Abstract
Resting-state functional magnetic resonance imaging (RS-FMRI) has been widely used to map brain functional connectivity, but it is unclear how to probe connectivity within and around lesions. In this study, we characterize RS-FMRI signal time course properties and evaluate different seed placements within and around hemorrhagic traumatic axonal injury (hTAI) lesions. RS-FMRI was performed on a 7 Tesla scanner in a patient who recovered consciousness after traumatic coma and in three healthy controls. Eleven lesions in the patient were characterized in terms of (1) temporal signal-to-noise ratio (tSNR); (2) physiological noise, through comparison of noise regressors derived from the white matter (WM), cerebrospinal fluid (CSF), and gray matter (GM); and (3) seed-based functional connectivity. Temporal SNR at the center of the lesions was 38.3% and 74.1% lower compared with the same region in the contralesional hemisphere of the patient and in the ipsilesional hemispheres of the controls, respectively. Within the lesions, WM noise was more prominent than CSF and GM noise. Lesional seeds did not produce discernable networks, but seeds in the contralesional hemisphere revealed networks whose nodes appeared to be shifted or obscured due to overlapping or nearby lesions. Single-voxel seed analysis demonstrated that placing a seed within a lesion's periphery was necessary to identify networks associated with the lesion region. These findings provide evidence of resting-state network changes in the human brain after recovery from traumatic coma. Furthermore, we show that seed placement within a lesion's periphery or in the contralesional hemisphere may be necessary for network identification in patients with hTAI.
Keywords: : brain connectivity, coma, functional MRI, lesion analysis, resting-state, traumatic axonal injury
Introduction
Hemorrhagic traumatic axonal injury (hTAI) is associated with disruption of neural networks and loss of cognitive function in patients with severe traumatic brain injury (TBI) (Bodien et al., 2017; Sharp et al., 2014). Yet, recent evidence from longitudinal imaging studies and histo-radiologic correlation studies suggests that hTAI lesions may not invariably destroy axons (Edlow et al., 2013, 2016). Rather, red blood cells may intercalate between partially injured axons, which have the potential to recover their structural integrity (Edlow et al., 2013). Functional connectivity derived from resting-state functional magnetic resonance imaging (RS-FMRI) enables investigation of how lesions disrupt brain networks, and how networks may compensate for injury. However, the optimal methodology for determining how hTAI lesions affect brain networks using RS-FMRI is unknown. Specifically, network maps may differ depending on whether a seed region-of-interest (ROI) is placed within or at the periphery of an hTAI lesion.
When attempting to identify neural network disruptions due to focal brain lesions, seed-based RS-FMRI (Biswal et al., 1995; Di Martino et al., 2008; Fox et al., 2005; van de Ven et al., 2004), as opposed to the data-driven independent component analysis (ICA) approach (Chen et al., 2008; McKeown et al., 2003; van de Ven et al., 2004), offers the opportunity for targeted studies of lesion-specific connectivity (or lack thereof). One potential challenge with the seed-based approach is that the T2* values within hTAI lesions tend to be shortened due to the presence of hemosiderin (Gomori et al., 1985). As a result, the signal-to-noise ratio (SNR) within a lesion may be too low to effectively identify correlations between signal time courses in other regions of the brain. Furthermore, loss of signal within a lesion may be associated with a local image intensity gradient, resulting in increased signal fluctuation due to dynamic partial volume effects driven by head motion and tissue pulsatility.
In prior RS-FMRI studies of patients with moderate-to-severe TBI, investigators typically placed seeds in well-established nodes of resting-state networks (Hillary et al., 2011; Palacios et al., 2013; Rigon et al., 2016; Venkatesan et al., 2015). Additional studies focused on altered functional connectivity within the default mode network (DMN) (Falletta Caravasso et al., 2016; Li et al., 2017; Sharp et al., 2011), salience network (Ham et al., 2014; Li et al., 2017), and somatomotor network (Falletta Caravasso et al., 2016). These studies used ICA and/or seed-based analysis, but none placed seeds within the lesion areas. Instead, prior studies applied ICA to compare functional connectivity between patients and controls, or performed ICA to identify network nodes that could be used as seeds in subsequent seed-based analysis. Several studies have also used a graph theory approach to analyze RS-FMRI results in TBI patients (Caeyenberghs et al., 2012; Nakamura et al., 2009; Pandit et al., 2013).
In contrast to these prior studies that focused on uninjured network nodes, our aim is to characterize RS-FMRI signal properties from seed regions within and around hTAI lesions. Specifically, we examine how signal time courses for ipsilesional and contralesional ROIs correlate with those from white matter (WM) and cerebrospinal fluid (CSF) regions, which are contaminated by physiological noise, as well as gray matter (GM). We also study the connectivity patterns derived from different seed placement strategies and compare with connectivity in the patient's contralesional side, as well as to connectivity in healthy controls. Ultimately, these investigations are intended to provide insights into how to use and interpret RS-FMRI to detect connectivity changes associated with hTAI.
Materials and Methods
Patient and healthy controls
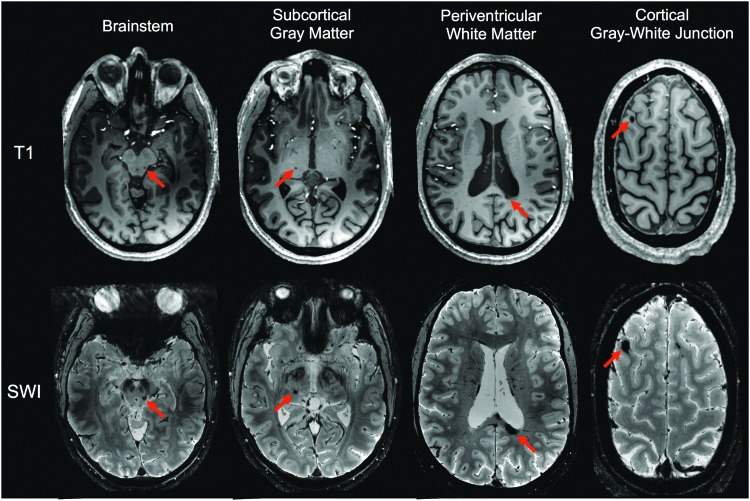
All study procedures were approved by the local institutional review board, and written informed consent was provided by all subjects. RS-FMRI was performed on a 27-year-old man who recovered from traumatic coma and three healthy controls (2F, 1 M, 25–26 years old). Six years and 7 months prior, the patient had experienced a severe TBI after he was struck by a car while riding his bicycle. He was in a coma for 14 days. A clinical MRI scan performed in the intensive care unit revealed grade 3 diffuse axonal injury involving the WM of the bilateral cerebral hemispheres, splenium of the corpus callosum, and dorsolateral midbrain. After emergence from coma, the patient underwent one and a half months of inpatient rehabilitation, followed by 1 year of outpatient rehabilitation. At the time of our imaging study, 11 hTAI lesions were still visible due to chronic hemosiderin deposition in the cortical gray/white junction, periventricular WM, subcortical GM, brainstem, and deep WM. Examples of hTAI lesions are shown in Figure 1. All hTAI lesions in the patient are shown in Supplementary Figure S1 (Supplementary Data are available online at www.liebertpub.com/brain).
