Abstract
It is estimated that nearly 2 billion people currently suffer from latent Mycobacterium tuberculosis infection. Although the key front-line antituberculosis drugs are effective in treating individuals with acute tuberculosis, these drugs are ineffective in eliminating M. tuberculosis during the persistent stages of latent infection. Consequently, therapeutics that directly target persistent bacilli are urgently needed. We have conducted a global analysis on a group of regulatory determinants that may play a role in M. tuberculosis virulence, and identified a two-component response regulator whose expression is required for entrance into and maintenance of persistent infection. Inactivation of this response regulator, Rv0981 (termed here mprA for mycobacterial persistence regulator), affected M. tuberculosis H37Rv growth in vivo in an organ- and infection stage-specific fashion. These results indicate that two-component systems are important for adaptation of the tubercle bacillus during stages of persistent infection.
Tuberculosis remains a serious health concern worldwide with more than one-third of the world's population being latently infected with Mycobacterium tuberculosis (1). The success of M. tuberculosis as a pathogen is caused, in part, by its ability to survive in macrophages and establish long-term, persistent infection in the host during periods of control by the cell-mediated immunity. The persistent, latent infection is one of the least understood aspects of tuberculosis despite the fact that latency plays a crucial role in the propagation of tuberculosis and maintenance of effective infectious cycles in human populations (2). Although several M. tuberculosis factors contributing to the initial stages of disease pathogenesis have been identified (2–6), little is known about factors that contribute to persistent infection. Recently, isocitrate lyase, an enzyme in the glyoxylate shunt, was shown to be required by M. tuberculosis for survival in macrophages in vitro and during persistent infection in vivo (7), implicating intermediary metabolism adjustments during latency periods. Cell-surface components also contribute to long-term persistence of M. tuberculosis in vivo (8). Other candidate determinants, including a family of glycine-rich proteins, Pro/Glu polymorphic repetitive sequences with unknown function are required for persistent infection and granuloma formation in poikilothermic animal models infected with another pathogenic mycobacterial species, Mycobacterium marinum (9).
Adaptive responses and virulence determinants in pathogenic organisms are frequently controlled by two-component signal transduction systems (10, 11). M. tuberculosis contains 11 complete two-component systems and several unlinked sensor kinase or response regulator homologs (12). The importance of two-component systems in M. tuberculosis physiology has already been suggested. For example, mtrA is an essential gene in the tubercle bacillus (13), and the prrA-prrB two-component system is expressed during growth of M. tuberculosis in human macrophages in vitro but not during growth in artificial media (14). Here we present a concerted effort to examine the role of two-component signal transduction systems in the physiology and virulence of M. tuberculosis. We show expression analysis of M. tuberculosis response regulator genes in infected macrophages by using global analysis with green fluorescent protein (GFP) technology, and present evidence indicating a role for these systems in persistence-related adaptation phenomena, including entrance into and maintenance of chronic infection stages.
Materials and Methods
Bacterial Strains and Growth Conditions.
M. tuberculosis H37Rv (ATCC 27294), Mycobacterium bovis bacillus Calmette–Guérin Pasteur (ATCC 27291), and Escherichia coli DH5α were used. Mycobacteria were grown under standard laboratory conditions in Middlebrook 7H9 broth and Middlebrook 7H10 or 7H11 agar medium (Difco Laboratories) supplemented with 0.5% glycerol, 10% albumin-dextrose-catalase or oleic acid-albumin-dextrose-catalase, and 0.05% Tween 80. E. coli was grown in LB medium. When required, media were supplemented with 25 or 50 μg of kanamycin sulfate, or 50 and 100 μg of hygromycin B for Mycobacterium and E. coli, respectively.
Genetic Analyses.
Homologs of M. tuberculosis H37Rv two-component systems and adjacent genes were determined from currently available mycobacterial genomes including Mycobacterium avium strain 104, Mycobacterium leprae, M. bovis, Mycobacterium smegmatis, and M. tuberculosis CSU#93 by using blast database searches available from the Sanger Center (www.sanger.ac.uk) or TIGR (www.tigr.org). Conservation of M. tuberculosis two-component systems in other mycobacteria was further confirmed from detailed genetic analyses of cosmid or genomic sequence information.
