Summary
The biotechnical platform strain Ralstonia eutropha H16 was genetically engineered to express a cox subcluster of the carboxydotrophic Oligotropha carboxidovorans OM5, including (i) the structural genes coxM, ‐S and ‐L, coding for an aerobic carbon monoxide dehydrogenase (CODH) and (ii) the genes coxD, ‐E, ‐F and ‐G, essential for the maturation of CODH. The cox Oc genes expressed under control of the CO 2‐inducible promoter PL enabled R. eutropha to oxidize CO to CO 2 for the use as carbon source, as demonstrated by 13 CO experiments, but the recombinant strains remained dependent on H2 as external energy supply. Therefore, a synthetic metabolism, which could be described as ‘carboxyhydrogenotrophic’, was established in R. eutropha. With this extension of the bacterium's substrate range, growth in CO‐, H2‐ and CO 2‐containing artificial synthesis gas atmosphere was enhanced, and poly(3‐hydroxybutyrate) synthesis was increased by more than 20%.
Introduction
The β‐proteobacterium Ralstonia eutropha H16, currently named as Cupriavidus necator H16, represents a model organism for autotrophic lifestyle and has been studied extensively for its efficient utilization of carbon dioxide and hydrogen as carbon and energy sources (Friedrich and Schwartz, 1993). By means of enzymes of the Calvin‐Benson‐Bassham (CBB) cycle, R. eutropha H16 fixes CO2 from the atmosphere as the sole source of carbon as well as from CO2, generated from its own intracellular oxidation of organic carbon compounds like formate (Kärst and Friedrich, 1984; Shimizu et al., 2015). When growing lithotrophically, R. eutropha H16 oxidizes molecular hydrogen by a soluble and a membrane‐bound hydrogenase, which are tolerant to oxygen, as well as to carbon monoxide (Buhrke et al., 2005; Burgdorf et al., 2005; Bürstel et al., 2016).
Several approaches have been made to exploit R. eutropha's efficient chemolithoautotrophic lifestyle to potentially capture CO2 from waste‐ or industrial exhaust gases for the conversion to various products of increased value. These so‐called second‐generation bioproducts include polymers, fuels and fine chemicals (Cook and Schlegel, 1978; Tanaka and Ishizaki, 1994; Müller et al., 2013; Lu and Yu, 2017). An alternative form of a gaseous feedstock that can be produced from fossil as well as from renewable resources, is the platform chemical synthesis gas (syngas), which has gained considerable attention for the use as cheap, energy‐rich and abundant feedstock for microbial processes in recent years (Drzyzga et al., 2015). Besides hydrogen and some CO2, a major component of syngas is carbon monoxide, which can be utilized as carbon and energy source by various bacteria. These include the so‐called aerobic carboxydotrophs, such as Oligotropha carboxidovorans and Alcaligenes carboxydus, which exhibit notable physiological similarities to R. eutropha (Cypionka and Meyer, 1982; King, 2003). Carboxydotrophic bacteria oxidize CO with H2O to CO2 and 2 H+ + 2 e− by a carbon monoxide dehydrogenase (CODH), thereby obtaining energy and carbon, that is usually fixed in the CBB cycle. The CODH‐associated proteins of the model organism for carboxydotrophy, O. carboxidovorans OM5, are encoded in a single 14.5‐kbp cox gene cluster, which is located on the organism's megaplasmid pHCG3 (Fig. S1; Fuhrmann et al., 2003). This cluster includes the structural genes for CODH, coxM, coxS and coxL, as well as genes that code for proteins mediating the post‐translational maturation of CODH (coxD, coxE, coxF, coxG; Schübel et al., 1995). The remaining cox Oc genes, which encode transmembrane complexes (coxB, coxK, coxI) and a putative signal transduction system (coxC, coxH), have yet to be further characterized.
Although a utilization of carbon monoxide as a source of carbon or energy has not been shown, R. eutropha H16 has several characteristics, which suggest that a possible exploitation of the energy‐rich substrate CO by this bacterium is not far off. Similarly to its two hydrogenases, examined cytochromes of R. eutropha H16 showed low affinity towards CO (Bernard et al., 1974). This has led to the assumption that the respiratory system of R. eutropha H16 is mostly CO‐insensitive, which was supported by the study of Cypionka and Meyer (1982), who observed a substantial inhibition of growth of R. eutropha only at CO concentrations that exceeded those of conventional syngas by far. Furthermore, the two chromosomes of R. eutropha H16 harbour putative genes with significant similarities to cox genes of O. carboxidovorans (Cramm, 2009). Recently, R. eutropha was genetically engineered to display a cell surface‐anchoring protein consisting of CODH subunits of different anaerobic bacteria that converted CO to CO2 (Hyeon et al., 2015). However, an intracellular utilization of CO by R. eutropha, resulting in growth, has not been shown to this date. In this study, cultivations of R. eutropha H16 with a standardized artificial syngas mixture or different, defined compositions of CO, CO2 and H2 were carried out and the effect of heterologously expressed cox genes of O. carboxidovorans OM5 on growth in CO‐containing gas mixtures by R. eutropha was examined.
Results and discussion
Cultivation of strains of R. eutropha in the presence of carbon monoxide
To assess and enhance growth of R. eutropha H16 in CO‐containing atmospheres, cells of the wild type and of recombinant strains were initially cultivated aerobically with 30% (by volume) of an artificial syngas mixture. This gas mixture consisted of (by volume) 40% CO, 40% H2, 10% CO2 and 10% N2, and resembled syngas compositions, which were previously used in academic studies and were actually obtained from gasification of biomass (Bridgewater, 1995; Heinrich et al., 2016; Revelles et al., 2016). To improve the utilization of syngas, of which CO is a major component, seven cox genes of O. carboxidovorans OM5 were heterologously expressed in R. eutropha H16. For this, coxM, ‐S, ‐L, ‐D, ‐E, ‐F and ‐G (locus tags: OCA5_RS17205‐260), which are the conserved cox genes among carboxydotrophs and are regarded as essential for carbon monoxide utilization (Santiago et al., 1999; King and Weber, 2007), were cloned to yield the construct pBBR1MCS‐3::coxMSLDEFG Oc. Furthermore, the CO2‐inducible promoter PL was cloned upstream of the cox Oc genes to exploit the gas exposure for enhanced expression. This promoter mediates gene transcription of the two homologous cbb operons of R. eutropha and binds the transcription activator CbbR (Jeffke et al., 1999). Syngas cultures of R. eutropha showed exponential growth after an initial lag phase of three to four days, which showed that all gluconate of the precultures had been successfully washed out from the medium prior to inoculation (Fig. 1). From day five onwards, the gas phase was renewed with every sample withdrawal to prevent a lack of carbon, or energy supply for these cultures. Recombinant R. eutropha strains, which harboured the coxMSLDEFG Oc cluster, showed substantially enhanced growth in particular during the first 10 days of cultivation (Table 1) when compared to the control strains, which carried the plasmids pBBR1MCS‐3 or pBBR1MCS‐3‐PL. The strain harbouring pBBR1MCS‐3‐PL::coxMSLDEFG Oc grew to the highest optical density (OD600nm: 5.48). The increased optical densities were not only a result of cell growth, as depicted by the measured cell dry weights (CDW), but also of the accumulated amounts of the carbon storage compound poly(3‐hydroxybutyrate) (poly[3HB]), which were increased by more than 20% concomitant with heterologous expression of the cox Oc genes (Table 1). The crystalline short chain length polymer poly(3HB) is known to considerably affect the transmission of light through the cell material (Schlegel et al., 1970). Due to the technical limitation of the applied proof‐of‐concept set‐up, the exact amounts of utilized CO and CO2 from applied syngas were not determined. However, by means of unpaired t‐tests, the impact of the heterologously expressed coxMSLDEFG Oc genes on growth and poly(3HB) synthesis of the respective R. eutropha strains (Table 1) was determined to be significant for all analysed parameters (P < 0.05), which suggested an increased utilization of carbon, CO and/or CO2, which was present in the artificial syngas mixture.
