Abstract
Background
Recently, 34 genes had been associated with differential expression relative to blood pressure (BP)/ hypertension (HTN). We hypothesize that some of the genes associated with BP/HTN are also associated with BP response to antihypertensive treatment with thiazide diuretics.
Methods
We assessed these 34 genes for association with differential expression to BP response to thiazide diuretics with RNA sequencing in whole blood samples from 150 hypertensive participants from the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) and PEAR-2 studies. PEAR white and PEAR-2 white and black participants (n = 50 for each group) were selected based on the upper and lower quartile of BP response to hydrochlorothiazide (HCTZ) and to chlorthalidone.
Results
FOS, DUSP1 and PPP1R15A were differentially expressed across all cohorts (meta-analysis p-value < 2.0 × 10− 6), and responders to HCTZ or chlorthalidone presented up-regulated transcripts. Rs11065987 in chromosome 12, a trans-eQTL for expression of FOS, PPP1R15A and other genes, is also associated with BP response to HCTZ in PEAR whites (SBP: β = − 2.1; p = 1.7 × 10− 3; DBP: β = − 1.4; p = 2.9 × 10− 3).
Conclusions
These findings suggest FOS, DUSP1 and PPP1R15A as potential molecular determinants of antihypertensive response to thiazide diuretics.
Trial registration
Electronic supplementary material
The online version of this article (10.1186/s12920-018-0370-x) contains supplementary material, which is available to authorized users.
Keywords: Pharmacogenomics, Hypertension, Thiazide diuretics, Personalized medicine, RNA-Seq, eQTL
Background
Hypertension (HTN) is the most important modifiable risk factor for cardiovascular diseases- coronary artery disease, myocardial infarction, heart failure, stroke and peripheral vascular diseases; controlling blood pressure (BP) is critical for reducing long-term mortality and morbidity rates [1]. Despite the plethora of therapeutic options, selection of the initial anti-HTN treatment remains empirical. Worldwide, 1 billion people suffer from HTN [2] but only about 50% of those under drug therapy achieve the treatment goal, which highlights that anti-HTN drug selection for a specific patient likely impacts therapy success [3, 4].
Thiazide diuretics (TD) are a centerpiece of anti-HTN therapy due to their effectiveness, and safety profile in the management of HTN. Among the available anti-HTN medications, HCTZ, chlorthalidone and other TD are considered first line options for most patients with uncomplicated essential HTN, and are highly recommended for patients requiring more than one anti-HTN therapy for control of BP [5]. However, TD have variable efficacy, and less than 50% of HCTZ-treated patients achieve BP control [3]. The inter-individual variability in BP response to TD is likely to contribute to suboptimal BP control.
Using a genome-wide association (GWAS) approach, two replicated regions, one in PRKCA (protein kinase C, alpha) and the other one near GNAS (G protein alpha subunit), were identified with clinically relevant effects on BP response to HCTZ [6]. Despite the successes, the GWAS approach provides only one dimension of molecular information about BP response to anti-HTN treatment. While it is a critical dimension, analyzing DNA variation alone is insufficient for achieving an understanding of the multidimensional complexity of BP response to TD. In this context, transcriptomics (gene expression profiling) has been described as an innovative approach that enables biomarker discovery associated with different diseases and traits [7–10].
Recently, Huan et al. [9] identified 34 genes associated with differential expression relative to BP/HTN, which in aggregate explain ~ 9% of inter-individual variability in BP. In addition, previous findings suggest that some signals from HTN GWAS may predict anti-HTN drug response [11]. This study tests the hypothesis that some of the differentially expressed genes associated with BP/HTN are also associated with BP response to antihypertensive treatment with TD. We assessed the association of these 34 genes with differential expression to BP response to TD by applying RNA sequencing data from the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) and PEAR-2 studies.
Methods
Study population and ethics statement
This study includes data from PEAR and PEAR-2 (NCT00246519, NCT01203852 www.clinicaltrials.gov), which were previously described in details [12]. Briefly, PEAR was a multicenter, randomized clinical trial with the primary aim of evaluating the role of genetic variability on BP response of HCTZ and/or atenolol treated patients. Study participants (n = 768) with uncomplicated HTN were randomized to receive monotherapy of either the thiazide diuretic HCTZ, or the beta-blocker atenolol for a period of 9 weeks. Fasting blood and urine samples were collected at baseline (untreated), after 9 weeks of monotherapy, and after 9 weeks of combination therapy. BP responses were assessed using office, home, and 24-h ambulatory BP and then a composite BP response was constructed [13].
The PEAR-2 clinical trial included a hypertensive population similar to the one in PEAR, and for which metoprolol, a beta-blocker, and chlorthalidone, a thiazide-like diuretic, were tested. Details of this prospective, clinical trial were previously published [14]. Briefly, 417 hypertensive participants were treated in a sequential monotherapy design with metoprolol and then chlorthalidone with at least 4 week washout periods prior to each active treatment. Data collected included home and clinic BP measurements, adverse metabolic effects, fasting whole blood, and urine samples.
Gene expression profile with RNA-Seq
PEAR whites and PEAR-2 white and black participants were selected for gene expression profiling with RNA-Seq based on the differences in their BP response to HCTZ and chlorthalidone treatment, respectively. A total of 150 patients with BP responses to either HCTZ or chlorthalidone in the top and bottom quartiles from each of the three cohorts were selected and classified as poor BP responders (non-responders) and good BP responders (responders). Sample size was selected based on the theoretical statistical calculations [15], which revealed that with 32 million reads per sample (average number of RNA-Seq reads generated), coefficient of variation (σ) = 0.8, two-sided α level = 0.05, and 25 samples per group (25 responders and 25 non-responders to thiazide diuretics) we have greater than 80% power to detect two-fold differences in expression.