FIG. 1.
T1-weighted images and susceptibility-weighted imaging of a patient with severe traumatic brain injury based on the lesion locations. Red arrows indicate examples of lesions in each of the anatomical categories: cortical gray/white junction, periventricular white matter, subcortical gray matter, brainstem, and deep white matter. SWI, susceptibility-weighted imaging. Color images available online at www.liebertpub.com/brain
Acquisition
Data were acquired using a 7T Siemens MRI scanner (Siemens Healthcare, Erlangen, Germany) equipped with a custom-built 32-channel RF receive coil and birdcage transmit coil (Keil et al., 2010). The BOLD-weighted acquisition consisted of a single-shot gradient-echo echo planar imaging (EPI) pulse sequence with echo time (TE)/repetition time (TR) = 25/3560 msec, 1.5 mm isotropic voxels, field of view (FOV) = 19.2 cm × 19.2 cm, 60 slices, partial Fourier = 7/8, BW = 1776 Hz/pixel, R = 2 inplane acceleration, online Generalized Autocalibrating Partially Parallel Acquisition (GRAPPA) reconstruction (Griswold et al., 2002) using Fast Low-angle Excitation EPI Technique (FLEET) autocalibration signal (ACS) acquisition (Polimeni et al., 2013, 2016), echo spacing = 0.33 msec, 180 time points, and total scan time = 11 min. To assist with lesion identification and registration to Montreal Neurological Institute (MNI) space, T1-weighted multiecho magnetization-prepared rapid acquisition gradient echo (ME-MPRAGE) images (van der Kouwe et al., 2008) were acquired with TE1/TE2/TE3/TE4/TR/TI = 1.72/3.55/5.38/7.21/2530/1100 msec, flip angle = 7°, 0.75 mm isotropic voxels, FOV = 25.6 × 25.6 cm, 192 slices, full Fourier, R = 2 (GRAPPA), and BW = 696 Hz/pixel. Susceptibility-weighted images (Haacke et al., 2004) were acquired with TE/TR = 21/1830 msec, 0.3 × 0.3 × 1 mm resolution, FOV = 19.2 cm × 19.2 cm, 40 slices, GRAPPA R = 2, and BW = 30 Hz/pixel.
Data analysis
Network analyses were conducted in each subject's native space. Preprocessing of the RS-FMRI data was performed using tools from the FMRIB Software Library (FSL, www.fmrib.ox.ac.uk/fsl) (Jenkinson et al., 2012; Smith et al., 2004; Woolrich et al., 2009). The preprocessing pipeline included creating a mask of the brain using the Brain Extraction Tool, regression of nuisance variables (WM, CSF, movement, and global signal), temporal high-pass filtering with 0.01 Hz cutoff using FMRIB Expert Analysis Tool (FEAT), motion correction using MCFLIRT (Jenkinson et al., 2002), and spatial smoothing with a 3 mm Gaussian kernel. The WM, CSF, and GM masks were created by applying FMRIB's Automated Segmentation Tool (FAST) (Zhang et al., 2001) to EPI volumes. To avoid effects of partial voluming, the WM, CSF, and GM masks were eroded using a 3 × 3 × 3 kernel to zero any nonzero voxels at the ROI boundaries. That is, whenever a zero was detected in the kernel, all of the voxels within the kernel were set to zero. To exclude lesional tissues in the WM, CSF, and GM masks, all the lesion ROIs were subtracted from the masks. WM and CSF masks were used to define regressors for RS-FMRI preprocessing and only ventricular CSF was included in the CSF regressor mask (no peripheral CSF). All the three masks were used for signal characterization.
After testing connectivity maps with and without smoothing (2 and 3 mm Gaussian kernel), a 3 mm Gaussian kernel was selected because it provided increased sensitivity without changing the connectivity patterns (Supplementary Fig. S2). The first six image volumes were discarded to avoid data acquired during the approach to steady-state magnetization. ICA was performed using FSL's MELODIC. Seed-based analysis was performed using the Analysis of Functional NeuroImages (AFNI, https://afni.nimh.nih.gov/) software package (Cox, 1996; Cox and Hyde, 1997; Gold et al., 1998). Temporal SNR (tSNR) was derived from the detrended, motion-corrected data and defined as the ratio of time-series mean over time-series standard deviation.
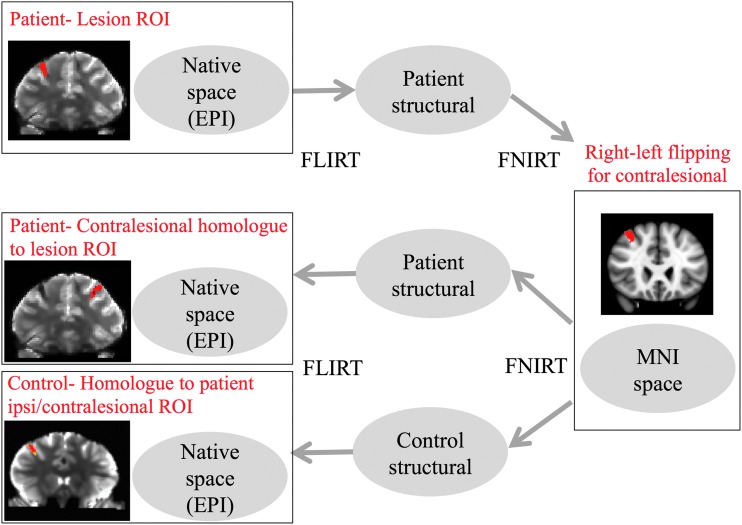
For seed-based analysis, seed ROIs were created using two approaches. First, a neurologist (B.L.E.) manually traced a multivoxel seed for each lesion that covered the entire hTAI lesion. All seeds were drawn directly on the EPI data set in native space, with reference to the susceptibility-weighted imaging (SWI) and T1 ME-MPRAGE data sets for neuroanatomic confirmation of lesion placement. Second, a series of single-voxel seeds were created along a straight line that bisected each multivoxel lesion in the axial plane. Each multivoxel and single-voxel seed ROI was then transformed from the patient's EPI data set to the healthy controls' EPI data sets. Image coregistration was performed using FMRIB's Linear Image Registration Tool (FLIRT) and FMRIB's non-FLIRT. EPI volumes were first warped to their respective T1-weighted images and then to MNI space. Equivalent multivoxel seeds were also generated in the contralesional hemisphere by performing a right/left flip in MNI space and then warping the contralesional seed back to native image space (i.e., the EPI data). The spatial normalization process is shown in Figure 2 and registered ROIs in the patient and each control are shown in Supplementary Figure S3. A neurologist (B.L.E.) blinded to the RS-FMRI data confirmed the neuroanatomic accuracy of the coregistrations and, if necessary, manually repositioned the ROIs based on anatomic landmarks. Of note, lesion K could not be identified on controls 2 and 3 due to distortions in the inferoanterior temporal lobe.
