Abstract
Although mechanisms for chromosomal instability in tumors have been described in animal and in vitro models, little is known about these processes in man. To explore cytogenetic evolution in human tumors, chromosomal breakpoint profiles were constructed for 102 pancreatic carcinomas and 140 osteosarcomas, two tumor types characterized by extensive genomic instability. Cases with few chromosomal alterations showed a preferential clustering of breakpoints to the terminal bands, whereas tumors with many changes showed primarily interstitial and centromeric breakpoints. The terminal breakpoint frequency was negatively correlated to telomeric TTAGGG repeat length, and fluorescence in situ hybridization with telomeric TTAGGG probes consistently indicated shortened telomeres and >10% of chromosome ends lacking telomeric signals. Because telomeric dysfunction may lead to formation of unstable ring and dicentric chromosomes, mitotic figures were also evaluated. Anaphase bridges were found in all cases, and fluorescence in situ hybridization demonstrated extensive structural rearrangements of chromosomes, with terminal transferase detection showing fragmented DNA in 5–20% of interphase cells. Less than 2% of cells showed evidence of necrosis or apoptosis, and telomerase was expressed in the majority of cases. Telomeric dysfunction may thus trigger chromosomal fragmentation through persistent bridge-breakage events in pancreatic carcinomas and osteosarcomas, leading to a continuous reorganization of the tumor genome. Telomerase expression is not sufficient for completely stabilizing the chromosome complement but may be crucial for preventing complete genomic deterioration and maintaining cellular survival.
Complex chromosome abnormalities occur in many malignancies, but the underlying mechanisms remain largely unknown. In many tumor types, the number of chromosomal aberrations increases in parallel to histological grade and the risk of metastasis, indicating a stepwise accumulation of cytogenetic changes during tumor progression (1–3). Several in vitro and animal models have shown that abrogation of DNA damage checkpoints or repair systems may lead to an elevated rate of chromosome mutations in cancer cells (4). Many human tumors also exhibit significantly shortened telomeric repeat sequences, which may trigger the formation of telomeric fusions between chromosome arms (5, 6). Such fusions lead to ring and dicentric chromosomes that at cell division form bridges, which may break and result in novel chromosome rearrangements by fusion of broken ends (7, 8). Some of these abnormal chromosomes may, in turn, form bridges at the next cell division. It has recently been demonstrated that such breakage-fusion-bridge (BFB) cycles triggered by telomeric dysfunction may play an important role for epithelial carcinogenesis in mice (9). If a similar relationship between BFB events and telomeric dysfunction is present also in human tumors is unclear.
The present study explores the evolution of chromosome aberrations in one epithelial and one mesenchymal tumor, pancreatic carcinoma (PC) and osteosarcoma (OS). These neoplasms are both characterized by complex chromosome aberrations and extensive genomic instability (10, 11). We used cytogenetic data to infer a temporal sequence of chromosomal breakpoints during tumor development. This model was correlated to telomere repeat status, BFB instability, and telomerase expression. The consequences of anaphase bridging for interphase chromosome structure and cellular survival were also investigated.
Materials and Methods
Cytogenetic Meta-Analyses.
Abnormal karyotypes of 169 PC and 106 OS were retrieved from the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman, December 2000). The OS were supplemented with 54 unpublished cases from Lund University Hospital (Sweden), and the University of Nebraska Medical Center. After removal of incomplete karyotypes with <5 resolved abnormalities and those with loss of the Y chromosome as the sole abnormality, 102 PC and 140 OS remained for analysis. For each case, net gains and losses of chromosome segments were calculated as previously described (12). As it has been shown that BFB cycles may result in structural as well as numerical changes (7, 8), the number of chromosome aberrations in a karyotype was measured as the sum of resolved structural and numerical changes plus the maximum number of unidentified ring and marker chromosomes.
Tissue Culture and Chromosome Preparation.
Peripheral blood lymphocytes and dermal fibroblasts were obtained from healthy donors. The osteosarcoma and the pancreatic carcinoma cell lines (OSA and LPC3, LPC6, LPC10, and LPC13) have been previously described (11, 13). Primary tumor cultures were initiated within 24 h after surgical excision. Harvesting and chromosome preparation were performed after 2 to 10 days (14). For analysis of mitotic figures, cells were harvested without Colcemid, washed in PBS for 5 min, fixed in methanol:acetic acid (3:1) for 30 min, air dried, and stained with hematoxylin & eosin. At least 10 anaphase cells were analyzed in each case. Long-term metaphase arrest was obtained by 0.03 μg/ml Colcemid treatment.