Construction of gfp Reporter Plasmids and Analysis of GFP Fluorescence.
Plasmids pmtrA-gfp, phsp60-gfp, and pMYGFP2 have been described (15). PCR was used to amplify promoter regions from the Rv0844c (narL), Rv0757 (phoP), Rv0903c (prrA), Rv0981 (mprA), Rv1033c (tcrR), Rv3133c (devR), Rv3765c, Rv0818, Rv1626, and Rv2884 M. tuberculosis H37Rv response regulators. PCR products were cloned into pMYGFP2. All constructs were sequenced to confirm the correct orientation of promoter fragments with gfp. Transformants were screened by semiquantitative PCR for the presence of gfp by using equal amounts of template chromosomal DNA. Infection of macrophage monolayers and fluorescence microscopic analyses have been described (13). When indicated, infected macrophage monolayers were stained with auramine-rhodamine to visualize intracellular bacilli present but not fluorescent by GFP.
Disruption and Complementation of the Rv0981 Response Regulator in M. tuberculosis H37Rv.
pTZ198, a derivative of pPR27 (16), was used for the disruption of Rv0981 and was constructed as follows. An NheI-XbaI fragment containing the xylE reporter gene from pHSX-1 (17) was first cloned into the XbaI site of pPR27 to create pTZ184. A 2.0-kb fragment containing the Rv0981 gene including upstream and downstream sequences was amplified from the H37Rv chromosome and cloned into pTZ184 to create pTZ186. Finally, an NheI-SpeI fragment encoding the Kmr cassette from pMV206 (18) was cloned into an engineered SpeI site present in the Rv0981 coding sequence from pTZ186 to create pTZ198. Disruption of Rv0981 was as described (16). xylE− colonies (white upon spraying with catechol) resistant to both Km and 2% sucrose were screened by PCR for legitimate double-crossover homologous recombination. Insertional inactivation was confirmed by Southern blot analysis with an Rv0981-specific probe generated by PCR. The Rv0981∷Kmr mutant was complemented by transformation with pTZ215, a site-specific integration vector carrying wild-type Rv0981-Rv0982. The Rv0981-Rv0982 coding sequence was PCR-amplified from M. tuberculosis H37Rv by using Pfu polymerase and primers containing engineered NotI sites at their 5′ ends. This fragment was cloned into pCR2.1, digested with NotI, and cloned into the NotI site of pTZ189, a derivative of pMV306 (18) containing the Hygr gene from pOLYG (19). pTZ215 was sequenced to confirm the absence of mutations in the Rv0981-Rv0982 coding sequence.
M. tuberculosis Survival in Murine and Human Macrophages.
Wild-type M. tuberculosis H37Rv, the isogenic Rv0981∷Kmr mutant, or Rv0981∷Kmr [attB∷pTZ215] were infected into J774 macrophages, bone marrow macrophages derived from C57BL/6 mice (20), or human macrophages derived from peripheral blood monocytes (21) as described (13). For some experiments, murine macrophages were activated 24 h before infection and during the course of infection by the addition of 500 units of mouse IFN-γ and 500 ng of bacterial lipopolysaccharide (LPS) to tissue culture medium. Bacterial survival was determined by diluting macrophage lysates in sterile water containing 0.05% Tween and plating onto 7H10 medium.
In Vivo Growth Competition Between M. tuberculosis H37Rv and Rv0981∷Kmr.
Virulence of the Rv0981∷Kmr mutant of M. tuberculosis was measured by using an in vivo growth competition assay. Groups of five 6- to 8-week-old BALB/c mice (Harlan Sprague, Indianapolis) were infected intravenously with a bollus of 5.7 × 104 single-cell tubercle bacilli containing a mixture of wild-type H37Rv and its isogenic mutant Rv0981∷Kmr. At various times after infection (t = 24 h, 14 days, 28 days, 119 days, and 147 days) mice were killed and the lung, spleen, and liver recovered. Organs were homogenized and diluted in PBS containing 0.05% Tween 80. Total colony-forming units were determined by plating onto 7H11 agar medium. The fraction of mutant bacilli was subsequently determined by patching onto 7H10 agar medium and 7H10 agar medium containing kanamycin. Before the 147-day point, a group of five mice were given two s.c. injections (spaced 5 days apart) of a 25-mg hydrocortisone acetate emulsion (22) to suppress the immune system during the persistent stage of infection.
