Short abstract
Objective
Long noncoding RNAs (lncRNAs) offer great potential as cancer biomarkers. This study was performed to assess the applicability of serum lncRNA urothelial carcinoma-associated 1 (UCA1) as a diagnostic and/or prognostic biomarker for hepatocellular carcinoma (HCC).
Methods
We examined UCA1 expression in serum samples from 105 patients with HCC, 105 patients with benign liver disease (BLD), and 105 healthy volunteers using reverse-transcription polymerase chain reaction and analyzed the relationship between serum UCA1 and clinicopathological parameters of HCC as well as survival.
Results
Expression of serum UCA1 was significantly higher in patients with HCC and allowed for discrimination of HCC from BLD and healthy controls. High expression of serum UCA1 was significantly associated with a high tumor grade, large tumor size, positive vascular invasion, and advanced TNM stage. Multivariate analysis revealed that a high serum UCA1 level was an independent unfavorable prognostic factor for HCC.
Conclusions
Our results confirm the upregulation of serum UCA1 expression in HCC and indicate its clinical value as a noninvasive biomarker for HCC screening and prognostic prediction.
Keywords: LncRNAs, UCA1, hepatocellular carcinoma, biomarkers, diagnosis, polymerase chain reaction
Introduction
Hepatocellular carcinoma (HCC) is one of the most aggressive carcinomas and the third major cause of cancer-related mortality worldwide.1 Despite recent progress in clinical treatment, the 5-year overall survival rate in patients with HCC is still far from satisfactory, largely because of delayed diagnosis, frequent cancer metastasis, and high recurrence rates.2 At present, the combination of serum alpha-fetoprotein measurement and an imaging technique such as computed tomography or magnetic resonance imaging is the most widely used strategy for evaluation of suspicious HCC nodules. However, the sensitivity of serum alpha-fetoprotein for the detection of early-stage HCC is only 39% to 65%.3 Additionally, upregulation of serum alpha-fetoprotein can also occur in patients with benign liver diseases such as hepatitis and cirrhosis. Thus, reliable noninvasive biomarkers for HCC are needed.
Long noncoding RNAs (lncRNAs) are a large family of transcripts longer than 200 bp with no protein-coding function.4 Previous studies have confirmed that lncRNAs play critical roles in tumorigenesis and cancer development.5 Moreover, lncRNAs are detectable and relatively stable in cell-free body fluids, indicating great potential of circulating lncRNAs for biomarker applications.6 In one study, for example, lncRNA H19 expression was upregulated in plasma samples from patients with gastric cancer and significantly associated with the TNM stage.7 The combination of serum lncRNA XIST and HIF1A-AS1 was successfully used for non-small cell lung cancer diagnosis in another study.8 Moreover, increased circulating lncRNA HOTAIR expression was shown to possibly serve as an unfavorable prognostic marker for colorectal cancer.9
LncRNA urothelial carcinoma-associated 1 (UCA1), located in chromosome 19p13.12, was originally identified in bladder cancer and suggested to induce cell proliferation and migration and confer drug resistance.10 Subsequent studies revealed UCA1 overexpression and its role as an oncogene in many malignancies, such as HCC,11 prostate cancer,12 gastric cancer,13 breast cancer,14 colorectal cancer,15 pancreatic cancer,16 and osteosarcoma.17 Three UCA1 isoforms have been reported: 1.4, 2.2 and 2.7 kb in length. The 1.4-kb isoform is contained in the 2.2-kb isoform, the biological function of which is unclear. High levels of UCA1 in HCC tumor tissues are closely related to large tumor size, vascular invasion, advanced TNM stage, and poor postoperative survival.11,18 UCA1 was recently found to be significantly upregulated in the serum/plasma of patients with osteosarcoma,19 lung cancer,20 and gastric cancer21 and might be used for discrimination between patients with cancer and healthy controls. However, the potential significance of serum UCA1 in HCC remains elusive. This prompted us to investigate the serum levels of UCA1 (1.4-kb isoform) in patients with HCC patients explore the clinical value of UCA1 as a noninvasive biomarker for early diagnosis and prognostic prediction of HCC.