We determined the mean changes of serum potassium concentrations and uric acid levels in non-responders before and after treatment with HCTZ and chlorthalidone with the premise that if the cause of the nonresponse for BP lowering was nonadherence, then these individuals would also not have any adverse metabolic responses that are typically seen with thiazide. We also compared changes from baseline to after treatment serum potassium and uric acid using paired t-tests. Potassium depletion and uric acid elevation are commonly observed secondary to treatment with TD [16–18], and were lab parameters with statistically significant change in the overall clinical study from PEAR participants [19, 20].
Using whole blood samples collected before HCTZ or chlorthalidone monotherapy, RNA was extracted using the PAXgene Blood RNA kit IVD (Qiagen, Valenica, CA). The selection of poly(A) mRNA from total RNA was performed using Sera-Mag Magnetic Oligo(dT) Beads (Illumina, San Diego,CA) according to the manufacturer’s protocol. 100 ng of RNA was then used as a template for cDNA synthesis. Libraries were prepared following the strand-specific protocol [21]. DNA clusters were generated using the Illumina cluster station, followed by 100 cycles of paired-end sequencing on the Illumina HiSeq 2000, performed at Baylor Human Genome Sequencing Center in Texas. For data quality control purposes, read duplicates removal was implemented using Picard (http://broadinstitute.github.io/picard/) MarkDuplicates option.
The 100 bp reads generated in the paired-end RNA sequencing were uniquely mapped to the human reference genome (hg19) using TopHat v2.0.10 [22] allowing for four reads mismatches, read edit distance of six, one mismatch in the anchor region of a spliced read, and a maximum of five multi-hits. Transcript assembly was performed using Cufflinks v2.2.1. Statistical analysis were carried out with Cuffdiff and gene expression levels are reported in fragments per kilobase per million reads (FPKM), considering reads mapped to exonic regions of the 34 genes previously associated with BP/HTN [9].
Additionally, we performed differential expression analysis using alternative tools in order to adjust the expression levels for age, sex and baseline diastolic BP because we observed, for these variables, statistically significant differences between participants classified as responders and non-responders to thiazide diuretics (Table 1). Other common covariates in association studies of hypertension, such as Body Mass Index (BMI) and smoking were not included as covariates because previous analysis of BP response in PEAR [23] established that these variables were not associated with BP response. By using BAM files from TopHat 2 alignments, we were able to count the number of reads for each known human genes (Gencode gene annotation release 18) applying the htseq-count function from the HTSeq bioconductor package [24]. Counts were modeled to a Negative Binomial distribution using a generalized linear model in edgeR [25].
Table 1.
Characteristics of PEAR and PEAR-2 participants classified as responder and non-responders for the RNA-Seq analysis
| Whites (n = 99) | Blacks (n = 50) | |||||
|---|---|---|---|---|---|---|
| HCTZ | Chlorthalidone | Chlorthalidone | ||||
| Characteristics | Responders (n = 24) | Non-responders (n = 25) | Responders (n = 25) | Non-responders (n = 25) | Responders (n = 25) | Non-responders (n = 25) |
| Age | 48 ± 12 | 48 ± 8 | 53 ± 8 | 48 ± 10 | 52 ± 8 | 50 ± 10 |
| Female, n (%) | 11 (44%) | 10 (40%) | 15 (75%)a | 5 (25%)a | 12 (48%) | 12 (48%) |
| BMI, kgam− 2 | 29 ± 5 | 32 ± 6 | 32 ± 5 | 30.5 ± 5 | 30 ± 6 | 31 ± 5 |
| Baseline DBP | 93 ± 5 | 94 ± 4 | 97 ± 6a | 93 ± 5a | 98 ± 6a | 93 ± 4a |
| Baseline SBP | 146 ± 10 | 144 ± 10 | 152 ± 11a | 144 ± 9a | 152 ± 10a | 146 ± 10a |
| DBP response to TD | −9 ± 6b | 0.06 ± 4b | − 14 ± 4b | − 0.2 ± 2b | − 17 ± 4b | − 1.4 ± 3b |
| SBP response to TD | −12 ± 6b | − 0.9 ± 6b | − 22 ± 7b | − 1.5 ± 5b | − 27 ± 7b | − 4.4 ± 5b |
Mean and Standard Deviation values for the continuous variables were presented
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, TD thiazide diuretics
aSignificant at the 0.05 probability level
bSignificant at the 0.001 probability level
Statistical methods
Based on the fact that the BP signature genes, selected for this analysis, were discovered in whites, the primary data analysis was also performed in whites treated with HCTZ or chlorthalidone. Associations of differences in expression levels of these genes in responders compared to non-responders to TD was evaluated using a t-test to quantify the statistical significance in the differences observed among the gene expression measurements (FPKM). Bonferroni corrected P values < 0.0015 (0.05/34) were considered statistically significant. In addition, we assessed the statistical significance (hypergeometric test) of the overlap between the 34 BP signature genes and the 29 genes associated with thiazide diuretics blood pressure response at the whole transcriptome level (FDR p-value < 0.05) [29].
For each differentially expressed gene in PEAR or in PEAR-2 whites (6 in total), we attempted replication in PEAR-2 blacks and the alternate group of whites in order to validate the association of the genes with BP response to TD. A strict approach was established for validation with Bonferroni corrected P value (< 0.05/6 = 0.008) and the same fold change direction (either up or down regulation) as the primary analysis in whites treated with HCTZ or chlorthalidone.
