Abstract
The archaellum, the rotating motility structure of archaea, is best studied in the crenarchaeon Sulfolobus acidocaldarius. To better understand how assembly and rotation of this structure is driven, two ATP-binding proteins, FlaI and FlaH of the motor complex of the archaellum of the euryarchaeon Pyrococcus furiosus, were overexpressed, purified and studied. Contrary to the FlaI ATPase of S. acidocaldarius, which only forms a hexamer after binding of nucleotides, FlaI of P. furiosus formed a hexamer in a nucleotide independent manner. In this hexamer only 2 of the ATP binding sites were available for binding of the fluorescent ATP-analog MANT-ATP, suggesting a twofold symmetry in the hexamer. P. furiosus FlaI showed a 250-fold higher ATPase activity than S. acidocaldarius FlaI. Interaction studies between the isolated N- and C-terminal domains of FlaI showed interactions between the N- and C-terminal domains and strong interactions between the N-terminal domains not previously observed for ATPases involved in archaellum assembly. These interactions played a role in oligomerization and activity, suggesting a conformational state of the hexamer not observed before. Further interaction studies show that the C-terminal domain of PfFlaI interacts with the nucleotide binding protein FlaH. This interaction stimulates the ATPase activity of FlaI optimally at a 1:1 stoichiometry, suggesting that hexameric PfFlaI interacts with hexameric PfFlaH. These data help to further understand the complex interactions that are required to energize the archaellar motor.
Keywords: Archaea, Motility, ATPase activity, Type IV pili
Introduction
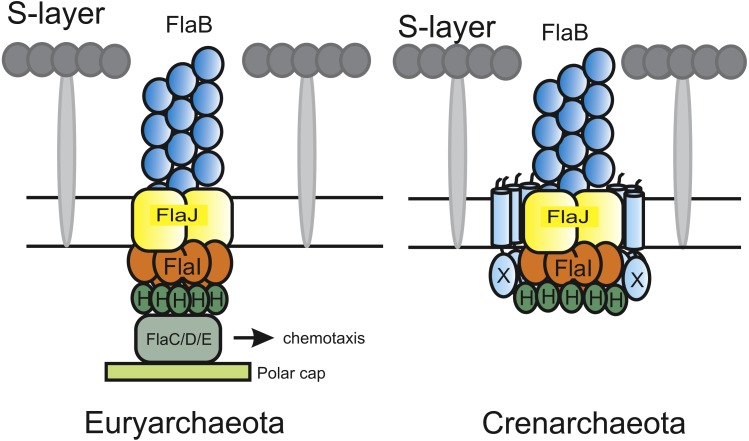
Motility in archaea is driven by a rotating cell surface appendage, called the archaellum (Jarrell & Albers, 2012; Albers & Jarrell, 2015). The archaellum is widely spread in archaea, and was identified in many of the archaeal phyla, e.g., in crenarchaeota, euryarchaeota, thaumarchaeota and nanoarchaeota (Makarova, Koonin & Albers, 2016). Although in function it resembles the bacterial flagellum, it is structurally different as it is evolutionarily related to archaeal and bacterial type IV pilus assembly systems (T4PSs) and Type II secretion systems (T2SS) (Jarrell & Albers, 2012; Berry & Pelicic, 2015; Albers & Jarrell, 2015). The archaellum consists of 7–13 proteins, which are all essential for the assembly and function (Patenge et al., 2001; Thomas, Pawson & Jarrell, 2001; Chaban et al., 2007; Lassak et al., 2012). Similar to the pilins of T4PSs, archaellins also possess N-terminal class III signal peptides, which are processed by a dedicated membrane bound aspartic acid peptidase (Albers, Szabó & Driessen, 2003; Bardy & Jarrell, 2003). After N-terminal cleavage, the mature archaellins are inserted into the growing archaellum filament. Recently, cryo EM structures of the euryarchaeotes Methanospirillum hungatei and Pyrococcus furiosus archaellum revealed that the archaellin monomer has two domains: an N-terminal domain, which forms a long hydrophobic α-helix, and a C-terminal domain with an eight-stranded anti-parallel β-barrel (Poweleit et al., 2016; Daum et al., 2017). The assembled archaellins showed different inter-subunit interactions than the assembled pilins of T4PSs (Craig et al., 2006; Wang et al., 2017) and the Iho670 adhesion filament of non-motile Ignicoccus hospitalis (Braun et al., 2016). Similar to the assembly of the pilus in T4PS and T2SSs (Jakovljevic et al., 2008; Chiang et al., 2008; Yamagata & Tainer, 2007), assembly of the archaellum is energized by ATP hydrolysis (Thomas, Pawson & Jarrell, 2001; Reindl et al., 2013). Operons encoding components of the archaellum contain two ATP binding proteins, FlaI and FlaH. FlaI of the crenarchaeon Sulfolobus acidocaldarius (SaFlaI) forms an ATP-dependent hexamer and the nucleotide bound crystal structure showed a conserved C-terminal ATPase domain (CTD) which is connected via a flexible linker to the variable N-terminal domain (NTD) (Ghosh et al., 2011; Reindl et al., 2013). In the hexameric crystal structure, an intrinsic twofold symmetry results in three unique subunit conformations, and superimposition of unique NTDs and CTDs shows that the individual NTD and CTD structures are similar, with only small changes in the NTD. The intra-subunit interface is largest between the CTDs. Indeed, SaFlaI lacking the NTD still exhibits 75% of the ATPase activity compared to the full-length FlaI (Reindl et al., 2013). In the crystal structures, the interaction between the NTD and CTD from one subunit is small compared to the interaction between one NTD and the neighboring subunit CTD (Reindl et al., 2013). Together with the hexameric structures of Type II secretion and Type IV pili systems (Yamagata & Tainer, 2007; Misic, Satyshur & Forest, 2010; Lu et al., 2013; McCallum et al., 2017) a model evolved in which successive rounds of ATP binding, ATP hydrolysis and ADP release in the three unique subunits result in conformational changes of the subunits. Based on the homology of FlaI and FlaJ with the T4PS proteins, it seems likely that FlaI interacts with FlaJ (Chiang, Habash & Burrows, 2005; Takhar et al., 2013; Bischof et al., 2016). Indeed, the flexible crown groove (residues 61–128) of the structure of SaFlaI contains negatively charged amino acid patches which were proposed to interact with the positively charged cytoplasmic loops of SaFlaJ (Reindl et al., 2013; Banerjee et al., 2013). A similar interaction was recently proposed between the PilB and PilC of Geobacter metallireducens (McCallum et al., 2017). Conformational changes in the membrane platform protein then might result in insertion or extrusion of the pilin or archaellin into the pilus/archaellum (Chang et al., 2016; McCallum et al., 2017). Indeed, the ATPase activity of Myxococcus xanthus PilB is stimulated by interaction with PilC (Bischof et al., 2016). Additional to its assembly, the archaellum also needs to rotate. How the switch between assembly and rotation occurs is currently unknown, but possibly FlaH, a second ATPase only identified in operons encoding the archaellum but not in operons encoding archaeal Type IV pili systems, is involved in this switch (Chaudhury et al., 2016). FlaH belongs to the RecA superfamily of ATPases. S. acidocaldarius FlaH (SaFlaH) can bind ATP, but is unable to hydrolyze it, most likely due to the presence of a non-canonical walker B motif. The crystal structures of SaFlaH and Methanocaldococcus jannaschii FlaH were solved (Meshcheryakov & Wolf, 2016; Chaudhury et al., 2016). SaFlaI and SaFlaH interact with each other in an ATP dependent manner (Chaudhury et al., 2016). SaFlaI and SaFlaH were also shown to interact with S. acidocaldarius FlaX (SaFlaX) (Banerjee et al., 2013). FlaX, which was only identified in crenarchaea, contains an N-terminal transmembrane domain and a C-terminal cytoplasmic domain which, for SaFlaX, forms a ring-like oligomeric structure with a diameter of 30 nm (Banerjee et al., 2012). Deletion of 57 amino acids which correspond to three helices from the C-terminus of SaFlaX abolished formation of the ring and interaction with FlaI in vitro (Banerjee et al., 2012; Banerjee et al., 2013). Electron microscopy revealed that, in vitro, SaFlaH could assemble as a second ring inside the SaFlaX ring (Chaudhury et al., 2016). Thus, it was proposed that the central core of the crenarchaeal archaellum is formed by FlaI together with FlaH, FlaX and FlaJ (Banerjee et al., 2013) (Fig. 1). In addition to these proteins, a minimal functional archaellum requires the FlaF and FlaG proteins. FlaF and FlaG are monotopic membrane proteins where SaFlaF has a β-sandwich fold and interacts with S-layer proteins suggesting that it might act as a stator that anchors the rotating archaellum (Banerjee et al., 2015).