Fluorescence in Situ Hybridization (FISH) Analyses.
Telomeric TTAGGG repeats were detected by using fluorescein-conjugated (CCCTAA)3 peptide nucleic acid probes (15). Signal intensity was directly quantified by Cytovision software (Applied Imaging, Newcastle, UK). Whole chromosome painting probes were purchased from Oncor and Cambio (Cambridge, U.K.). Yeast artificial chromosome clones were obtained from Centre d'Étude du Polymorphisme Humain (Paris). Clones 939b4, 914c5, and 931a9 were located in 13q12, q14, and q31, respectively, clones 868h8, 427g11, 961f10, and 935a6 in 17p13.3, 17p13.1, 17p12, and 17p11, respectively, and clone 956d4 in 16q12. Clone positions were determined according to the Whitehead Institute radiation hybrid maps (www.wi.mit.edu). Probe preparation, labeling, hybridization, and detection were as described before (16).
Detection of Fragmented DNA.
Cells on chamber slides were washed in PBS and fixed in methanol for 20 min. DNA breaks were detected by incorporation of fluoroscein–dUTP by terminal deoxynucleotidyl transferase (In Situ Cell Death detection kit, Boehringer Mannheim). Enzyme treatment was at 37°C for 1 h. The cells were then washed in PBS for 3 × 2 min, dehydrated in ethanol, and counterstained with diamidinophenylindole.
Detection of Cell Death.
Methanol-fixed cells were rinsed in water, air-dried, and immersed in 100% ethanol for 3 min, followed by 1 min in 70% ethanol, 2 × 1 min in water, and 15 min in 0.06% potassium permanganate. The slides were then rinsed in water, immersed in 0.001% Fluoro-Jade (Histo-Chem, Jefferson, AR)/0.1% acetic acid for 30 min, stained 5 min in 2 μg/ml Hoechst 33342, and mounted in xylene. As a positive control for cell death, OSA cultures were treated with 2 μM calcium ionophore A23187 (Sigma) for 2 days.
Expression Analysis of the Telomerase Regulatory Subunit.
Total RNA extraction and cDNA synthesis were according to standard procedures. The reverse transcriptase subunit of telomerase (TERT) was coamplified with an ACTB fragment in a PTC-225 temperature cycler (MJ Research, Cambridge, MA). An initial 5-min step at 96°C was followed by 35 cycles at 96°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. Primers for ACTB (17) were added after 10 cycles. Amplified products were separated on a 1.8% agarose gel and visualized by using Vista Green nucleic acid gel stain (Amersham Pharmacia). A 145-bp segment outside the alternatively spliced region was amplified by using the primers TERT F1784 (CGGAAGAGTGTCTGGAGCAA) and TERT R1928 (GGATGAAGCGGAGTCTGGA), and alternative splice products of the TERT transcript were amplified by using the primers TERT F2164 (GCCTGAGCTGTACTTTGTCAA) and TERT R2620 (CGCAAACAGCTTGTTCTCCATGTC) as described by Ulaner et al. (18).
Results
Telomeric Breakpoints Occur Early in Chromosomal Evolution.