FIG. 2.
A schematic diagram illustrating the image space normalization process. EPI, echo planar imaging; MNI, Montreal Neurological Institute; ROI, region-of-interest. Color images available online at www.liebertpub.com/brain
Lesion signal characterization
To characterize the physiological noise properties of the lesions, we calculated the square of the Pearson correlation coefficient, R2, between each ROI and the WM, CSF, and GM masks.
Connectivity maps from multivoxel seeds
Connectivity maps were derived for all ipsilesional and contralesional multivoxel seed ROIs. Each map was evaluated in terms of (1) identifiable networks, (2) the presence of distinct clusters/nodes, (3) similarity of ipsilesionally and contralesionally seeded maps, and (4) symmetry of connectivity maps. For connectivity maps generated using the contralesional seed, we looked for network nodes that were potentially displaced or shifted on the lesional side. For each potentially affected node that was identified, we calculated a distance associated with the shift by thresholding, clustering, and binarizing the left and right nodes in question and then performing a right/left flip in the MNI space to reflect the contralesional node onto the lesional node. The distance between the centroid of the lesional node and the contralesional node (reflected onto the lesional node) was then calculated.
Connectivity maps from single-voxel seeds
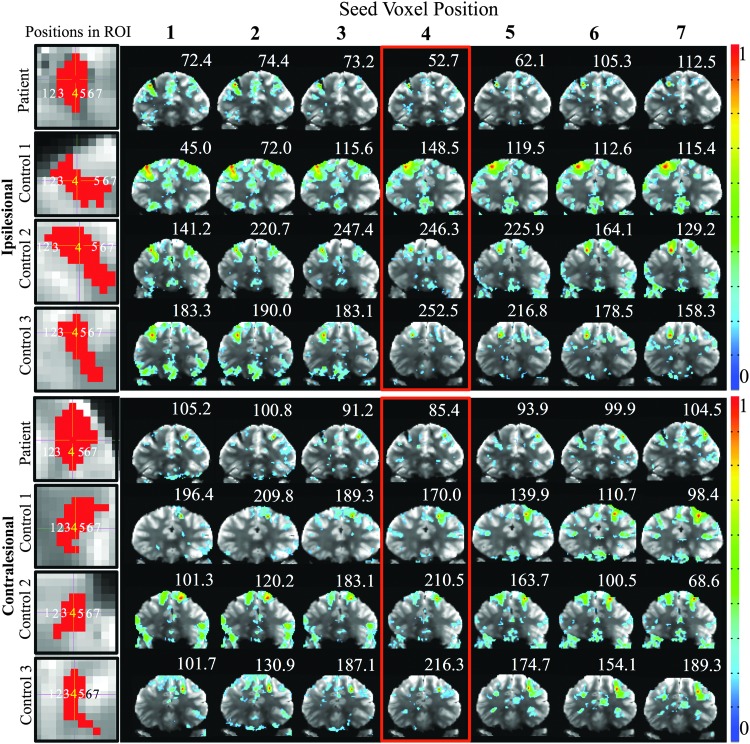
To evaluate how connectivity maps change in relation to the proximity of a seed voxel to the lesion ROI, we derived connectivity maps for seven separate single-voxel seeds placed along a straight line oriented medial/laterally through the center of lesion A in the axial plane. Temporal SNR was determined for each of the seven seed voxels. The seven single-voxel seed positions were placed from lateral to medial across the lesion as follows: #1: two voxels away from the lesion edge in the lateral direction, #2: one voxel away from the lesion edge in the lateral direction, #3: at the lateral edge of the lesion, #4: at the center of the lesion, #5: at the medial edge of the lesion, #6: one voxel away from the lesion edge in the medial direction, and #7: two voxels away from the lesion edge in the medial direction. Due to the different shapes and sizes of lesion ROIs after mapping them into each subject's native space, the distance between seed voxels #3, 4, and 5 varied across subjects. For example, a larger lesion diameter causes a greater distance between the center and edges of the lesion (i.e., the distances between seed voxels #3 and #4 and between #4 and #5). Across all of the lesion ROIs, the resulting distance between the center and edge lesion seed voxels varied between one and three voxels.
Results
Lesion signal characterization
Table 1 reports R2 values between the ipsilesional and contralesional ROI correlations with the WM, CSF, and GM masks. On average across all lesions, the R2 value between the lesion signal and the signal within WM, CSF, and GM masks was 141.2% higher, 85.1% higher, and 52.9% lower compared with the same regions on the contralesional side. Therefore, this provides some evidence that WM noise is more prevalent within the lesion than CSF or GM noise in our patient. We explored the WM noise component of the lesions by performing a principal component analysis (Behzadi et al., 2007) on the WM signal to generate the top 10 components of the WM signal. We calculated R2 value between the ten WM component signals and each of the lesions. The top two components of the WM signal showed significant correlations. The first WM component explained a substantial (R2 = 0.17–0.60) component of the variance for all but two lesions (i.e., lesions C and G). The second WM component explained a significant (R2 = 0.14–0.41) component of the variance for 7 of the 11 lesions (i.e., not lesions E, F, H, or J). Changes in WM, CSF, and GM correlations, compared with the equivalent contralesional regions, varied substantially for different lesions. No trends in noise properties were found for lesions with similar anatomical locations. Signal time courses for the lesions and the WM, CSF and GM signals were plotted in an attempt to gain additional insight from their visualization. Unfortunately, the correlations, while meaningful, proved too small to provide additional insight by eye.
Table 1.