Figure 1. Current models of the euryarchaeal and crenarchaeal archaellum motor complex.
In both shown archaellum motor complexes, FlaH, FlaI and FlaJ are conserved. FlaB, the archaellin, builds the fiament in the archaeal cell envelope (S-layer). The euryarchaeal archaellum motor complex contains FlaC/D/E which are thought to interact with the chemotaxis system. Additionally, a polar cap structure was identified very recently. However, its function is so far unknown. In the crenarchaeal archaellum motor complex FlaX forms a ring-like structure and is thought to act as a scaffold protein for motor protein assembly.
Although the archaellum is widely spread among archaea, the archaellum has mainly been biochemically characterized in the crenarchaeote S. acidocaldarius. Several differences have been observed between the archaellum systems of crenarchaea and euryarchaea. For example, whereas most species of crenarchaeota contain only one archaellin, euryarchaeota may possess up to five different archaellins (Jarrell & Albers, 2012). In Methanococcus maripaludis, the archaellum showed a hook-like structure which was not observed in a deletion mutant of the minor archaellin flaB3 (Chaban et al., 2007). Furthermore, FlaX was only identified in crenarchaea (Ghosh & Albers, 2011), whereas euryarchaeota contain the flaC, flaD and flaE genes which conversely are not found in crenarchaea (Jarrell & Albers, 2012; Albers & Jarrell, 2015) (Fig. 1). Finally, many euryarchaea exhibit chemotaxis systems, which have not been identified in crenarchaea till date (Wuichet, Cantwell & Zhulin, 2010). In Halobacterium salinarum and Haloferax volcanii, it has been demonstrated that the FlaC, FlaD and FlaE proteins link rotation of the archaellum to the CheY signal transduction cascade and thus to the chemotaxis system (Schlesner et al., 2009; Quax et al., 2018).
Recently, a low resolution image of the archaellar basal body of the euryarchaeote Thermococcus kodakaraensis was obtained by cryo-tomography (Briegel et al., 2017). This structure shows similarities to the structures of bacterial T4P (Chang et al., 2016), but also shows several unique features. For example, a large conical frustum of up to 500 nm in diameter was observed at the cytosolic base of the structure. The resolution of this structure is however not high enough to distinguish or identify individual components (Briegel et al., 2017). The cryo-EM structure of the P. furiosus archaellum allowed a more detailed view of the archaellum motor complex (Daum et al., 2017). As in T. kodakaraensis, a cone structure is present below the archaellum motor complexes. After modelling the structures of SaFlaI and SaFlaH in the densities close to the membrane, several remaining densities were observed. These probably contain the FlaCDE proteins.
In this study, we set out to characterize the motor subunits of the archaellum of Pyrococcus furiosus, an anaerobic, heterotrophic hyperthermophilic euryarchaeote that can grow at temperatures between 70 °C and 103 °C, and a pH between 5 and 9 (Fiala & Stetter, 1986). P. furiosus contains monopolar polytrichous archaella (Fiala & Stetter, 1986), which it does not only use to swim, but also to form cable-like cell–cell connections to adhere to solid surfaces (Näther et al., 2006). Here, we continue our previous studies on the biochemical characterization of P. furiosus FlaI (PfFlaI) and FlaH (PfFlaH).
Materials & Methods
Strains and plasmids
Escherichia coli strains NEB 10-beta (New England BioLabs) and Rosetta (DE3) (Novagen) were used for cloning purposes and overexpression respectively. Genomic DNA of Pyrococcus furiosus DSM 3638 (Robb et al., 2001) was used as a template for PCR reactions. Plasmids and their construction are described in Supplementary Table S1. Primers used are described in Supplementary Table S2. All plasmids sequences were confirmed by PCR and sequencing.
Overproduction and purification
Overproduction and purification of His-tagged P. furiosus FlaI (PfFlaI), PfFlaI-NTD, PfFlaI-CTD, and P.furiosus FlaH (PfFlaH) and the PfFlaH (K39A) and PfFlaH (D126N) mutants were performed as described previously (Chaudhury et al., 2016). Overproduction and purification of StrepII-tagged PfFlaI (E336A) were essentially performed the same except that Streptactin column material (IBA GmbH, Göttingen, Germany) was used, and that the protein was eluted with 2.5 mM d-desthiobiotin. Samples were stored at −80 °C until use.
Analytical gel filtration
PfFlaI, PfFlaI-NTD and PfFlaI-CTD were concentrated to 1 mg/ml in buffer containing 20mM Tris HCl pH 8.0, 150 mM NaCl (buffer A) using Amicon concentrators (Millipore) with a 10 kDa cut-off. 500 µl of the concentrated samples or 250 µl PfFlaI-NTD mixed with 250 µl PfFlaI-CTD and applied to Superdex 200 10/300 GL or Superdex 75 10/300 GL size exclusion columns equilibrated with buffer A. Fractions were analyzed on SDS-PAGE. Thyroglobulin (669 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.35 kDa) were used as size standards.
MANT-ATP binding
Binding of the fluorescent ATP analog 2′-/3′-O-(N-methylanthraniloyl) adenosine 5′-triphosphate (MANT-ATP, JENA biosciences) was detected by titrating PfFlaI (20 nM, 100 nM and 5 µM in buffer A containing 5 mM MgCl2) with increasing concentrations of MANT-ATP in a 150 µl cuvette at 20 °C in a Fluoromax-4 fluorimeter (HORIBA Scientific, Irvine, CA, USA). Excitation and emission wavelengths were set to 285 and 450 nm respectively with slit widths of 10 nm. Fluorescence was corrected for MANT-ATP fluorescence in the absence of protein. To determine the binding affinity of ATP, competition assays between MANT-ATP and ATP were performed. Total fluorescence was determined under the conditions described above after addition of increasing amounts of ATP to a solution containing 20 nM PfFlaI and 10 nM of MANT-ATP. The data were fitted with the Hill equation: , where F = Fluorescence, Fmin = minimal fluorescence, Fmax = maximal fluorescence, [ATP] is the ATP concentration, IC50 is the ATP concentration where the fluorescence is reduced by half, and n = Hill coefficient.