Figure 1.
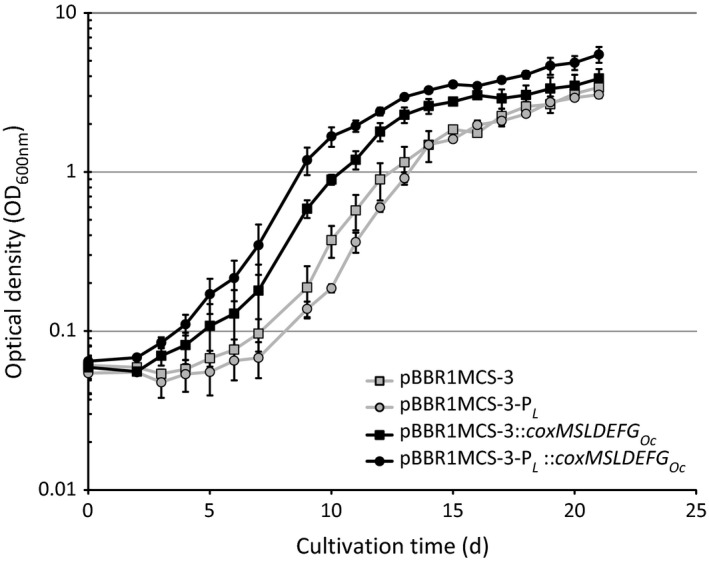
Cultivation of strains of Ralstonia eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐P L, pBBR1MCS‐3::coxMSLDEFG Oc, or pBBR1MCS‐3‐PL::coxMSLDEFG Oc, in 50 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and an agitation of 130 r.p.m. Cells were grown under oxic conditions in 1 L Duran flasks with an atmosphere of 30% of an artificial synthesis gas mixture (by volume 40% CO, 40% H2, 10% CO 2, 10% N2). From day five onwards, gas atmospheres were renewed with each sampling procedure. Standard deviations of optical densities (OD 600nm) are shown by error bars. Data were obtained from duplicate flasks of each strain over the course of two biological experiments. d, days.
Table 1.
Growth rates, cell dry weights and poly(3HB) accumulation of recombinant strains of Ralstonia eutropha H16, cultivated for 21 days. Cells were grown in 50 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and 130 r.p.m. under oxic conditions in 1 L Duran flasks with an atmosphere of 30% of an artificial synthesis gas mixture (by volume 40% CO, 40% H2, 10% CO2, 10% N2). From day five onwards, gas atmospheres were renewed with each sampling procedure. Data were obtained from duplicate flasks of each strain over the course of two biological experiments. d, days; CDW, cell dry weight
| R. eutropha pBBR1MCS‐3:: | Growth rate (d−1; d 1–10) | Cell density (g CDW/L) | Poly(3HB) content (%, wt/wt, of CDW) |
|---|---|---|---|
| (Only vector) | 0.18 ± 0.03 | 1.84 ± 0.18 | 40.8 ± 0.5 |
| PL | 0.12 ± 0.02 | 1.75 ± 0.13 | 39.0 ± 0.2 |
| coxMSLDEFG Oc | 0.27 ± 0.04 | 2.05 ± 0.23 | 46.4 ± 0.8 |
| PL::coxMSLDEFG Oc | 0.33 ± 0.03 | 2.62 ± 0.30 | 49.7 ± 0.4 |
To verify this assumed utilization of CO, respective strains were cultivated in a similar set‐up, in which cells were cultivated with syngas for 5 days, harvested and washed with medium and then transferred to flasks containing fresh media with an aerobic atmosphere containing (i) 10% CO with 10% H2, (ii) 10% CO, or (iii) 10% H2 (by volume). To gain sufficient amounts of cell matter for poly(3HB) analysis without extending the cultivation time of the previous growth experiment (Fig. 1), the cells were initially inoculated to a higher OD600nm. The difference in growth during the first 5 days of cultivation between R. eutropha pBBR1MCS‐3::coxMSLDEFG Oc and its corresponding control strain (empty vector) appeared to be less substantial than during the first days of the previous syngas cultivation (Fig. 1). Taking the higher concentrations of the inocula into account, it could be assumed that the positive effect of the vector pBBR1MCS‐3::coxMSLDEFG Oc on growth with syngas was less noticeable with increasing OD600nm (Fig. 1, d 10–15; Fig. 2, d 0‐5). At these higher cell densities, the comparably lower amount of CO substrate available for each individual cell, combined with the presumably low concentration of recombinant intracellular CODHOc resulting from basal gene expression through Plac, might have led to a relatively low rate of conversion of CO to CO2 for growth.
Figure 2.
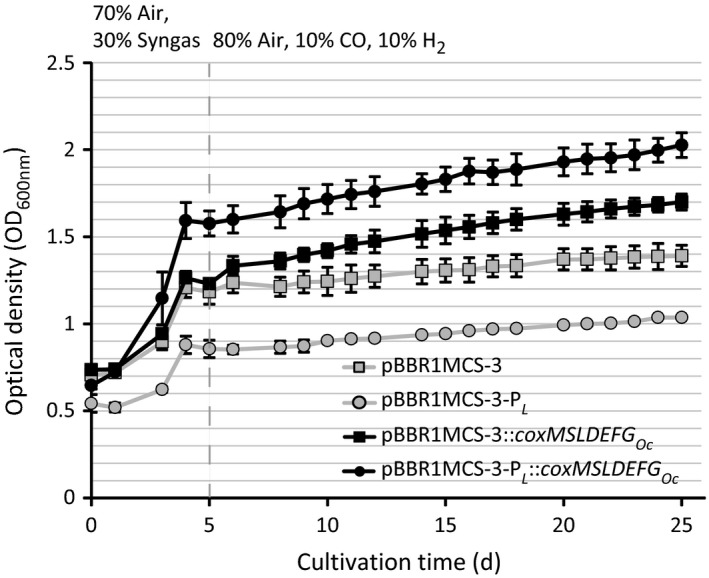
Cultivation of strains of Ralstonia eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐P L, pBBR1MCS‐3::coxMSLDEFG Oc, or pBBR1MCS‐3‐P L::coxMSLDEFG Oc, in 1 L Duran flasks filled with 50 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and an agitation of 130 r.p.m. Initially, cultures were grown under oxic conditions in an atmosphere of 30% of an artificial synthesis gas mixture (by volume 40% CO, 40% H2, 10% CO 2, 10% N2). After five days of cultivation (vertical dashed line), cells were washed and transferred to fresh media. Cultures were then set to an atmosphere of (by volume) 80% air, 10% CO and 10% H2. Atmospheres were renewed after every five days. Standard deviations of optical densities (OD 600nm) are represented by error bars. Data were obtained from triplicate flasks of each strain. d, days.