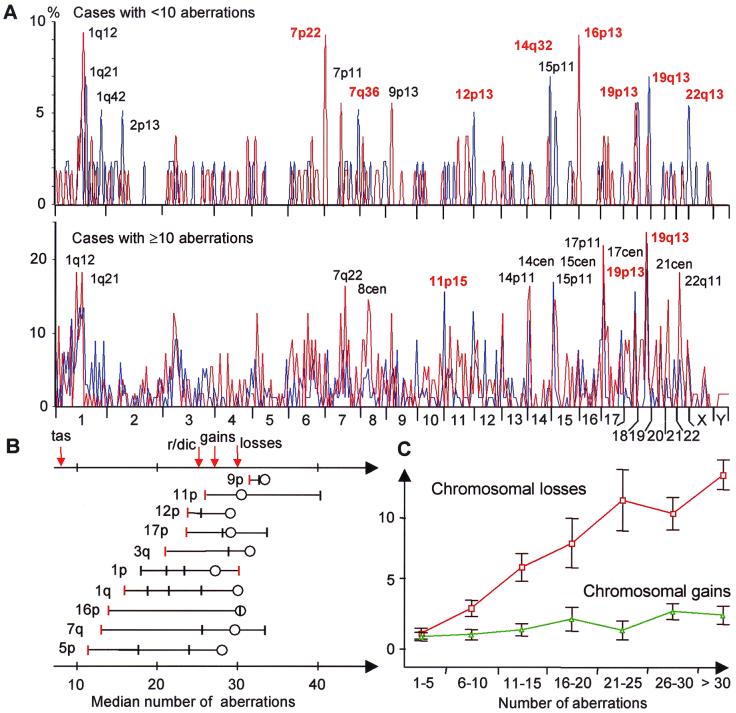
Chromosomal breakpoint profiles were constructed from 102 PC and 140 OS. The most common breakpoint in both tumor types was 19q13, occurring in 10% and 17% of the cases, respectively, followed by 1q21 (9%) and 17p11 (9%) in PC, and 19p13 (13%) and 17p11 (12%) in OS. All other breakpoints occurred at lower frequencies, with an extensive variation in breakpoint pattern among the tumors. However, the majority of karyotypes (60% of PC and 69% of OS) showed one or more breakpoints in telomeric bands. To investigate changes in breakpoint distribution during the accumulation of cytogenetic abnormalities, karyotypes were then divided into two groups: those with less than 10 aberrations and those with 10 or more aberrations (Fig. 1A). In the first group, telomeric breakpoints occurred more frequently than expected from a random distribution in both OS and PC (χ2 test; P = 0.01 and 0.007, respectively). Of the breakpoints occurring in >5% of cases, approximately half (43% in PC and 55% in OS) occurred in terminal bands. The second, more complex, group of karyotypes showed a less distinct breakpoint pattern. Only one-fourth or less (16% in PC and 25% in OS) of common (>5%) breakpoints were telomeric among these tumors.
Figure 1.
(A) Chromosome breakpoint profiles of 102 PC (blue line) and 140 OS (red line). Cases with <10 aberrations preferentially showed breakpoints in telomeric bands (red type), whereas cases with ≥10 aberrations preferentially had breakpoints in interstitial bands (black type); only the 15 most frequent breakpoints have been indicated. (B) Evolution of breakpoints along chromosomes in OS. For each chromosome arm, the median of the total number of aberrations per case has been indicated for tumors with terminal breakpoints (red bars), interstitial breakpoints (black bars), and chromosome arm losses (circles). In all arms, except 1p, terminal breaks occurred at lower levels of cytogenetic complexity than interstitial breaks. Telomeric associations (tas) occurred in tumors with few aberrations, followed by rings (r) and dicentrics (dic), gains, and finally losses of chromosome arms. (C) The mean number of whole chromosome losses (red) increased in parallel to the total cytogenetic complexity in OS, whereas the number of whole chromosome gains (green) remained constant; vertical bars represent confidence intervals.
To study the relative order of breakage events along single chromosomes, OS karyotypes were used to identify chromosome arms with at least two breakpoints, each occurring in at least five cases (Fig. 1B). Cases with terminal breakpoints had fewer changes than those with breakpoints occurring in nonterminal bands (Wilcoxon paired-difference test; P = 0.007). In all chromosome arms analyzed, except 1p, the terminal band broke first, commonly (5 of 10 arms) followed by breaks in the centromere or the pericentric bands (p11 or q11). Because of the more scattered distribution of breakpoints in PC, a similar analysis could not be performed here.
Abnormal Distribution of Telomeric Repeats in PC and OS.
The high rate of terminal breakpoints, especially in cases with few other chromosomal changes, suggests early disruption of telomeric integrity in the development of PC and OS. When comparing the terminal breakpoint frequencies to previously published measurements of the relative TTAGGG repeat lengths of chromosome arms (19), a negative correlation was found in both PC (r = −0.39, P = 0.007; Spearman rank-order correlation) and OS (r = −0.46, P = 0.001), indicating that the tendency for involvement in chromosomal rearrangements was partly related to telomere repeat length.