R2 Correlation Values Between Resting-State Functional Magnetic Resonance Imaging Signal Time Courses Extracted from Each Lesion Region-of-Interest and (1) the White Matter; (2) Cerebrospinal Fluid; and (3) Gray Matter Masks Compared with Those from Contralesional Regions-of-Interest
| Cortical gray/white junction | Periventricular WM | Subcortical GM | Brainstem | Deep WM | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | D | I | K | B | C | E | F | H | G | J | All lesions | |||||||||||
| Lesion | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | Contra | Ipsi | |
| WM | 0.147 | 0.173 | 0.010 | 0.214 | 0.155 | 0.102 | 0.001 | 0.002 | 0.086 | 0.011 | — | 0.002 | 0.054 | 0.062 | 0.005 | 0.000 | 0.016 | 0.076 | 0.058 | 0.000 | 0.180 | 0.132 |
| CSF | 0.093 | 0.184 | 0.062 | 0.066 | 0.089 | 0.218 | 0.119 | 0.060 | 0.136 | 0.128 | — | 0.080 | 0.038 | 0.163 | 0.052 | 0.118 | 0.004 | 0.005 | 0.042 | 0.079 | 0.161 | 0.022 |
| GM | 0.084 | 0.192 | 0.016 | 0.287 | 0.053 | 0.088 | 0.053 | 0.005 | 0.019 | 0.011 | — | 0.002 | 0.027 | 0.002 | 0.001 | 0.004 | 0.050 | 0.016 | 0.000 | 0.003 | 0.171 | 0.021 |
For each lesion, larger correlation values between ipsi- and contralesional ROIs are shown in bold.
WM, white matter; CSF, cerebrospinal fluid; GM, gray matter.
Independent component analysis
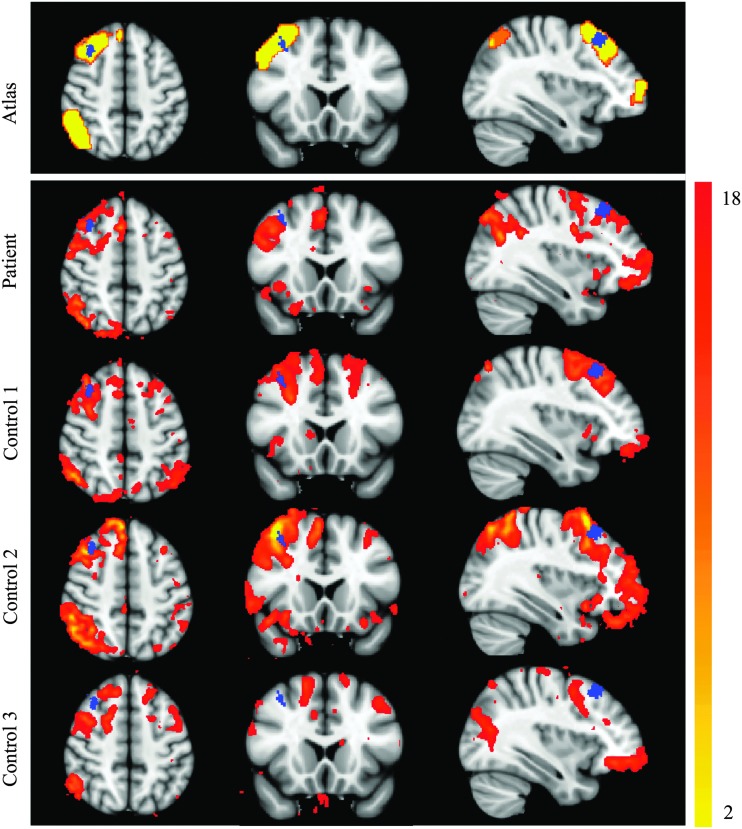
ICA applied to the RS-FMRI data revealed all of the networks shown in the FIND Lab atlas (Shirer et al., 2012) except for the basal ganglia network. Table 2 shows specifically which networks were found in which subject. The only three networks from the FIND Lab atlas (Shirer et al., 2012) that were not found in the patient were the left executive control network (ECN) and the anterior and posterior salience networks. However, the salience networks were not identified consistently in the controls. Specifically, the anterior salience network was not found in controls 1 and 3 and the posterior salience network was not found in controls 2 and 3. Identification of the posterior salience network in control 1 was only possible by increasing the smoothing level to 6 mm FWHM. Additional networks that could only be identified with an increased smoothing level of 6 mm FWHM include visuospatial (patient) and left ECN (control 2), auditory (control 2) and precuneus (control 1 and 2). Lesion A is the only lesion in the patient that overlaps with a known network node (the dorsolateral prefrontal cortex [DLPFC] node of the right ECN, Fig. 3), although several lesions are close to other network node boundaries (i.e., lesion D is very close to left parahippocampal DMN and lesion E is close to posterior salience network). Qualitatively, the ICA maps did not show any decrease in connectivity within lesion A compared to the surrounding tissue in the DLPFC node. Of the networks that were identified, no qualitative differences were observed between the patient and controls. Maps of the right ECN from the patient and controls, registered to MNI space, are shown in Figure 3 with the location of lesion A indicated in blue. Lesion A overlaps with the DLPFC node of the ECN from the atlas and in the ECN of controls 1 and 2. For control 3, the right ECN is not well demarcated. In the patient's coronal plane, it appears that the DLPFC node is located at a more inferior position compared with the location of the same node in the atlas or in the networks of controls 1 or 2.
Table 2.
Networks Found in the Patient and Controls Using Independent Component Analysis
| Network | Patient | Control 1 | Control 2 | Control 3 |
|---|---|---|---|---|
| Left ECN | X | V | 6 mm | V |
| Right ECN | V | V | V | V |
| Dorsal DMN | V | V | V | V |
| Visuospatial | 6 mm | V | V | V |
| Ventral DMN | V | V | V | V |
| Auditory | V | V | 6 mm | V |
| Precuneus | V | 6 mm | 6 mm | X |
| Higher visual | V | V | V | V |
| Motor | V | V | V | V |
| Primary visual | V | V | V | V |
| Anterior salience | X | X | V | X |
| Posterior salience | X | 6 mm | X | X |
| Language | V | V | V | V |
| Total number of networks found | 10 | 12 | 12 | 10 |
In the table, “V” indicates that the network is found in both 6 and 3 mm smoothed data, “6 mm” indicates that the network is found only in 6 mm smoothed data, and “X” indicates that the network is not found in the data.
DMN, default mode network; ECN, executive control network.
FIG. 3.