ATPase assay
Release of inorganic phosphate after ATP hydrolysis was determined using the Malachite green assay (Lanzetta et al., 1979) by determining the colorimetric change at 620 nm using a Clariostar plate reader (BMG labtech). ATP hydrolysis at different temperatures was determined by incubating 12.5 µg/ml PfFlaI in buffer A containing 5 mM MgCl2 and1 mM ATP for 5 min at different temperatures. ATP hydrolysis at different pHs was determined by incubating 12.5 µg/ml PfFlaI in different buffers (20 mM citrate (pH 3.0), 20 mM 2-(N-morpholino)ethane sulfonic acid (MES, pH 6.0), 20 mM 4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid (HEPES, pH 7.2), 20 mM 2-amino-2-hydroxymethyl-propane-1,3-diol (TRIS, pH 8.0/ 9.5), 20 mM sodium carbonate-bicarbonate (pH 10.0)) containing 5 mM MgCl2 and1 mM ATP for 5 min at 70 °C. To determine the ATPase activity of PfFlaI at different ATP concentrations, 12.5 µg/ml PfFlaI was incubated for 5 min at 70°C in buffer A containing 5mM MgCl2 and different ATP concentrations. The curve was fitted to the Michaelis–Menten equation (v = vmax∗[ATP]∕(Km + [ATP])). The Hill coefficient was determined from the slope of a plot of log ([ATP]) vs log (v∕(vmax − v)). ATPase activity of PfFlaI, PfFlaI-NTD, PfFlaI-CTD and the stoichiometric mixture of PfFlaI-NTD and PfFlaI-CTD was determined by incubating the proteins at a concentration of 12.5 µg/ml in buffer A containing 5 mM MgCl2 at 70 °C.
Microscale thermophoresis
Microscale thermophoresis was performed as described previously (Chaudhury et al., 2016). To determine the binding affinity of CTD and NTD of PfFlaI, 28 nM labeled PfFlaH in buffer A containing 0.05% (v/v) Tween-20 was titrated with increasing concentrations (1.3 nM–22.5 µM) of Pf-CTD and (5.6 nM–11.6 µM) Pf-NTD of PfFlaI on a Nano Temper Monolith NT.115 Pico instrument. The data were fitted as described previously (Chaudhury et al., 2016).
Results
Overproduction, purification and characterization of P. furiosus FlaI
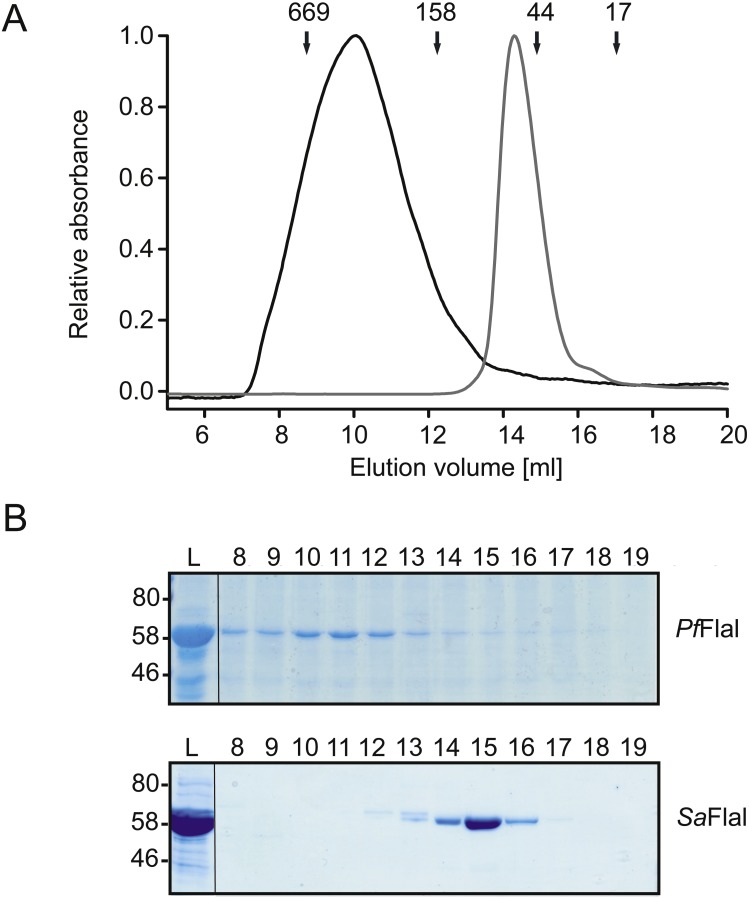
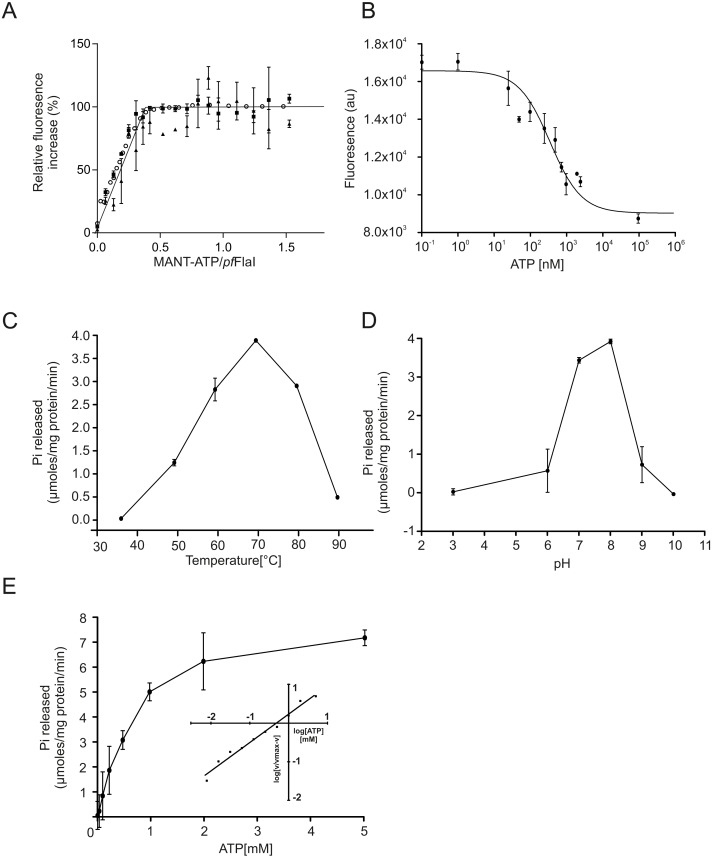
To compare the crenarchaeal and euryarchaeal archaellum, we set out to biochemically characterize the archaellum of Pyrococcus furiosus. The genetic region that encodes the components of the archaellum of P. furiosus encodes the major archaellin (flaB0), followed by two minor archaellins (flaB1, flaB2) and the flaCDFGHIJ genes (Robb et al., 2001; Näther et al., 2006; Näther-Schindler et al., 2014). Here, we focus on the FlaI and FlaH proteins. Similar to SaFlaI, Pyrococcus furiosus FlaI (PfFlaI) contains both an N-terminal and a C-terminal domain, which show respectively 36% and 57% identity with SaFlaI. PfFlaI was previously overexpressed and purified (Chaudhury et al., 2016), and is here characterized further. Analysis of the oligomeric state of PfFlaI using size exclusion chromatography showed that PfFlaI eluted at a position corresponding to a hexamer (Figs. 2A and 2B). Thus, in this respect, PfFlaI differs from SaFlaI, which after purification elutes as a monomer and formed a hexamer in the presence of the non-hydrolysable ATP analog adenylyl-imidodiphosphate (AMP-PNP) (Ghosh et al., 2011) (Figs. 2A and 2B). The Abs260/Abs280 ratio of 0.3 suggested that PfFlaI was isolated in the nucleotide free form. Indeed, further purification steps, dialysis or ammonium sulphate precipitation did not result in a change in the Abs260/Abs280 ratio. To test whether PfFlaI could bind ATP, titrations with the fluorescent ATP analog MANT-ATP were performed. Similar experiments were performed to determine the nucleotide binding affinity of SaFlaI (Ghosh et al., 2011). For PfFlaI, the fluorescence increased linearly upon addition of MANT-ATP, until a maximum was reached after which no further increase was observed. This saturation was dependent on the protein concentration, and when 20 nM, 100 nM or 5 µM protein PfFlaI (Fig. 3A) was used, the maximum was reached for all three concentrations at 1/3 of the PfFlaI concentration used, demonstrating that only two of the ATP binding sites in the hexamer are available for binding of MANT-ATP. Indeed, SaFlaI crystallized as a hexameric ring with an intrinsic twofold symmetry resulting in three different conformations of the monomers (Reindl et al., 2013), suggesting that also for PfFlaI such a twofold symmetry with three unique subunits occurs. To determine the affinity for ATP, bound MANT-ATP was competed with ATP