Statistical Analyses.
All statistical analysis (ANOVA and Fisher's protected least significant difference) was performed with ANOVA (Version 1.11; Abacus Software, Abacus Concepts, Berkley, CA).
Results and Discussion
Global Analysis of M. tuberculosis Two-Component Systems.
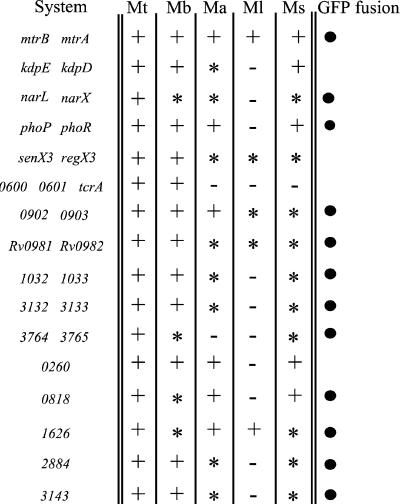
Fig. 1 shows comparative genomic analysis of two-component systems in M. tuberculosis and other mycobacterial species. It also indicates the response regulator genes where it was possible to generate promoter-gfp fusions because of paired arrangements with cognate sensor kinase genes. Transcriptional fusions between response regulator promoters and gfp were introduced into M. tuberculosis H37Rv or M. bovis bacillus Calmette–Guérin Pasteur and analyzed for expression levels by epifluorescence microscopy upon infection of macrophages. Two types of cells were used: the murine macrophage-like cell line J774, and human peripheral blood monocyte-derived macrophages (Fig. 2). M. tuberculosis response regulators could be grouped into five classes based on expression patterns: (i) those constitutively expressed in vivo and in vitro (data not shown) in both M. bovis bacillus Calmette–Guérin and M. tuberculosis H37Rv (Fig. 2, class I; e.g., phoP and Rv0818); (ii) those expressed in M. bovis bacillus Calmette–Guérin and in M. tuberculosis H37Rv during growth in macrophages with signs of induction of expression in vivo (Fig. 2, class II; e.g., mtrA; ref. 13); (iii) those expressed in M. bovis bacillus Calmette–Guérin but not expressed in M. tuberculosis H37Rv under the conditions tested (Fig. 2, class III; e.g., Rv0981 and Rv3143); (iv) those with no detectable expression in M. bovis bacillus Calmette–Guérin but induced for expression in M. tuberculosis H37Rv during growth in macrophages (Fig. 2, class IV; e.g., Rv3133c); and (v) those with no detectable expression by the method used in our study (narL, Rv0903c, Rv1033c, Rv3765c, Rv1626, and Rv2884).
Figure 1.
Conservation of M. tuberculosis H37Rv two-component signal transduction systems. Homologs of these genes in other mycobacterial species were: +, present with conserved genetic arrangement; ✻, present with differences in genetic arrangement; or −, absent from the genomes of M. avium (Ma), M. bovis (Mb), M. leprae (Ml), M. smegmatis (Ms), and M. tuberculosis CSU#91 (Mt). When indicated (●), promoter fragments from response regulators were fused to the promoterless gfp gene. Note that not all response regulator promoters could be fused to gfp because of paired arrangements with sensor kinase genes.
Figure 2.
In vivo expression of M. tuberculosis response regulators in macrophages. Response regulator promoter fusions were transformed into M. tuberculosis H37Rv or M. bovis bacillus Calmette–Guérin Pasteur and analyzed for expression during growth in macrophages by epifluorescence microscopy. Semiquantitative PCR was used to analyze differences in reporter plasmid copy number between strains. M. tuberculosis response regulator fusions not shown exhibited no detectable fluorescence in M. bovis bacillus Calmette–Guérin or in M. tuberculosis during growth in vitro or during growth in macrophages. +, 0–100 relative fluorescence units (RFU); ++, 100–225 RFU; +++, 225–450 RFU.
