ctnwt/fl; Rosa26YFP (heterozygous floxed p120ctn, KYp120ctnfl/wt) (cell line ID: 363, 732, 866) and LSL-KrasG12D/wt; p120ctnfl/fl; Rosa26YFP pancreata (homozygous floxed p120ctn, KYp120ctnfl/fl) (cell line ID: 288, 690, 791) (Deramaudt et al., 2006; Reichert et al., 2013a). Lentiviral-mediated, Cre recombinase in vitro followed by FACS for YFP positive (YFP+) cells was utilized to establish stable, recombined cell lines (Figure 4A). The in vitro recombination approach was necessary since bi-allelic p120ctn loss in combination with KrasG12D is lethal in utero.
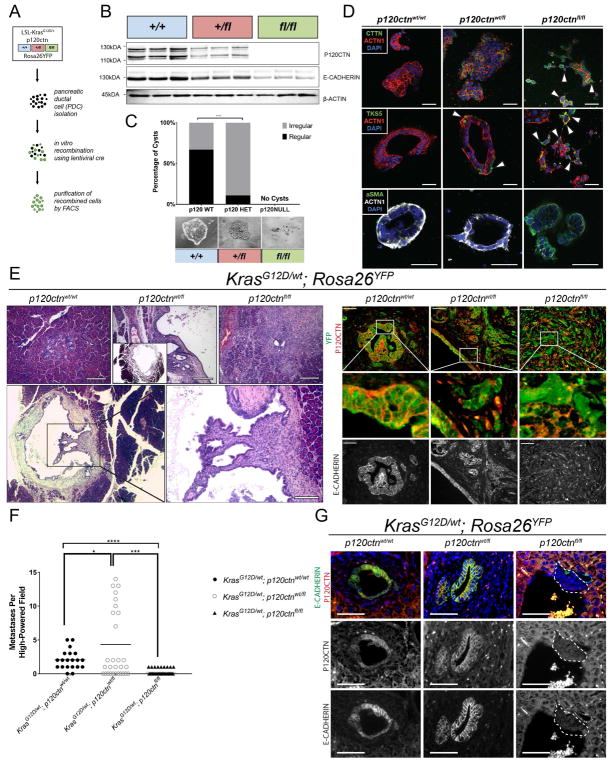
Figure 4. A single allele of p120ctn is sufficient to stabilize E-CADHERIN and to colonize the liver.
A. Ex-vivo recombination of KrasG12D/wt; Rosa26YFP (KY) cells with either wild-type p120ctn or mono-/bi-allelic loss. B. Confirmation of efficiency of recombination with ex-vivo cre in three replicates per genotype. C. 3D organotypic culture demonstrates that normal cyst formation is dependent on P120CTN. Cells were plated in chamber slides in quadruplicate in bovine collagen. Cyst formation was quantified 5 days later. KYp120ctnwt/wt cells formed normal cysts a majority of the time (94 regular versus 47 irregular), whereas KYp120ctnwt/fl cells showed dramatically impaired regular cystogenesis (9 regular versus 70 irregular). KYp120ctnfl/fl cells formed no cysts in 4 technical replicates. Fisher’s Exact Test. D. 3D organotypic cultures were stained for Tks5, cortactin (CTTN), α-smooth muscle actin (α-SMA), actin (ACTN1), and DAPI. Arrowheads indicates protrusions reminiscent of invadopodia. E. Nude athymic mice were orthotopically injected with 500,000 cells and harvested three weeks post-injection (n = 3 per genotype). H&E on the left and immunofluorescence (P120CTN in red, YFP in green, E-CADHERIN lower panel]. F. Athymic nude mice were injected with 750,000 cells and harvested two weeks post-injection. Ten high power fields were analyzed per mouse (n = 2 wildtype, n = 3 heterozygous/null). Welch’s T-test. G. Immunofluorescence of liver metastasis in intraportal vein injection model. (P120CTN in red, E-CADHERIN in green]. 50 μm scale bar. See Supplemental Figure 4.
We confirmed the recombination efficiency in YFP+ cells by FACS (Figure S4A) and western blot (Figure 4B), which indicated a gene dose-dependent reduction of P120CTN levels that correlated with E-CADHERIN levels. 3D culture reveals that wild-type p120ctn cells form multicellular organoids with hollow lumens (Figure 4C). Mono-allelic p120ctn loss disrupts the symmetry of these structures with a loss of hollow lumens; and leads to invadopodia-like protrusions. Bi-allelic loss of p120ctn completely prevents cells from forming organized structures (Figure 4C). To characterize these 3D structures, we used the established markers of invadapodia Tks5 (Blouw et al., 2015; Seals et al., 2005) and cortactin (Clark et al., 2007), in addition to α-smooth muscle actin (α-SMA). We observed higher expression of these invadapodia markers and αSMA with p120ctn loss (Figure 4D). To validate this phenotype in vivo, we next injected these cell lines orthotopically into the pancreata of athymic nude mice (Figure 4E). KYp120ctnwt/wt cells form PanIN-like structures within 3 weeks (21 days). Surprisingly, KYp120ctnwt/fl cells establish large cystic structures reminiscent of IPMN/MCN lesions with highly organized regions, especially at the epithelial lining of the cyst, as well as more invasive areas (Figure 4E, middle upper panel and two lower panels). The epithelial lining of the cysts displayed membranous expression of P120CTN and E-CADHERIN whereas the invasive parts of the tumor showed varying localization of P120CTN and E-CADHERIN. Finally, KYp120ctnfl/fl cells, characterized by a spindle shape and lack of membranous E-CADHERIN, generate invasive undifferentiated tumors.
MET is required for metastatic outgrowth in the liver but not in the lung
The liver is the most common site of metastasis in PDAC (Yachida and Iacobuzio-Donahue, 2009). Thus, we next asked the question whether the remaining p120ctn allele is required for metastatic outgrowth in the liver. In order to focus on steps within the metastatic cascade that do not require primary tumor growth, invasion and intravasation, we employed intraportal vein injections. We again used the cell lines described above harboring mutant KrasG12D, a YFP reporter and one of three p120ctn genotypes: p120ctnwt/wt, p120ctnwt/fl, or p120ctnfl/fl. Fourteen days post-injection, no macroscopic liver metastases were observed. Histologically, KYp120ctnwt/wt cells show sparse metastases in the livers (2.05 ± 0.32 lesions per HPF, mean ± SD) (Figure 4F). KYp120ctnwt/fl cells colonize the liver significantly more frequently (4.33 ± 1.03 lesions per HPF, mean ± SD). KYp120ctnfl/fl cells demonstrated the least capacity to form liver metastases (0.3 ± 0.09 lesions per HPF, mean ± SD). Additionally, immunofluorescence confirmed that P120CTN and E-CADHERIN expression track with each other (Figure 4G). These data suggest that bi-allelic p120ctn loss abrogates the ability of pancreatic cancer cells to establish liver metastases.
To test whether p120ctn levels alter the frequency of lung metastasis, we utilized the retro-orbital injection model. Due to transpulmonary passage, this model system preferentially metastasizes to the lung (Galaup et al., 2006; Hollern et al., 2014). Interestingly, KrasG12D cells form lung metastases irrespective of their p120ctn status (Figure S4B). As expected, p120ctn wild-type cells establish metastases with high E-CADHERIN levels. Cells with mono-allelic p120ctn loss show intermediate E-CADHERIN expression whereas E-CADHERIN is absent in metastases formed by cells with bi-allelic p120ctn loss (Figure S4B).