Upon the exchange of atmospheres, none of the four tested strains of R. eutropha was able to grow, when solely CO or H2 were applied (Fig. S2). In contrast, cells showed a slight, but significant increase in optical density, when 10% H2 was added to 10% of CO (Fig. 2). Again, R. eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc appeared to be the best‐performing strain and gained nearly 0.5 units of OD600nm within 20 days of cultivation with CO/H2.
These cultivations suggested that the strains of R. eutropha expressing the seven cox genes of O. carboxidovorans OM5 were able to utilize carbon monoxide as carbon source but were unable to channel the resulting electrons into the respiratory chain for the generation of energy, as provision of H2 was essential for an increase of the OD. Surprisingly, also the control strains harbouring the respective vectors without cox Oc genes showed a slight increase in OD600nm, however, to a much lower degree as compared to the recombinant strains. In these two‐phase cultivation experiments, the cells accumulated considerably less poly(3HB) when compared to the previous cultivations, in which syngas was applied over the entire course of the cultivation (Table 1). As syngas contained 10% of CO2, which in contrast to CO can be immediately assimilated through the CBB cycle, the amount of available carbon, crucial for poly(3HB) accumulation in R. eutropha, was likely higher when the strains were cultivated in syngas atmosphere (Anderson and Dawes, 1990). Furthermore, strains of R. eutropha, which harboured the cox Oc genes, accumulated only slightly more poly(3HB) (coxMSLDEFG Oc: 18.1% ± 1.1%; PL::coxMSLDEFG Oc: 18.9% ± 0.9% [wt/wt of CDW]) than the corresponding control strains (empty vector: 17.0% ± 0.3%; pBBR1MCS‐3‐PL: 16.7% ± 0.5% [wt/wt of CDW]). This implied that the increased optical density of these recombinant strains was not only due to light scattering of the accumulated polymer granules, but also to cell division.
Remarkably, no growth was detected when cells were directly exposed to the aerobic CO/H2‐containing atmosphere after inoculating the cells from the heterotrophically grown precultures. An explanation for this might be the display of cytochrome patterns by R. eutropha, which evidently vary according to different growth phases and conditions (Kömen et al., 1991). No experiments on the cytochrome CO sensitivity of heterotrophically grown cells of R. eutropha H16 have been carried out; however, Probst and Schlegel (1976) reported a moderate affinity of cytochromes towards CO in CO2/H2‐cultivated cells of this bacterium. Therefore, one could speculate that cells of R. eutropha that were grown in syngas prior to the switch to CO/H2 atmosphere built a cytochrome pattern, which was more adapted to CO exposure (Fig. 2).
In both cultivation experiments, the applied PL promoter had a positive impact on growth of the recombinant R. eutropha strain, which harboured the cox Oc genes. This was most likely due to an increased level of expression of the episomal heterologous genes that was caused by the induced PL promoter. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) protein patterns of separated soluble supernatants of disrupted syngas‐grown cells did not show notable differences in occurrence or abundancy of proteins between the recombinant cox Oc strains and the R. eutropha H16 wild type, harbouring the empty vector (Fig. S3). However, the protein pattern of the membrane‐containing pellet of R. eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc (Fig. 3) displayed a distinct spot that matched the size of the large CODH subunit CoxLOc (88.7 kDa; Dobbek et al., 1999). Applying matrix‐assisted laser desorption/ionization‐time‐of‐flight‐tandem‐mass spectrometry (MALDI‐TOF‐MS/MS) and subsequent data analysis with the Mascot search engine, this protein spot was clearly identified as CoxLOc (Protein score, 105.5; Confidence interval [C.I.%], 100%). Other Cox subunits or accessory proteins could not be detected; this may be due to (i) an overlay of proteins, (ii) weakspecific binding of the Coomassie dye or (iii) partial 3′‐terminal degradation of mRNA of a respective gene, downstream of coxL Oc. Moreover, the expression of cox Oc genes under control of the pBBR1MCS‐3 standard Plac promoter, which cannot be induced but exhibits basal activity in R. eutropha strains, presumably occurred at a low level. This led to protein concentrations that cannot be detected in SDS PAGE protein patterns.
Figure 3.
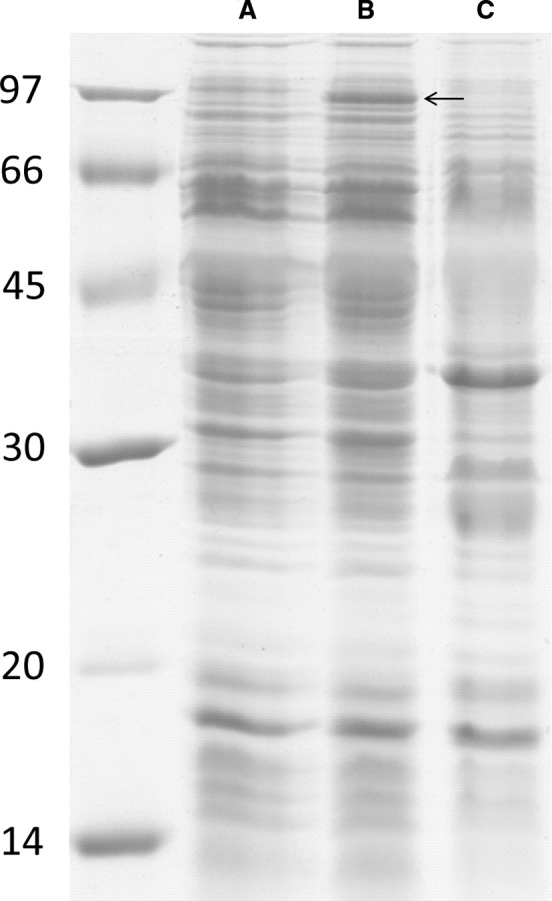
Protein patterns of the non‐soluble pellet of lysed cells of Ralstonia eutropha, harbouring pBBR1MCS‐3::coxMSLDEFG Oc (A), pBBR1MCS‐3‐P L::coxMSLDEFG Oc (B), or pBBR1MCS‐3 (empty vector; C). Cells were obtained after six days of cultivation in mineral salts medium (Schlegel et al., 1961) at 30 °C, an agitation of 130 r.p.m. and an atmosphere of (by volume) 70% air and 30% of an artificial syngas mixture that contained 40% CO, 40% H2, 10% CO 2, 10% N2. Proteins of 2 mg of cell pellet were separated in an SDS polyacrylamide gel and subsequently stained with Coomassie blue. Molecular masses of proteins (in kilodaltons) are displayed on the left margin. An arrow indicates a protein band, identified as CoxL Oc in lane ‘B’.