resulting in an IC50 of 260 nM at 20 °C (Fig. 3B). The hydrolysis of ATP by PfFlaI was tested at different temperatures and at different pHs (Figs. 3C and 3D) and the highest activity was found at 70 °C and pH 8.0. Even though P. furiosus can live up to 103 °C, the ATPase activity in vitro decreased above 70 °C suggesting that at temperatures above 70 °C PfFlaI is unstable. At 70 °C and pH 8.0, ATP was hydrolyzed with a vmax of 8 µmoles mg−1 min−1 (Chaudhury et al., 2016) and a Km of 580 µM (Fig. 3E). ATP hydrolysis did not show cooperativity with increasing ATP concentrations (Hill coefficient = 0.9) (Fig. 3E, inset). The maximum activity observed for PfFlaI was 250-fold higher than the maximum activity (at pH 6.5 and 75 °C) observed for SaFlaI (Chaudhury et al., 2016), and equals a turn-over of ∼500 ATP min−1.
Figure 2. PfFlaI forms a stable hexamer.
(A) Relative absorbance at 280 nm of size exclusion chromatography of PfFlaI (64 kDa) shown in black line and SaFlaI (59 kDa) is shown in grey line using Superdex 200 10/300GL column. Elution positions of molecular mass standards (kDa) were indicated with arrows. (B) Coomassie stained SDS-PAGE analysis of the different elution fractions.
Figure 3. ATP binding and hydrolysis of PfFlaI.
(A) Fluorescence increase at increasing concentrations of MANT-ATP upon addition of 20 nM, 100 nM and 5 µM PfFlaI. Lines depict the linear fits of the two observed phases. The lines cross at a MANT-ATP concentration of 1.7 µM. (B) Total fluorescence after addition of increasing amounts of ATP to a solution containing 20 nM PfFlaI and 10 nM of MANT-ATP. The data were fitted with the Hill equation: resulting in a best fit (R2 = 0.98) with IC50 = 260 nM and n = 0.67. (C, D) ATP hydrolysis by 12.5 µg/ml PfFlaI at different temperatures and at different pHs respectively. (E) ATPase activity of PfFlaI at different ATP concentrations. The curve was fitted to the Michaelis-Menten equation (V = Vmax∗[ATP]∕(Km + [ATP])), resulting in a Km of 580 nM. The inset shows the same data plotted according to the Hill equation (Hill coefficient = 0.9). Experiments were performed with at least two biological and three technical replicates. Error bars depict the standard error obtained from the technical replicates.
The N-terminal domains of PfFlaI interact with each other and with the C-terminal domains to stimulate ATP hydrolysis
The crystal structures of SaFlaI revealed that, similar to other described T2SS and T4PS ATPases (Robien et al., 2003; Yamagata & Tainer, 2007; Satyshur et al., 2007; Misic, Satyshur & Forest, 2010; Lu et al., 2013; McCallum et al., 2017), SaFlaI consists of a variable NTD and a CTD that binds and hydrolyses ATP (Reindl et al., 2013). These domains are connected via a short flexible linker (Reindl et al., 2013). The hexameric structure of SaFlaI was only observed in the crystal structure after incubation with AMP-PNP. Since PfFlaI forms a much more stable hexamer, we set out to study the interactions between the NTDs and CTDs of PfFlaI.
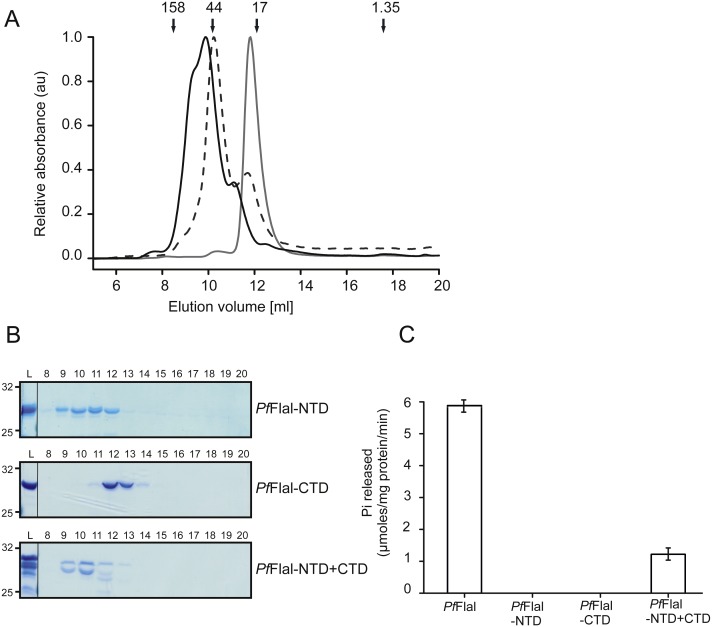
Both the NTD (PfFlaI-NTD; 29 kDa) and the CTD (PfFlaI-CTD; 31 kDa) of PfFlaI were overexpressed in E. coli, purified, and were then analyzed on analytical size exclusion chromatography (Figs. 4A and 4B). The PfFlaI-CTD eluted as a monomer whereas, contrary to what was expected, the PfFlaI-NTD eluted not only as monomer but also as dimer and possibly a higher order oligomer. This demonstrates that the NTDs of PfFlaI interact with each other and might play a role in the formation of the hexameric ring. Since in the crystal structure of the SaFlaI hexamer the NTDs interact with the CTDs of the neighboring subunit, it was tested whether also the PfFlaI-NTD and the PfFlaI-CTD interacted. Equimolar concentrations of PfFlaI-NTD and PfFlaI-CTD were mixed, analyzed on analytical size exclusion chromatography. This resulted in a co-elution of the PfFlaI-NTD and PfFlaI-CTD and a shift of especially the elution position of the PfFlaI-CTD, demonstrating an interaction between the PfFlaI-NTD and the PfFlaI-CTD. To test whether interaction between the PfFlaI-NTD and the PfFlaI-CTD influenced the ATPase activity, ATP hydrolysis of the single domains and of the mixed domains was determined (Fig. 4C). In these experiments, ATP hydrolysis was only observed when both the PfFlaI-NTD and PfFlaI-CTD were present, demonstrating that the PfFlaI-NTD can stimulate ATP hydrolysis by the PfFlaI-CTD.