Long-term orthotopic transplantation of KYp120ctnwt/fl cells results in both macroscopic solid tumors and fluid-filled cystic structures
We next orthotopically injected 5 × 105 KYp120ctnwt/fl cells into 12 nude mice and aged them for up to 180 days. Intriguingly, we saw two distinct paths reminiscent of the PanIN versus IPMN/MCN phenotype in the KCYp120ctnwt/fl model. Specifically, 3 mice exhibited macroscopic solid tumors whereas 4 harbored macroscopic cystic lesions. 1 mouse showed a mixed solid/cystic phenotype, and 4 showed no evidence of tumor (Figure S5A). The difference between these two pathways is evident both macroscopically and histologically (Figure S5B) where immunohistochemistry was used to confirm YFP+ expression in solid tumor cells and the epithelial lining of the cysts. Retention of P120CTN and E-CADHERIN was seen in the ductal structures of the solid tumor and in the epithelial lining of the cysts (Figure S5B).
Restoration of MET shifts metastatic organotropism from the lung back to the liver
We next tested whether we could restore liver metastatic ability in KYp120ctnfl/fl cells by reintroducting p120ctn. To investigate this, we rescued our KYp120ctnfl/fl cells with the full-length p120ctn isoform p120ctn1A. KYp120ctnfl/fl cells were stably transfected with either an empty control construct or p120ctn1A and validated by western blot (Figure 5A). 5 × 105 cells were orthotopically injected into the pancreata of athymic nude mice and aged for between 10–14 weeks. Parental KrasG12D/wt; p120ctnfl/fl; Rosa26YFP cells formed macroscopic lung metastases in a higher number of mice than macroscopic liver metastases (Figures 5B and 5C). Cells without the ability to modulate E-cadherin and undergo MET bypass the liver altogether and preferentially go to the lung. We were able to restore macroscopic liver metastasis in 8/17 mice orthotopically injected with p120ctn1A-rescued cells (Figures 5B and 5D). These mice demonstrated significant macroscopic liver and lung metastatic burden; however, metastatic lesions derived from the same primary tumor in the p120ctn1A-recused orthotopics showed a distinct appearance based upon site of metastasis. Specifically, lesions in the liver demonstrated an almost entirely epithelial phenotype characterized by membranous E-CADHERIN and well-organized ductal structures (Figure 5D and 5F). Conversely, lung metastases were disorganized and de-differentiated. 12/18 liver lesions were well-whereas 27/30 lesions were de-differentiated (Figure 5E). It is interesting to note that the invasive front of these metastatic liver lesions is characterized by well-differentiated ductal structures and not individual cells (Figure 5E). We also noted that this difference was not entirely due to size, as even large lung lesions, comparable in size to some liver lesions, were de-differentiated. Additionally, even small liver lesions were well-differentiated. This supports our observation that P120CTN/E-CADHERIN expression levels are strongly membranous in KPCY liver metastasis (Figure 2G).
Figure 5. KrasG12D; p120ctnfl/fl; Rosa26YFP (KYp120ctnfl/fl) cells readily colonize the lung, but not the liver, unless rescued with p120ctn1A isoform.
A. Western blot demonstrates p120ctn1A restoration in KYp120ctnfl/fl cells. B. Nude mice orthotopically injected with 500,000 KYp120ctnfl/fl cells (n = 10) show lung (n = 6/10), but not liver (n = 0/10), metastasis. p120ctn1A rescue (n = 17) restores liver metastasis (n = 8/17) to the liver while still permitting lung metastasis (n = 9/14 [3 lungs excluded from this analysis due to lack of gross fluorescence images]). Fisher’s Exact Test. C and D. Representative gross pathology and histology from nude mice orthotopically injected with KYp120ctnfl/fl cells stably transfected with an empty vector (C) or p120ctn1A-expressing vector (D) and aged out for up to 102 days post-injection. The examples represented here were aged for 70- (D) and 98-days (E) post-implantation. E. 18 liver and 30 lung metastatic lesions in mice injected with p120ctn1A-rescue cells were classified as either well-differentiated or moderately/poorly-differentiated on the basis of the presence or absence of ductal structures. 12/18 liver lesions were well-differentiated compared to only 3/30 lung lesions classified as well-differentiated. Fisher’s Exact Test. F. Immunofluorescence for P120CTN, E-CADHERIN, and YFP in p120ctn1A-rescued liver metastasis. 1 mm scale bar for gross pathology. 50 μm scale bar for histology. *p < .05, ****p < .0001. See Supplemental Figure 5 and Supplemental Tables 5 and 6.
Epithelial integrity mediated by P120CTN and E-CADHERIN is critical in human PDAC-derived liver metastases
We next determined the expression pattern of P120CTN and E-CADHERIN in human PDAC and associated liver metastasis using a human tumor microarray. In total, we analyzed 21 and 20 primary tumors for E-CADHERIN and P120CTN, respectively (Figure 6A). All primary and metastatic PDAC tissues with artifacts or necrosis were excluded. For matched liver metastases, we analyzed 14 patients for E-CADHERIN and 13 for P120CTN. Each sample was assigned to one of four groups for staining pattern: membranous (M), membranous and cytoplasmic (M+C), cytoplasmic (C), or negative (N). Remarkably, liver metastases were significantly more epithelial than paired primary tumors based upon E-CADHERIN localization: only 9 out of 21 primary tumors (42.9%) demonstrated membranous or membranous and cytoplasmic staining whereas 11 out of 14 liver metastases (78.6%) did so (Figure 6A). When we examined only these liver metastases with paired primary tumors, we found that of 8 out 12 paired samples showed a shift from cytoplasmic E-CADHERIN staining at the primary site to membranous and cytoplasmic staining in the liver. Of the other 4 patients, 2 exhibited membranous E-CADHERIN in both the primary and metastatic site; 1 was predominantly cytoplasmic in both locations; and only 1 showed a shift from membranous to cytoplasmic E-CADHERIN from primary tumor to liver metastasis. These results support the premise that MET is important in liver colonization and outgrowth. Analysis of P120CTN revealed that, while there was no significant difference between P120CTN at the primary and metastatic site in terms of localization (Figure 6A), there was a significant increase in membranous staining intensity from primary tumor to liver metastasis (Figure 6B). 17/20 primary tumors showed a maximum of weak staining whereas 6/12 liver metastases expressed moderate or higher membranous staining intensity. This trend is consistent with the observed increase in membranous E-CADHERIN localization observed in liver metastases.
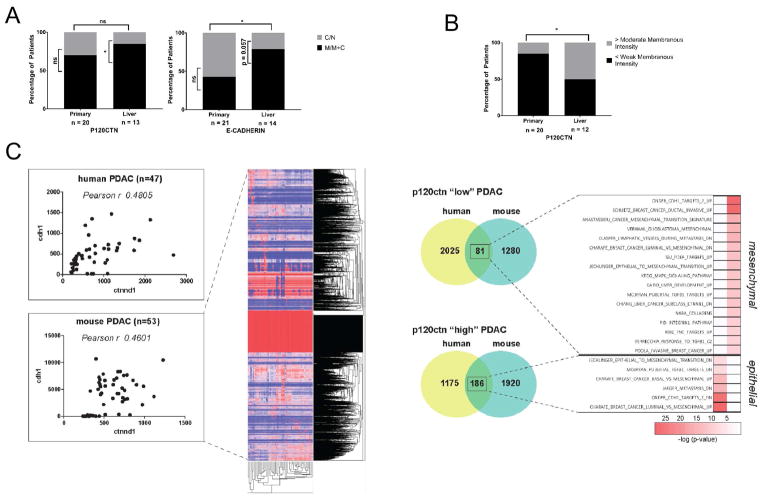
Figure 6. P120CTN and E-CADHERIN expression in human PDAC.