The detection of at least the CoxLOc subunit in disrupted cells of R. eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc together with the increased growth in CO‐containing atmospheres of this strain indicated that the applied PL is a suitable promoter for heterologous gene expression in syngas cultivations with R. eutropha. The CO2, which was responsible for induction of the PL promoter, was solved in the culture broth and additionally emerged from the assumed intracellular conversion of CO to CO2. In contrast, the presence of the PL promoter on the control plasmid pBBR1MCS‐3‐PL leads to decreased growth (Figs 1 and 2). As episomal copies of PL could capture the regulating CbbR proteins, which are required for transcription of the cbb genes of Ralstonia eutropha, CO2 fixation of the respective strain might have been hampered (Bowien and Kusian, 2002).
As an additional observation, the positive impact of the cox Oc genes on growth of the recombinant R. eutropha strains in CO‐containing atmospheres was more substantial in syngas atmosphere (Fig. 1) than in the two‐phase cultivation, where CO was the sole carbon source (Fig. 2). Possibly, the presence of CO2, which is a natural substrate of R. eutropha, leads to an increased fitness of the recombinant strains as compared to cultivations, where cells solely relied on carbon monoxide. As a mere hypothesis, the recombinant CODHOc could not only provide limited amounts of CO2 as carbon source for R. eutropha, but as a side‐effect could also dispose the medium of accumulating CO, which would be partially inhibiting growth at otherwise increasingly higher concentrations (Cypionka and Meyer, 1982). Despite their inability to gain sufficient amounts of energy from carbon monoxide, this possible detoxification effect could explain the enhanced productivity of the recombinant R. eutropha cox Oc strains in the conducted syngas cultivations, as compared to the wild type.
In vitro analysis of carbon monoxide oxidation by strains of R. eutropha H16
To demonstrate the ability of the recombinant R. eutropha strains to oxidize CO to CO2 in vitro, an altered protocol of the photometric assay of Meyer and Schlegel (1978), in which the oxidation of CO is coupled to the reduction of methylene blue, was applied. For this, duplicate flasks with cells of the different generated strains of R. eutropha were cultivated in 30% of syngas (by volume) to ODs600nm of 0.8–1.0. The wild type of O. carboxidovorans OM5, which was cultivated under oxic conditions with (by volume) 30% CO for 10 days to an OD600nm of 1.0, was used as a reference. Following the disruption of cells, the soluble supernatant and the non‐soluble pelleted fraction were analysed for CODH activity. Of the tested cox Oc ‐ and control strains of R. eutropha H16, cell extracts of the two strains, which expressed the coxMSLDEFG Oc cluster, showed an oxidation of carbon monoxide in the photometric enzyme assay. However, CODH activity of the applied recombinant R. eutropha cell extract was found exclusively in the pellet, containing cytoplasmic membranes of lysed cells. This was in accordance with the detection of CoxLOc, in protein patterns, prepared from the non‐soluble cell pellet of R. eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc (Fig. 3). Due to the constitutive episomal expression of coxG Oc, which codes for the CODH membrane‐anchoring protein, the recombinant CODH might be attached to the cytoplasmic membrane of R. eutropha during all stages of growth. In contrast, the native CO dehydrogenases of O. carboxidovorans OM5 are attached to the cytoplasmic membrane and diffused in the cytoplasm at different ratios, depending on the respective growth phase (Rohde et al., 1984). Consequently, the soluble cell fraction of O. carboxidovorans OM5 catalysed the oxidation of 28.5 nanomoles CO·min−1·mg protein−1, whereas the pellet of lysed cells oxidized CO at a rate of 4.8 nanomoles·min−1·mg dried cell pellet−1. The activity of CO oxidation of the R. eutropha cox Oc strains appeared to be clearly lower than for the reference strain O. carboxidovorans OM5, as pellets of lysed R. eutropha cells harbouring the plasmids pBBR1MCS‐3::coxMSLDEFG Oc and pBBR1MCS‐3‐PL::coxMSLDEFG Oc oxidized CO at rates of 0.7 and 0.9 nanomoles·min−1·mg dried cell pellet‐1. The recorded increase in the rate of CO oxidation resulting from application of the PL promoter was approximately 23%, although the cellular abundance of at least CoxLOc, which appeared to be much higher for this strain (Fig. 3), would have suggested an even more increased activity of respective cell extract. A potential approach to further increase in CO oxidation by the recombinant strains could be the further fine tuning of their cox Oc gene expression, aiming at the equal abundance of all CODHOc subunits, which could result in an increased amount of functional cellular CODH enzymes.
Cultivation of strains of R. eutropha H16 with 13CO
To demonstrate that the R. eutropha cox Oc strains were not only able to oxidize carbon monoxide but also to assimilate the CO‐derived carbon, the strains were cultivated with 13C‐labelled carbon monoxide as the sole C source. For this purpose, a shortened two‐stage cultivation was carried out, which comprised 5 days of growth with the artificial syngas mixture followed by 10 days of exposure to (by volume) 10% 13CO + 10% H2 (Fig. S4). Along with the cell numbers of withdrawn samples, conversion of 13CO into poly(3HB) was determined by gas chromatography–mass spectrometry (GC‐MS), as poly(3HB) derives from the central metabolite acetyl coenzyme A (acetyl‐CoA). Therefore, 13C enrichment in the characteristic m/z 103 fragment of methanolyzed poly(3HB) was determined (Table 2). The recombinant cox Oc strains of R. eutropha H16 again showed only a slight increase in optical density, but incorporated substantial amounts of 13C‐labelled carbon, which derived from the applied 13CO, into the backbone of the accumulated poly(3HB) after 10 days of cultivation with 13CO (Table 2). Taking the natural abundance of carbon, oxygen and hydrogen isotopes into account, most of these labelled monomers appeared to contain three 13C atoms (m/z: 106), whereas only a small amount of monomers containing one 13C atom (m/z: 104) and no monomers containing two 13C (m/z 105) atoms were detected (Fig. S5). As cultures were supplied with exclusively 12C containing syngas (d 0–5) or 13CO (d 5–15), the occurrence of polymer constituents containing both 12C and 13C atoms could have resulted from 12C which was mobilized from the central metabolism, biomass or poly(3HB) into poly(3HB) precursors during growth with 13CO. The amount of incorporated 13C was higher than one could expect from the small increase in accumulated poly(3HB) during cultivation with 13CO/H2 (Table 2). This may be due to the simultaneous synthesis and degradation of poly(3‐hydroxyalkanoate) by R. eutropha H16, first reported by Doi et al. (1990). The 3HB constituents originating from non‐labelled carbon compounds of syngas were thereby steadily replaced by constituents derived from 13CO. Furthermore, the cell number of the strains, harbouring cox Oc genes slightly increased over the course of the exposure to 13CO and H2 (Table 2), which suggested that carbon from CO was utilized for cell growth. A minor increase in cell number was also recorded for the R. eutropha control strains harbouring the empty pBBR1MCS‐3(‐PL), which, however, did not meet statistical significance (P > 0.05). The pH of the cultures after 15 days of cultivation with 13CO/H2 was in the range of 6.6 (± 0.2), whereas cultures displayed an approximate pH of 6.7 (± 0.1) after the five‐day syngas cultivation phase, which implied that this parameter did not affect growth with CO and H2. These results, combined with the foregoing growth experiments and the photometric enzyme assay, provided unequivocal evidence that the recombinant R. eutropha strains generated in this study were able to oxidize and utilize CO as the sole carbon source by CODHOc‐mediated conversion to CO2.