Right ECN of the patient and controls from ICA. Right ECN from the FIND Lab atlas and those of the patient and controls obtained from ICA (red-yellow color) are superimposed on top of MNI space. Blue label represents lesion A ROI. ICA, independent component analysis; ECN, executive control network. Color images available online at www.liebertpub.com/brain
Connectivity maps from multivoxel seeds
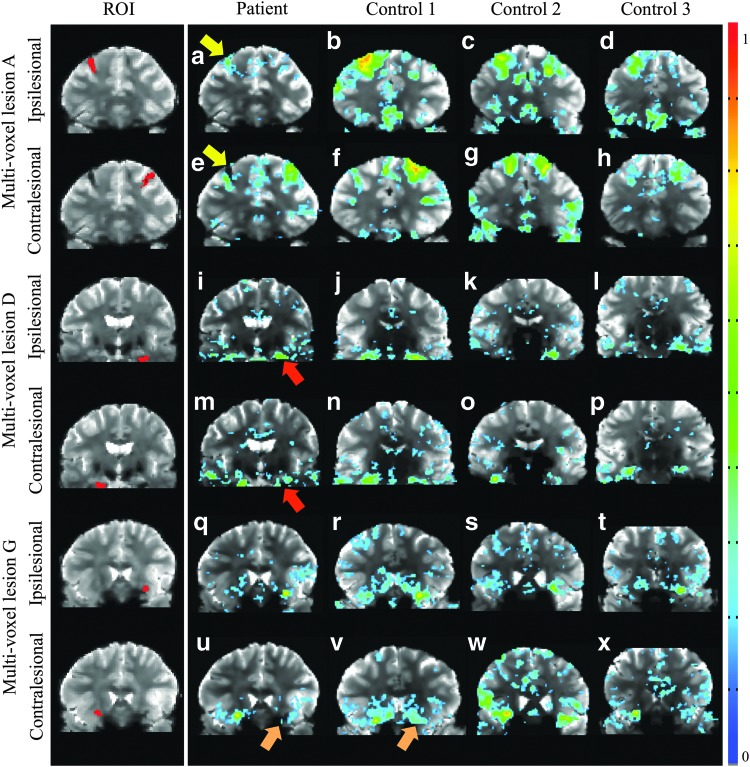
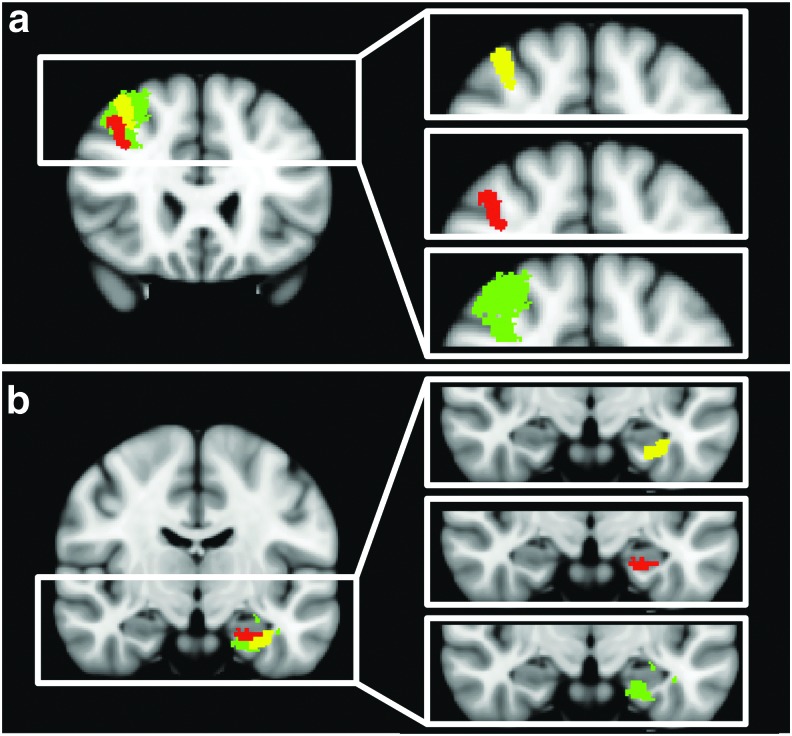
A summary of the connectivity map results is presented in Table 3. Connectivity maps from lesion ROIs do not show recognizable network nodes. Even lesion A, located within the DLPFC node of the right ECN, did not produce the expected identifiable network (i.e., the right ECN). However, multivoxel seeds placed in the lesion-equivalent region within the contralesional hemisphere did produce a node located adjacent to lesion A and nodes in the left and right temporal lobe. Furthermore, the connectivity clusters/nodes found for contralesional multivoxel seeds for lesions A, D, and G were visibly different between the patient and controls. The differences between the patient and controls for connectivity maps derived from contralesional multivoxel seeds for lesions A, D, and G are summarized as follows. The lesion A seed showed weaker correlations and smaller clusters in the patient compared with controls (Fig. 4a) and the DLPFC node of the ECN on the ipsilesional side of the patient is smaller and centered more inferolateral compared with the contralesional side or compared with controls (Fig. 4e). The lesion D seed produced similar connectivity maps for patient and controls with the exception of a node near the inferior part of the brain on the ipsilesional side that appeared shifted superiorly in the patient (Fig. 4m). The lesion G seed showed similar connectivity patterns for the patient and the controls, but overall smaller clusters (Fig. 4u). Figure 5 displays the nodes that were potentially shifted or obscured on the lesional side compared with the reflected contralesional node. Specifically, the right DLPFC appears to be partially obscured by lesion A (Fig. 5a) and the left parahippocampal DMN node appears to be shifted superiorly (Fig. 5b). For lesion A, the distance between the centroid of the right (lesional) DLPFC node and the reflected left (contralesional) DLPFC node is 18 mm and for lesion D, the distance between the centroid of the left (lesional) parahippocampal DMN node and the reflected right (contralesional) parahippocampal DMN node is 8 mm.
Table 3.
A Summary of the Seed-Based Connectivity Results
| Lesion ID | A | B | C | D | E |
|---|---|---|---|---|---|
| Location | Right frontal | Left splenium corpus callosum | Body corpus callosum | Left hippocampus | Right thalamus |
| Category | Cortical gray/white junction | Periventricular white matter | Periventricular white matter | Cortical gray/white junction | Subcortical gray matter |
| Size of ROI (no. of voxels) | 156 | 121 | 96 | 124 | 24 |
| Connectivity using ipsilesional ROI | Smaller cluster and not symmetric | No network nodes | No network nodes | No network nodes | No network nodes |
| Connectivity using contralesional ROI | Shifted connectivity | No network nodes | — | Shifted connectivity and not symmetric | No network nodes |
| Lesion ID | F | G | H | I | J |
|---|---|---|---|---|---|
| Location | Right cerebral peduncle | Left uncinate fasciculus | Left midbrain tegmentum | Left frontal | Right external capsule |
| Category | Brainstem | Deep white matter | Brainstem | Cortical gray/white junction | Deep white matter |
| Size of ROI (no. of voxels) | 8 | 29 | 19 | 11 | 28 |
| Connectivity using ipsilesional ROI | No network nodes | Not symmetric | No network nodes | Smaller cluster and not symmetric | No network nodes |
| Connectivity using contralesional ROI | No network nodes | Smaller cluster and not symmetric | No network nodes | Not symmetric | Not symmetric |
Each lesion's location and anatomic category are listed and the characteristics of the connectivity map of the patient using ipsilesional and contralesional ROIs are described compared with controls. Since lesion C is in the middle of brain, there is no contralesional ROI. “No network nodes” indicates that there are no visible clusters of connectivity in the patient nor in the controls; “Not symmetric” indicates that maps of controls show symmetric patterns, however, the corresponding map in the patient does not.