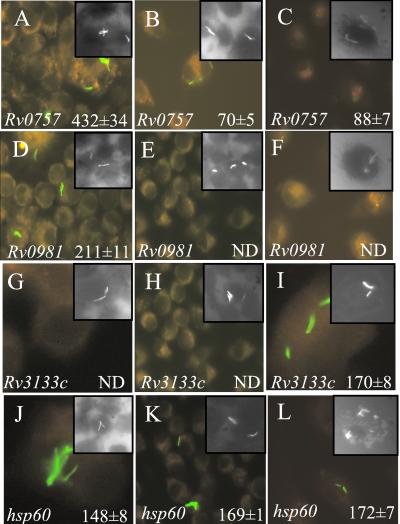
Expression of reporter constructs from class I was consistently higher in M. bovis bacillus Calmette–Guérin than expression levels observed in M. tuberculosis H37Rv during growth in vitro (data not shown) and during growth in macrophages (Fig. 2). For example, after 3 days of growth in J774 macrophages, the mean relative fluorescence intensity from the pphoP-gfp reporter plasmid in M. bovis bacillus Calmette–Guérin was 432 ± 34, whereas the mean relative fluorescence intensity from the same reporter construct in M. tuberculosis H37Rv was 70 ± 5 (Fig. 3 A and B; P < 0.0001; ANOVA). Similar results were obtained for pphoP-gfp after growth of M. tuberculosis H37Rv in macrophages derived from human peripheral blood monocytes (Fig. 3C; P < 0.0001; ANOVA). Differences in expression levels were not simply caused by variation in plasmid copy number or loss of reporter constructs during infection, because similar amounts of gfp were amplified from strains before infection by using semiquantitative PCR (Fig. 2), and expression from strains containing the phsp60-gfp control plasmid did not vary between M. tuberculosis H37Rv and M. bovis bacillus Calmette–Guérin (Fig. 2, row g; Fig. 3 J–L).
Figure 3.
Epifluorescence microscopy of macrophage-grown M. tuberculosis H37Rv or M. bovis bacillus Calmette–Guérin representative response regulator promoter fusions from class I (A–C), class III (D–F), class IV (G–I), or the hsp60-gfp control (J–L) are shown. Mean fluorescence intensity was determined from M. bovis bacillus Calmette–Guérin-infected J774 monolayers (A, D, G, and J), M. tuberculosis H37Rv-infected J774 monolayers (B, E, H, and K), or M. tuberculosis H37Rv-infected human peripheral blood monocyte-derived macrophages (C, F, I, and L). Insets show intracellular bacilli stained with rhodamine-auramine.
Expression of Rv0981 Is Repressed in M. tuberculosis H37Rv but Not in M. bovis Bacillus Calmette–Guérin During Growth in Macrophages.
We next focused on the response regulators that we hypothesized might play a role in persistent infection. We reasoned that class III response regulators may be candidate regulators of M. tuberculosis virulence factors that play a role in later stages of infection, because their expression was silenced in virulent M. tuberculosis H37Rv, whereas their expression was deregulated and detectable in the avirulent M. bovis bacillus Calmette–Guérin strain. The response regulator Rv0981 fits this pattern because it showed a strong difference in expression between M. bovis bacillus Calmette–Guérin, where it was inducible in macrophages, and M. tuberculosis H37Rv, where its expression in macrophages was not detectable by monitoring gfp transcriptional fusion activity (Fig. 2, row d). Fluorescence of M. bovis bacillus Calmette–Guérin containing the pRv0981-gfp reporter construct was induced after growth in J774 macrophages (Fig. 2, row d) and showed fluorescence of 211 ± 11 relative units (Fig. 3D). Increased Rv0981 expression was also quantitated by flow cytometric analysis of M. bovis bacillus Calmette–Guérin in macrophages and changed from 9% to 22% in the high fluorescence gate depending on time after infection. In contrast, fluorescence from the pRv0981-gfp reporter construct in M. tuberculosis H37Rv was undetectable during growth in vitro (data not shown) and during growth in J774 macrophages at all times examined (Fig. 2, row d; Fig. 3E). Rv0981 expression was also undetectable in M. tuberculosis (pRv0981-gfp)-infected human monocyte-derived macrophages (Fig. 2, row d; Fig. 3F). The inability to detect fluorescence from pRv0981-gfp