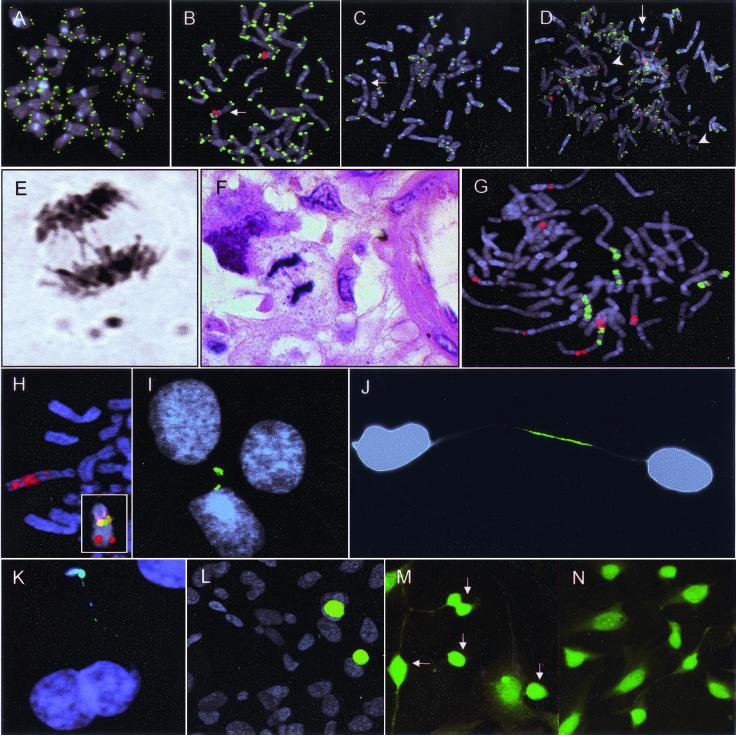
To visualize telomeric sequences, peptide nucleic acid probes for TTAGGG repeats were hybridized to metaphase cells. As controls, fibroblasts from one and lymphocytes from six individuals, age 19–60 years, as well as a myxoid liposarcoma with a 12;16-translocation as the only chromosome abnormality, were used. In all controls, >98% of chromosome termini showed signals of approximately equal intensity (Fig. 2 A and B). Metaphase cells from four PC (LPC3, LPC6, LPC10, and LPC13) and one OS cell line (OSA), as well as short-term cultures from 10 OS, were then analyzed (Table 1). The cell lines showed complex chromosomal aberrations by G-banding and lacked telomeric signals at 9–36% of chromosome termini in every cell (Fig. 2C). All but one of the 10 OS cultures showed complex karyotypes (Table 1). Because these cultures did not contain pure tumor cell populations, a centromere probe for chromosome 17 was cohybridized with the telomeric probes to identify abnormal metaphase cells. This chromosome is known to show copy-number alterations in OS at a high frequency (12). In eight of nine cases, cells with >2 copies of centromere 17 had 10–30% of chromosome termini lacking TTAGGG signals (Fig. 2D), compared with <2% in cells that were disomic. One case (OS10) showed strong signals, covering all chromosome ends, also in abnormal cells. The tumor with a simple karyotype (OS1) had large supernumerary ring chromosomes as the only abnormality. Aberrant cells were easily identified by counterstaining and showed a telomeric hybridization pattern identical to OS2–9.
Figure 2.
Hybridization with TTAGGG probes to (A) normal fibroblast metaphase cells resulted in signals of similar intensity at all chromosome termini. (B) Identical pattern in a liposarcoma cell with a t(12;16); the der(16) is identified by a 16q12 probe and diamidinophenylindol banding (red; arrow). (C) LPC6 exhibited several TTAGGG-negative chromosome ends and dicentric chromosomes (arrow). (D) Similar telomeric hybridization pattern in OS3 with ring (arrow) and dicentric (arrowhead) chromosomes, and with aneusomy for chromosome 17 (centromeric probes in red). (E and F) Anaphase bridges in LPC6 and OS4. (G) Whole chromosome paint shows extensive structural rearrangements of chromosomes 13 (green) and 17 (red) in OS7. (H) Four-color FISH with probes in 13q (Inset) demonstrates amplification of 13q31 sequences (red) in a marker chromosome in OSA. (I) Fragmented DNA (green) in a nuclear bleb and a micronucleus in LPC6, shown by the terminal transferase reaction. (J and K) Broken DNA in chromatin bridges in OSA and LPC10, respectively. (L) Apoptotic cells in A23187-exposed OSA cultures with fragmentation of whole nuclear DNA content (green). (M) Multiple dying cells (arrows) in A23187-exposed OSA cultures shown by Fluoro-Jade staining. (N) Low prevalence of dying cells in cultures not exposed to A23187.
Table 1.