Materials and methods
Sample collection
The research protocol was approved by the ethics committee of Xijing Hospital Affiliated to The Fourth Military Medical University (No. 2013036), and each participant provided signed, written informed consent.
From June 2008 to July 2012, a total of 105 patients with histologically confirmed HCC were included in this study. Patients with a history of previous cancer were excluded. Serum samples were drawn before surgery. No chemotherapy, radiotherapy, or targeted therapy was used prior to blood collection. For each patient, 8 mL of peripheral blood was obtained by venous puncture, followed by centrifugation at 3000 × g for 10 min at 4°C. Cell-free serum was then stored at −80°C until RNA extraction. Control samples were obtained from 105 patients with benign liver disease (BLD) (75 patients with alcoholic liver disease and 30 patients with nonalcoholic fatty liver disease, all without hepatitis B infection) and 105 healthy volunteers (both age- and sex-matched). Clinical follow-up data were available for all patients with HCC. Patients with incomplete medical records or prior chemotherapy/radiotherapy were excluded from this study.
RNA isolation and quantitative reverse-transcription polymerase chain reaction
Total RNA in serum was extracted using a miRNeasy Micro Kit (QIAGEN, Valencia, CA, USA). The isolated RNA (100 ng) was reverse-transcribed into complementary DNA (cDNA) using the High Capacity cDNA Reverse Transcription Kit (Takara, Dalian, China). Next, 2 µL of cDNA was used as a template, and polymerase chain reaction (PCR) was performed with the iTaq Universal SYBR Green One-Step Kit (Bio-Rad, Hercules, CA, USA) on a CFX96 Real-Time PCR Detection System (Bio-Rad). The cycling conditions were 95°C for 30 s (predenaturation), followed by 40 cycles at 95°C for 10 s (denaturation) and 65°C for 30 s (annealing/extension). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was employed as an endogenous control. The sequences of the PCR primers for UCA1 and GAPDH were as follows: UCA1, 5′-TTC CTT ATT ATC TCT TCTG-3′ (forward) and 5′-TCC ATC ATA CGA ATA GTA-3′ (reverse); GAPDH, 5′-CTC GCT TTG GCA GCA CA-3′ (forward) and 5′-AAC GCT TCA CGA ATT TGC GT-3′ (reverse). The relative quantitative value was determined by the 2−ΔΔCt method.
Statistics
All data were processed with SPSS 18.0 software (IBM Corp., Armonk, NY, USA). The serum levels of UCA1 were compared between the groups using the Mann–Whitney U-test. Categorical data were analyzed using the chi-square test. Receiver-operating characteristic (ROC) curves were used to evaluate the diagnostic value of serum UCA1 for HCC. Overall survival was compared by the Kaplan–Meier method. Univariate and multivariate Cox regression analyses were performed to examine the relationships between patient survival and prognostic variables. A P-value of <0.05 was considered statistically significant.