Table 2.
Changes in optical density, cell number and poly(3HB) accumulation as well as incorporation of 13C into poly(3HB) of generated strains of Ralstonia eutropha H16 after ten days of cultivation with 80% air, 10% 13CO and 10% H2 (by volume). Cells were initially grown in 100 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and 130 r.p.m. under oxic conditions in 1 L Duran flasks with an atmosphere of 30% of an artificial synthesis gas mixture (by volume 40% CO, 40% H2, 10% CO2, 10% N2). After five days of cultivation, 50 ml of culture broth was harvested, and the remaining 50 ml was washed and transferred to fresh media. Cultures were then set to an atmosphere of 80% air, 10% 13CO and 10% H2 (by volume), which was renewed after five further days of cultivation, before the remaining cultures were harvested. Data were obtained from duplicate flasks. M0 to M3 display the m + 0 to m + 3 enrichments of the 3HB‐methyl ester′s m/z 103 fragment with 13C, detected by GC‐MS as described by Tan et al. (2016). The fractions of 13C‐enriched 3HB constituents were determined after correcting for natural isotope abundances
| R. eutropha pBBR1MCS‐3:: | Δ OD600nm | Δ Cell number (108 ml−1) | Δ poly(3HB) (wt/wt of CDW) | Incorporation of 13C (M0/M1/M2/M3) |
|---|---|---|---|---|
| (Only vector) |
+0.08 ±0.02 |
+0.21 ±0.11 |
+0.2% ±0.1% |
99.8/0.2/–/– ±0.2/0.2/–/– |
| PL |
+0.06 ±0.00 |
+0.09 ±0.05 |
−0.1% ±0.1% |
100/–/–/– ±0.0/–/–/– |
| coxMSLDEFG Oc |
+0.22 ±0.00 |
+0.95 ±0.13 |
+1.6% ±0.4% |
92.6/0.8/–/6.6 ±0.6/0.2/–/0.4 |
| PL::coxMSLDEFG Oc |
+0.23 ±0.04 |
+1.04 ±0.19 |
+2.0% ±0.5% |
88.6/2.2/–/9.2 ±0.8/1.0/–/0.2 |
To this point, the cause for the inability to sufficiently exploit the reducing power of CO for cell growth remains unclear but cannot be experimentally solved within this study. A critical point when discussing the transfer of electrons from the CODHOc to the respiratory chain is the identification of the responsible electron acceptor. Although cytochrome b 561 was widely considered to accept the reducing equivalents from the CODH of O. carboxidovorans (Cypionka and Meyer, 1983), recent experiments suggested that the common redox carrier ubiquinone (coenzyme Q10) initially interacts with the CODH flavin site (Wilcoxen et al., 2011). Furthermore, sequence analysis revealed that R. eutropha H16 possesses a cytochrome b large subunit (HoxZ) with a high identity to cytochrome b 561 of O. carboxidovorans OM5 (BLASTP expect value, 4e−83; identities, 147/242 [60%]; positives, 185/242 [76%]; gaps, 3/242 [1%]; Altschul et al., 1997). Therefore, R. eutropha could potentially transfer reducing equivalents from CO to its respiratory chain either way. Possibly, an imperfect insertion of the recombinant CODH into the cytoplasmic membrane sterically hindered the transmission of reducing equivalents to their acceptor in the R. eutropha cox Oc strains. In this case, the fate of the electrons of the reduced CODH would be unclear at this point. A simpler explanation would be that the amount of CO, which is oxidized by the recombinant CODH, is insufficient to feed the energy demand of the cell, for instance, CO2 fixation through the CBB cycle. This was implied by the considerably lower CO oxidation rates of the recombinant R. eutropha strains as compared to the reference O. carboxidovorans OM5. Several bacteria, e.g. of the genus Mesorhizobium, can oxidize CO but are unable to utilize the resulting CO2 for cell growth, due to the absence of CO2‐fixing mechanisms and are thus referred to as ‘carboxydovores’ (King, 2003). In contrast to these lithoheterotrophic ‘carboxydovores’, the recombinant strains of R. eutropha cultivated in this study metabolized CO as source of carbon, but both were dependent on molecular hydrogen for cell growth. Consequently, the CO‐related lifestyle of the engineered strains of R. eutropha could be referred to as ‘carboxyhydrogenotrophic’.
No in vitro CO oxidation was detected for the R. eutropha control strains harbouring the empty pBBR1MCS‐3 or pBBR1MCS‐3‐PL, which consequently were therefore either unable to oxidize CO under the applied conditions, or their reactions were below the detection level respectively. Previous studies by King and Weber (2007), as well as Cramm (2009) reported the presence of multiple coxL homologues in the genome of R. eutropha H16. Of these, one putative coxL gene (locus tag: H16_RS02155) is located in a coxSLMDEGI cluster, which was shown to be constitutively transcribed, at least under heterotrophic growth conditions, in foregoing studies by Peplinski et al. (2010). This cluster was annotated to putatively encode a xanthine oxidase, an enzyme that has diverse substrate specificities and is phylogenetically close to CODHs of, e.g. O. carboxidovorans (King, 2003; Hille, 2005). Still, a possible activity of this putatively formed enzyme towards carbon monoxide remains to be investigated in further studies.
Due to its metabolic versatility, R. eutropha H16 has been exploited to synthesize biobased products from various heterotrophic and autotrophic carbon sources (Volodina et al., 2016). Yet CO has been widely ignored as substrate for this bacterium, despite the importance of utilizing all carbon compounds of waste gases for a desirable circular economy. In this study, the utilization of CO as part of the economically and ecologically sustainable feedstock syngas by R. eutropha H16 through heterologous expression of the coxMSLDEFG genes of O. carboxidovorans OM5 has been demonstrated both in vivo and in vitro. A conceivable benefit of this CODHOc‐mediated oxidation of CO over the recently developed cell surface‐attached CODH (Hyeon et al., 2015) could be the intracellularly nascent CO2, that is readily available to be incorporated into the CBB cycle by the ribulose 1,5 bisphosphate carboxylase. In contrast, extracellular CO2 enters the cell as HCO3 − and thus initially must be converted by the R. eutropha carbonic anhydrase Cag (Gai et al., 2014). By oxidation of toxic CO to CO2 and the resulting supply of additional accessible carbon, potential syngas processes involving R. eutropha H16, could exhibit enhanced productivity and efficiency, as shown for the commercially applicable biopolymer poly(3HB) in this study.