Clinical data and results from telomere and telomerase analyses
| Case* | Age/sex | Grade†§ | Number of aberrations | TTAGGG negative termini (%)‡ | Bridges, %§ | TERT expression§ |
|---|---|---|---|---|---|---|
| Pancreatic carcinoma | ||||||
| LPC3 | 53/F | WD | 15 | 9–11 | 14 | Yes |
| LPC6 | 52/M | MD | 13 | 14–20 | 50 | Yes |
| LPC10 | 61/F | MD | 13 | 12–16 | 18 | Yes |
| LPC13 | 80/M | MD | 5 | 32–36 | 25 | No |
| Osteosarcoma | ||||||
| OSA | 19/M | — | 17 | 9–16 | 7 | Yes |
| OS1 | 44/M | II | 2 | 20–32 | 50 | — |
| OS2 | 61/F | IV | 13 | 14–19 | 40 | — |
| OS3 | 56/F | III–IV | 21 | 28–29 | 33 | — |
| OS4 | 48/F | III–IV | 8 | 17–27 | 48 | — |
| OS5 | 18/M | III–IV | 7 | 10–25 | 48 | — |
| OS6 | 15/F | III–IV | 43 | 14–16 | — | — |
| OS7 | 15/F | III–IV | 37 | 9–13 | — | — |
| OS8 | 38/M | III–IV | 17 | 13–21 | — | — |
| OS9 | 45/M | IV | 37 | 22–33 | — | — |
| OS10 | 34/M | IV | 20 | 0–2 | — | — |
Short-term tumor cultures in bold type.
WD, well differentiated; MD, moderately differentiated; osteosarcomas were graded from I–IV.
Range from 2 to 15 metaphase cells.
—, not determined.
Telomeric Dysfunction Is Associated with Breakage-Fusion-Bridge Events.
When anaphase cells were analyzed in the PC and OS cell lines and five of the OS short-term cultures, bridges were found in all cases (Fig. 2E) at frequencies between 7% and 50% (Table 1). To exclude the possibility that such bridges were in vitro artefacts, open biopsies taken before treatment from three of the OS were also analyzed. These showed bridges in 33–50% of anaphase cells (Fig. 2F). Another seven, randomly selected OS biopsies from untreated patients all showed bridges at frequencies between 10% and 60%. One of these cases was a low-grade tumor with five aberrations by G-banding, whereas the others were classical high-grade OS exhibiting karyotypes with at least eight abnormalities (data not shown). No bridges were found in normal fibroblasts or the myxoid liposarcoma with t(12;16).
Whole Chromosome Losses May Reflect Extensive Chromosome Fragmentation.
BFB events have previously been shown to cause complex alterations in chromosome structure (20, 21). Nondisjunction of chromosomes suspended in anaphase bridges could also lead to gains and losses of genetic material (7). Frequent bridging should thus lead to a progressive increase in structural rearrangements on the one hand, and chromosome gains and losses in equal numbers on the other. Indeed, in PC the number of whole chromosome gains and losses increased with the total number of aberrations in approximately equal proportions (data not shown). However, in OS, whole chromosome losses increased far more than whole chromosome gains (Fig. 1C). Chromosomes 13 and 17 showed the highest frequencies of loss (50%). Approximately 80% of OS karyotypes contained unidentified marker chromosomes that could potentially harbor material from these seemingly lost chromosomes. To investigate whether this was the case, metaphase cells from two high-grade OS (OS5 and OS7) and the OSA cell line, all showing −13 by G-banding, were hybridized with whole chromosome 13 paint and a panel of single-copy probes for 13q. In all cases, FISH demonstrated that chromosome 13 was involved in multiple unbalanced structural rearrangements with other chromosomes (Fig. 2 G and H). OS7 also showed loss of chromosome 17 by G-banding. At hybridization with a panel of four probes located along 17p, complex rearrangements leading to gain of sequences from 17p11 (probe 935a6) and 17p13 (probe 868h8) were revealed. Thus, a proportion of the whole chromosome losses in OS may actually reflect structural rearrangements leading to gain or loss of discrete chromosome segments rather than of whole chromosomes.
Fragmented DNA Is Retained in Interphase Nuclei with Little Impairment of Cellular Survival.