ROI, region-of-interest.
FIG. 4.
Connectivity maps for the patient and controls from the multivoxel seed ROIs covering the entirety of lesions A (a–d), D (i–l), and G (q–t) and the equivalent contralesional regions A (e–h), D (m–p), and G (u–x). Maps are thresholded at p < 0.05. The color bar represents the Pearson correlation coefficient R. Arrows highlight differences between the contra- and ipsilesional ROI-derived maps for lesions A and D and between patient and controls for lesion G. Color images available online at www.liebertpub.com/brain
FIG. 5.
Coronal images in MNI space depicting the location of lesions (yellow), ipsilesional nodes (red), and contralesional nodes reflected back onto the lesional hemisphere (green). Lesion A, which overlaps with the right dorsolateral prefrontal cortex of the ECN, is shown in (a) and lesion D, which is close to the left parahippocampal node of the default mode network, is shown in (b). Color images available online at www.liebertpub.com/brain
Connectivity maps from single-voxel seeds
The connectivity maps and tSNR values for the single-voxel seeds within and surrounding lesion A and ipsilesional and contralesional hemispheres are shown in Figure 6. For the patient, the tSNR for the voxel at the center of lesion A is 38.3% lower compared with the contralesional side and 74.1% ± 8.3% [mean ± SD] lower compared with controls. For the three controls, the ECN appears for all voxel positions through lesion A. For the patient, the voxel at the center of the lesion ROI (voxel #4) shows only a few small, nondescript clusters, whereas voxel #1 located two voxels laterally shifted from the lesion boundary shows the ECN most reliably. Contralesional connectivity maps in the patient show the ECN for voxel #4, #5, #6, and #7. The average tSNR across all seven ipsilesional voxel positions in the patient is 18.9% lower compared with the contralesional side and 47.8 ± 20.5 [mean ± SD] lower compared with the controls. Of note, the same single-voxel analysis was tested along straight lines running along other orientations (diagonal) and similar connectivity patterns were found. We also note that the whole-brain nonlesional tissue within the patient had a lower mean tSNR (92.3) compared with the three controls (120.2, 149.9, and 144.6).
FIG. 6.
Connectivity maps for patients and controls from single-voxel seeds. The red voxels in the first column represent the lesion A ROI for patient and controls and ipsilesional and contralesional sides. Each number superimposed on lesion A in the first column corresponds to single-voxel seed placements across the ROIs and subsequent columns show corresponding connectivity maps thresholded at p < 0.05. The number above each image is the temporal signal-to-noise ratio at the seed voxel position. Color bars represent the correlation coefficient. Color images available online at www.liebertpub.com/brain
Discussion
In this study, we investigated brain network functional connectivity in a patient who recovered from traumatic coma and compared these results with those of three healthy controls. We evaluated the effects of seed placement, the signal time course properties within lesions, and the potential for detecting network changes. Our results reveal several important considerations for lesion-specific RS-FMRI analyses.
First, single-voxel seeds demonstrated the importance of probing the periphery of a lesion and the equivalent region in the contralesional hemisphere to identify lesion-specific networks. This observation is likely attributable to the low tSNR at the lesion center, which was 38.3% and 74.1% lower compared with the contralesional hemisphere in the patient and ipsilesional hemisphere in controls, respectively.
Second, we found that WM noise was more strongly elevated within the lesions than CSF and GM noise. This observation is consistent with prior studies suggesting that lesions are more strongly influenced by cardiac pulsatility than they are by breathing effects (Van de Moortele et al., 2002; van Gelderen et al., 2007). Pulsatile movement of brain tissue may cause the lesions to move slightly, and since lesions typically have lower signal intensity compared to surrounding tissue, a sharp lesion boundary could cause temporal signal modulation due to dynamic partial volume effects. When grouping lesions according to their anatomical locations, CSF noise was more strongly elevated within lesions located at the cortical GM-WM junction and within periventricular WM and deep WM, whereas WM noise was more strongly elevated within lesions in the subcortical GM and brainstem. There was, however, in some cases, significant variation in noise properties for lesions within similar anatomical locations.
Third, when evaluating the ability to detect network changes, our data show that aside from the ECN, connectivity maps derived from ICA did not demonstrate a difference in the number or qualitative appearance of identified brain networks in the patient versus the controls. However, our seed-based analysis did reveal potential changes of brain networks. For example, the right DLPFC node appears to have been partially obscured by lesion A (i.e., only the lower portion of the node, inferior to the lesion, is present). Furthermore, a network node near the hippocampus was potentially shifted to a slightly superior location due to lesion D (Fig. 5).
Our results offer new insight into the potential utility of RS-FMRI as a marker of brain function, and possibly brain plasticity, in patients who recover from traumatic coma. Prior RS-FMRI studies of patients with TBI have attempted to elucidate recovery mechanisms but have focused on ICA or seed-based analysis using nonlesion areas as seeds. For example, Hillary et al. (2011) observed increased resting-state connectivity in the DMN during the first 6 months of recovery in patients with moderate-to-severe TBI. However, in contrast to our study, these prior lesion connectivity studies did not specifically investigate connectivity between the lesion area and other parts of the brain. Rather, many RS-FMRI studies of patients with brain injuries have masked and eliminated the lesion area before identifying functional networks (Hayes et al., 2012; van Meer et al., 2010). Other studies included visible lesion areas as a small part of a much larger seed-ROI (Alstott et al., 2009; Zhang et al., 2009). Alstott et al. (2009) simulated the effect of lesions on brain functional connectivity as measured by RS-FMRI and found that the modeled lesions altered brain functional connectivity in both the ipsilateral and contralateral hemispheres. Along similar lines, Zhang et al. (2009) studied sensorimotor mapping in brain tumor patients. The Zhang et al. (2009) study placed seeds in the hemisphere contralateral to the tumor and found that in the hemisphere ipsilateral to the tumor, the equivalent network node was displaced anteriorly and laterally to the tumor. In our study, we observed two instances of recognizable network nodes (the DLPFC node and the parahippocampal DMN node) that were potentially spatially shifted or obscured due to lesions (Figs. 4e, m, and 5). Our use of seeds within the contralesional hemisphere to identify this potential remapping of a network node aligns well with the Zhang et al. study. However, our study is further complicated by the fact that there are many lesions spread across both hemispheres that could be impacting the spatial location of network nodes. Furthermore, it is important to note that not all brain regions have a functional contralateral homologue and this is a fundamental limitation of our approach of seeding in the contralesional hemisphere. Even for lateralized function, however, there is the possibility of cross-hemispheric plasticity. Therefore, consideration of the lesion location is critically important for interpreting the lesion connectivity results. Furthermore, it is unclear why the patient's nonlesional tissue had a lower mean tSNR compared with the controls and this may have decreased our sensitivity to some measures of connectivity in the patient compared with the controls.