Experimental procedures
Microorganisms, plasmids and oligonucleotides
Bacterial strains, plasmids and oligonucleotides, which were used in this study, are listed in Table S1. Escherichia coli TOP10 was used for cloning procedures that included the propagation and isolation of generated plasmids. E. coli C41 was used as a negative reference strain in the photometric CODH assay.
Cultivation of bacteria
Escherichia coli strains were grown in 5 ml of lysogeny broth (Sambrook et al., 1989) in test tubes at 37 °C and an agitation speed of 130 r.p.m. O. carboxidovorans OM5 and strains of R. eutropha H16 were grown at 30 °C in mineral salts medium (Schlegel et al., 1961), which contained (per litre) 4.5 g Na2HPO4·2 H2O, 1.5 g KH2PO4, 1.0 g NH4Cl, 0.2 g MgSO4·7 H2O, 0.02 g CaCl2, 1.2 mg NH4Fe(III) citrate and 10 μl of 10 000‐fold SL6 (Pfennig, 1974). For growth on solid media and for precultures, 1% (wt/vol) of sodium gluconate for R. eutropha or 0.3% (wt/vol) of sodium acetate for O. carboxidovorans OM5 was added to the medium. For precultures, cells were grown in 20 ml of medium in 100 ml Erlenmeyer flasks and an agitation of 130 r.p.m. for 24–32 h. Cells were washed with carbon‐free mineral salts medium by centrifugation for 15 min at 4000 g before inoculating the main culture with 1 ml of a concentrated cell suspension. Cultivations of cells of R. eutropha with different gas mixtures were carried out at a volume of 50 ml in baffled 1 L Duran flasks, which were sealed with butyl rubber plugs. Upon evacuation of the respective defined amount of air, flasks were filled with an artificial syngas mixture (by volume, 40% CO, 40% H2, 10% CO2 and 10% N2; Air Liquide, Bottrop, D) to a final concentration of 30%. Similarly, atmospheres of (by volume) 10% (13)CO, 10% H2 and 10% CO2 (Air Liquide, Bottrop D/Eurisotop, Saarbrücken, D) or different combinations of these gas concentrations were established. Autotrophic cultivations of O. carboxidovorans OM5 were carried out in an atmosphere of (by volume) 70% air and 30% CO. Optical densities of samples were determined at 600 nm (OD600nm). The cell count (·ml−1) was determined from samples, diluted between 1:10 and 1:50, using a Thoma counting chamber. Cell harvest was carried out by centrifugation (15 min, 4000 g, 4 °C). Cultures of recombinant strains contained tetracycline at concentrations of 25 μg ml−1 for R. eutropha and 12.5 μg ml−1 for E. coli. During gas cultivations, tetracycline was added repeatedly after 7 days of cultivation with recombinant strains of R. eutropha to maintain plasmid stability.
Construction of vectors and generation of recombinant strains of E. coli and R. eutropha
Nucleic acids were processed according to Sambrook et al. (1989). DNA fragments were amplified from genomic DNA of O. carboxidovorans OM5 and R. eutropha H16 by applying the Phusion High‐Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA) with oligonucleotides, listed in Table S1. FastDigest restriction enzymes (Thermo Scientific, Waltham, MA, USA) were used to digest DNA fragments, before ligation into a likewise digested target plasmid applying T4 DNA ligase (Thermo Scientific, Waltham, MA, USA). To propagate, isolate and validate generated hybrid plasmids, chemically competent cells of E. coli TOP10 were transformed, applying the method of Hanahan (1983). Re‐isolated plasmids were then transferred into R. eutropha H16 by electroporation (Aneja et al., 2009). The vector pBBR1MCS‐3::coxMSLDEFG Oc was generated by digesting the amplified coxMSLDEFG Oc DNA fragment with SpeI and SacI, and subsequent ligation into a likewise digested pBBR1MCS‐3 vector. SpeI‐digested PL fragments were then inserted into the corresponding site to yield the vector pBBR1MCS‐3‐PL::coxMSLDEFG Oc.
Preparation of cell extract and analysis of protein patterns
Harvested cells were washed and resuspended in 50‐mm KH2PO4‐KOH buffer (pH 7.0) with added protease inhibitor (cOmplete ULTRA Tablets, Roche, Basel, CH), and cell extracts of R. eutropha were obtained by sonication and subsequent centrifugation (10 min, 13 000 g, 4 °C). Protein concentrations were determined employing the method of Bradford (1976), and 40 μg of protein were heated in denaturing buffer for 10 min at 95 °C. Upon separation by SDS PAGE (Laemmli, 1970), proteins were stained with Coomassie brilliant blue R‐250. Proteins were identified by MALDI‐TOF‐MS/MS. For this, respective protein spots were excised from SDS gels and transferred to 1.5 ml reaction tubes containing 50 μl 10% (vol/vol) acetic acid. Protein samples were subjected to MALDI‐TOF‐MS/MS analysis as described by Wolf et al. (2008). Applying a 5800 Proteomics Analyzer (AB Sciex, Framingham, MA, USA), the spectra were recorded in a reflector mode in a mass range from 900 to 3700 Da with a focus mass of 200 Da. Acquired MS data were compared to the proteome database of O. carboxidovorans OM5 and R. eutropha H16 using the Mascot engine (version 2.1.0.4).
Determination of CODH activity in cell extracts
CODH activity in cell lysates was determined with a method modified according to Meyer and Schlegel (1978). In the present protocol, the oxidation of glucose by a glucose oxidase/catalase mix to dispose residual oxygen was omitted, as it affected the reduction of methylene blue from CO in the applied experimental set‐up. The reaction was started by adding 20 μl of soluble cell extract (2–8 mg ml−1 of protein) or 1–3 mg of resuspended wet cell pellet to 1.8 ml of the CO–saturated 50 mm methylene blue solution.
Analysis of poly(3HB) content of cells
Upon cell harvest, cells were lyophilized, and 5–10 mg of each sample was subjected to acidic methanolysis as described by Brandl et al. (1988). Synthesized 3HB‐methyl esters were then quantified by gas chromatography (Timm et al., 1990), using an Agilent 6850 gas chromatograph (GC), which was equipped with a BP21 polyethylene glycol capillary column (50 m by 0.22 mm; 250 nm film thickness) and a flame ionization detector (Agilent Technologies, Waldbronn, D). To determine the content of 13C, obtained 3HB‐methylesters were analysed by GC‐MS according to Tan et al. (2016). Applying an Agilent 6890 GC, which was connected to an Agilent HP 5973 mass spectrometer (MS), the samples were separated on a BPX35 polyethylene glycol capillary column (60 m by 0.22 mm; 250 nm film thickness; SGE Deutschland GmbH, Darmstadt, D). For this, the temperature programme described by Andreeßen et al. (2010) was applied.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Table S1. Microorganisms, target vectors and oligonucleotides, which were applied in this study.