A. Primary PDAC (n = 20 for P120CTN, n = 21 for E-CADHERIN) and paired liver metastases (n = 13 for P120CTN, n = 14 for E-CADHERIN) were analyzed for localization patterns. Whereas 9/21 primary tumors demonstrated M or M+C E-CADHERIN, 11/14 liver metastases demonstrated M or M+C staining. Fisher’s Exact Test for comparison between organs. Binomial Test for comparison within organs. B. Comparison of P120CTN staining intensity between primary PDAC and liver metastases. Whereas 17/20 primary tumors showed, at most, weak membranous staining, 6/12 liver metastases showed moderate staining or higher. Fisher’s Exact Test for comparison between organs. Binomial Test for comparison within organs. C.Correlation of P120CTN (CTNND1) and E-CADHERIN (CDH1) expression in human PDAC (n=47) (www.broadinstitute.org) (Pearson r = 0.4805). Correlation of p120ctn (Ctnnd1) and E-cadherin (Cdh1) expression in murine PDAC (n=53) (Pearson r = 0.4601). Gene expression profiles were divided according to p120ctn expression levels. Genes associated with a p120 high status (> 90 percentile) and a p120 low status (< 10 percentile) shared in human and mouse are enriched in gene sets associated with an epithelial or mesenchymal signature, respectively. Scale bar: 50 μm. *p < .05, **p < .01, ***p < .001, ****p < .0001. See Supplemental Table 7.
p120ctn is associated with epithelial-to-mesenchymal transition in human and mouse PDAC
To evaluate the possible correlation of P120CTN and E-CADHERIN expression in human PDAC (n=47), we utilized publicly available databases (www.broadinstitute.org). In addition, we have generated a unique library of gene expression profiles of 53 primary mouse PDAC cell lines isolated from several different mouse models of PDAC and a variety of PDAC phenotypes. Interestingly, both human (Pearson r = 0.4805) (Collisson et al., 2011) and mouse (Pearson r = 0.4601) PDAC cell lines show a significant correlation of p120ctn (CTNND1/ctnnd1) and E-cadherin (cdh1) expression (Figure 6C). We analyzed gene sets that are enriched in mouse and humans depending upon their p120ctn expression levels (p120ctn “low” and “high” PDAC). Indeed, p120ctn “low” genes are enriched in gene sets that are associated with a mesenchymal signature whereas p120ctn “high” PDAC genes are found in gene sets with an epithelial signature (Figure 6C). These data suggest that p120ctn in human and mouse PDAC influences the epithelial or mesenchymal signature.
P120CTN and E-CADHERIN status in human IPMN
We analyzed a cohort of IPMN patients from across a range of subtypes for P120CTN (Figure S6A and S6B). Overall, human IPMN demonstrated robust epithelial staining with some subtype specific differences. We next looked at E-CADHERIN intensity levels in IPMN patients, also across multiple subtypes (Figure S6C and S6D). While we did not observe statistical differences in overall localization, we did observe statistically significant greater E-CADHERIN intensity in the membranous than in the cytoplasmic compartment.
P120CTN and E-CADHERIN status in human pancreatic neuroendocrine tumors
We evaluated NET samples along with normal adjacent tissue from 2 patients. As we showed in Figure S1, the endocrine compartment is not affected by p120ctn loss in the Pdx1-cre; p120ctnfl/fl mouse. Interestingly, no PNET samples showed membranous P120CTN (Figure S6E). Although 6 samples showed both membranous and cytoplasmic E-CADHERIN localization, no samples showed only membranous staining, and, the intensity of membranous E-CADHERIN staining was less than that in the cytoplasmic compartment (Figure S6F). Lastly, we confirmed that the normal human pancreas expresses both P120CTN and E-CAHDERIN at the cell membrane (Figure S6G), in contrast to PNET (Figure S6H). Therefore, PNET demonstrates dispensability of epithelial properties, which is consistent with the ability of islet cells to retain normal architecture in Pdx1-cre; p120ctnfl/fl mice.
Unbiased analysis of KRASG12D-transformed pancreatic epithelial cells with varying p120ctn status reveals a differential EMT and metastasis signature
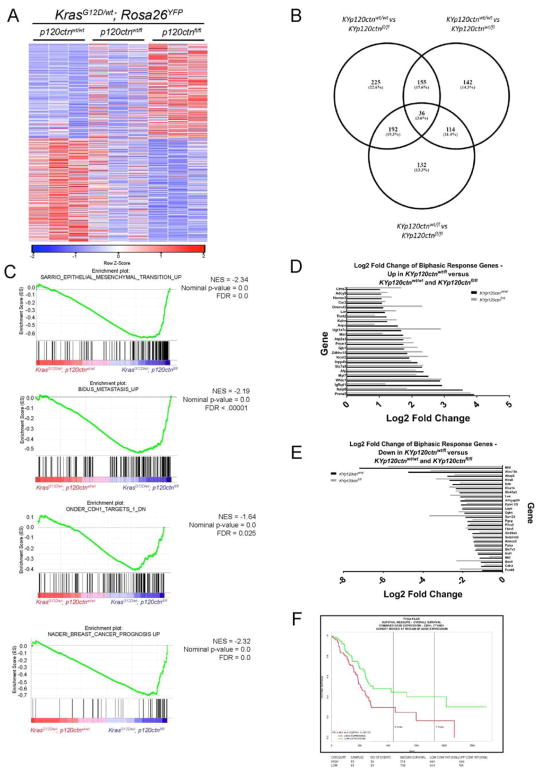
We next performed RNA-Seq on KYp120ctnwt/wt, KYp120ctnwt/fl, and KYp120ctnfl/fl primary cell lines. Homozygous deletion of p120ctn significantly altered 608 genes (Figure 7A; P<0.05 and fold-change>2). Cells with wild-type p120ctn and bi-allelic p120ctn loss showed the largest difference, with 225 differentially expressed genes. Additionally, there was significant overlap between lists of differentially expressed genes – 191 genes were common in the comparison between mono-allelic or bi-allelic loss and wild-type p120ctn (Figure 7B). Gene Set Enrichment Analysis (GSEA) of these data validated our in vivo results by showing that cells lacking p120ctn are enriched for EMT and metastasis gene sets (Figure 7C). Of note, EMT and metastasis gene sets that were enriched in comparisons of KYp120ctnwt/wt and KYp120ctnfl/fl cells were not enriched in comparisons of KYp120ctnwt/wt and KYp120ctnwt/fl cells, suggesting that one allele of p120ctn is sufficient to restrain EMT programs. In addition to restrained EMT, certain genes exhibit a biphasic response to mono-allelic p120ctn loss, with both KYp120ctnwt/wt and KYp120ctnfl/fl cells having a statistically significant difference in the same direction compared to KYp120ctnwt/fl cells. It is possible that the phenotypes described are mediated by some of the genes upregulated (Figure 7D) and downregulated (Figure 7E) in KYp120ctnwt/fl cells compared to both p120ctn wild-type and p120ctn null states. Interestingly, we noted by GSEA that KYp120ctnfl/fl cells were enriched for a positive-prognosis gene signature seen in breast cancer. This observation was consistent with interrogation of TCGA, which revealed that higher p120ctn and Cdh1 levels correlate with worse survival (Figure 7F). Patients with p120ctn and Cdh1 expression below the 50th percentile have median survival of 518 days compared to 684 days for patients with expression above the 50th percentile. Our work may provide a biological rationale for why TCGA data demonstrates a worse outcome with higher epithelial markers as they are conducive for liver colonization and outgrowth. However, tumor cells in a fixed mesenchymal state form undifferentiated tumors but fewer liver metastases.
Figure 7. A single allele of p120ctn restrains EMT programs, and p120ctn and E-cadherin are negative predictors of outcomes.
A. Heatmap depicting 608 significantly altered genes (P<0.05 and fold-change>2) identified through RNA-Seq analysis comparing KYp120ctnfl/wt and KYp120ctnfl/fl cells. B. There is significant overlap in differentially induced genes between comparisons of KYp120ctnwt/wt cells with KYp120ctnfl/wt and KYp120ctnfl/fl cells. C. GSEA plots EMT and metastasis pathways significantly enriched in KYp120ctnfl/fl cells compared to either KYp120ctnwt/wt or KYp120ctnfl/wt cells. Note that mono-allelic p120ctn loss restrains transcriptional programs associated with EMT. D. Genes that are significantly upregulated in KYp120ctnwt/fl cells compared to both KYp120ctnwt/wt and KYp120ctnfl/fl cells. E. Genes that are significantly downregulated in KYp120ctnwt/fl cells compared to both KYp120ctnwt/wt and KYp120ctnfl/fl cells. F. Interrogation of TCGA reveals that P120CTN and E-CADHERIN are negative predictors of outcomes together (shown) or independently (not shown). See Supplemental Figure 7.