It is important to consider that the results from one patient who recovered from coma cannot be generalized to the entire population of patients with traumatic coma, and the relevance of our chronic lesion mapping approach to patients with acute lesions remains to be determined. Nevertheless, the patient in our study provided a unique opportunity to study the functional connectivity of multiple lesions located in different brain regions and to generate patient-specific brain network connectivity maps. This individualized approach to brain mapping is especially relevant to patients with traumatic coma, since hTAI lesions vary in their number and neuroanatomic locations between patients. Furthermore, a prior study that correlated histopathologic and structural connectivity data in a patient who died after traumatic coma showed that even hTAI lesions within the same brain may have different effects on tissue microstructure (Edlow et al., 2013). A recent review article by Aerts et al. (2016) highlighted that lesion effects on brain connectivity depend critically on lesion location, with lesions located in network hubs causing the most significant alterations. This may be why prior studies such as Roy et al. (2017) have not found measures of total lesion burden to be predictive of network changes. Automated or semiautomated lesion detection would likely enhance the translational potential of our methods and possibly also increase their reliability for patient-specific analysis. Lesion detection software is continually improving and recent efforts have focused specifically on detection of TBI lesions (Roy et al., 2017; van den Heuvel et al., 2016).
A strength of this study is that the patient was scanned 6 years and 7 months postinjury, and therefore, his brain structure and function were not confounded by the acute effects of edema. Our study also benefits from data acquired with state-of-the art equipment and acquisition technology, including a 7T MRI scanner equipped with a 32-channel coil and an acquisition that utilized GRAPPA with FLEET-ACS to control motion sensitivity of the parallel imaging calibration data acquisition (Polimeni et al., 2013, 2016). The use of a 7T scanner benefits our study in two key ways. First, lesion identification is enhanced on SWI, T1-weighted and T2-weighted images at 7T compared with 3T (Madai et al., 2012; Moenninghoff et al., 2015). We illustrate an example of this in Supplementary Figure S4. All lesions can be visualized on the 3T data set, but they can be identified more clearly on the 7T images. Second, RS-FMRI measurements are more sensitive at 7T compared with 3T (De Martino et al., 2011; Yacoub et al., 2001). We point out, however, that the performance of our 7T protocol was far from the limits of the 7T scanner and similar protocols can be achieved with modern 3T scanners. In addition, advanced RF array coils can provide additional sensitivity at 3T. Development of a 3T RS-FMRI protocol that could be translated to clinical MRI scanners is therefore likely feasible within the near future. Last, the implementation of 7T scanners has accelerated in recent years and the recent Food and Drug Administration approval of 7T MRI for clinical use may also make 7T imaging more accessible in the near future.
In conclusion, we show that RS-FMRI may detect functional connectivity related to hTAI lesion areas. We also show that RS-FMRI may enable identification of network nodes that have been shifted or obscured as a result of an hTAI lesion overlapping with that node. Finally, we demonstrate the importance of seeding in the contralesional hemisphere or a larger perilesional region or systematically shifting ROI placement to the lesion periphery to identify network changes. This lesion-specific approach represents a new way to assess brain function and plasticity in traumatic coma patients and may provide important diagnostic and prognostic information to clinicians and families.
Supplementary Material
Acknowledgments
We thank the patient for his participation and support of this work. The authors also thank Thomas Witzel for assistance with data acquisition, Michael D. Greicius for helping with data preprocessing and templates, and Gary Glover and Jingyuan Chen for helpful discussions. Support for this work was provided by the NIH: K23-NS094538, R01-NS095985, R01-EB019437, and R01-MH111444. Funding was also provided by the Charles A. Dana Foundation and the James S. McDonnell Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- Aerts H, Fias W, Caeyenberghs K, Marinazzo D. 2016. Brain networks under attack: robustness properties and the impact of lesions. Brain 139:3063–3083 [DOI] [PubMed] [Google Scholar]
- Alstott J, Breakspear M, Hagmann P, Cammoun L, Sporns O. 2009. Modeling the impact of lesions in the human brain. PLoS Comput Biol 5:e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. 2007. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage 37:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med 34:537–541 [DOI] [PubMed] [Google Scholar]
- Bodien YG, Chatelle C, Edlow BL. 2017. Functional networks in disorders of consciousness. Semin Neurol 37:485–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caeyenberghs K, Leemans A, Heitger MH, Leunissen I, Dhollander T, Sunaert S, et al. . 2012. Graph analysis of functional brain networks for cognitive control of action in traumatic brain injury. Brain 135:1293–1307 [DOI] [PubMed] [Google Scholar]
- Chen S, Ross TJ, Zhan W, Myers CS, Chuang KS, Heishman SJ, et al. . 2008. Group independent component analysis reveals consistent resting-state networks across multiple sessions. Brain Res 1239:141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29:162–173 [DOI] [PubMed] [Google Scholar]
- Cox RW, Hyde JS. 1997. Software tools for analysis and visualization of fMRI data. NMR Biomed 10:171–178 [DOI] [PubMed] [Google Scholar]
- De Martino F, Esposito F, van de Moortele PF, Harel N, Formisano E, Goebel R, et al. . 2011. Whole brain high-resolution functional imaging at ultra high magnetic fields: an application to the analysis of resting state networks. Neuroimage 57:1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Scheres A, Margulies DS, Kelly AM, Uddin LQ, Shehzad Z, et al. . 2008. Functional connectivity of human striatum: a resting state FMRI study. Cereb Cortex 18:2735–2747 [DOI] [PubMed] [Google Scholar]
- Edlow BL, Copen WA, Izzy S, van der Kouwe A, Glenn MB, Greenberg SM, et al. . 2016. Longitudinal diffusion tensor imaging detects recovery of fractional anisotropy within traumatic axonal injury lesions. Neurocrit Care 24:342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlow BL, Haynes RL, Takahashi E, Klein JP, Cummings P, Benner T, et al. . 2013. Disconnection of the ascending arousal system in traumatic coma. J Neuropathol Exp Neurol 72:505–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falletta Caravasso C, de Pasquale F, Ciurli P, Catani S, Formisano R, Sabatini U. 2016. The default mode network connectivity predicts cognitive recovery in severe acquired brain injured patients: a longitudinal study. J Neurotrauma 33:1247–1262 [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold S, Christian B, Arndt S, Zeien G, Cizadlo T, Johnson DL, et al. . 1998. Functional MRI statistical software packages: a comparative analysis. Hum Brain Mapp 6:73–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomori JM, Grossman RI, Goldberg HI, Zimmerman RA, Bilaniuk LT. 1985. Intracranial hematomas: imaging by high-field MR. Radiology 157:87–93 [DOI] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, et al. . 2002. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 47:1202–1210 [DOI] [PubMed] [Google Scholar]