Results
Increased serum UCA1 expression in patients with HCC and its diagnostic value
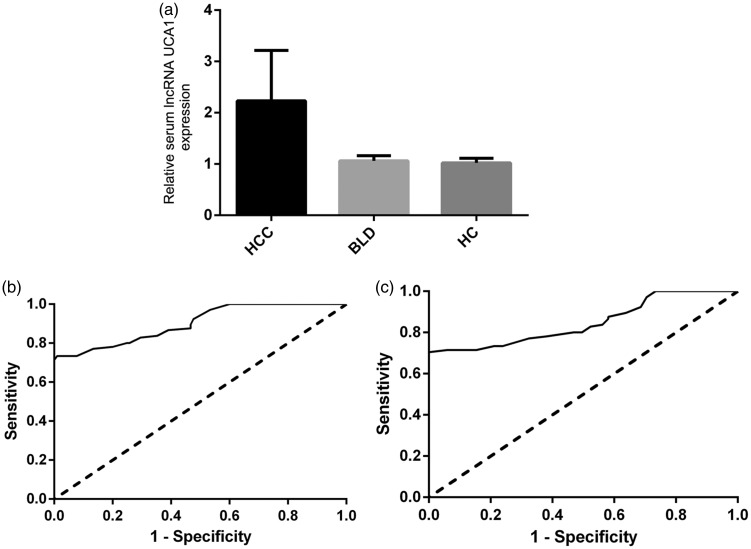
The relative expression of serum UCA1 was detected and analyzed in each sample using reverse-transcription PCR. We found that the serum UCA1 levels in the 105 patients with HCC were significantly higher than those in the 105 patients with BLD and 105 healthy volunteers (both P < 0.01) (Figure 1(a)). There was no significant difference between the BLD group and healthy volunteers with respect to serum UCA1 expression (Figure 1(a)).
Figure 1.
Relative serum urothelial carcinoma-associated 1 (UCA1) expression levels and their diagnostic value in patients with hepatocellular carcinoma (HCC). (a) The expression of serum UCA1 was significantly higher in patients with HCC than in patients with benign liver disease (BLD) and healthy controls (HCs). (b) Receiver operating characteristic (ROC) curve analysis of serum UCA1 for discriminating patients with HCC from HCs. (c) ROC curve analysis of serum UCA1 for discriminating patients with HCC from patients with BLD.
We performed ROC curve analyses to evaluate whether serum UCA1 can be used as a potential diagnostic biomarker for HCC. The results showed that serum UCA1 could differentiate patients with HCC from healthy controls, with an area under the ROC curve of 0.902 (95% confidence interval, 0.862–0.942). At the cut-off value of 1.85, the sensitivity and specificity were 73.3% and 99.0%, respectively (Figure 1(b)). Serum UCA1 could also differentiate patients with HCC from those with BLD, with an area under the curve of 0.848 (95% confidence interval, 0.795–0.902). At the cut-off value of 1.99, the sensitivity and specificity were 71.4% and 94.3%, respectively (Figure 1(c)).
Relationship of serum UCA1 with clinicopathological characteristics and patients’ prognosis
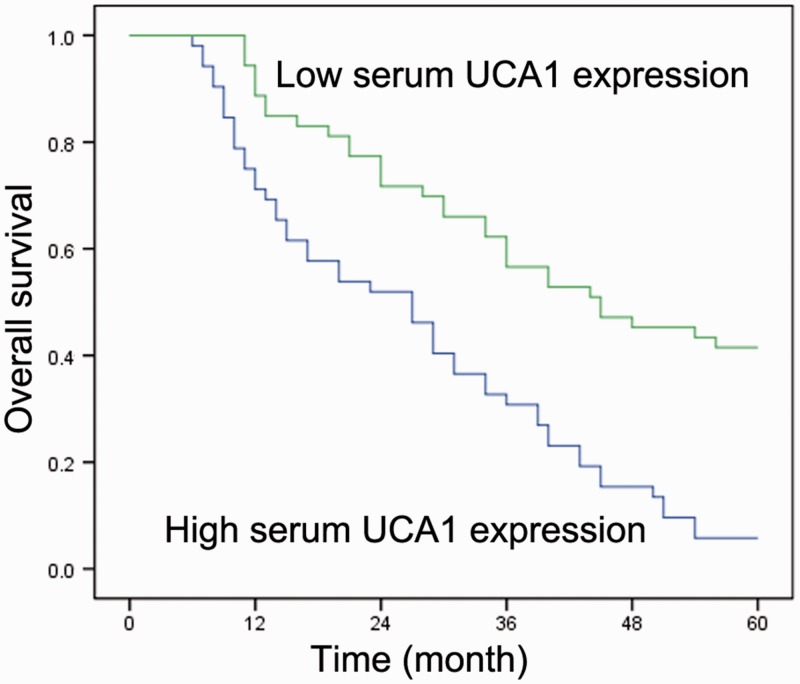
Associations between the serum UCA1 level and clinicopathological features of the patients with HCC are summarized in Table 1. All patients were subdivided into high- and low-expression groups according to the median serum UCA1 level. High serum UCA1 expression was found to be significantly associated with a high tumor grade (P = 0.011), large tumor size (P = 0.01), positive