Discussion
P120CTN and E-CADHERIN mediate epithelial plasticity and PDAC metastatic organotropism
The ability of epithelial cells to shift between epithelial and mesenchymal identities is critical to fundamental biological processes, such as embryonic development, differentiation, regeneration and carcinogenesis (Thiery et al., 2009). Epithelial cells acquire a mesenchymal phenotype in a gene dose-dependent manner in vitro and in vivo based upon our findings. We show that pancreatic cancer cells lacking both alleles of p120ctn fail to establish any sort of contact with neighboring epithelial cells in 3D culture.
We use multiple independent genetic and transplantation mouse models to highlight the mechanistic basis underlying metastatic organotropism. We demonstrate that liver metastasis is highly dependent upon P120CTN-mediated stabilization of membranous E-CADHERIN or MET, whereas the lung appears permissive to colonization by cells that are not MET-capable. Of note, PDAC patients with recurrent pulmonary metastases demonstrate significantly longer overall survival compared to patients with recurrence at other sites (40.3 vs. 20.9 months) (Yamashita et al., 2015). Along with our data it is tempting to speculate whether the aggressiveness of PDAC is dependent upon epithelial plasticity and not exclusively upon a mesenchymal/de-differentiated phenotype, although we certainly acknowledge the importance of this as well (Genovese et al. 2017).
Introduction of a mutant KrasG12D allele in combination with heterozygous p120ctn loss accelerates PanIN formation and PDAC progression. Importantly, in this PDAC progression model, we observed the classical PanIN-PDAC sequence as well as the IPMN/MCN-PDAC sequence when both mutations were targeted to all (acinar, ductal and endocrine) lineages with Pdx1-cre. However, in our orthotopic transplantation experiments in which we utilized purified pancreatic ductal cells harboring the same genetic mutations, the IPMN/MCN pathway to PDAC was also present, suggesting that the ductal compartment may represent the cell of origin of cystic neoplasms (IPMNs, MCNs) as suggested previously (von Figura et al., 2014). During PDAC progression, in all our model systems (autochthonous and orthotopic mouse models) we observed that at least one p120ctn allele is expressed in liver metastasis. Interestingly, the retained expression of P120CTN is sufficient to stabilize E-CADHERIN at the cell membrane. Removing both alleles of p120ctn results in the absence of liver metastases while lung metastases prevail. This is also true in an E-CADHERIN-dependent mouse model of PDAC (Pdx1-Flp; FSF-KrasG12D/wt; FSF-R26GAG-CreERT2;CDH1fl/fl). These observations were confirmed by analysis of P120CTN and E-CADHERIN localization patterns in lung and liver metastases from two previously published PDAC models – the KPCY mouse (Rhim et al., 2012) and inducible tet-on Elastase-KRASG12V mouse (Guerra et al., 2007).
The crucial role of P120CTN-mediated stabilization in driving liver metastasis was observed and underscored in two new genetic mouse models. First, we demonstrated that the metastatic switch from liver to lung in KPCYp120ctnwt/fl mice is characterized by P120CTN-deficient lung metastases arising from a mixed epithelial-mesenchymal primary tumor. Second, we demonstrated that in a mosaic E- CADHERIN knockout model of PDAC, the liver metastases are characterized by overwhelming E- CADHERIN -positivity whereas the lung metastases are overwhelmingly E-CADHERIN-negative. Third, analysis of human liver metastases shows a shift to membranous E-CADHERIN and stronger P120CTN membranous staining compared to primary PDAC.
Epithelial identity and plasticity are important as a new principle in PDAC metastatic organotropism
For a tumor cell to form a metastatic lesion, it must undergo several steps, including delamination from the primary epithelial layers, intravasation, survival in circulation, extravasation, survival at the metastatic site, and reactivation and outgrowth (Giancotti, 2013; Massagué and Obenauf, 2016; Massagué et al., 2017). Our data show that cells with or without epithelial plasticity can accomplish all the steps up to and including survival in circulation since we observe lung metastasis independent of membranous P120CTN or E-CADHERIN localization. We used an unbiased RNA-seq approach and demonstrated that p120ctn deletion in pancreatic tumor cells may lead to the induction of pathways associated with metastasis based upon GSEA. Furthermore, in comparing bi-allelic p120ctn loss and mono-allelic p120ctn loss, GSEA reveals induction of EMT in the former, but not in the latter.
We and others have published on the paradoxical observation that metastasis may be enhanced by certain epithelial properties, including E-CADHERIN expression. Tail vein injection of breast cancer cells with knockdown of the EMT transcription factors PRRX1 and Twist1 showed increased lung metastatic burden (Ocaña et al., 2012). In a skin cancer model, persistent induction of Twist was seen to inhibit metastatic outgrowth, whereas turning off Twist expression promoted it (Tsai et al., 2012). In pancreatic cancer, liver metastatic outgrowth was associated with the acquisition of an epithelial phenotype (Aiello et al., 2016a). Additionally, we have demonstrated that two isoforms of the transcription factor Prrx1 (Reichert et al., 2013b), Prrx1A and Prrx1B, promote MET and EMT, respectively (Takano et al., 2016). PRRX1A, the MET-promoting isoform, is significantly upregulated in human PDAC liver metastases compared to primary tumor. In addition, using orthotopic transplantation of Prrx1A- or Prrx1B-inducible mouse pancreatic cancer cells, we found that either induction of Prrx1A (i.e., MET) or repression of Prrx1B (i.e., turning off EMT) promoted both the incidence of liver metastasis as well as the size of the lesions. The microenvironment of the liver may therefore demand acquisition of an epithelial phenotype for reasons that do not exist in the lung. It is possible that a proclivity towards acquiring an epithelial state in cancer cells occurs prior to actual metastatic colonization (del Pozo Martin et al., 2015; Korpal et al., 2011; Tsai et al., 2012). Intriguingly, in this issue, Aiello et al. demonstrate that pancreatic cancer cells undergo either “classical” EMT or “partial” (p-EMT), with the latter being characterized by retained epithelial gene expression. This p-EMT predisposes to invasion in a collective manner. In light of our observation that EMT-MET plasticity is involved in regulating metastatic organotropism, this work informs our understanding of how disseminated tumor cells regulate their position on the EMT-MET spectrum.
We believe that epithelial plasticity under the governance of P120CTN is critical to the establishment of pancreatic cancer cellular colonization in the liver or lung (Figure S7). Subsequent to initial colonization and further outgrowth, maintenance of epithelial identity is critical in the liver but not required in the lung. Our delineation of this mechanism underlying pancreatic cancer metastatic organotropism provides a window of opportunity to tackle new perspectives in therapeutic innovation and customization.
Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Anil K. Rustgi (anil2@pennmedicine.upenn.edu).
Experimental Model and Subject Details
General Animal Surgery
All animal work was approved by the University of Pennsylvania Institutional Animal Care and Use Committee under protocol #804959. For all procedures, animals are anesthetized using isoflurane (1–4%) delivered through an isoflurane vaporizer and face mask. Adequate anesthesia is determined by loss of hind limb retraction to stimulation. Buprenorphine was provided for analgesia. Athymic female nude mice between the ages of 8–12 weeks were used for all transplantation studies (Taconic Cat#NCRNU-F).