- Haacke EM, Xu Y, Cheng YC, Reichenbach JR. 2004. Susceptibility weighted imaging (SWI). Magn Reson Med 52:612–618 [DOI] [PubMed] [Google Scholar]
- Ham TE, Bonnelle V, Hellyer P, Jilka S, Robertson IH, Leech R, Sharp DJ. 2014. The neural basis of impaired self-awareness after traumatic brain injury. Brain 137:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes SM, Salat DH, Verfaellie M. 2012. Default network connectivity in medial temporal lobe amnesia. J Neurosci 32:14622–14629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillary FG, Slocomb J, Hills EC, Fitzpatrick NM, Medaglia JD, Wang J, et al. . 2011. Changes in resting connectivity during recovery from severe traumatic brain injury. Int J Psychophysiol 82:115–123 [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. 2002. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841 [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. 2012. FSL. Neuroimage 62:782–790 [DOI] [PubMed] [Google Scholar]
- Keil B, Triantafyllou C, Hamm M, Wald LL. 2010. Design optimization of a 32-channel head coil at 7T. Proceedings of the International Society for Magnetic Resonance in Medicine 18 [Google Scholar]
- Li J, Gao L, Xie K, Zhan J, Luo X, Wang H, et al. . 2017. Detection of functional homotopy in traumatic axonal injury. Eur Radiol 27:325–335 [DOI] [PubMed] [Google Scholar]
- Madai VI, von Samson-Himmelstjerna FC, Bauer M, Stengl KL, Mutke MA. Tovar-Martinez E, et al. . 2012. Ultrahigh-field MRI in human ischemic stroke—a 7 Tesla study. PLoS One 7:e37631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Hansen LK, Sejnowsk TJ. 2003. Independent component analysis of functional MRI: what is signal and what is noise? Curr Opin Neurobiol 13:620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenninghoff C, Kraff O, Maderwald S, Umutlu L, Theysohn JM, Ringelstein A, et al. . 2015. Diffuse axonal injury at ultra-high field MRI. PLoS One 10:e0122329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Hillary FG, Biswal BB. 2009. Resting network plasticity following brain injury. PLoS One 4:e8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios EM, Sala-Llonch R, Junque C, Roig T, Tormos JM, Bargallo N, Vendrell P. 2013. Resting-state functional magnetic resonance imaging activity and connectivity and cognitive outcome in traumatic brain injury. JAMA Neurol 70:845–851 [DOI] [PubMed] [Google Scholar]
- Pandit AS, Expert P, Lambiotte R, Bonnelle V, Leech R, Turkheimer FE, Sharp DJ. 2013. Traumatic brain injury impairs small-world topology. Neurology 80:1826–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimeni JR, Bhat H, Benner T, Feiweier T, Inati SJ, Witzel T, et al. . 2013. Sequential-segment multi-shot auto-calibration for GRAPPA EPI: maximizing temporal SNR and reducing motion sensitivity. Proceedings of the International Society for Magnetic Resonance in Medicine 21:2646 [Google Scholar]
- Polimeni JR, Bhat H, Witzel T, Benner T, Feiweier T, Inati SJ, et al. . 2016. Reducing sensitivity losses due to respiration and motion in accelerated echo planar imaging by reordering the autocalibration data acquisition. Magn Reson Med 75:665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigon A, Duff MC, McAuley E, Kramer AF, Voss MW. 2016. Is traumatic brain injury associated with reduced inter-hemispheric functional connectivity? A Study of Large-Scale Resting State Networks following traumatic brain injury. J Neurotrauma 33:977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Bernier RA, Wang J, Benson M, French JJ, Jr., Good DC, Hillary FG. 2017. The evolution of cost-efficiency in neural networks during recovery from traumatic brain injury. PLoS One 12:e0170541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Beckmann CF, Greenwood R, Kinnunen KM, Bonnelle V, De Boissezon X, et al. . 2011. Default mode network functional and structural connectivity after traumatic brain injury. Brain 134:2233–2247 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, Scott G, Leech R. 2014. Network dysfunction after traumatic brain injury. Nat Rev Neurol 10:156–166 [DOI] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. 2012. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22:158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. . 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 Suppl 1:S208–S219 [DOI] [PubMed] [Google Scholar]
- Van de Moortele PF, Pfeuffer J, Glover GH, Ugurbil K, Hu X, 2002. Respiration-induced B0 fluctuations and their spatial distribution in the human brain at 7 Tesla. Magn Reson Med 47:888–895 [DOI] [PubMed] [Google Scholar]
- van de Ven VG, Formisano E, Prvulovic D, Roeder CH, Linden DE. 2004. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum Brain Mapp 22:165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel TL, van der Eerden AW, Manniesing R, Ghafoorian M, Tan T, Andriessen TM, et al. . 2016. Automated detection of cerebral microbleeds in patients with traumatic brain injury. Neuroimage Clin 12:241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kouwe AJ, Benner T, Salat DH, Fischl B. 2008. Brain morphometry with multiecho MPRAGE. Neuroimage 40:559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen P, de Zwart JA, Starewicz P, Hinks RS, Duyn JH. 2007. Real-time shimming to compensate for respiration-induced B0 fluctuations. Magn Reson Med 57:362–368 [DOI] [PubMed] [Google Scholar]
- van Meer MP, van der Marel K, Wang K, Otte WM, El Bouazati S, Roeling TA, et al. . 2010. Recovery of sensorimotor function after experimental stroke correlates with restoration of resting-state interhemispheric functional connectivity. J Neurosci 30:3964–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan UM, Dennis NA, Hillary FG. 2015. Chronology and chronicity of altered resting-state functional connectivity after traumatic brain injury. J Neurotrauma 32:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, et al. . 2009. Bayesian analysis of neuroimaging data in FSL. Neuroimage 45:S173–S186 [DOI] [PubMed] [Google Scholar]
- Yacoub E, Shmuel A, Pfeuffer J, Van De Moortele PF, Adriany G, Andersen P, et al. . 2001. Imaging brain function in humans at 7 Tesla. Magn Reson Med 45:588–594 [DOI] [PubMed] [Google Scholar]
- Zhang D, Johnston JM, Fox MD, Leuthardt EC, Grubb RL, Chicoine MR, et al. . 2009. Preoperative sensorimotor mapping in brain tumor patients using spontaneous fluctuations in neuronal activity imaged with functional magnetic resonance imaging: initial experience. Neurosurgery 65:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S. 2001. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20:45–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.