Figure 4. Analysis of the interaction between the N- and C-terminal domains of PfFlaI.
(A) Relative absorbance at 280 nm of size exclusion chromatography at 280 nm of size exclusion chromatography of PfFlaI-NTD (black line), PfFlaI-CTD (grey line) or a stoichiometric mixture of PfFlaI-NTD and PfFlaI-CTD (dashed line) using Superdex 75 10/300GL column. Elution positions of molecular mass standards (kDa) are indicated with arrows. (B) SDS-PAGE analysis of the elution fractions described in A. (C) ATPase activity at 70 °C of the main elution fraction after size exclusion chromatography of PfFlaI, PfFlaI-NTD, PfFlaI-CTD and the stoichiometric mixture of PfFlaI-NTD and PfFlaI-CTD at a concentration of 12.5 µg/ml. For A and B, a representative experiment is shown. For C, graph shows the average of two biological replicates with two technical replicates.
PfFlaH stimulates the ATPase activity of PfFlaI
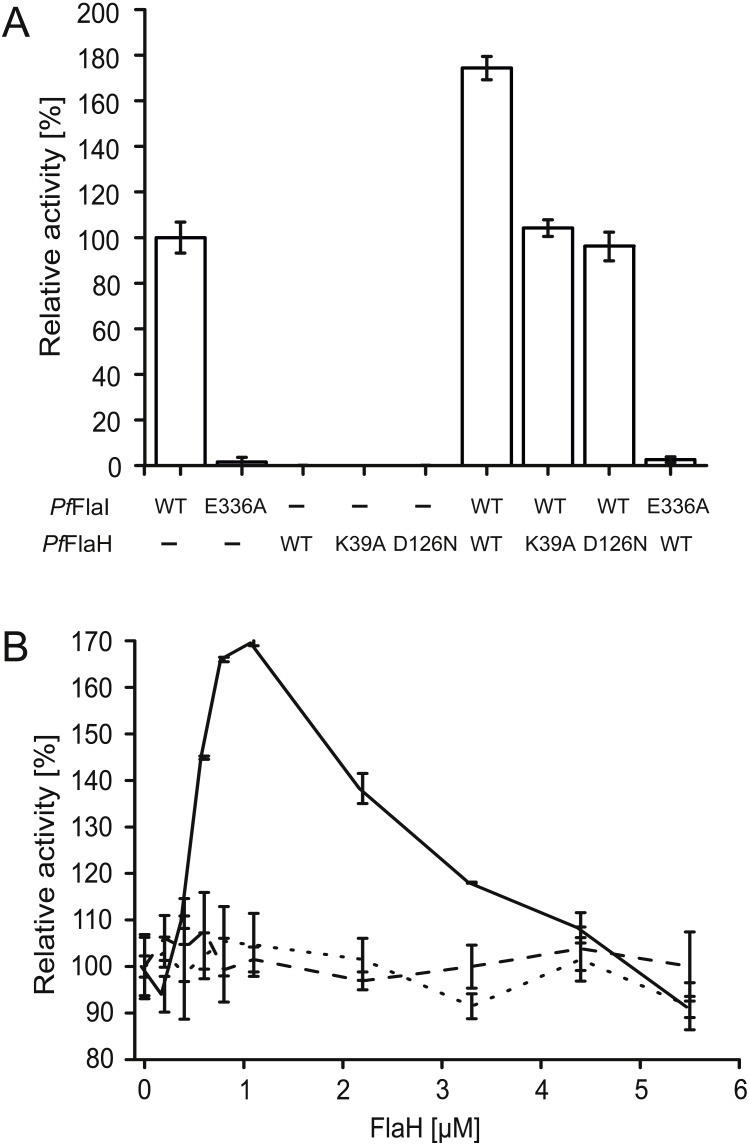
We have overexpressed and purified P. furiosus FlaH (PfFlaH) and used microscale thermophoresis (MST) to show that nucleotide-bound PfFlaH bound to PfFlaI with a KD of 1 µM (Chaudhury et al., 2016). PfFlaH containing mutations in the Walker A (PfFlaH K39A) and Walker B motifs (PfFlaH D126N) had a strongly reduced affinity for nucleotides, and for PfFlaI, demonstrating that nucleotide binding to PfFlaH is important for its interaction with PfFlaI (Chaudhury et al., 2016). Here, we further investigated the influence of this interaction on the ATPase activity of PfFlaI. As observed, PfFlaI hydrolyzed ATP, whereas for PfFlaH, no ATPase activity could be observed (Fig. 5A, Chaudhury et al., 2016). Also, no ATP hydrolysis was observed for PfFlaI with an E336A mutation in the Walker B motif, and for the PfFlaHK39A and PfFlaHD126N proteins. Addition of PfFlaH to PfFlaI stimulated the total ATPase activity. To test whether the stimulation of the ATPase is derived from an increase of the activity of PfFlaH or of PfFlaI, different combinations of mutants in PfFlaH and PfFlaI with WT proteins were tested (Fig. 5A). This demonstrated unequivocally that binding of nucleotide bound PfFlaH stimulates the ATPase activity of PfFlaI. To test the stoichiometry of this interaction, 1 µM PfFlaI was incubated with increasing concentrations of PfFlaH. A maximal stimulation was observed when PfFlaH was present in stoichiometric amounts to PfFlaI (Fig. 5B). The stimulation decreased when further increasing amounts of PfFlaH were added. Thus PfFlaI and PfFlaH interact in a 1:1 stoichiometry.
Figure 5. Nucleotide bound PfFlaH stimulates the ATPase activity of PfFlaI.
(A) The ATPase activity at 70 °C was determined for PfFlaI, PfFlaH and these proteins with mutations in their respective Walker A PfFlaH(K39A) and walker B motifs PfFlaI(E336A), PfFlaH(D126N) and of different combinations of these proteins. Proteins were added to a final concentration of 1 µM. (B) The ATPase activity at 70 °C was determined for 1 µM PfFlaI and increasing amounts of PfFlaH, PfFlaH(K39A) and PfFlaH(D126N) shown in solid line, dashed line and dotted line respectively. The graphs show the average of two biological replicates with two technical replicates.