Orthotopic Transplantation
After anesthesia and proper sterile preparation of the abdomen, a small (5–10mm) incision was made over the left upper quadrant of the abdomen. After exposure of the peritoneal cavity, the spleen was located and exteriorized onto a sterile field surrounding the incision site. 500,000 cells suspended in 50 μl DMEM (10 or 20% FBS) were then injected into the tail of the pancreas via an insulin syringe. A cotton can be held over the injection site for 1 min to ensure that no cancer cells leak into the peritoneal cavity, though this is not necessary with the cells used in this study. Successful injection of the tail of the pancreas was be confirmed by the appearance of a liquid bleb in the pancreas, without leakage of contents into the peritoneum. Afterwards, the spleen and pancreas as be placed back into the peritoneal cavity and ventral incision closed with 4-0 sterile absorbable suture in a double layer closure. A useful published protocol can be found in Aiello, Rhim, and Stanger (2016).
Intraportal Vein Injection
After anesthesia and proper sterile preparation of the abdomen, a small (10–15mm) incision will be made on the upper median of the abdomen, and followed by mobilization of the duodenum to liberate the portal vein. Tumor cells suspended in 200 μl 20% FBS DMEM will then be injected into the portal vein using a needle. After removal of the needle, a cotton swab will then be held over the injection site for 3 min to stop bleeding. The injection procedure will be considered successful if there is no post-injection bleeding from the puncture site or recoil of tumor cells within the injection canal or tumor cell spread into the abdominal cavity. After tumor cell injection the intestine will be repositioned and the ventral incision will then be closed with 4-0 absorbable suture in a double layer closure using a running suture technique.
Retroorbital Injection
Generally anesthetized mice are given local ophthalmic anesthesia (0.5% proparacaine hydrochloride) on the eye that will receive the bolus of cells. Cells are injected at a 30° angle on the medial side and the needle is slowly and smoothly retracted. 7.5 × 105 tumor cells suspended in 200 μl 20% FBS DMEM were injected into the retro-orbital sinus of 10 weeks old athymic nude mice. Lungs were harvested 21 days after injection.
Pancreatitis
Acute pancreatitis was induced as described previously (Reichert et al., 2013c). Mice were starved starting 18 hours prior to induction while given access to water ad libitum. On days -1 and day 0, mice were intraperitoneally injected once an hour for eight hours with either 200 μl of sterile PBS (control group) or 200 μl of 10 μg/mL cerulein (Sigma Cat#C9026) (experimental group). On days 1, 3, and 7, pancreata were harvested and fixed.
Human Tissues
De-identified and IRB-exempt human pancreatic tissue was obtained from the Technical University of Munich Institute of Pathology. In detail, 26 cases of resected pancreatic adenocarcinomas and matched liver metastases were included in the cohort. Mean patient age was 68.4 years (range: 46 to 82). 13 patients were female 11 were male. According to the 7th TNM classification, one tumor was pT1, 24 were pT3 and one was pT4. 6 patients were pN0, 18 were pN1, per definition all cases were pM1. 5 patients received neoadjuvant chemotherapy. For the creation of tissue microarrays, tumor cores (2 cores of 1.5mm diameter) were punched out of formalin-fixed paraffin embedded (FFPE) tissue blocks and arranged in two newly generated paraffin block using a tissue microarrayer (Beecher Instruments, Sun Praierie, USA) after tumor areas were marked by a board certified pathologist. Use of the tissue was approved by the institutional review board of the Technical University Munich (N° 403/17 S).
De-identified and IRB-exempt human IPMN slides were obtained from Dr. Volkan Adsay at Emory University. Deidentified and IRB-exempt human PNET slides were obtained from Dr. Andrea Califano at Columbia University.
Method Details
Primary Pancreatic Cell Line Generation and Culture
Primary pancreatic mouse cells were generated as follows: pancreata were harvested from mice and placed in G solution (1 L HBSS [Invitrogen Cat#14175079] + 10 mL penicillin-streptomycin [Invitrogen Cat#15140122] + 10 mL Fungizone [Invitrogen Cat#15290018] + 400 μl 0.11M CaCl2]. Pancreata were physically dissociated with scissors and then underwent three washes (allow cells to settle, aspirate G solution and floating tissues, and the resuspend in 25 mL G solution). In the last wash, all G solution was aspirated save for a small amount with cells and remaining tissue was resuspended in 25 mL of a 1 mg/mL solution of collagenase type V (Sigma Aldrich Cat#9263) diluted in DMEM/F12 (Gibco Cat#11330-032). This mixture was incubated in a bottle with a magnet on a stir plate at 37 C for 20–30 minutes. Following digestion, the mixture was centrifuged, trypsinzed, resuspended, filtered through a 40 um mesh, and plated on collagen gels (type I rat tail collagen [Corning Cat#354236] mixed with 10X PBS and 1 N NaOH) in primary pancreatic cell media (see below). All tools, bottles, and magnets were autoclaved prior to use, and the G solution and collagenase mix were filter sterilized.
Primary pancreatic cells were grown in a 12-component special media composed of (amounts are for a 500 mL bottle) 500 mL DMEM/F12 (Gibco Cat#11330-032), 25 mL Nu-Serum (BD Biosciences Cat#355104), 2.5 g D-Glucose (Sigma-Aldrich Cat#G5400), 0.66 g Nicotinamide (Sigma-Aldrich Cat#N3376), 5 mL penicillin-streptomycin (Invitrogen Cat#15140122), 2.5 mL ITS+ Premix (BD Biosciences Cat#354352), 100 ng/mL cholera toxin (Sigma-Aldrich Cat#C8052), 20 ng/mL mouse recombinant EGF (Gibco Cat#PMG8043), 1 μM dexamethasone (Sigma-Aldrich Cat#1756), 5nM T3 (Sigma-Aldrich Cat#T6397), 900 μl bovine pituitary extract (Gemini BioProducts Cat#500-102), and 0.5 g STI (Gibco Cat#17075029).
Passage of primary pancreatic cells cultured on collagen plates was performed by digestion of collagen gels in Collagenase Type IV (Worthington Biochemical Cat#LS004189) diluted in DMEM/F12 (Gibco Cat#11330-032).
A useful methods paper for the generation and culture of primary pancreatic cells can be found in Reichert et al. (2013).
Cells were infected with lentiviral-Cre-SD (see “Lentiviral Virus Generation and Transduction” below) and sorted for YFP. All nine lines used in this paper (KrasG12D/wt; p120ctnwt/wt; Rosa26YFP: 611, 793, 801; KrasG12D/wt; p120ctnwt/fl; Rosa26YFP: 363, 732, 866; KrasG12D/wt; p120ctnfl/fl; Rosa26YFP: 288, 690, 791) are detailed in reagent table.
Lentiviral Virus Generation and Transduction
LV-Cre-SD (Addgene #12105, gift from Inder Verma) (Pfeifer et al., 2001) was made using pCMVR8.74 (Addgene #22036, gift from Didier Trono) and pMD2.G (Addgene #12259, gift from Didier Trono) packaging. HEK293T cells were plated at 6×106 in a 10 cm plate in 10% FBS (Sigma-Aldrich Cat#F2442) in DMEM (Corning #MT10013CV) the day prior to transfection. For transfection, two cocktails were made (quantities are per 10 cm plate). First, 10 μg of LV-Cre-SD was mixed with 6.5 μg pCMV dR8.74 and 3.5 μg pMD2 VSVG in 1 mL of Opti-MEM-I (Gibco #31985070). Second, 1 mL of Opti-MEM-I with 30 μl of Lipofectamine LTX (Invitrogen Cat#15338100). After 5 minutes of incubation at room temperature, the latter was added to the former and left at room temperature for 30 minutes. 2 mL of the mix was then added to each 10 cm plate. After either a 3–6 hour incubation or overnight incubation, media was replaced and harvested 24 or 48 hours later with a second 72-hour incubation. For transduction, 400 μl of viral supernatant was placed on each collagen-coated well of a 6-well plate of PDCs in two sequential spin infections. YFP sorted cells were continued to be cultured on collagen.