For those genes that passed the validation criteria, the differential expression results from each study cohort were combined in a meta-analysis, using standardized p-values to follow the assumption of the Fisher p-value combination method implemented by the R package MetaRNASeq [26]. We considered that genes with meta-analysis p values < 2.0 × 10− 6 (0.05/25,000) achieved transcriptome-wide association with BP response to TD.
To evaluate whether FOS, DUSP1 and PPP1R15A robustly predict BP response to TD, PEAR participants were assigned into the derivation cohort for logistic regression model building. PEAR-2 whites constituted the validation cohort, in which area under the receiver operator curve was calculated in the R ROCR package [27], for model evaluation. The TD prediction model was compared to logistic regression model including randomly selected genes from whole transcriptome analysis. Twenty randomly selected genes were sampled (R function “sample”, 20 rounds), and each random signature was fitted to a logistic regression model to assess the probability of random gene signature performing better than the TD genes. Gene expression measures in FPKM were used for this analysis.
Genomics analysis
Previous studies have explored the genome-wide genotyping results for the PEAR and PEAR-2 studies in much more detail [6, 11]. GWAS data for chlorthalidone in PEAR-2 will be reported separately. Briefly, DNA samples were genotyped using Illumina Human Omni-1Million Quad BeadChip and 2.5 M-8 BeadChip (Illumina, San Diego CA) for PEAR and PEAR-2, respectively. Genotypes were called using GenTrain2 clustering algorithm (GenomeStudio, Illumina, San Diego CA). MaCH software (version 1.0.16) was used to impute SNPs based on HapMapIII haplotypes.
In order to identify SNPs potentially regulating the expression of the genes differentially expressed in the RNA-Seq data, we consulted the Blood eQTL browser [28]. The SNPs identified as eQTL for the differentially expressed genes were then evaluated in the PEAR and PEAR2 GWAS data, to test for a genetic association with BP response to TD. SNP associations with BP response were evaluated using previously conducted GWAS analyses [6] that included data on systolic and diastolic BP responses to HCTZ in 228 whites participants from PEAR, and responses to chlorthalidone in 185 white and 142 black participants from PEAR-2. PLINK software was used to run the analysis with adjustment for age, sex, pre-HCTZ/chlorthalidone BP and population substructure by considering the first and second principal components (PC1 and PC2) in all our analysis.
Results
Table 1 summarizes baseline and demographic characteristics from PEAR white participants treated with HCTZ and PEAR-2 white and black participants treated with chlorthalidone who were selected for RNA-Sequencing. For PEAR, age, body mass index (BMI), sex and baseline BP were not statistically different between participants classified as responders and non-responders to HCTZ. However, in PEAR-2 white participants, differences in sex and baseline BP were statistically significant between responders and non-responders to chlorthalidone. Differences in baseline BP were also observed in PEAR-2 blacks between responders and non-responders to chlorthalidone.
After treatment with HCTZ and chlorthalidone, there were significant reductions on serum potassium concentrations and significant increases serum uric acid levels in participants classified as non-responders (Additional file 1: Table S1). These changes are consistent with previously reported metabolic effects after treatment with TD [19, 20], and suggest high treatment adherence in the group of BP non-responders to TD.
In order to identify genes with differential expression involved in BP response to TD, whole transcriptome sequences were generated from 149 participants treated with HCTZ or chlorthalidone. One of the samples from HCTZ responders did not achieve enough library yield for adequate performance in sequencing. On average, 32 million reads per sample were mapped to the human reference genome (hg 19) and about 93% were uniquely mapped (Additional file 1: Figure S1). Whole transcriptome analyses from the PEAR and PEAR2 studies were previously published [29].
At a Bonferroni corrected alpha (0.0015), six genes were differentially expressed in whites treated with HCTZ or chlorthalidone (Additional file 1: Table S2). Of those GPR56, FOS and FGFBP2 were common between the gene lists as significant for BP response to TD at the whole transcriptome level (FDR p-value < 0.05 in PEAR or PEAR-2 whites) [29] and the BP signature genes (Additional file 1: Table S2). A hypergeometric test (p = 9.0 × 10− 7) showed that this overlap between the 29 genes associated to TD BP response at the whole transcriptome level [29] and the 34 BP signature genes is statistically significant. For each gene differentially expressed (P < 0.0015) in PEAR or PEAR-2 white participants, we attempted replication in the other group of white study participants and in black participants from PEAR-2 (Additional file 1: Table S2). Of the six genes identified, FOS and DUSP1 were differentially expressed and showed consistent fold change direction in all 3 cohorts (Table 2), passing the stringent Bonferroni corrected alpha at 0.008 for validation. PPP1R15A showed consistent directional fold change in all three cohorts, and met the Bonferroni threshold p value in PEAR whites given HCTZ (Fold Change (Responders/non-responders): 1.27, p = 1.15 × 10− 3) and PEAR-2 blacks given chlorthalidone (Fold Change: 1.29, p = 1.75 × 10− 3), while only achieving nominal significance in PEAR-2 whites (Fold Change: 1.19, p = 3.61 × 10− 2) (Table 2). The meta-analysis of all participants with RNA-Seq data included FOS, DUSP1 and PPP1R15A, and confirmed transcriptome-wide associations that far exceeded transcriptome wide (and genome wide) significance for FOS (p = 2 × 10− 12), DUSP1 (p = 9.5 × 10− 12) and PPP1R15A (p = 3.6 × 10− 8) expression and BP response to TD (Table 2). Even though the statistical strength of the association lessened after the adjustment for age, sex and baseline BP, the fold change direction remains consistent across PEAR whites and PEAR-2 whites and blacks regardless of the statistical methods used (Additional file 1: Table S3).