FlaH interacts at the C-terminal domain of FlaI
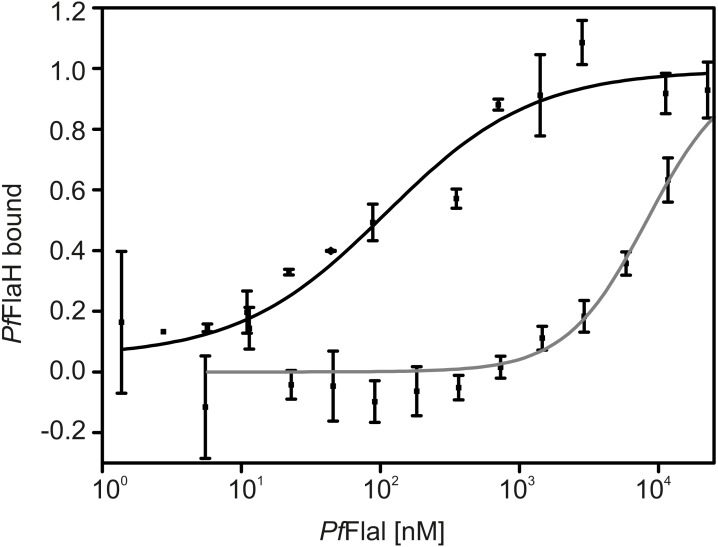
Nucleotide-bound PfFlaH can interact with PfFlaI with an affinity of 1 µM (Chaudhury et al., 2016). To test whether FlaH interacts with the NTD or CTD of FlaI, interaction assays using microscale thermophoresis were performed using PfFlaH and the P. furiosus NTD and CTD of FlaI (Fig. 6). The PfFlaI-CTD and the PfFlaI-NTD interacted with PfFlaH with affinities of 100 nM and >8 µM, respectively. Thus, it was concluded that PfFlaH interacts specifically with the PfFlaI-CTD. This interaction occurs with a 10-fold higher affinity than the interaction between full length hexameric PfFlaI and PfFlaH, showing that PfFlaH can interact in different manners with the hexameric PfFlaI and the monomeric PfFlaI-CTD.
Figure 6. PfFlaH interacts with higher affinity with PfFlaI-CTD than PfFlaI-NTD.
The binding of 28 nM fluorescently labeled Pf FlaH to increasing concentrations of 1.4 nM–22.5 µM for PfFlaI-CTD (black) and 5.7 nM–11.7 µM for PfFlaI-NTD (grey) was studied by microscale thermophoresis. Binding is depicted as the fraction bound and binding curves were fitted to the Hill equation. Curves were obtained from at least two independent experiments.
Discussion
Many archaeal cell surface structures are homologous to bacterial T4PSs, which function in cell attachment to surfaces, DNA transport, biofilm formation and motility (Jarrell et al., 2013; Makarova, Koonin & Albers, 2016; Chaudhury, Quax & Albers, 2018). Similar to the T2SSs and T4PSs, most of these systems possess a pilin protein, a prepilin peptidase which cleaves at the N-terminus of the prepilin, an ATPase of the superfamily of traffic ATPases, and a membrane platform protein (Peabody, 2003; Nishida & Chen, 2004). The traffic ATPase was suggested to interact with the membrane platform protein for several systems (Reindl et al., 2013; Banerjee et al., 2013; Bischof et al., 2016; Takhar et al., 2013), and it has been proposed that interactions between the processed pilin, the traffic ATPase protein and the platform protein drive the assembly of the pilus (Chang et al., 2016; McCallum et al., 2017). FlaI, the traffic ATPase of the archaellum differs from the other traffic ATPases in the fact that it not only energizes the assembly of the archaella, but also should drive its rotation. Comparison of the different crystal structures of traffic ATPases showed many different conformations of the NTD relative to the CTD, but in general, binding of ATP results in large domain movements bringing the NTD and CTD closer together (Reindl et al., 2013; Lu et al., 2013; Satyshur et al., 2007). Comparison of the conformations in SaFlaI and Vc GspE, the traffic ATPase of the T2SS of Vibrio cholerae, showed that in Vc GspE, ATP hydrolysis results in an up and down movement of the domains, whereas, SaFlaI shows a more rotating movement of the domains, possibly explaining the differences between SaFlaI and other traffic ATPases (Reindl et al., 2013). The rotating movement of the SaFlaI hexamer is the result of three different alternating conformations which mostly differ in the position of the NTD relative to the CTD.
Here the euryarchaeal PfFlaI was characterized, and compared to the well characterized crenarchaeal SaFlaI. Firstly, it was observed that PfFlaI forms a stable hexamer whereas SaFlaI was a monomer in solution. The SaFlaI hexamer was observed only after incubation with a non-hydrolyzable ATP analogue and at high protein concentrations in the crystal structure. In the ADP-bound SaFlaI crystal structure, three different alternating conformations were observed, which all contained a nucleotide. ATP binding assays with PfFlaI showed that only two of the six positions in the PfFlaI hexamer were accessible for MANT-ATP, suggesting that PfFlaI also contains alternating conformations, and that four of the six subunits of the hexamer are not accessible to fluorescently labelled nucleotides. ATP hydrolysis by SaFlaI was highly co-operative, whereas ATP hydrolysis by PfFlaI did not show any cooperativity, suggesting that cooperatively occurs during the assembly of the SaFlaI hexamer, while no cooperativity occurs in the assembled PfFlaI hexamer. Indeed, the PfFlaI hexamer is, contrary to SaFlaI, formed in an ATP independent manner. This suggest that PfFlaI and SaFlaI might differ structurally. Another suggestion that PfFlaI and SaFlaI differ in their structures comes from the experiments with the isolated NTD and CTD. Whereas in the different SaFlaI crystal structures obtained, interactions are found between the CTDs and between the CTDs and the NTD, no strong interactions were observed between the NTDs. Indeed, SaFlaI lacking the NTD still exhibits 75% of the ATPase activity compared to the full-length FlaI (Reindl et al., 2013), suggesting that the NTD plays no important role in oligomerization. In PfFlaI however, deletion of the NTD strongly reduces or abolishes oligomerization and ATPase activity. The activity and oligomerization can be partly recovered by the addition of the isolated NTD, suggesting that the NTD for PfFlaI also plays an intrinsic role in oligomerization. Based on the crystal structure of SaFlaI, it could be expected that this is caused by the stabilization of the interaction between two CTDs by an NTD, but our observation that the isolated NTDs of PfFlaI also oligomerize suggests that interactions between NTDs might also play a role in the oligomerization and activity of PfFlaI. Strong interactions between NTDs of traffic ATPases, have currently only been observed for the HP0525 traffic ATPase of the Helicobacter pylori type IV secretion system (Yeo et al., 2000).