vascular invasion (P = 0.014), and advanced TNM stage (P = 0.003). Kaplan–Meier curve analysis revealed that the 5-year overall survival in patients with HCC with high serum UCA1 levels was inferior to that in patients with low serum UCA1 levels (P < 0.001) (Figure 2). Univariate Cox regression analysis also showed a statistically significant correlation between overall survival and tumor size, vascular invasion, and clinical stage (Table 2). The multivariate Cox regression analysis confirmed the independent effects of serum UCA1 (P = 0.016), tumor size (P = 0.037), vascular invasion (P = 0.028), and TNM stage (P = 0.001) on the prognosis of patients with HCC (Table 2).
Table 1.
Correlations between serum long noncoding RNA UCA1 and clinicopathological variables of hepatocellular carcinoma
| Clinicopathological features | Cases (n) |
Serum UCA1 expression |
P-value | |
|---|---|---|---|---|
| Low (n, %) | High (n, %) | |||
| Age (years) | ||||
| <60 | 51 | 28 (54.9) | 23 (45.1) | 0.437 |
| ≥60 | 54 | 25 (46.3) | 29 (53.7) | |
| Sex | ||||
| Male | 78 | 38 (48.7) | 40 (51.3) | 0.656 |
| Female | 27 | 15 (55.6) | 12 (44.4) | |
| Tumor grade | ||||
| G1 | 34 | 23 (67.6) | 11 (32.4) | 0.011 |
| G2 + G3 | 71 | 30 (42.3) | 41 (57.7) | |
| AFP (ng/L) | ||||
| ≥400 | 59 | 26 (44.1) | 33 (55.9) | 0.170 |
| <400 | 46 | 27 (58.7) | 19 (41.3) | |
| Tumor diameter (cm) | ||||
| <5 | 67 | 40 (59.7) | 27 (40.3) | 0.010 |
| ≥5 | 38 | 13 (34.2) | 25 (65.8) | |
| Tumor nodes | ||||
| Multiple | 35 | 14 (40.0) | 21 (60.0) | 0.151 |
| Single | 70 | 39 (55.7) | 31 (44.3) | |
| Cirrhosis | ||||
| Negative | 24 | 11 (45.8) | 13 (54.2) | 0.648 |
| Positive | 81 | 42 (51.9) | 39 (48.1) | |
| Venous infiltration | ||||
| Present | 36 | 12 (33.3) | 24 (66.7) | 0.014 |
| Absent | 69 | 41 (59.4) | 28 (40.6) | |
| TNM stage | ||||
| I + II | 50 | 33 (66.0) | 17 (34.0) | 0.003 |
| III | 55 | 20 (36.4) | 35 (63.6) | |
UCA1, urothelial carcinoma-associated 1; AFP, alpha-fetoprotein.
Figure 2.
Kaplan–Meier overall survival curves by serum urothelial carcinoma-associated 1 (UCA1) level. Patients with hepatocellular carcinoma with high serum UCA1 expression showed lower overall survival than patients with low serum UCA1 expression (log-rank test; P < 0.001).
Table 2.