in M. tuberculosis during intracellular growth was not a result of inefficient infection into macrophage monolayers, because intracellular bacilli were detected in all macrophage infections by rhodamine-auramine staining (Fig. 3 D–F Insets), and the presence of pRv0981-gfp in M. tuberculosis H37Rv was verified by PCR (Fig. 2). However, the absence of a discernible GFP signal with Rv0981 in M. tuberculosis H37Rv does not preclude low-level expression of this gene. For example, another response regulator, Rv0903c, not expressed at high levels in our experiments (i.e., being classified with five other response regulators in the category of genes with no detectable signals by using gfp fusions) has been reported as having detectable expression by a PCR-based method (14). Furthermore, inactivation of Rv0981 in M. tuberculosis H37Rv had a phenotypic effect on growth in macrophages (see next section), supporting the notion that this gene is most likely expressed in the intracellular environment of phagocytic cells, albeit at low levels. Nevertheless, it is possible to conclude that expression of Rv0981 is differentially regulated in avirulent M. bovis bacillus Calmette–Guérin where it is expressed upon macrophage infection, relative to virulent M. tuberculosis H37Rv, where its expression was not detectable by GFP for the duration of in vitro macrophage infection.
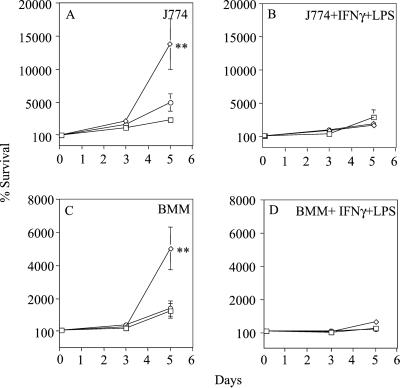
Inactivation of Rv0981 Enhances M. tuberculosis H37Rv Survival in Resting Macrophages.
The high-level expression of Rv0981 in the avirulent strain M. bovis bacillus Calmette–Guérin and its silencing or low-level expression in the virulent M. tuberculosis strain H37Rv during growth in macrophages at first seems counterintuitive if a gene is to be associated with virulence. However, the pattern of Rv0981 expression observed in M. tuberculosis and M. bovis bacillus Calmette–Guérin during growth in macrophages in vitro fit the profile of the type of regulator that we rationalized may play a role in persistent infection. To test this hypothesis, we generated an Rv0981∷Kmr mutant of M. tuberculosis H37Rv by allelic exchange (see Materials and Methods). First, we tested survival of the Rv0981∷Kmr mutant in macrophages infected in vitro. Growth of the Rv0981∷Kmr mutant was significantly higher than that observed with wild-type H37Rv after infection into J774 and murine bone marrow-derived macrophages (Fig. 4 A and C). For example, the number of Rv0981∷Kmr bacilli recovered 5 days after infection was significantly higher than the number of wild-type H37Rv recovered from macrophages (Fig. 4 A and C; P < 0.0001; ANOVA). Growth of the Rv0981∷Kmr mutant was also higher after infection of human macrophages derived from peripheral blood monocytes (data not shown). The difference in survival between the Rv0981∷Kmr mutant and wild-type H37Rv was not simply caused by differences in growth rate, because no significant differences in growth were observed in 7H9 medium during the same period. The improved survival in macrophages of the Rv0981∷Kmr mutant relative to wild-type H37Rv was also not caused by a polar effect of the Kmr insertion on downstream gene expression, because complementation of the Rv0981∷Kmr mutant with wild-type Rv0981-Rv0982, Rv0981∷Kmr [attB∷pTZ215], reversed the improved growth of the mutant in macrophages to wild-type levels (Fig. 4 A and C). We also examined survival of the Rv0981∷Kmr mutant during infection of macrophages activated with IFN-γ and LPS. The enhanced growth of the Rv0981∷Kmr mutant in resting macrophages was abrogated upon activation with IFN-γ an LPS (Fig. 4 B and D).