Table 2.
Genes differentially expressed between responders and non-responders to HCTZ and chlorthalidone in all 3 cohorts, with consistent direction and transcriptome-wide statistical significance when meta-analyzed
| HCTZ Whites | Chlorthalidone Whites | Chlorthalidone Blacks | Meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Non-resp. | Resp. | Fold Change | P value | Non-resp. | Resp. | Fold Change | P value | Non-resp. | Resp. | Fold Change | P value | P value |
| FOS | 39.2 | 49.5 | 1.26 | 2.90E-03 | 29.4 | 38.0 | 1.29 | 1.15E-03 | 24.6 | 35.9 | 1.46 | 5.00E-05 | 2.08E-12 |
| DUSP1 | 76.0 | 105.2 | 1.38 | 1.50E-04 | 71.5 | 92.8 | 1.30 | 1.35E-03 | 63.3 | 81.7 | 1.29 | 3.55E-03 | 9.50E-12 |
| PPP1R15A | 38.3 | 48.7 | 1.27 | 1.15E-03 | 29.9 | 35.5 | 1.19 | 3.61E-02 | 27.6 | 35.6 | 1.29 | 1.75E-03 | 3.64E-08 |
Fold change corresponds to gene expression levels in responders divided by levels in non-responders, in fragments per kilobase per million reads (FPKM)
The combination of FOS, DUSP1 or PPP1R15A gene expression in a logistic regression model was statistically significant (P = 0.02), and explained 23.3% of the variability in drug response to TD in the derivation cohort (PEAR). For independent assessment of this model in PEAR-2, the area under the curve was 0.71, indicative of the model’s good prediction for BP response to TD (Fig. 1). Additionally, this model performed better than randomly selected signature genes of identical size, re-sampled 20 times (P range: = 0.045–0.96) (Additional file 1: Figure S2).
Fig. 1.
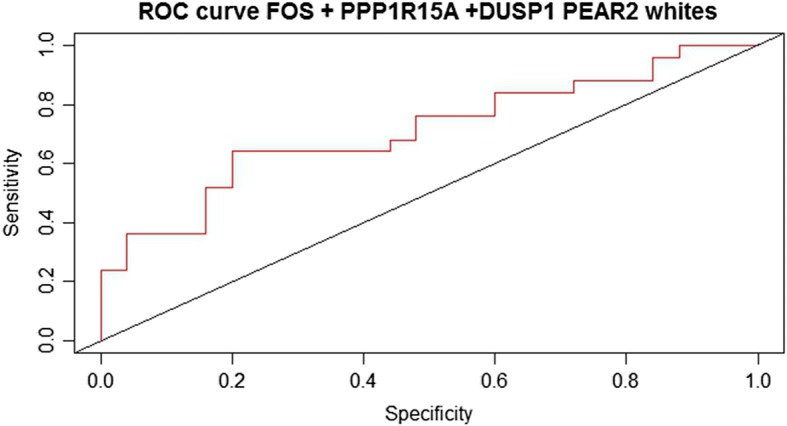
Receiver operator curve for assessment of logistic regression model prediction in PEAR-2. Statistical model including thiazide diuretic (TD) genes FOS, DUSP1 and PPP1R15A showed area under the curve was 0.71, indicative of the model’s good prediction for blood pressure response to TD. Gene expression measures reported in Fragments per Kilobase of Exon per Million mapped (FPKM)
Based on data in the Blood eQTL browser [28], we identified 4 trans-eQTLs (rs11065987, rs653178, rs10774625 and rs11066301) associated with reduced expression of both FOS and PPP1R15A (Additional file 1: Table S4). Because of the high linkage disequilibrium between these SNPs (Additional file 1: Figure S3), we selected a representative SNP (rs11065987) to test for an association with BP response with TD. Rs11065987 was associated with SBP and DBP response to HCTZ in PEAR whites (SBP: β = − 2.1; p = 1.7 × 10–3; DBP: β = − 1.4; p = 2.9 × 10− 3) (Fig. 2) and showed consistent directional association in PEAR-2 whites that did not reach statistical significance in PEAR-2 whites or blacks treated with chlorthalidone (Additional file 1: Table S4).
Fig. 2.
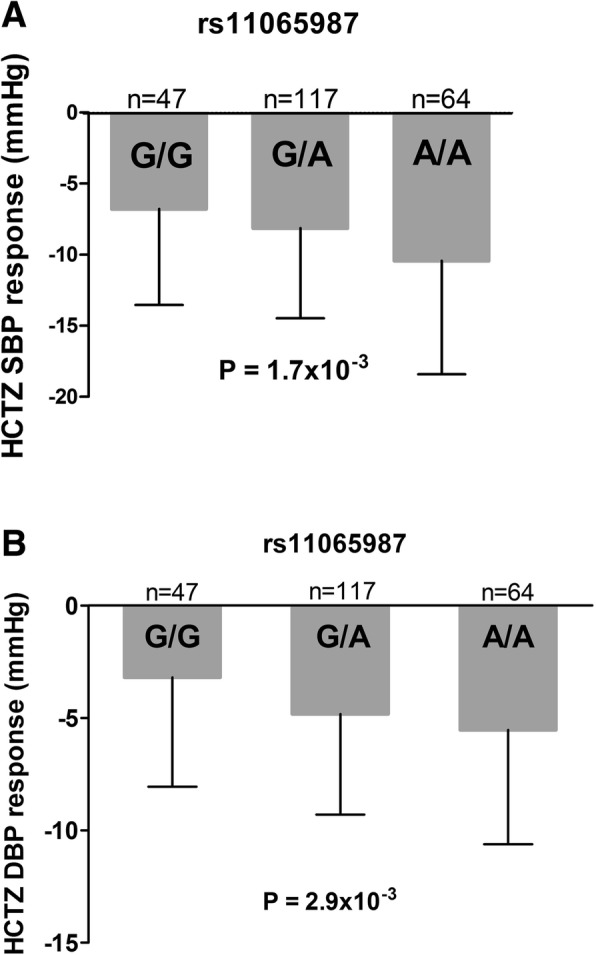
The effect of rs11065987 polymorphism on the blood pressure response of Whites treated with HCTZ in PEAR. Blood pressure responses were adjusted for baseline blood pressure, age, sex, and population substructure. P-values represent the contrast of adjusted means between different genotype groups in the PEAR white participants. Error bars represent standard error of the mean. a systolic blood pressure response to HCTZ in PEAR whites. b diastolic blood pressure response to HCTZ in PEAR whites
In order to investigate FOS and PPP1R15A co-expression, we calculated the Pearson correlation coefficient between the log transformation of expression levels of these genes. FOS and PPP1R15A showed strong positive correlation with r2 = 0.9 in PEAR whites treated with HCTZ and r2 = 0.8 in PEAR-2 whites treated with chlorthalidone. This indicates potential common co-regulatory mechanism involving the expression of these genes and driven by rs11065987 or its proxy SNPs.