3D Culture and Immunofluorescence
Primary pancreatic epithelium was seeded into 4-well chamber slides (Fisher Scientific Cat#12-565-21) and allowed to grow for 5 days. First, a collagen solution was prepared as follows (amounts are for a full 4-well chamber slide): 294 μl L-Glutamine (Gibco Cat#25030) + 77 μl sodium bicarbonate (Gibco Cat#25080) + 245 μl MEM (Invitrogen Cat#11430030) + 816 μl bovine collagen (Advanced BioMatrix Cat#5010-50ML) + 49 μl HEPES (Sigma-Aldrich Cat#H0887) + 1020 μl water. Second, 225 μl of this solution was allowed to solidify on the bottom the chamber slides at 37°C for 30–60 minutes in an oven at atmospheric conditions. Third, a mixture of 350 μl of collagen solution was mixed with 5000 cells in 50 μl of primary pancreatic media and placed on top of the acellular collagen layer with another 30–60 minutes of solidification in a 37°C oven at atmospheric con ditions. Fourth, 500 μl primary pancreatic media was placed on top and replaced every 2–3 days.
For 3D culture immunofluorescence, samples were fixed with 4% PFA (room temperature for 30 minutes) and permeabilized (gently rocking at 30 minutes room temperature or 4 C overnight) with a solution composed of 0.35 g fish skin gelatin (Sigma Cat#G7765) and 250 μl Triton X-100 diluted in 50 mL of PBS with Ca2+ and Mg2+ (PBS+) (Invitrogen Cat#14040). Primary antibodies were diluted in permeabilization solution and incubated for 3 hours at room temperature of 4 C overnight. Wash samples with permeabilization solution 3 times for 10 minutes and incubate in secondary antibodies/DAPI diluted in permeabilization solution (3 hours at room temperature of 4 C overnight). Following additional permeabilization solution and PBS+ washes, incubate for 30 minutes in 4% PFS. Wash with PBS+; remove chambers that surround samples; mount coverslip; and image.
A useful, step-by-step protocol can be found in Reichert et al. (2013) for both 3D culture growth and immunofluorescence staining.
Antibodies used can be found in “Primary and Secondary Antibody Dilutions.”
CTC Isolation and Immunofluorescence
1 ml of blood from KPCY mice was obtained using a heparinized insulin syringe. Blood was transferred immediately to a 10 cm plate containing PBS. Single and clustered CTCs were identified under fluorescent microscopy and handpicked using a 10ul pipette and tip. Each cell was placed into 25 μl of a 1:1 Matrigel:PBS solution on glass coverslip bottom plate. After allowing Matrigel/PBS mix to harden for 1 hour at 37 C, cells were fixed with 4% PFA and then washed 3 times with PBS.
Staining of these Matrigel-embedded CTCs was done as follows. First, 0.5% Triton (diluted in PBS) was used to permeabilize for 10 minutes at 4 C followed by 3 PBS washes. Second, 10% donkey serum was used to block. Third, primary antibodies were diluted in 10% donkey serum and incubated overnight at 4 C. On day 2, following further washes in 10% donkey serum, secondary antibodies were incubated overnight. On day 3, two further washes in 10% donkey serum and two further PBS washes were done before DAPI staining.
Antibodies used can be found in “Primary and Secondary Antibody Dilutions.”
Cloning
p120ctn1A was PCR amplified from of a Mammalian Gene Collection clone (Cat# MMM1013-202859190) with primers having NHEI and EcoRI enzyme restriction sites and then ligated into PiggyBac plasmid EF1α-MCS-IRES-Neo (Systems Bioscience Cat#PB533A-2). The sequence was validated through the University of Pennsylvania Genomics Analysis Core.
PiggyBac Transfection
Transfections were performed using Lipofectamine 3000 (Invitrogen Cat#L3000008) per manufacturer’s instruction with changes to ratio of DNA to transfection components. Briefly, cells were plated to 70–90% confluency. On day of transfection, media was replenished and 125 μl of mixture A (3 μg PiggyBac [Systems Bioscience Cat#PB533A-2] vector [either empty control vector or p120ctn1A rescue vector], 0.6 μg Hyperactive Piggybac Transposase, and 7.2 μl P3000 Reagent [part of Invitrogen Cat#L3000008] diluted in 125 μl Opti-MEM) was added to mixture B (16.2 μl Lipofectamine 3000 diluted in 125 μl Opti-MEM [Gibco #31985070]) and allowed to incubate for 5 minutes. Fresh media was placed on each well and transfection solution added. Media was replaced 24 hours later, and G-418 antibiotic selection (Gold Biotechnology Cat#G-418-5]) started 48 hours post-transfection at 100 μg/mL. Cells were propagated indefinitely in G418 at 70 μg/mL.
Primary and Secondary Antibody Dilutions
For western blots, primary antibody dilutions were as follows: 1:1000 P120CTN (BD Biosciences Cat#610134), 1:1000 E-CADHERIN (Cell Signaling Cat#3195), 1:5000 β-actin (Sigma Cat#A5316). Secondary antibody dilutions were 1:5000-1:15000 for anti-mouse (LiCor Cat#925-68070) and anti-rabbit (LiCor Cat#926-32211).
For tissue immunohistochemistry, primary antibody dilutions were as follows: 1:400 P120CTN (BD Biosciences Cat#610134), 1:400 E-CADHERIN (Cell Signaling Cat#3195), and 1:2500 GFP (Abcam Cat#13970). Secondary antibody dilutions were 1:200 for biotin-conjugated anti-mouse (Vector Labs Cat#BA-2001), anti-rabbit (Vector Labs Cat#BA1000), and anti-chicken (Vector Labs Cat#BA-9010).
For tissue immunofluorescence, primary antibody dilutions were as follows: 1:250 to 1:500 P120CTN (BD Biosciences Cat#610134), 1:50 to 1:500 E-CADHERIN (Cell Signaling Cat#3195), 1:250 GFP (Abcam Cat#13970), 1:100 amylase (Santa Cruz Cat#12821). Secondary antibody dilutions were 1:600 for Cy-2 (usually anti-chicken, Jackson ImmunoResearch Cat#703-225-155) and Cy-3 (usually anti-mouse, Jackson ImmunoResearch Cat#715-165-150) conjugated secondary antibodies and 1:300 for Cy-5 (usually anti-rabbit, Jackson ImmunoResearch Cat#711-175-152) conjugated secondary antibodies.
For 3D culture immunofluorescence, primary antibody dilutions were as follows: 1:200 CTTN (Abcam Cat#81208), 1:100 TKS5 (Merck Cat#09-268), 1:200 αSMA (Sigma Cat#A2547), and 1:100 ACTIN-1 (Thermo-Fisher Cat#A22287).
For CTC immunofluorescence, primary antibody dilutions were as follows: 1:200 P120CTN (BD Biosciences Cat#610134), 1:250 E-CADHERIN (Clonetech Cat#M108), 1:200 β-CATENIN (Cell Signaling Cat#8480), 1:1000 CK-19 synthesized as described (Ito et al., 2000; Tanimizu et al., 2003; Zong et al., 2009), and 1:250 YFP (Abcam Cat#6673). All secondary antibody dilutions used for CTC immunofluorescence were 1:200.
Western Blot
Equal amounts of protein were run in reducing conditions on SDS-Page gels and transferred on Immobilon-FL PVDF membrane (Millipore Cat#IPFL00010). After blocking in Odyssey Blocking Buffer (Li-Cor Biosciences Cat#927-40003), membranes were incubated overnight at 4° in antibody diluted in Odyssey Blocking Buffer. After PBS-Tween washes, membranes were incubated in the dark at room temperature in the appropriate secondary antibody conjugated to an infrared fluorophore. Following another round of PBS-Tween washes and then PBS washes, membranes were imaged on Li-Cor Odyssey.