Figure 4.
Survival of M. tuberculosis strains growing in macrophages in vitro. Wild-type H37Rv (squares), the Rv0981∷Kmr mutant (diamonds), or Rv0981∷Kmr [attB∷pTZ215] (circles) were used to infect J774 macrophages (J774; A and B) and murine bone marrow-derived macrophages (BMM; C and D). When indicated, macrophages were activated with IFN-γ and LPS 24 h before and during the course of infection (B and D). Bacterial survival has been normalized to the 2-h time point and set to 100%. No growth of M. tuberculosis was observed in tissue culture medium alone. Values represent the mean and standard error of at least three independent experiments. Statistical significance for Rv0981∷Kmr relative to H37Rv or Rv0981∷Kmr [attB∷pTZ215] at each time point: ✻✻, P < 0.01 (ANOVA).
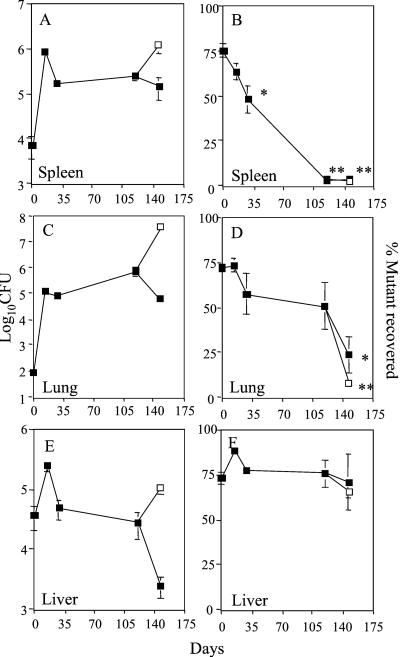
Response Regulator Rv0981 Is Important for Persistent Infection in an Organ- and Infection Stage-Specific Fashion.
To examine the profile of the Rv0981∷Kmr mutant for survival in vivo, we performed growth experiments by using a modification of a low-infection-dose murine model of tuberculosis (23). Mice infected with M. tuberculosis by this method develop a persistent infection (24) that can be reactivated during the chronic stage on suppression of the immune system (22). We chose to examine differences in virulence between the Rv0981∷Kmr mutant and its parental strain with an in vivo growth competition assay (25–27), because this method is highly sensitive and results in a low variation in inoculum dosage between strains (25–29). After i.v. infection of BALB/c mice, the Rv0981∷Kmr mutant was evaluated in lungs, spleens, and livers of infected animals at different stages of infection. The infection presented a typical course in total colony-forming units recovered from different organs at various times after inoculation (Fig. 5 A, C, and E; ref. 30). In the spleen, the Rv0981∷Kmr mutant was attenuated for growth during the acute phase of infection and failed to establish a persistent infection (Fig. 5B). For example, although the mutant comprised 75% ± 3% of total colony-forming units in the spleen 1 day after infection, the Rv0981∷Kmr mutant comprised only 2% of total colony-forming units in the spleen 147 days after infection (Fig. 5B; P < 0.0001; ANOVA). The decline in Rv0981∷Kmr survival was initiated during the acute phase of infection where 64% ± 4% of the bacteria recovered 14 days after infection were mutant (Fig. 5B; P = 0.141; ANOVA), and became significant by 28 days after infection following the onset of acquired immunity (Fig. 5A), when only 48% ± 8% of total colony-forming units recovered were the Rv0981∷Kmr mutant (Fig. 5B; P = 0.0017, ANOVA). In the lungs, the Rv0981∷Kmr mutant was not, however, attenuated for growth during acute infection. Instead, the Rv0981∷Kmr mutant was unable to maintain viability and persist during the latent stage of infection. For example, no statistically significant decline in Rv0981∷Kmr mutant growth was observed in the lungs of infected animals during acute infection (Fig. 5D), even though total colony-forming units in the lung increased 1,000-fold (Fig. 5C). However, after the emergence of cell-mediated immunity and entrance of the tubercle bacillus into a persistent state of infection, the mutant was unable to remain in the lungs of latently infected mice. A significant reduction in the fraction of Rv0981∷Kmr mutant recovered was observed in the lungs of infected mice at 147 days after infection (Fig. 5D; P = 0.0001; ANOVA). The inability to maintain persistent infection in the lungs of infected mice was tissue-specific, because no significant difference in survival between wild-type H37Rv and the Rv0981∷Kmr mutant was observed in the liver during the course of infection (Fig. 5F). In addition, the inability to establish infection in the spleen or maintain persistent infection in the lung was partially independent of host immunity, because only a minor additional drop in Rv0981∷Kmr mutant bacilli was observed between 119 and 147 days after infection in the lungs and spleens of mice treated with hydrocortisone relative to control mice (Fig. 5 B and D). Immune suppression and reactivation of bacteria from persistent infection in these experiments was efficient, as evidenced by a significant increase in total colony-forming units observed in hydrocortisone-treated animals (Fig. 5 A and C). Taken together, these results support a role for two-component systems, specifically Rv0981, in the establishment and maintenance of persistent infection by M. tuberculosis.