Discussion
Despite the widespread use of TD, there is large inter-individual variability in BP or drug response, which has motivated the identification of genetic markers with the potential to optimize antihypertensive treatment selection. GWAS results have definitely contributed to enlarge the current knowledge on the potential role of genetics in inter-individual variability in drug response in general and also to thiazide BP response [6, 11]. However, this approach provides only one dimension of molecular information in thiazide BP response, which may not be sufficient to understand the complexity of this phenotype. Gene expression has been shown to have predictive and prognostic value in disease genomics studies, applying transcriptomics tools for the characterization of novel cancer subtypes [30–32] or the identification of signature genes for hypertension [9, 33], heart failure [34–36] and other diseases. Recent studies presented a remarkable effect of whole transcriptome gene expression data in enriching for drug responders to docetaxel and cisplatin treatment of breast cancer [37], and to erlotinib in non-small cell lung cancer [38]. Each of these studies highlights the potential scientific insights that can be gained through experimental approaches that apply gene expression data. In this study, we investigated differences in gene expression underlying extreme BP response to thiazides in white and black participants from PEAR and PEAR-2. Such approaches have the potential to provide methods for precision medicine, but additionally may provide previously unrecognized insights into BP regulation and responses to antihypertensive drugs.
Herein, we have shown that applying transcriptome sequencing data helped us to identify molecular markers potentially implicated in BP response to TD. Among the 34 genes previously documented to influence BP/HTN, FOS, DUSP1 and PPP1R15A mRNAs were differentially expressed between responders and non-responders in three different cohorts treated with TD, with consistent directional fold change in whites treated with HCTZ and whites and blacks treated with chlorthalidone.
Among these three genes, only FOS has been associated previously with the pathophysiology of HTN. Expression of FOS (FBJ murine osteosarcoma viral oncogene homolog, also known as AP-1 transcription factor subunit), a leucine zipper protein that when dimerized with JUN forms a transcription factor complex, is linked to neuronal activation of vasomotor areas in mice [39]. Also, the blockade of FOS expression with oligonucleotides attenuates high BP in HTN-induced and spontaneously HTN mice [40].
We did not find any direct evidence in the literature of the involvement of DUSP1 and PPP1R15A that could account mechanistically for a potential susceptibility for HTN and/or BP response to thiazides. However, we found that these genes are involved in biological processes related to BP regulatory mechanisms. For instance, DUSP1 has shown consistent inhibition of ERK 1/2 (Extracellular Regulated Kinases) signaling in vitro and in vivo [41], with potential attenuation on the effects of angiotensin II-mediated vascular smooth muscle cell (VSMC) proliferation and vasoconstriction [42].
PPP1R15A is a regulatory subunit for phosphatase protein (PP) 1 [43]. PP1 is the catalytic subunit for myosin phosphatases, a key convergence point on contractility pathways in VSMC, that dephosphorylates the myosin light chain and initiates the relaxation process for vasodilation [44]. Of relevance, PP1 has a highly specific inhibitor 1 (I-1) which, when activated by protein kinase A, forms a heterotrimeric complex with PP1 and PPP1R15A [43]. This specific interaction of PPP1R15A with the C-terminal region of I-1 engenders strong PP1 inhibition [43] and a potential amplification of contractile response in VSMC [45]. In addition, recent research shows that I-1 regulates thiazide-sensitive NaCl cotransporter (NCC) activity and phosphorylation in the distal convoluted tube (DCT), and loss of I-1 expression in mice lowers arterial BP [46]. Since there is no concrete evidence of the consequences of I-1/PPP1R15A interaction in the regulation of contractile signaling, in VSMC, or in the regulation of NCC activity in DCT, we can only speculate that PPP1R15A may be important for BP regulatory mechanisms. Further experimental validation will be crucial to close the link between PPP1R15A interactions with I-1 for the regulation of PP1 and NCC activity in VSMC and DCT.
In addition, we found rs11065987 associated with both systolic and diastolic BP responses to HCTZ in PEAR whites, and it is also associated in trans with decreased expression of two genes in our top list of BP signature genes: FOS and PPP1R15A, which are co-expressed in the whole blood samples tested in this study. rs11065987, the leading SNP in a small haplotype block, is an intergenic SNP in chromosome 12, where the closest gene is BRCA1 associated protein and previous cardiovascular disease GWA studies identified 12q4 as a risk locus for coronary artery disease and HTN [47]. Further experiments will be valuable to understand the mechanisms involved in gene expression regulation in the chromosome 12q4 region that could potentially affect BP regulation as well.