Fig. S1. Cluster of coxgenes in Oligotropha carboxidovorans OM5.
Fig. S2. Cultivation of strains of R. eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐PL, pBBR1MCS‐3::coxMSLDEFG Oc or pBBR1MCS‐3‐PL::coxMSLDEFG Oc in 1L Duran flasks filled with 50 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and 130 r.p.m.
Fig. S3. Protein patterns of the soluble supernatant of lysed cells of R. eutropha, harbouring pBBR1MCS‐3::coxMSLDEFG Oc (A), pBBR1MCS‐3‐PL::coxMSLDEFG Oc (B); or pBBR1MCS‐3 (empty vector; C).
Fig. S4. Cultivation of strains of R. eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐PL, pBBR1MCS‐3::coxMSLDEFG Oc, or pBBR1MCS‐3‐PL::coxMSLDEFG Oc, in 1L Duran flasks at 30 °C and an agitation of 130 r.p.m.
Fig. S5. Mass spectra of 3‐hydroxybutyrate methyl esters, extracted from cells of Ralstonia eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc.
Acknowledgements
Partial funding of this study by the European Union′s Seventh Framework Program for research, technological development and demonstration under grant agreement no:311815 is gratefully acknowledged. We also thank Dr. Dirk Albrecht, Dr. Daniela Zühlke and Prof. Dr. Katharina Riedel at the Institute of Microbiology of the Ernst‐Moritz‐Arndt University Greifswald for MALDI‐TOF‐MS/MS analysis of the CoxL protein.
Microbial Biotechnology (2018) 11(4), 647–656
Funding Information
Seventh Framework Programme (311815).
References
- Altschul, S.F. , Madden, T.L. , Schaffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. , and Lipman, D.J. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, A.J. , Dawes, E.A . (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54: 450‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeßen, B. , Lange, A.B. , Robenek, H. , and Steinbüchel, A. (2010) Conversion of glycerol to poly(3‐hydroxypropionate) in recombinant Escherichia coli . Appl Environ Microbiol 76: 622–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneja, K.K. , Ashby, R.D. , and Solaiman, D.K.Y. (2009) Altered composition of Ralstonia eutropha poly(hydroxyalkanoate) through expression of PHA synthase from Allochromatium vinosum ATCC 35206. Biotechnol Lett 31: 1601–1612. [DOI] [PubMed] [Google Scholar]
- Bernard, U. , Probst, I. , and Schlegel, H.G. (1974) The cytochromes of some hydrogen bacteria. Arch Microbiol 95: 29–37. [DOI] [PubMed] [Google Scholar]
- Bowien, B. , and Kusian, B. (2002) Genetics and control of CO2 assimilation in the chemoautotroph Ralstonia eutropha . Arch Microbiol 178: 85–93. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976) A rapid and sensitive method for the quantification of microgram‐quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brandl, H. , Gross, R.A. , Lenz, R.W. , and Fuller, R.C. (1988) Pseudomonas oleovorans as a source of poly(β‐hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol 54: 1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgewater, A.V. (1995) The technical and economic feasibility of biomass gasification for power generation. Fuel 74: 631–653. [Google Scholar]
- Buhrke, T. , Lenz, O. , Krauss, N. , and Friedrich, B. (2005) Oxygen tolerance of the H2‐sensing [NiFe] hydrogenase from Ralstonia eutropha H16 is based on limited access of oxygen to the active site. J Biol Chem 280: 23791–23796. [DOI] [PubMed] [Google Scholar]
- Burgdorf, T. , Lenz, O. , Buhrke, T. , van der Linden, E. , Jones, A.K. , Albracht, S.P. , and Friedrich, B. (2005) [NiFe]‐hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen‐tolerant biological hydrogen oxidation. J Mol Microbiol Biotechnol 10: 181–196. [DOI] [PubMed] [Google Scholar]
- Bürstel, I. , Siebert, E. , Frielingsdorf, S. , Zebger, I. , Friedrich, B. , and Lenz, O. (2016) CO synthesized from the central one‐carbon pool as source for the iron carbonyl in O2‐tolerant [NiFe]‐hydrogenase. Proc Natl Acad Sci USA 113: 14722–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, A.M. , and Schlegel, H.G. (1978) Metabolite concentrations in Alcaligenes eutrophus H16 and a mutant defective in poly‐β‐hydroxybutyrate synthesis. Arch Microbiol 119: 231–235. [Google Scholar]
- Cramm, R. (2009) Genomic view of energy metabolism in Ralstonia eutropha H16. J Mol Microbiol Biotechnol 16: 38–52. [DOI] [PubMed] [Google Scholar]
- Cypionka, H. , and Meyer, O. (1982) Influence of carbon monoxide on growth and respiration of carboxydobacteria and other aerobic organisms. FEMS Microbiol Lett 15: 209–214. [Google Scholar]
- Cypionka, H. , and Meyer, O. (1983) Carbon monoxide‐insensitive respiratory chain of Pseudomonas carboxydovorans . J Bacteriol 156: 1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbek, H. , Gremer, L. , Meyer, O. , and Huber, R. (1999) Crystal structure and mechanism of CO dehydrogenase, a molybdo iron‐sulfur flavoprotein containing S‐selanylcysteine. Proc Natl Acad Sci USA 96: 8884–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, Y. , Segawa, A. , Kawaguchi, Y. , and Kunioka, M. (1990) Cyclic nature of poly(3‐hydroxyalkanoate) metabolism in Alcaligenes eutrophus . FEMS Microbiol Lett 67: 165–170. [DOI] [PubMed] [Google Scholar]
- Drzyzga, O. , Revelles, O. , Durante‐Rodríguez, G. , Díaz, E. , García, J.L. , and Prieto, A. (2015) New challenges for syngas fermentation: towards production of biopolymers. J Chem Technol Biotechnol 90: 1735–1751. [Google Scholar]
- Friedrich, B. , and Schwartz, E. (1993) Molecular biology of hydrogen utilization in aerobic chemolithotrophs. Annu Rev Microbiol 47: 351–383. [DOI] [PubMed] [Google Scholar]
- Fuhrmann, S. , Ferner, M. , Jeffke, T. , Henne, A. , Gottschalk, G. , and Meyer, O. (2003) Complete nucleotide sequence of the circular megaplasmid pHCG3 of Oligotropha carboxidovorans: function in the chemolithoautotrophic utilization of CO, H2 and CO2 . Gene 322: 67–75. [DOI] [PubMed] [Google Scholar]
- Gai, C.S. , Lu, J. , Brigham, C.J. , Bernardi, A.C. , and Sinskey, A.J. (2014) Insights into bacterial CO2 metabolism revealed by the characterization of four carbonic anhydrases in Ralstonia eutropha H16. AMB Express 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580. [DOI] [PubMed] [Google Scholar]