Next to the characterization PfFlaI, the interaction of PfFlaI with PfFlaH was further analyzed. Binding of nucleotide bound PfFlaH to PfFlaI stimulated its ATPase activity twofold, further demonstrating the importance of this interaction. Maximum stimulation was found at a 1:1 stoichometery, suggesting that hexameric PfFlaH interacts with hexameric PfFlaI. Previously, it was demonstrated that both SaFlaI and SaFlaH interact with SaFlaX (Banerjee et al., 2013). In vitro assembly of the C-terminal domain of SaFlaX resulted in ring-like structures with 15- to 23- fold symmetry with widely different diameters (Banerjee et al., 2012). After incubation with SaFlaH, monomeric SaFlaH particles were observed inside these SaFlaX rings (Chaudhury et al., 2016). The amount of FlaH bound inside the rings varied with the size of the ring, but in the most occurring rings with a 20- fold symmetry, 9–10 SaFlaH monomers could be observed (Chaudhury et al., 2016). The size of the in vivo FlaX ring in the S. acidocaldarius archaellum complex is currently still unknown. However, since SaFlaX also interacts with SaFlaI, and a FlaI hexamer interacts with hexameric FlaH, it seems likely that the motor complex in S. acidocaldarius consists of hexameric SaFlaI, bound to hexameric SaFlaH, surrounded by a SaFlaX ring. Like all euryarchaea, P. furiosus does not encode a FlaX homolog, but encodes the FlaC and FlaD proteins. However, it is currently not known whether they also form a ring-like structure and whether possible interactions with FlaI or FlaH exist. Remarkably, P. furiosus does not encode a chemotaxis system (Maeder et al., 1999) and thus the FlaCDE proteins should not be related to the CheY signal transduction cascade.
It was proposed that the NTD of FlaI interacts with FlaJ (Reindl et al., 2013; Banerjee et al., 2013), making it likely that the CTD domain of FlaI would interact with FlaH, and we found that PfFlaH interacts with the PfFlaI-CTD. This interaction occurs with a 10-fold higher affinity than the interaction between full length PfFlaI and PfFlaH. This suggest that the affinity of the interaction between FlaH and FlaI might be modulated by factors that increase or decrease the accessibility of the CTD of FlaI to FlaH, and this might facilitate a switch between assembly and rotation of the archaellum.
Conclusions
Our results showed that FlaI of P. furiosus differs significantly from the extensively studied FlaI of S. acidocaldarius. Contrary to FlaI of S. acidocaldarius, which only forms a hexamer after binding of nucleotides, FlaI of P. furiosus forms a stable hexamer in a nucleotide independent manner. The presence of the stable hexamer allowed us to study nucleotide binding to the hexamer. This showed that only 2 of the 6 ATP binding sites were available for binding of the fluorescent ATP-analog MANT-ATP, suggesting a twofold symmetry in the hexamer and further suggesting that individual proteins in the hexamer alternate between the empty, ATP and ADP bound states. We also identified strong interactions between the N-terminal domains S. acidocaldarius FlaI not identified before for S. acidocaldarius FlaI. These interactions played a role in oligomerization and activity, suggesting a conformational state of the hexamer not observed previously. We further showed that interaction between FlaI and FlaH stimulates the ATPase activity of FlaI. This occurs optimally at a 1:1 stoichiometry, suggesting that a FlaI hexamer can interact with six FlaH proteins or with a FlaH hexamer. Further interaction studies showed that FlaH interacts with the C-terminal domain of PfFlaI.
Supplemental Information
Tables listing the primers, plasmids and strains used in this study.
All SDS-PAGE images and excel tables for all shown graphs.
Acknowledgments
We thank Prof. Carola Hunte for giving access to Nanotemper Monolith NT.115 Pico instrument and Dr. Rashmi Kumariya for providing the data of the SaFlaI gel filtration.
Funding Statement
Paushali Chaudhury and Sonja-Verena Albers were supported by a starting grant from the European Reserach Council (Nr. 311523, Archaellum). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Sonja-Verena Albers is an Academic Editor for PeerJ.
Author Contributions
Paushali Chaudhury conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Chris van der Does conceived and designed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Sonja-Verena Albers conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The raw data are provided in a Supplemental File.
References
- Albers, Szabó & Driessen (2003).Albers SV, Szabó Z, Driessen AJM. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. Journal of Bacteriology. 2003;185:3918–3925. doi: 10.1128/JB.185.13.3918-3925.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers & Jarrell (2015).Albers S-V, Jarrell KF. The archaellum: how Archaea swim. Frontiers in Microbiology. 2015;6:23. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee et al. (2012).Banerjee A, Ghosh A, Mills DJ, Kahnt J, Vonck J, Albers S-VV. FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI. Journal of Biological Chemistry. 2012;287:43322–43330. doi: 10.1074/jbc.M112.414383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee et al. (2013).Banerjee A, Neiner T, Tripp P, Albers S-V. Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS Journal. 2013;280:6141–6149. doi: 10.1111/febs.12534. [DOI] [PubMed] [Google Scholar]
- Banerjee et al. (2015).Banerjee A, Tsai CL, Chaudhury P, Tripp P, Arvai AS, Ishida JP, Tainer JA, Albers SV. FlaF is a β-sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S-layer protein. Structure. 2015;23:863–872. doi: 10.1016/j.str.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy & Jarrell (2003).Bardy SL, Jarrell KF. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Molecular Microbiology. 2003;50:1339–1347. doi: 10.1046/j.1365-2958.2003.03758.x. [DOI] [PubMed] [Google Scholar]
- Berry & Pelicic (2015).Berry J-LJL, Pelicic V. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiology Reviews. 2015;39:1–21. doi: 10.1093/femsre/fuu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof et al. (2016).Bischof LF, Friedrich C, Harms A, Søgaard-Andersen L, Van der Does C. The type IV pilus assembly ATPase PilB of Myxococcus xanthus interacts with the inner membrane platform protein PilC and the nucleotide-binding protein PilM. Journal of Biological Chemistry. 2016;291:6946–6957. doi: 10.1074/jbc.M115.701284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun et al. (2016).Braun T, Vos MR, Kalisman N, Sherman NE, Rachel R, Wirth R, Schröder GF, Egelman EH. Archaeal flagellin combines a bacterial type IV pilin domain with an Ig-like domain. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:10352–10357. doi: 10.1073/pnas.1607756113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel et al. (2017).Briegel A, Oikonomou CM, Chang Y, Kjær A, Huang AN, Kim KW, Ghosal D, Nguyen HH, Kenny D, Orgorzalek Loo RR, Gunsalus RP, Jensen GJ. Morphology of the archaellar motor and associated cytoplasmic cone in Thermococcus kodakaraensis. EMBO Reports. 2017;18:1660–1670. doi: 10.15252/embr.201744070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban et al. (2007).Chaban B, Ng SYM, Kanbe M, Saltzman I, Nimmo G, Aizawa SI, Jarrell KF. Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Molecular Microbiology. 2007;66:596–609. doi: 10.1111/j.1365-2958.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Chang et al. (2016).Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ. Architecture of the type IVa pilus machine. Science. 2016;351(6278):aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury et al. (2016).Chaudhury P, Neiner T, D’Imprima E, Banerjee A, Reindl S, Ghosh A, Arvai AS, Mills DJ, Van der Does C, Tainer JA, Vonck J, Albers SV. The nucleotide-dependent interaction of FlaH and FlaI is essential for assembly and function of the archaellum motor. Molecular Microbiology. 2016;99:674–685. doi: 10.1111/mmi.13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, Quax & Albers (2018).Chaudhury P, Quax TEF, Albers S-V. Versatile cell surface structures of archaea. Molecular Microbiology. 2018;107:298–311. doi: 10.1111/mmi.13889. [DOI] [PubMed] [Google Scholar]