Univariate and multivariate regression analyses of parameters associated with prognosis of patients with hepatocellular carcinoma
| Variables |
Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| Hazard ratio | P-value | Hazard ratio | P-value | |
| Age, years (≥60/<60) | 1.173 | 0.383 | – | – |
| Sex (male/female) | 1.154 | 0.406 | – | – |
| Tumor grade (G1/G2+G3) | 1.855 | 0.097 | – | – |
| AFP, ng/L (≥400/<400) | 1.261 | 0.214 | – | – |
| Tumor diameter, cm (≥5/<5) | 3.247 | 0.011 | 2.387 | 0.037 |
| Tumor nodes (multiple/single) | 1.498 | 0.122 | – | – |
| Venous infiltration (present/absent) | 3.866 | 0.004 | 3.126 | 0.028 |
| TNM stage (I–II/III) | 4.457 | 0.002 | 4.783 | 0.001 |
| Serum UCA1 expression (low/high) | 4.891 | <0.001 | 3.649 | 0.016 |
AFP, alpha fetoprotein; UCA1, urothelial carcinoma-associated 1.
Discussion
Despite recent progression in cancer research, the pathogenesis of HCC remains largely unknown. Recent studies have demonstrated that dysregulation of lncRNAs is involved in tumorigenesis and progression of HCC. For example, ectopic expression of SNHG6-003 in HCC cells promotes cell proliferation and induces drug resistance.22 The knockdown of nuclear enriched abundant transcript 1 reduces HCC cell invasion and migration.23 A high level of BRAF-activated non-protein coding RNA expression in HCC tissues is correlated with poor cancer differentiation and advanced TNM stages and predicts unfavorable patient survival.24
Early diagnosis of HCC can improve clinical outcomes. Because of their easy accessibility, high stability, and crucial roles in carcinogenesis and cancer progression, circulating lncRNAs have been regarded as promising candidate biomarkers for cancer detection and/or prognosis. In the present study, we showed that increased serum UCA1 expression is correlated with high tumor grade, large tumor size, positive vascular invasion, and advanced TNM stage. Notably, serum UCA1 was found to be a potential diagnostic biomarker and independent prognostic factor for HCC. To the best of our knowledge, this is the first study to detect serum UCA1 expression and evaluate its clinical significance in patients with HCC.
Interestingly, upregulation of serum/plasma UCA1 also occurs in several other malignancies. Serum UCA1 expression in patients with osteosarcoma is closely related to clinical stage and metastasis and might serve as a prognostic biomarker.19 Plasma UCA1 is also upregulated in patients with non-small cell lung cancer and gastric cancer and shows good diagnostic value.20,21 Thus, UCA1 does not appear to be a specific marker for HCC, and the clinical significance of serum UCA1 in other human malignancies is worthy of further investigation.
Previous studies have shown that UCA1 can promote tumor progression through multiple mechanisms in various types of cancer. UCA1 may function as a sponge for several tumor suppressor microRNAs, such as miR-184,12 miR-204-5p,15 and miR-182.25 Some downstream pathways have also been identified, including the AKT/mTOR,26 p27Kip1/CDK2,27 KLF4-KRT6/13,28 and Wnt signaling pathways.29 Xiao et al.11 revealed that upregulation of UCA1 increased epithelial–mesenchymal transition in HCC via sponging to miR-203 and thereby activating the expression of transcription factor Snail2. Wang et al.18 reported that UCA1 overexpression promoted HCC progression through inhibition of miR-216b and activation of the FGFR1/ERK signaling pathway. Taken together, these findings indicate that UCA1 might be involved in an extensive regulatory network, and more potential targets should be identified to further clarify the mechanisms of how UCA1 acts as an oncogene in HCC.
The current study indicates that upregulation of serum UCA1 might be a valuable biomarker for HCC screening and prognostic prediction. Prospective studies with large sample sizes are encouraged to confirm our conclusions.