Figure 5.
Response regulator Rv0981 (mprA) is required for persistent M. tuberculosis infection. Growth competition experiments were performed between wild-type M. tuberculosis H37Rv and the Rv0981∷Kmr mutant. At various times after infection, colony-forming units (CFU) were determined in the spleen (A), lung (C), and liver (E) of infected mice. Resulting colonies were picked and patched onto selective medium to determine the fraction of Rv0981∷Kmr mutant bacilli recovered (B, D, and F). A subset of infected mice (open squares) received a s.c. injection of hydrocortisone 10 and 5 days before the 147-day time point. Values represent the mean and standard error of total colony-forming units or percent of mutant bacteria recovered from organs. Statistical significance compared percent of mutant recovered at 14, 28, 119, and 147 days after infection relative to the percent of mutant recovered 1 day after infection: ✻, P < 0.05; ✻✻, P < 0.01 (ANOVA).
Conclusion.
This work has established a role for two-component signal transduction systems in the virulence of M. tuberculosis. The data presented show that Rv0981, now designated mprA (mycobacterium persistence regulator), is required for maintenance of persistent lung infection by M. tuberculosis in a murine model of tuberculosis. The inability of the mprA∷Kmr (Rv0981∷Kmr) mutant to maintain persistent infection in the lung is likely associated with altered adaptation phenomena under conditions of latent infection to which the mprA mutant cannot adjust.
Several conditions present in the host have been suggested to affect persistence of M. tuberculosis. Low oxygen, believed to exist in granulomatous lesions, has been suggested to contribute to persistence of M. tuberculosis in vivo (31). For example, isocitrate lyase is induced 4-fold upon entry of M. tuberculosis into a low-oxygen-induced state in vitro (32), is required for M. tuberculosis survival in activated macrophages in vitro, and is important for intermediary metabolism processes during the persistent stage of infection in the murine model of tuberculosis in vivo (7). In addition to isocitrate lyase, the 16-kDa α-crystalline-like small-heat-shock protein is also induced on oxygen limitation in vitro and is required for survival of M. tuberculosis during intracellular growth in macrophages (33, 34). Apart from genes regulated in response to microaerophilic environments, other genetic determinants may also be required for persistent infection, including genes encoding cell-surface components (8), and genes from the Pro/Glu polymorphic repetitive sequence family of proteins (9). The identification of the mprAB system as critical during stages of persistent infection is, to our knowledge, the first report of a regulatory system in the tubercle bacillus required for latency-related phenomena. Although the growth defect of the M. tuberculosis mprA∷Kmr mutant observed in the lungs of infected animals during the persistent stage of infection is less overt than the growth defect of the mprA∷Kmr mutant observed in the spleen, the degree of attenuation in virulence is significant and is similar to that observed with other “persistence” mutants (7, 8). However, unlike other persistence mutants that have been identified to date, the disruption of mprA in M. tuberculosis H37Rv is tissue-specific and affects processes required for growth of the tubercle bacillus during multiple stages of the infection process. Further characterization of this two-component system should provide additional insights into the genes and conditions required for the establishment and maintenance of persistent infection by M. tuberculosis.