Although it is not clear how FOS, DUSP1 and PPP1R15A are involved in BP regulation, the differences in gene expression documented in this study taken together with evidence of gene expression regulatory mechanism with trans-eQTLs associated with BP response to HCTZ suggest that these genes may be markers of response to TD. Further functional studies may provide additional insights to the field.
This study presents some limitations. First, the number of samples with RNA-Seq data may have limited the power to identify additional genes differentially expressed as well as to validate some of the transcriptomics signals; however, we enhanced the power of the number of samples tested by taking an extreme phenotype approach. Second, using RNA from whole blood for RNA-Seq data analysis may have limited the detection of the expression of some genes/regulatory mechanisms that might be cell type-specific. However, it may be challenging to select only one tissue in order to investigate gene expression as a marker of BP regulation since drug response to anti-HTN might arise from a variety of target tissues such as heart, brain, kidney or vasculature. Not only are these tissues difficult to access in relatively healthy patients, as hypertensive patients are, but it is also not obvious which tissue should be used. Thus we are using whole blood as a surrogate for multiple tissues. Moreover, the original study that served as the basis for selection of BP signature genes also used whole blood samples for that transcriptome-wide gene expression study due to the convenience to identify biomarkers using easily accessible body fluids [9].
Conclusions
For over half century, thiazide diuretics have been a centerpiece of antihypertensive therapy with more than 100 million prescriptions annually in the US alone. Its large inter-individual variability in BP response emphasizes the need for molecular predictors of drug response that hold potential for improving the antihypertensive therapy. Results of the present study suggest that whole transcriptome data can provide insights into genes potentially involved in the pharmacogenetic phenotype of antihypertensive drug response. We were able to demonstrate that genes previously identified through BP/HTN transcriptome profiling that are also relevant determinants of BP response to TD. Specifically, FOS, DUSP1 and PPP1R15A, through their differential expression, may be involved in the response to TD. To strengthen the finding, through use of a publicly available eQTL database, we found an eQTL (SNP) of FOS and PPP1R15A that associated with BP response to TD and other SNPs with evidence of gene expression regulatory mechanisms. Further work is needed to understand the mechanistic basis by which differential expression of FOS, DUSP1 and PPP1R15A may influence BP regulation and response to TD.
Additional file
Figure S1. Mapping statistics for PEAR and PEAR-2 RNA-Seq data. Table S1. Potassium, glucose and uric acid mean changes in participants classified as non-responders after treatment with HCTZ and chlorthalidone. Table S2. Genes previously associated with BP/HTN (34 BP signature genes) and the expression measurements in PEAR and PEAR-2. Table S3. Differences in baseline expression levels for FOS, DUSP1 and PPP1R15A between thiazide diuretics responders and non-responders in PEAR and PEAR-2 with adjustment for age, gender and baseline blood pressure. Figure S2. P-value distribution for association with thiazide diuretics blood pressure response for 20 randomly selected genes. Table S4. Representative trans eQTL for top differentially expressed genes. Figure S3. Linkage disequilibrium plots between rs10655987, rs653178, rs10774625 and rs11066301. (DOCX 5615 kb)
Acknowledgements
We thank the valuable contributions of the Pharmacogenomics Evaluation of Antihypertensive Responses (PEAR) study participants, support staff, and study physicians. We also thank University of Florida Research Computing (http://www.rc.ufl.edu/) for providing computational resources and support as well as BCM-HGSC personnel including Viktoriya Korchina, HarshaVardhan Doddapaneni, Donna Muzny and Richard Gibbs that have contributed to the research results reported in this publication.
Funding
The Pharmacogenomics Evaluation of Antihypertensive Responses (PEAR) study was supported by the National Institute of Health Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida), UL1 TR000454 (Emory University), and UL1 TR000135 (Mayo Clinic). The PEAR study was also supported by funds from the Mayo Foundation. RNA-Seq data production was supported by the National Institutes of Health Pharmacogenetics Research Network grants U19-GM061388 and U19-GM061390.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviation
- BP
Blood Pressure
- DBP
Diastolic Blood Pressure
- eQTL
Expression Quantitative Trait Loci
- ERK
Extracellular Regulated Kinases
- FDR
False Discovery Rate
- GWAS
Genome-wide Association Studies
- HCTZ
Hydrochlorothiazide
- HTN
Hypertension
- PEAR
Pharmacogenomics Evaluation of Antihypertensives Response
- PP
Phosphatase Protein
- RNA-Seq
RNA Sequencing
- SBP
Systolic Blood Pressure
- SNP
Single Nucleotide Polymorphis
- VSMC
Vascular Smooth Muscle Cell
Authors’ contributions
ACCS drafted the manuscript and prepared the figures and tables. ACCS and AW performed the RNA-Seq data analyses. SD, MHS, YG and CWM developed the analysis plan for analysis and quality control of the RNA-Seq data and the statistical analysis of the data, and assisted in interpretation of the data. TYL and SES performed the laboratory work for sequencing, and assisted in the analysis and interpretation of the data. JAJ, STT, EB, RMC, WS, ABC, SES, JGG and ALB conceptualized the study and study design and secured funding. JAJ, RMC, STT, ABC, JGG conducted the clinical trial. All authors provided critical review of manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All study participants from PEAR and PEAR-2 (NCT00246519, NCT01203852 www.clinicaltrials.gov) provided written informed consent. The Institutional Review Boards at participating clinical trial sites including the University of Florida, Mayo Clinic, and Emory University approved both PEAR and PEAR-2. The studies were conducted in accordance with the principles of the Declaration of Helsinki and the US Code of the Federal Regulations for Protection of Human Subjects.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12920-018-0370-x) contains supplementary material, which is available to authorized users.