To test whether broken chromosomes were present at interphase in cell populations undergoing BFB events, the terminal deoxynucleotidyl transferase enzyme was used to incorporate fluorescently labeled nucleotides at sites of DNA fragmentation (22). Cultured fibroblasts and myxoid liposarcoma cells showed <0.1% positive cells. In contrast, both OSA and the PC cell lines showed chromatin regions positive for broken DNA in 5–20% of interphase cells. These areas were typically located in micronuclei, nuclear blebs, and fragmented chromatin bridges, but also occurred in the central parts of the nuclei (Fig. 2 I–K).
To compare this staining pattern to that in apoptotic cells, also known to harbor broken DNA, OSA cells were treated with the toxic calcium ionophore A23187. This resulted in a strong staining pattern for broken DNA covering the entire nucleus in ≈10% of cells, a pattern clearly different from that observed in unexposed cultures and most probably representing apoptotic cells (Fig. 2L). By the Fluoro-Jade assay, sensitive for both necrosis and apoptosis (23), 40% of the exposed OSA cells showed strong cytoplasmic and nuclear staining (Fig. 2M). In contrast, the prevalence of Fluoro-Jade-positive cells in unexposed OS and PC lines was low (0.4–1.5%, Fig. 2N) and comparable to that in normal fibroblast cultures (0.1%).
If the broken DNA in interphase resulted from BFB events, the prevalence of terminal deoxynucleotidyl transferase positive cells should be dependent on the separation of sister chromatids by the mitotic machinery. When normal spindle-fiber contraction in OSA was inhibited by Colcemid treatment for 8 days, the frequency of cells exhibiting broken DNA indeed decreased from 10% to 5.0% (Mann–Whitney U test; P < 0.05). The frequency of binucleated cells increased from 1.4% to 82%. Many of these cells exhibited intact chromatin strings between two or more nuclei, most probably representing bridges that had failed to undergo anaphase breakage.
Breakage-Fusion-Bridge Instability Occurs in the Presence of TERT Expression.
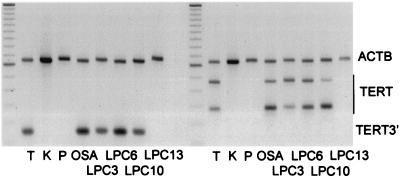
Human telomerase activity is determined by the expression of its reverse transcriptase subunit, TERT. However, the TERT transcript is alternatively spliced, and only the expression of full-length messages correlates with enzyme activity (18). Reverse transcriptase–PCR analysis of OSA and the PC lines revealed expression of full-length TERT in OSA, LPC3, LPC6, and LPC10 but not in LPC13 (Fig. 3). Full-length TERT was also found in thymus, used as a positive control, but not in normal kidney and pancreas, used as negative controls.
Figure 3.
TERT RNA expression measured by reverse transcriptase–PCR with primers outside the alternatively spliced region (TERT3′, Left) and primers for four alternative splice products (TERT, Right). TERT3′ expression as well as two major (239 and 421 bp) and two minor (275 and 457 bp) TERT products are present in thymus (T) and the cell lines OSA, LPC3, LPC6, and LPC10, but not in kidney (K), pancreas (P), or LPC13; β-actin (ACTB) was used as internal control.
Discussion
Chromosomal abnormalities in tumors may be caused by alterations in the untranscribed architectural elements of chromosomes. Data from mice deficient in the RNA subunit of telomerase have indicated that telomere dysfunction plays an important role in epithelial carcinogenesis by promoting unbalanced chromosome rearrangements (9). The present study demonstrates that similar mechanisms may play a role in the development of human PC and OS. FISH with TTAGGG-specific probes demonstrated an extensive heterogeneity in telomere signal intensity as well as chromosome ends lacking signals in the majority of cases. Analysis of anaphase figures in cell lines and tumor biopsies from untreated patients showed frequent BFB events in both high- and low-grade tumors. These processes were also reflected in the karyotypic patterns. The terminal breakpoint frequency in PC and OS was negatively correlated to telomere repeat length. Furthermore, tumors with a relatively low number of chromosomal abnormalities showed a high frequency of terminal breakpoints, whereas those with more complex karyotypes had preferentially interstitial and centromeric breaks. This is consistent with a model in which BFB cycles are initiated by telomere fusion, followed by a gradual shift of breakpoints from the chromosomal termini toward central regions as repeated anaphase breakage erodes the chromosome arms (8). The finding that the frequent whole chromosome losses in OS actually reflected cryptic structural rearrangements is also in accordance with this scenario.