Acknowledgments
We thank V. Vishwanath and M. Mudd for preparation of human peripheral blood monocyte-derived macrophages, and E. Pagan-Ramos for helpful discussions. B. Gicquel kindly provided plasmid pPR27. This work was supported by National Research Service Award AI10278 (to T.C.Z.), and by National Institute of Allergy and Infectious Disease Grants AI35217, AI42999, and AI45148 (to V.D.).
Abbreviations
- GFP
green fluorescent protein
- LPS
lipopolysaccharide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. J Am Med Assoc. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 2.Manabe Y C, Bishai W R. Nat Med. 2000;6:1327–1329. doi: 10.1038/82139. [DOI] [PubMed] [Google Scholar]
- 3.Camacho L R, Ensergueix D, Perez E, Gicquel B, Guilhot C. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 4.Berthet F X, Lagranderie M, Gounon P, Laurent-Winter C, Ensergueix D, Chavarot P, Thouron F, Maranghi E, Pelicic V, Portnoi D, et al. Science. 1998;282:759–762. doi: 10.1126/science.282.5389.759. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Kelley C, Collins F, Rouse D, Morris S. J Infect Dis. 1998;177:1030–1035. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]
- 6.Cox J S, Chen B, McNeil M, Jacobs W R., Jr Nature (London) 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 7.McKinney J D, Honer zu Bentrup K, Munoz-Elias E J, Miczak A, Chen B, Chan W T, Swenson D, Sacchettini J C, Jacobs W R, Jr, Russell D G. Nature (London) 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 8.Glickman M S, Cox J S, Jacobs W R., Jr Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 9.Ramakrishnan L, Federspiel N A, Falkow S. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 10.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 11.Rappuoli R, Scarlato V, Arico B. Signal Transduction and Bacterial Virulence. Austin, TX: R. G. Landes Co.; 1995. [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Zahrt T C, Deretic V. J Bacteriol. 2000;182:3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham J E, Clark-Curtiss J E. Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 16.Pelicic V, Jackson M, Reyrat J M, Jacobs W, Jr, Gicquel B, Guilhot C. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curcic R, Dhandayuthapani S, Deretic V. Mol Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 18.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, et al. Nature (London) 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 19.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D B. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 20.Via L E, Fratti R A, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. J Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 21.Boyum A. Scand J Clin Lab Invest Suppl. 1968;97:7. [PubMed] [Google Scholar]
- 22.North R J, Izzo A A. J Exp Med. 1993;177:1723–1733. doi: 10.1084/jem.177.6.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orme I M. Am Rev Respir Dis. 1988;137:716–718. doi: 10.1164/ajrccm/137.3.716. [DOI] [PubMed] [Google Scholar]
- 24.Flynn J L, Scanga C A, Tanaka K E, Chan J. J Immunol. 1998;160:1796–1803. [PubMed] [Google Scholar]
- 25.Merrell D S, Camilli A. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 26.Ho T D, Slauch J M. J Bacteriol. 2001;183:611–620. doi: 10.1128/JB.183.2.611-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lestrate P, Delrue R M, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang C M, Letesson J J. Mol Microbiol. 2000;38:543–551. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 28.Monack D M, Mecsas J, Bouley D, Falkow S. J Exp Med. 1998;188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beuzon C R, Meresse S, Unsworth K E, Ruiz-Albert J, Garvis S, Waterman S R, Ryder T A, Boucrot E, Holden D W. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orme I M, Collins F M. In: Tuberculosis: Pathogenesis, Protection, and Control. Bloom B R, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 113–134. [Google Scholar]
- 31.Segal W. In: The Mycobacteria: A Sourcebook. Kaplan G P, Wayne L G, editors. New York: Dekker; 1984. pp. 547–573. [Google Scholar]
- 32.Wayne L G, Lin K Y. Infect Immun. 1982;37:1042–1049. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham A F, Spreadbury C L. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan Y, Crane D D, Simpson R M, Zhu Y Q, Hickey M J, Sherman D R, Barry C E., III Proc Natl Acad Sci USA. 1998;95:9578–9583. doi: 10.1073/pnas.95.16.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]