Contributor Information
Ana Caroline C. Sá, Email: carolsa@ufl.edu
Amy Webb, Email: Amy.Webb@osumc.edu.
Yan Gong, Email: gong@cop.ufl.edu.
Caitrin W. McDonough, Email: cmcdonough@cop.ufl.edu
Mohamed H. Shahin, Email: mhossam@cop.ufl.edu
Somnath Datta, Email: somnath.datta@ufl.edu.
Taimour Y. Langaee, Email: Langaee@cop.ufl.edu
Stephen T. Turner, Email: Turner.Stephen@mayo.edu
Amber L. Beitelshees, Email: abeitels@medicine.umaryland.edu
Arlene B. Chapman, Email: achapman1@medicine.bsd.uchicago.edu
Eric Boerwinkle, Email: Eric.Boerwinkle@uth.tmc.edu.
John G. Gums, Email: jgums@ufl.edu
Steven E. Scherer, Email: sscherer@bcm.edu
Rhonda M. Cooper-DeHoff, Email: Dehoff@cop.ufl.edu
Wolfgang Sadee, Email: Wolfgang.Sadee@osumc.edu.
Julie A. Johnson, Phone: 352-273-6309, Email: johnson@cop.ufl.edu
References
- 1.Oparil S, Schmieder RE. New approaches in the treatment of hypertension. Circ Res. 2015;116(6):1074–1095. doi: 10.1161/CIRCRESAHA.116.303603. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Executive summary: heart disease and stroke Statistics-2016 update a report from the American Heart Association. Circulation. 2016;133(4):447–454. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 3.Materson BJ. Variability in response to antihypertensive drugs. Am J Med. 2007;120(4):10–20. doi: 10.1016/j.amjmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on antihypertensive agents. N Engl J Med. 1993;328(13):914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 5.James PA. 2014 Evidence-based guideline for the Management of High Blood Pressure in adults: report from the panel members appointed to the eighth joint National Committee (JNC 8) (vol 311, pg 507, 2014) Jama-J Am Med Assoc. 2014;311(17):1809. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 6.Turner ST, Boerwinkle E, O'Connell JR, Bailey KR, Gong Y, Chapman AB, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62(2):391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chepelev I, Wei G, Tang QS, Zhao KJ. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009;37(16):e106. [DOI] [PMC free article] [PubMed]
- 8.Himes BE, Jiang XF, Wagner P, Hu RX, Wang QY, Klanderman B, et al. RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells. PLoS One. 2014;9(6) [DOI] [PMC free article] [PubMed]
- 9.Huan T, Esko T, Peters MJ, Pilling LC, Schramm K, Schurmann C, et al. A meta-analysis of gene expression signatures of blood pressure and hypertension. PLoS Genet. 2015;11(3):e1005035. doi: 10.1371/journal.pgen.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng ZY, Cheng YB, Tan BCM, Kang L, Tian ZJ, Zhu YK, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30(3):253. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 11.Gong Y, McDonough CW, Wang Z, Hou W, Cooper-DeHoff RM, Langaee TY, et al. Hypertension susceptibility loci and blood pressure response to antihypertensives: results from the pharmacogenomic evaluation of antihypertensive responses study. Circ Cardiovasc Genet. 2012;5(6):686–691. doi: 10.1161/CIRCGENETICS.112.964080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: rationale and design of the Pharmacogenomic evaluation of antihypertensive responses (PEAR) study. Am Heart J. 2009;157(3):442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, et al. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamadeh IS, Langaee TY, Dwivedi R, Garcia S, Burkley BM, Skaar TC, et al. Impact of CYP2D6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin Pharmacol Ther. 2014;96(2):175–181. doi: 10.1038/clpt.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. J Comput Biol. 2013;20(12):970–978. doi: 10.1089/cmb.2012.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafi T, Appel LJ, Miller ER, 3rd, Klag MJ, Parekh RS. Changes in serum potassium mediate thiazide-induced diabetes. Hypertension. 2008;52(6):1022–1029. doi: 10.1161/HYPERTENSIONAHA.108.119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosfield E., Jr Thiazide-induced hyperuricemia. N Engl J Med. 1963;268:562. doi: 10.1056/NEJM196303072681026. [DOI] [PubMed] [Google Scholar]
- 18.Duarte JD, Cooper-DeHoff RM. Mechanisms for blood pressure lowering and metabolic effects of thiazide and thiazide-like diuretics. Expert Rev Cardiovasc Ther. 2010;8(6):793–802. doi: 10.1586/erc.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith SM, Anderson SD, Wen S, Gong Y, Turner ST, Cooper-Dehoff RM, et al. Lack of correlation between thiazide-induced hyperglycemia and hypokalemia: subgroup analysis of results from the pharmacogenomic evaluation of antihypertensive responses (PEAR) study. Pharmacotherapy. 2009;29(10):1157–1165. doi: 10.1592/phco.29.10.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith SM, Gong Y, Turner ST, Cooper-DeHoff RM, Beitelshees AL, Chapman AB, et al. Blood pressure responses and metabolic effects of hydrochlorothiazide and atenolol. Am J Hypertens. 2012;25(3):359–365. doi: 10.1038/ajh.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat Methods. 2010;7(9):709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-DeHoff RM, et al. Plasma renin activity predicts blood pressure responses to beta-blocker and thiazide diuretic as monotherapy and add-on therapy for hypertension. Am J Hypertens. 2010;23(9):1014–1022. doi: 10.1038/ajh.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rau A, Marot G, Jaffrezic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics. 2014;15:91. doi: 10.1186/1471-2105-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 28.