Because telomere shortening occurs at all chromosome ends, telomere-based instability would be expected to have a broad effect on the chromosome complement. Yet, the distribution of chromosome changes should not be entirely random. It is known that the length of terminal repeat sequences varies among chromosome arms, but the number of TTAGGG repeats on specific arms is similar in different tissues from the same individual and varies little between individuals (19). Hence, if telomere shortening would be the dominant mechanism for generating cytogenetic changes, one would expect considerable similarities in the aberration patterns in tumors from different tissues as well as from different patients. Indeed, similar profiles of chromosomal breakpoints and imbalances have been found among many tumor entities with complex karyotypes, including not only PC and OS but also lung, breast, colon, head and neck, and ovarian carcinomas (ref. 12; see also http://cgap.nci.nih.gov/Chromosomes/Mitelman). This indicates that histogenetic origin is not a strong determinant of chromosomal complexity in these tumors. Rather, the cytogenetic pattern may reflect the tendency of different chromosomes to enter BFB cycles. However, the correlation coefficient between breakpoint frequency and telomere length was low in both OS and PC. Undoubtedly, selection pressure and other factors, such as site-directed chemical mutagenesis and the induction of common fragile sites, may also influence the breakpoint pattern.
Breakage of ring and dicentric chromosomes at anaphase has so far not been characterized at the molecular level. The classical model for BFB events implies that anaphase bridging typically leads to one break in each chromatid (7, 8). Alternatively, the kinetochore detaches from the mitotic spindle, resulting in nondisjunction. However, the present study demonstrated multiple sites of chromosomal breakage per cell in populations undergoing BFB events. Pools of fragmented DNA were detected in abnormal nuclear structures, including blebs, micronuclei, and strings. Blebs and micronuclei can be remnants of broken anaphase bridges, whereas the strings probably represent chromatin bridges not fully disentangled after anaphase (24). This indicates that the traditional model for BFB instability is not directly applicable to OS and PC; instead of a few isolated breaks, anaphase bridging often leads to extensive fragmentation of chromosomal DNA. When tumor cells were prevented from entering anaphase, the frequency of broken DNA declined significantly. A possible explanation is that cells with broken chromosomes were gradually eliminated by apoptosis. However, the lower frequency of dying cells than cells with fragmented DNA in all five cell lines with BFB instability did not support this explanation. Instead, repair of broken ends may have occurred.
One mechanism for stabilizing broken chromosome ends could be the addition of novel TTAGGG repeats by telomerase (25). The majority of human cancers express the telomerase catalytic subunit TERT (18), and so did four of five PC and OS cell lines in this study. Nevertheless, these cell lines all exhibited frequent BFB events and several chromosome ends that were negative by TTAGGG probes. The absence of TTAGGG signals could either signify a total loss or a low copy number of telomeric repeats (15). The presence of fragmented DNA in interphase nuclei makes it likely that broken chromosome ends, totally lacking telomeric repeats, did in fact persist in some OS and PC cells. However, the TERT-negative cell line LPC13 showed a higher frequency of TTAGGG-negative ends than the TERT-positive lines. Taken together with other studies (26), this indicates that telomerase may still play a crucial role in karyotypic evolution by limiting the extent of chromosomal instability. It remains questionable whether this occurs only through elongation of remaining TTAGGG sequences, or whether telomerase acts on interstitial chromosome breaks as well. It is also possible that broken chromosome ends could heal by a mechanism related to nonhomologous end-joining, leading to fusion of broken chromosome fragments (8, 25). Indeed, the karyotypic pattern in PC and OS is well in accordance with fusion and reshuffling of broken ends into abnormal chromosomes with a highly complex architecture (10, 11, 16). The ends of these structures could be stabilized by parts of broken chromosomes still carrying TTAGGG sequences of sufficient length. On the other hand, fusion could result in the formation of mitotically dysfunctional ring and dicentric chromosomes that would proceed through further BFB events. The finding of BFB events and TTAGGG-negative chromosome ends at a high frequency in OS and PC, independent of tumor grade and cytogenetic complexity, indicates that telomeric dysfunction occurs at an early point in the development of these tumors and that the resulting BFB cycles are maintained during disease progression. Stabilizing factors such as telomerase expression and fusion of broken ends probably play vital roles in this process by keeping genomic instability at a moderate level and maintaining a high rate of cellular survival.