- Chiang, Habash & Burrows (2005).Chiang P, Habash M, Burrows LL. Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. Journal of Bacteriology. 2005;187:829–839. doi: 10.1128/JB.187.3.829-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang et al. (2008).Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. Functional role of conserved residues in the characteristics secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology. 2008;154:114–126. doi: 10.1099/mic.0.2007/011320-0. [DOI] [PubMed] [Google Scholar]
- Craig et al. (2006).Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Daum et al. (2017).Daum B, Vonck J, Bellack A, Chaudhury P, Reichelt R, Albers S-V, Rachel R, Kühlbrandt W. Structure and in situ organisation of the Pyrococcus furiosus archaellum machinery. Elife. 2017;6:e27470. doi: 10.7554/eLife.27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala & Stetter (1986).Fiala G, Stetter KO. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Archives of Microbiology. 1986;145:56–61. doi: 10.1007/BF00413027. [DOI] [Google Scholar]
- Ghosh & Albers (2011).Ghosh A, Albers S-V. Assembly and function of the archaeal flagellum. Biochemical Society Transactions. 2011;39:64–69. doi: 10.1042/BST0390064. [DOI] [PubMed] [Google Scholar]
- Ghosh et al. (2011).Ghosh A, Hartung S, Van der Does C, Tainer JA, Albers S-V. Archaeal flagellar ATPase motor shows ATP-dependent hexameric assembly and activity stimulation by specific lipid binding. Biochemical Journal. 2011;437:43–52. doi: 10.1042/BJ20110410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovljevic et al. (2008).Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. Journal of Bacteriology. 2008;190:2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell & Albers (2012).Jarrell KF, Albers SV. The archaellum: an old motility structure with a new name. Trends in Microbiology. 2012;20:307–312. doi: 10.1016/j.tim.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Jarrell et al. (2013).Jarrell K, Ding Y, Nair D, Siu S. Surface appendages of archaea: structure, function, genetics and assembly. Life. 2013;3:86–117. doi: 10.3390/life3010086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta et al. (1979).Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Analytical Biochemistry. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Lassak et al. (2012).Lassak K, Neiner T, Ghosh A, Klingl A, Wirth R, Albers SV. Molecular analysis of the crenarchaeal flagellum. Molecular Microbiology. 2012;83:110–124. doi: 10.1111/j.1365-2958.2011.07916.x. [DOI] [PubMed] [Google Scholar]
- Lu et al. (2013).Lu C, Turley S, Marionni S, Park Y-J, Lee KK, Patrick M, Shah R, Sandkvist M, Bush MF, Hol WG. Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure. 2013;21:1707–1717. doi: 10.1016/j.str.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder et al. (1999).Maeder DL, Weiss RB, Dunn DM, Cherry JL, González JM, DiRuggiero J, Robb FT. Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics. 1999;152(4):1299–1305. doi: 10.1093/genetics/152.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, Koonin & Albers (2016).Makarova KS, Koonin EV, Albers S-V. Diversity and evolution of type IV pili systems in archaea. Frontiers in Microbiology. 2016;7:667. doi: 10.3389/fmicb.2016.00667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum et al. (2017).McCallum M, Tammam S, Khan A, Burrows LL, Lynne Howell P. The molecular mechanism of the type IVa pilus motors. Nature Communications. 2017;8:15091. doi: 10.1038/ncomms15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshcheryakov & Wolf (2016).Meshcheryakov VA, Wolf M. Crystal structure of the flagellar accessory protein FlaH of Methanocaldococcus jannaschii suggests a regulatory role in archaeal flagellum assembly. Protein Science. 2016;25:1147–1155. doi: 10.1002/pro.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic, Satyshur & Forest (2010).Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT structures with and without nucleotide reveal a dynamic Type Iv pilus retraction motor. Journal of Biological Chemistry. 2010;400:1011–1021. doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther et al. (2006).Näther DJ, Rachel R, Wanner G, Wirth R. Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell-cell contacts. Journal of Bacteriology. 2006;188:6915–6923. doi: 10.1128/JB.00527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näther-Schindler et al. (2014).Näther-Schindler DJ, Schopf S, Bellack A, Rachel R, Wirth R. Pyrococcus furiosus flagella: biochemical and transcriptional analyses identify the newly detected flaB0 gene to encode the major flagellin. Frontiers in Microbiology. 2014;5:695. doi: 10.3389/fmicb.2014.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida & Chen (2004).Nishida T, Chen DG. Incorporating spatial autocorrelation into the general linear model with an application to the yellowfin tuna (Thunnus albacares) longline CPUE data. Fisheries Research. 2004;70:265–274. doi: 10.1016/j.fishres.2004.08.008. [DOI] [Google Scholar]
- Patenge et al. (2001).Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D. The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Molecular Microbiology. 2001;41:653–663. doi: 10.1046/j.1365-2958.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- Peabody (2003).Peabody CR. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology. 2003;149:3051–3072. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- Poweleit et al. (2016).Poweleit N, Ge P, Nguyen HH, Loo RRO, Gunsalus RP, Zhou ZH. CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nature Microbiology. 2016;2:16222. doi: 10.1038/nmicrobiol.2016.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax et al. (2018).Quax TEF, Altegoer F, Rossi F, Li Z, Rodriguez-Franco M, Kraus F, Bange G, Albers SV. Structure and function of the archaeal response regulator CheY. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E1259–E1268. doi: 10.1073/pnas.1716661115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl et al. (2013).Reindl S, Ghosh A, Williams GJ, Lassak K, Neiner T, Henche A-L, Albers SV, Tainer JA. Insights on FlaI functions in archaeal motor assembly and motilify from structures, conformations and genetics. Molecular Cell. 2013;49(6):1069–1082. doi: 10.1016/j.molcel.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb et al. (2001).Robb FT, Maeder DL, Brown JR, DiRuggiero J, Stump MD, Yeh RK, Weiss RB, Dunn DM. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods in Enzymology. 2001;330:134–157. doi: 10.1016/S0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- Robien et al. (2003).Robien MA, Krumm BE, Sandkvist M, Hol WGJ. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. Journal of Molecular Biology. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Satyshur et al. (2007).Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesner et al. (2009).Schlesner M, Miller A, Streif S, Staudinger WF, Müller J, Scheffer B, Siedler F, Oesterhelt D. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiology. 2009;9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhar et al. (2013).Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. Journal of Biological Chemistry. 2013;288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, Pawson & Jarrell (2001).Thomas NA, Pawson CT, Jarrell KF. Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non-flagellated cells. Molecular Genetics. 2001;265:596–603. doi: 10.1007/s004380100451. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2017).Wang F, Coureuil M, Osinski T, Orlova A, Altindal T, Gesbert G, Nassif X, Egelman EH, Craig L. Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV pili at sub-nanometer resolution. Structure. 2017;25:1423–1435. doi: 10.1016/j.str.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet, Cantwell & Zhulin (2010).Wuichet K, Cantwell BJ, Zhulin IB. Evolution and phyletic distribution of two-component signal transduction systems. Current Opinion in Microbiology. 2010;13:219–225. doi: 10.1016/j.mib.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata & Tainer (2007).Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO Journal. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo et al. (2000).Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Molecular Cell. 2000;6:1461–1472. doi: 10.1016/S1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables listing the primers, plasmids and strains used in this study.
All SDS-PAGE images and excel tables for all shown graphs.
Data Availability Statement
The following information was supplied regarding data availability:
The raw data are provided in a Supplemental File.