Tissue Immunohistochemistry
Tissue was fixed in zinc formalin followed by 70% ethanol dehydration. Following paraffin embedding, sectioning, and mounting, slides were incubated at 60 degrees for 30 minutes followed by xylene and ethanol washes. For antigen retrieval, slides were placed in pH 6 citrate buffer and microwaved for 15 minutes followed by 15 minutes cool down. Washes were performed as follows: 5 minutes PBS; 15 minutes 3% hydrogen peroxide; 5 minutes water; 15 minutes avidin D blocking; fast PBS wash; 15 minutes biotin blocking; fast PBS wash; 15 minutes in T-20 blocking agent (Fisher Cat#PI-37539); incubation overnight at 4°C in antibody diluted in PBT [43.5 mL water + 5 mL 10X PBS + 1 mL 10% TritonX-100 + 500 μl 10% BSA]. On day 2, slides were processed as follows: two 5-minute washes in PBS; 30 minutes in biotin-conjugated secondary antibody (anti-mouse [Vector Labs Cat#BA-2001], anti-rabbit [Vector Labs Cat#BA-1000], anti-mouse [Vector Labs Cat#BA-2001]) at 37°C; 2 5-minute washes in PBS; 30 minutes in ABC Solution (Vector Labs Cat#PK-6100) at 37°C; 5 minute PBS wash; 5 minute water wash; DAB exposure (Vector Labs Cat#SK-4100); 5 minutes under running tap water; hematoxylin counterstaining; ethanol dehydration and xylene washes followed by cover slipping.
Tissue Immunofluorescence
Tissue was fixed in zinc formalin followed by 70% ethanol dehydration. Following paraffin embedding, sectioning, and mounting, slides were incubated at 60 degrees for 15–30 minutes followed by xylene and ethanol washes. For antigen retrieval, slides were placed in ph 6 citrate buffer and either microwaved for 15 minutes (followed by 15 minutes cool down) or put in a pressure cooker for 2 hours. Following antigen retrieval, slides were washed in water and blocked at room temperature for 1 hour with 0.3% TritonX-100 PBS w/5% donkey serum (Jackson ImmunoResearch Laboratories Cat#017-000-121). Antibodies were diluted in 0.3% TritonX-100PBS w/5% donkey serum and incubated overnight a 4°C. On day 2, slides were washed with .05% TBS-Tween 3 times for 10 minutes and then incubated at room temperature for 2 hours in secondary antibody (1:600 for Cy2- and Cy3-conjugated antibodies, 1:300 for Cy-5 conjugated). After a final set of 3 10 minute washes in .05% TBS-Tween and TBS once for 5 minutes,
Relative Amylase Area
Relative amylase area was scored on a similar basis to a previously-published study (Morris et al., 2010). Immunofluorescence staining was performed using anti-amylase antibody (sc-12821) and DAPI (Life Technologies). The intensity of amylase staining and DAPI positive nuclei across all specimen was measured by iVision-Mac software (BioVision Technologies, Exton, PA, USA). A ratio of amylase intensity and DAPI intensity was calculated. Overall, 3 high power fields per mouse were used across 3 mice per genotype (Pdx1-cre; p120ctnwt/wt, Pdx1-cre; p120ctnwt/fl, Pdx1-cre; p120ctnfl/fl) per age group (10–20, 30–40, 50–60 weeks of age).
Pancreatitis Scoring
Pancreatitis was scored on the basis of a previously-published criteria covering edema, inflammation, vacuolization, and necrosis (Rongione et al., 1997). Three mice per genotype (Pdx1-cre; p120ctnwt/wt, Pdx1-cre; p120ctnwt/fl, Pdx1-cre; p120ctnfl/fl) were randomized to 4 groups: a control group that received sterile PBS and three experimental groups that were harvested at days 1, 3, and 7 post-induction (see “Pancreatitis” in Experimental Model and Subject Details). 3 high-powered fields were analyzed per mouse per condition and averaged. One mouse in the day 1 and day 3 group of the Pdx1-cre; p120ctnfl/fl genotype were excluded from final analysis. Statistical comparison was performed using multiple T-tests per row with a Holm-Sidak correction and assumed same standard deviation.
Human Tissue Scoring
Stained microarrays were scanned on an Aperio ScanScope slide scanner. Images were transferred to the Institute of General Pathology and Pathological Anatomy (Technical University of Munich, Munich, Germany) for analysis. Each specimen was graded for localization status. For P120CTN, samples were classified as very weak, weak, weak/moderate, moderate/strong, or strong. A specimen was classified in the highest group it fell in.
RNA-Sequencing Differential Expression Analysis
RNA was isolated using the Qiagen RNEasy Mini Kit (Cat#74104) from in vitro lentiviral-Cre recombined KrasG12D; p120ctnwt/wt, KrasG12D; p120ctnwt/fl, and KrasG12D; p120ctnfl/fl cell lines and analyzed by RNA-sequencing analysis at Hudson BioAlpha (Birmingham, Alabama). The FastQC software was applied on raw fastq files to examine the sequence quality. Paired-end reads were aligned to the mouse genome NCBIM37 using Tophat software. Alignments were performed using default parameters. Transcript expression quantification was performed using cufflinks. Differential gene expression analysis was performed using cuffdiff 6. Counts for each gene were computed by means of HTSeq Python package, using the annotation of the Ensembl 66 mouse genes and only reads that mapped to exons. Gene set enrichment analysis (GSEA) was performed on RNA-seq data using Broad Institute guidelines as previously established. Significantly associated gene sets had nominal p-values≤0.05 and false discovery rates (FDR)≤0.25 with 1000 permutations and weighted enrichment scoring (Subramanian et al., 2005). The RNA-seq data have been deposited at Gene Expression Omnibus accession number GSE96729: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE96729.
Quantification and Statistical Analysis
Replicates, statistical tests, and experimental results (quantification, mean, standard deviation) are indicated in figure legends for the respective experiments.
Data and Software Availability
Statistical tests were performed in Graphpad Prism. Immunofluorescence and immunohistochemical images were adjusted for contrast, color, and tone in Adobe Creative Cloud Photoshop. Western blots were imaged on the Odyssey Li-Cor software and manipulated using the built-in image processing software. Relative amylase area quantification was performed on iVision. No specific portion of any given image for immunofluorescence, immunohistochemistry, H&E’s, or Western blot was ever manipulated differently than other portions of the same image.
Supplementary Material
Highlights.