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45(10):1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sa ACC, Webb A, Gong Y, McDonough CW, Datta S, Langaee TY, et al. Whole transcriptome sequencing analyses reveal molecular markers of blood pressure response to thiazide diuretics. Sci Rep. 2017;7(1):16068. doi: 10.1038/s41598-017-16343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agnelli L, Bicciato S, Mattioli M, Fabris S, Intini D, Verdelli D, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23(29):7296–7306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 32.Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J, et al. Intrinsic subtypes of gastric cancer, based on gene Expr pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141(2):476–485. doi: 10.1053/j.gastro.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glastonbury CA, Vinuela A, Buil A, Halldorsson GH, Thorleifsson G, Helgason H, et al. Adiposity-dependent regulatory effects on multi-tissue transcriptomes. Am J Hum Genet. 2016;99(3):567–579. doi: 10.1016/j.ajhg.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.di Salvo TG, Yang KC, Brittain E, Absi T, Maltais S, Hemnes A. Right ventricular myocardial biomarkers in human heart failure. J Card Fail. 2015;21(5):398–411. doi: 10.1016/j.cardfail.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Salvo TG, Guo Y, Su YR, Clark T, Brittain E, Absi T, et al. Right ventricular long noncoding RNA expression in human heart failure. Pulm Circ. 2015;5(1):135–161. doi: 10.1086/679721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Morley M, Brandimarto J, Hannenhalli S, Hu Y, Ashley EA, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105(2):83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geeleher P, Cox NJ, Huang RS. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014;15(3):R47. doi: 10.1186/gb-2014-15-3-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wheeler HE, Aquino-Michaels K, Gamazon ER, Trubetskoy VV, Dolan ME, Huang RS, et al. Poly-omic prediction of complex traits: OmicKriging. Genet Epidemiol. 2014;38(5):402–415. doi: 10.1002/gepi.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minson J, Arnolda L, LlewellynSmith I, Pilowsky P, Chalmers J. Altered c-fos in rostral medulla and spinal cord of spontaneously hypertensive rats. Hypertension. 1996;27(3):433–441. doi: 10.1161/01.HYP.27.3.433. [DOI] [PubMed] [Google Scholar]
- 40.Rao F, Zhang L, Wessel J, Zhang K, Wen G, Kennedy BP, et al. Tyrosine hydroxylase, the rate-limiting enzyme in catecholamine biosynthesis - discovery of common human genetic variants governing transcription, autonomic activity, and blood pressure in vivo. Circulation. 2007;116(9):993–1006. doi: 10.1161/CIRCULATIONAHA.106.682302. [DOI] [PubMed] [Google Scholar]
- 41.Duff JL, Monia BP, Berk BC. Mitogen-activated protein (map) kinase is regulated by the map kinase phosphatase (Mkp-1) in vascular smooth-muscle cells. J Biol Chem. 1995;270(13):7161–7166. doi: 10.1074/jbc.270.13.7161. [DOI] [PubMed] [Google Scholar]
- 42.Touyz RM, Deschepper C, Park JB, He G, Chen X, Neves MF, et al. Inhibition of mitogen-activated protein/extracellular signal-regulated kinase improves endothelial function and attenuates Ang II-induced contractility of mesenteric resistance arteries from spontaneously hypertensive rats. J Hypertens. 2002;20(6):1127–1134. doi: 10.1097/00004872-200206000-00024. [DOI] [PubMed] [Google Scholar]
- 43.Connor JH, Weiser DC, Li S, Hallenbeck JM, Shenolikar S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol Cell Biol. 2001;21(20):6841–6850. doi: 10.1128/MCB.21.20.6841-6850.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429(6993):780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 45.Lipskaia L, Bobe R, Chen J, Turnbull IC, Lopez JJ, Merlet E, et al. Synergistic role of protein phosphatase inhibitor 1 and sarco/endoplasmic reticulum Ca2+ -ATPase in the acquisition of the contractile phenotype of arterial smooth muscle cells. Circulation. 2014;129(7):773–785. doi: 10.1161/CIRCULATIONAHA.113.002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picard N, Trompf K, Yang CL, Miller RL, Carrel M, Loffing-Cueni D, et al. Protein phosphatase 1 inhibitor-1 deficiency reduces phosphorylation of renal NaCl cotransporter and causes arterial hypotension. J Am Soc Nephrol. 2014;25(3):511–522. doi: 10.1681/ASN.2012121202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikram MK, Sim X, Jensen RA, Cotch MF, Hewitt AW, Ikram MA, et al. Four novel loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet. 2010;6(10):e1001184. doi: 10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mapping statistics for PEAR and PEAR-2 RNA-Seq data. Table S1. Potassium, glucose and uric acid mean changes in participants classified as non-responders after treatment with HCTZ and chlorthalidone. Table S2. Genes previously associated with BP/HTN (34 BP signature genes) and the expression measurements in PEAR and PEAR-2. Table S3. Differences in baseline expression levels for FOS, DUSP1 and PPP1R15A between thiazide diuretics responders and non-responders in PEAR and PEAR-2 with adjustment for age, gender and baseline blood pressure. Figure S2. P-value distribution for association with thiazide diuretics blood pressure response for 20 randomly selected genes. Table S4. Representative trans eQTL for top differentially expressed genes. Figure S3. Linkage disequilibrium plots between rs10655987, rs653178, rs10774625 and rs11066301. (DOCX 5615 kb)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