Acknowledgments
We are grateful to Professor Julia Bridge (University of Nebraska Medical Center) for providing unpublished reviewed karyotypes. This study was supported by the Swedish Cancer Society, the Swedish Child Cancer Fund, the IngaBritt and Arne Lundberg Research Foundation, and the John and Augusta Persson Foundation. D.G. was also supported by the Swedish Medical Society, the Blanceflor Foundation, a Lennander Scholarship, and a Young Investigator's Award from the American Cancer Society.
Abbreviations
- BFB
breakage-fusion-bridge
- FISH
fluorescence in situ hybridization
- OS
osteosarcoma
- PC
pancreatic carcinoma
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12331.
References
- 1.Nishizaki T, DeVries S, Chew K, Goodson W H, Ljung B M, Thor A, Waldman F M. Genes Chromosomes Cancer. 1997;19:267–272. doi: 10.1002/(sici)1098-2264(199708)19:4<267::aid-gcc9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mandahl N, Mertens F. Cancer Genet Cytogenet. 1997;95:1–8. doi: 10.1016/s0165-4608(96)00252-x. [DOI] [PubMed] [Google Scholar]
- 3.Al-Mulla F, Keith W N, Pickford I R, Going J J, Birnie G D. Genes Chromosomes Cancer. 1999;24:306–314. doi: 10.1002/(sici)1098-2264(199904)24:4<306::aid-gcc3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Nature (London) 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 6.Counter C M, Hirte H W, Bacchetti S, Harley C B. Proc Natl Acad Sci USA. 1994;91:2900–2904. doi: 10.1073/pnas.91.8.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClintock B. Genetics. 1938;23:215–376. doi: 10.1093/genetics/23.4.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClintock B. Genetics. 1940;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Artandi S E, Chang S, Lee S L, Alson S, Gottlieb G J, Chin L, DePinho R A. Nature (London) 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher J A, Gebhardt M C, Kozakewich H P. Cancer Genet Cytogenet. 1994;77:81–88. doi: 10.1016/0165-4608(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 11.Gorunova L, Höglund M, Andrén-Sandberg A, Dawiskiba S, Jin Y, Mitelman F, Johansson B. Genes Chromosomes Cancer. 1998;23:81–99. doi: 10.1002/(sici)1098-2264(199810)23:2<81::aid-gcc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 12.Mertens F, Johansson B, Höglund M, Mitelman F. Cancer Res. 1997;57:2765–2780. [PubMed] [Google Scholar]
- 13.Roberts W M, Douglass E C, Peiper S C, Houghton P J, Look A T. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- 14.Mandahl N. In: Human Cytogenetics: A Practical Approach. Malignancy and Acquired Abnormalities. Rooney D E, editor. Oxford: Oxford Univ. Press; 2001. [Google Scholar]
- 15.Landsdorp P M, Verwoerd N P, van de Rijke F M, Dragowska W, Little M-T, Dirks R W, Raap A K, Tanke H J. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 16.Gisselsson D, Mandahl N, Pålsson E, Gorunova L, Höglund M. Genes Chromosomes Cancer. 2000;28:347–352. [PubMed] [Google Scholar]
- 17.Raff T, van der Giet M, Endemann D, Wiederholt T, Paul M. BioTechniques. 1997;23:456–460. doi: 10.2144/97233st02. [DOI] [PubMed] [Google Scholar]
- 18.Ulaner G A, Hu J F, Vu T H, Giudice L C, Hoffman A R. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 19.Martens U M, Zijlmans J M, Poon S S, Dragowska W, Yui J, Chavez E A, Ward R K, Lansdorp P M. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- 20.Saunders W S, Shuster M, Huang X, Gharaibeh B, Enyenihi A H, Petersen I, Gollin S M. Proc Natl Acad Sci USA. 2000;97:303–308. doi: 10.1073/pnas.97.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gisselsson D, Pettersson L, Höglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. . (First Published April 18, 2000; 10.1073/pnas.090013497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmued L C, Hopkins K J. Toxicol Pathol. 2000;28:91–99. doi: 10.1177/019262330002800111. [DOI] [PubMed] [Google Scholar]
- 24.Gisselsson D, Björk J, Höglund M, Mertens F, Dal Cin P, Åkerman M, Mandahl N. Am J Pathol. 2001;158:199–206. doi: 10.1016/S0002-9440(10)63958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong K K, Chang S, Weiler S R, Ganesan S, Chaudhuri J, Zhu C, Artandi S E, Rudolph K L, Gottlieb G J, Chin L, et al. Nat Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 26.Counter C M, Avilion A A, LeFeuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]