Acknowledgements
None.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Diaz-Gonzalez A, Forner A, Rodriguez de Lope C, et al. New challenges in clinical research on hepatocellular carcinoma. Revista espanola de enfermedades digestivas: organo oficial de la Sociedad Espanola de Patologia Digestiva. 2016; 108: 485–493. [DOI] [PubMed] [Google Scholar]
- 2.Eggert T, McGlynn KA, Duffy A, et al. Epidemiology of fibrolamellar hepatocellular carcinoma in the USA, 2000-10. Gut. 2013; 62: 1667–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collier J, Sherman M. Screening for hepatocellular carcinoma. Hepatology. 1998; 27: 273–278. [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Wang Z, Wang D, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 2013; 41: D983–D986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Li W, Liang B, et al. Identification of cancer risk lncRNAs and cancer risk pathways regulated by cancer risk lncRNAs based on genome sequencing data in human cancers. Sci Rep 2016; 6: 39294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng C, Hao H, Chen L, et al. Long noncoding RNAs as novel serum biomarkers for the diagnosis of hepatocellular carcinoma: a systematic review and meta-analysis. Clin Transl Oncol 2017; 19: 961–968. [DOI] [PubMed] [Google Scholar]
- 7.Hashad D, Elbanna A, Ibrahim A, et al. Evaluation of the Role of Circulating Long Non-Coding RNA H19 as a Promising Novel Biomarker in Plasma of Patients with Gastric Cancer. J Clin Lab Anal 2016; 30: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tantai J, Hu D, Yang Y, et al. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol 2015; 8: 7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 9.Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014; 35: 1510–1515. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Li X, Xie X, et al. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett 2008; 582: 1919–1927. [DOI] [PubMed] [Google Scholar]
- 11.Xiao JN, Yan TH, Yu RM, et al. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Onco 2017; 143: 981–990. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Wang X, Zhang J, et al. Artesunate suppresses the viability and mobility of prostate cancer cells through UCA1, the sponge of miR-184. Oncotarget. 2017; 8: 18260–18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo ZK, Gong Y, Chen XH, et al. TGFbeta1-Induced LncRNA UCA1 Upregulation Promotes Gastric Cancer Invasion and Migration. DNA Cell Biol 2017; 36: 159–167. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Wang G, Yang L, et al. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/beta-Catenin Pathway. PLoS One. 2016; 11: e0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian Z, Jin L, Zhang J, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep 2016; 6: 23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P, Wan D, Zheng D, et al. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016; 83: 1220–1226. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol 2016; 5: 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015; 6: 7899–78917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen JJ, Ma YD, Yang GS, et al. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci 2017; 21: 498–503. [PubMed] [Google Scholar]
- 20.Wang HM, Lu JH, Chen WY, et al. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med 2015; 8: 11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Cao R, Mu H. Long non-coding RNA UCA1 may be a novel diagnostic and predictive biomarker in plasma for early gastric cancer. Int J Clin Exp Pathol 2015; 8: 12936–12942. [PMC free article] [PubMed] [Google Scholar]
- 22.Cao C, Zhang T, Zhang D, et al. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene 2017; 36: 1112–1122. [DOI] [PubMed] [Google Scholar]
- 23.Mang Y, Li L, Ran J, et al. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther 2017; 10: 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J Surg Oncol 2016; 14: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Z, Wang Y, Huang G, et al. The lncRNA UCA1 interacts with miR-182 to modulate glioma proliferation and migration by targeting iASPP. Arch Biochem Biophys 2017; 623–624: 1–8. [DOI] [PubMed] [Google Scholar]
- 26.Cheng N, Cai W, Ren S, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015; 6: 23582–23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu JJ, Song W, Zhang SD, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep 2016; 6: 23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Na XY, Liu ZY, Ren PP, et al. Long non-coding RNA UCA1 contributes to the progression of prostate cancer and regulates proliferation through KLF4-KRT6/13 signaling pathway. Int J Clin Exp Med 2015; 8: 12609–12616. [PMC free article] [PubMed] [Google Scholar]
- 29.Fan Y, Shen B, Tan M, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J 2014; 281: 1750–1758. [DOI] [PubMed] [Google Scholar]