Independent mouse models reveal that liver metastasis requires P120CTN/E-CADHERIN
P120CTN/E-CADHERIN interaction is not required for lung metastasis in these models
p120ctn isoform 1A restoration in p120ctn null cells reinstates liver tropism
Human liver metastases exhibit more epithelial properties relative to primary PDAC
Acknowledgments
We express our gratitude to the NIH/NIDDK P30-DK050306 Center for Molecular Studies in Digestive and Liver Diseases (and Molecular Pathology and Imaging, H-MARC, Cell Culture, and Transgenic and Chimeric Mouse Cores) as well as the Flow Cytometry/Sorting Core. We thank the following funding sources: F30 CA180601 (BB and AKR), R01 DK060694 (MR and AKR), American Cancer Society (AKR), National Pancreas Foundation (MR), German Cancer Aid Foundation (Max Eder Program, Deutsche Krebshilfe 111273, MR), KKF Program (School of Medicine, Klinikum rechts der Isar, Technical University Munich, MR), AGA-Actavis Research Award in Pancreatic Disorders (MR), Asociación Española Contra el Cáncer (Programa Avanzado en Oncología 2016, LM), Societat Catalatana de Digestologia (Beca d’estada a l’estranger 2015, LM), and F32CA221094 (JP). Part of this work was funded by the German Consortium for Translational Cancer Research (DKTK) and the BMBF-funded PANC-STRAT consortium (grant no. 01ZX1305 and 01ZX1605 to WW). We wish to thank Dr. Nicole Aiello (University of Pennsylvania, Laboratory of Dr. Ben Stanger) for generously providing us tissue from KPCY mice; Dr. Andrea Califano (Columbia University) for generously providing us human PNET samples; and Dr. Carmen Guerra (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain) for generously providing us Elastase-driven inducible KRASG12V PDAC tissue.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Author Contributions
MR, BB, and AKR conceived the experiments and wrote the manuscript. MR, BB, LM, JP, KF, LS, KS, RM, and MW performed experiments and analyzed data. AR, NVA, MK, WW, KS, DS, and GS provided reagents. AKS, AMS BP, BM, and NVA analyzed data. AKR secured funding and supervised the design, execution, and analysis of all work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aiello, Bajor DL, Norgard RJ, Sahmoud A, Bhagwat N, Pham MN, Cornish TC, Iacobuzio-Donahue CA, Vonderheide RH, Stanger BZ. Metastatic progression is associated with dynamic changes in the local microenvironment. Nat Commun. 2016a;7:12819. doi: 10.1038/ncomms12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello NM, Rhim AD, Stanger BZ. Orthotopic Injection of Pancreatic Cancer Cells. Cold Spring Harb Protoc. 2016b doi: 10.1101/pdb.prot078360. pdb.prot078360. [DOI] [PubMed] [Google Scholar]
- Aiello NM, Brabletz T, Kang Y, Nieto MA, Weinberg RA, Stanger BZ. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547:E7–E8. doi: 10.1038/nature22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello, et al. THIS ISSUE OF DEV CELL [Google Scholar]
- AmericanCancerSociety. American Cancer Society. Cancer Facts & Figures 2015. Atlanta: American Cancer Society; 2015. [Google Scholar]
- Blouw B, Patel M, Iizuka S, Abdullah C, You WK, Huang X, Li JL, Diaz B, Stallcup WB, Courtneidge SA. The Invadopodia Scaffold Protein Tks5 Is Required for the Growth of Human Breast Cancer Cells In Vitro and In Vivo. PLoS One. 2015;10:e0121003. doi: 10.1371/journal.pone.0121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin Is an Essential Regulator of Matrix Metalloproteinase Secretion and Extracellular Matrix Degradation in Invadopodia. Cancer Res. 2007;67:4227–4235. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- Davis MA, Reynolds AB. Blocked Acinar Development, E-Cadherin Reduction, and Intraepithelial Neoplasia upon Ablation of p120-Catenin in the Mouse Salivary Gland. Dev Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- del Pozo Martin Y, Park D, Ramachandran A, Ombrato L, Calvo F, Chakravarty P, Spencer-Dene B, Derzsi S, Hill CS, Sahai E, et al. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. Cell Rep. 2015;13:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deramaudt TB, Takaoka M, Upadhyay R, Bowser MJ, Porter J, Lee A, Rhoades B, Johnstone CN, Weissleder R, Hingorani SR, et al. N-cadherin and keratinocyte growth factor receptor mediate the functional interplay between Ki-RASG12V and p53V143A in promoting pancreatic cell migration, invasion, and tissue architecture disruption. Mol Cell Biol. 2006;26:4185–4200. doi: 10.1128/MCB.01055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Figura G, Fukuda A, Roy N, Liku ME, Morris JP, IV, Kim GE, Russ HA, Firpo MA, Mulvihill SJ, Dawson DW, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong STC, Choi H, El Rayes T, Ryu S, Troeger J, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaup A, Cazes A, Le Jan S, Philippe J, Connault E, Le Coz E, Mekid H, Mir LM, Opolon P, Corvol P, et al. Angiopoietin-like 4 prevents metastasis through inhibition of vascular permeability and tumor cell motility and invasiveness. Proc Natl Acad Sci U S A. 2006;103:18721–18726. doi: 10.1073/pnas.0609025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancotti FG. Mechanisms Governing Metastatic Dormancy and Reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic Pancreatitis Is Essential for Induction of Pancreatic Ductal Adenocarcinoma by K-Ras Oncogenes in Adult Mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Hidalgo M. Pancreatic Cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol Cell Biol. 2014;34:3229–3243. doi: 10.1128/MCB.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, et al. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N, Lee SH, Liu S, Li GY, Smith MJ, Reichardt LF, Ikura M. Dynamic and Static Interactions between p120 Catenin and E-Cadherin Regulate the Stability of Cell-Cell Adhesion. Cell. 2010;141:117–128. doi: 10.1016/j.cell.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JP, Pan FC, Akiyama H, Wright CVE, Jensen K, et al. Identification of Sox9-Dependent Acinar-to-Ductal Reprogramming as the Principal Mechanism for Initiation of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KM, Ward JM, Yew CCK, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Chang DK, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Batlle E, Gomis RR. Understanding the molecular mechanisms driving metastasis. Mol Oncol. 2017;11:3–4. doi: 10.1002/1878-0261.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J, Cano DA, Sekine S, Wang SC, Hebrok M, Schmid R. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120:508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocaña OH, Córcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-catenin mediates inflammatory responses in the skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M, Song W, Pasolli HA, Williams SE, Fuchs E. Loss of p120 catenin and links to mitotic alterations, inflammation, and skin cancer. Proc Natl Acad Sci. 2008;105:15399–15404. doi: 10.1073/pnas.0807301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: Efficient gene targeting in vivo. Proc Natl Acad Sci. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest. 2011;121:4572–4578. doi: 10.1172/JCI57131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert Takano, Heeg, Bakir B, Botta GP, Rustgi AK. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc. 2013a;8:1354–1365. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert Takano, Burstin V, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013b;27:288–300. doi: 10.1101/gad.204453.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Takano S, von Burstin J, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013c;27:288–300. doi: 10.1101/gad.204453.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongione A, Kusske A, Kwan K, Ashley S, Reber H, McFadden D. Interleukin 10 reduces the severity of acute pancreatitis in rats. Gastroenterology. 1997;112:960–967. doi: 10.1053/gast.1997.v112.pm9041259. [DOI] [PubMed] [Google Scholar]
- Schönhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, Eser S, Feyerabend TB, Paul MC, Eser P, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals DF, Azucena EF, Pass I, Tesfay L, Gordon R, Woodrow M, Resau JH, Courtneidge SA. The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell. 2005;7:155–165. doi: 10.1016/j.ccr.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Short SP, Kondo J, Smalley-Freed WG, Takeda H, Dohn MR, Powell AE, Carnahan RH, Washington MK, Tripathi M, Payne DM, et al. p120-Catenin is an obligate haploinsufficient tumor suppressor in intestinal neoplasia. J Clin Invest. 2017;127:4462–4476. doi: 10.1172/JCI77217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley-Freed WG, Efimov A, Burnett PE, Short SP, Davis MA, Gumucio DL, Washington MK, Coffey RJ, Reynolds AB. p120-catenin is essential for maintenance of barrier function and intestinal homeostasis in mice. J Clin Invest. 2010;120:1824–1835. doi: 10.1172/JCI41414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stairs DB, Bayne LJ, Rhoades B, Vega ME, Waldron TJ, Kalabis J, Klein-Szanto A, Lee JS, Katz JP, Diehl JA, et al. Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene. Cancer Cell. 2011;19:470–483. doi: 10.1016/j.ccr.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano S, Reichert M, Bakir B, Das KK, Nishida T, Miyazaki M, Heeg S, Collins MA, Marchand B, Hicks PD, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 2016;30:233–247. doi: 10.1101/gad.263327.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa M, Saito H, Tsujimura T, Miyajima A. Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J Cell Sci. 2003;116:1775–1786. doi: 10.1242/jcs.00388. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J, Abel EL, Angel JM, Kiguchi K, DiGiovanni J, Batlle E, et al. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Iacobuzio-Donahue CA. The Pathology and Genetics of Metastatic Pancreatic Cancer. 2009 doi: 10.5858/133.3.413. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Miyamoto A, Hama N, Asaoka T, Maeda S, Omiya H, Takami K, Doki Y, Mori M, Nakamori S. Survival Impact of Pulmonary Metastasis as Recurrence of Pancreatic Ductal Adenocarcinoma. Dig Surg. 2015;32:464–471. doi: 10.1159/000439545. [DOI] [PubMed] [Google Scholar]
- Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, Yang J, Weinberg RA. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547:E1–E3. doi: 10.1038/nature22816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.