- Heinrich, D. , Raberg, M. , Fricke, P. , Kenny, S.T. , Morales‐Gamez, L. , Babu, R.P. , et al (2016) Synthesis gas (Syngas)‐derived medium‐chain‐length polyhydroxyalkanoate synthesis in engineered Rhodospirillum rubrum . Appl Environ Microbiol 82: 6132–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, R. (2005) Molybdenum‐containing hydroxylases. Arch Biochem Biophys 433: 107–116. [DOI] [PubMed] [Google Scholar]
- Hyeon, J.E. , Kim, S.W. , Park, C. , and Han, S.O. (2015) Efficient biological conversion of carbon monoxide (CO) to carbon dioxide (CO2) and for utilization in bioplastic production by Ralstonia eutropha through the display of an enzyme complex on the cell surface. Chem Commun 51: 10202–10205. [DOI] [PubMed] [Google Scholar]
- Jeffke, T. , Gropp, N.H. , Kaiser, C. , Grzeszik, C. , Kusian, B. , and Bowien, B. (1999) Mutational analysis of the cbb operon (CO2 assimilation) promoter of Ralstonia eutropha . J Bacteriol 181: 4374–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärst, U. , and Friedrich, C.G. (1984) Mixotrophic capabilities of Alcaligenes eutrophus . J Gen Microbiol 130: 1987–1994. [Google Scholar]
- King, G.M. (2003) Molecular and culture‐based analyses of aerobic carbon monoxide oxidizer diversity. Appl Environ Microbiol 69: 7257–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, G.M. , and Weber, C.F. (2007) Distribution, diversity and ecology of aerobic CO‐oxidizing bacteria. Nat Rev Microbiol 5: 107–118. [DOI] [PubMed] [Google Scholar]
- Kömen, R. , Zannoni, D. , Ingledew, W.J. , and Schmidt, K. (1991) The electron transport system of Alcaligenes eutrophus . Arch Microbiol 155: 382–390. [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. II , and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- Lu, Y. , and Yu, J. (2017) Gas mass transfer with microbial CO2 fixation and poly(3‐hydroxybutyrate) synthesis in a packed bed bioreactor. Biochem Eng J 122: 13–21. [Google Scholar]
- Meyer, O. , and Schlegel, H.G. (1978) Reisolation of the carbon monoxide utilizing hydrogen bacterium Pseudomonas carboxidovorans (Kistner) comb. nov. Arch Microbiol 137: 118–143. [DOI] [PubMed] [Google Scholar]
- Meyer, O. , and Schlegel, H.G. (1979) Oxidation of carbon monoxide in cell extracts of Pseudomonas carboxydovorans . J Bacteriol 137: 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J. , MacEachran, D. , Burd, H. , Sathitsuksanoh, N. , Bi, C. , Yeh, Y.C. , et al (2013) Engineering of Ralstonia eutropha H16 for autotrophic and heterotrophic production of methyl ketones. Appl Environ Microbiol 79: 4433–4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplinski, K. , Ehrenreich, A. , Döring, C. , Bömeke, M. , Reinecke, F. , Hutmacher, C. , and Steinbüchel, A. (2010) Genome‐wide transcriptome analyses of the ‘Knallgas’ bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology (SGM) 156: 2136–2152. [DOI] [PubMed] [Google Scholar]
- Pfennig, N. (1974) Rhodopseudomonas globiformis, sp. n., a new species of the Rhodospirillaceae . Arch Microbiol 100: 197–206. [Google Scholar]
- Probst, I. , and Schlegel, H.G. (1976) Respiratory components and oxidase activities in Alcaligenes eutrophus . Biochim Biohys Acta 440: 412–428. [DOI] [PubMed] [Google Scholar]
- Revelles, O. , Beneroso, D. , Menéndez, J.A. , Arenillas, A. , García, J.L. , and Prieto, M.A. (2016) Syngas obtained by microwave pyrolysis of household wastes as feedstock for polyhydroxyalkanoate production in Rhodospirillum rubrum . Microb Biotechnol 10: 1406–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, M. , Mayer, F. , and Meyer, O. (1984) Immunocytochemical localization of carbon monoxide oxidase in Pseudomonas carboxydovorans. The enzyme is attached to the inner aspect of the cytoplasmic membrane. J Biol Chem 259: 14788–14792. [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Santiago, B. , Schübel, U. , Egelseer, C. , and Meyer, O. (1999) Sequence analysis, characterization and CO‐specific transcription of the cox gene cluster on the megaplasmid pHCG3 of Oligotropha carboxidovorans . Gene 236: 115–124. [DOI] [PubMed] [Google Scholar]
- Schlegel, H.G. , Kaltwasser, H. , and Gottschalk, G. (1961) Ein Submersverfahren zur Kultur Wasserstoff oxydierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Microbiol 38: 209–222. [Google Scholar]
- Schlegel, H.G. , Lafferty, R. , and Krauss, I. (1970) The isolation of mutants not accumulating poly‐β‐hydroxybutyric acid. Arch Microbiol 71: 283–294. [DOI] [PubMed] [Google Scholar]
- Schübel, U. , Kraut, M. , Mörsdorf, G. , and Meyer, O. (1995) Molecular characterization of the gene cluster coxMSL encoding the molybdenum‐containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans . J Bacteriol 177: 2197–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, R. , Dempo, Y. , Nakayama, Y. , Nakamura, S. , Bamba, T. , Fukusaki, E. , and Fukui, T. (2015) New insight into the role of the Calvin cycle: reutilization of CO2 emitted through sugar degradation. Sci Rep 5: 11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, G.Y.A. , Ge, L. , Pan, C. , Tan, S.N. , and Wang, J.Y. (2016) Current and emerging advanced technologies for biopolyesters characterization In Recent Advances in Biotechnology, vol 2, Koller M. (ed). Sharjah, UAE: Bentham Science Publishers, pp. 303–402. [Google Scholar]
- Tanaka, K. , and Ishizaki, A. (1994) Production of poly‐D‐3‐hydroxybutyric acid from carbon dioxide by a two‐stage culture method employing Alcaligenes eutrophus ATCC 17697T . J Ferment Bioeng 77: 425–527. [Google Scholar]
- Timm, A. , Byrom, D. , and Steinbüchel, A. (1990) Formation of blends of various poly(3‐hydroxyalkanoic acids) by a recombinant strain of Pseudomonas oleovorans . Appl Microbiol Biotechnol 33: 296–301. [Google Scholar]
- Volodina, E. , Raberg, M. , and Steinbüchel, A. (2016) Engineering the heterotrophic carbon sources utilization range of Ralstonia eutropha H16 for applications in biotechnology. Crit Rev Biotechnol 36: 978–991. [DOI] [PubMed] [Google Scholar]
- Wilcoxen, J. , Zhang, B. , and Hille, R. (2011) Reaction of the molybdenum‐ and copper‐containing carbon monoxide dehydrogenase from Oligotropha carboxydovorans with quinones. Biochemistry 50: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, C. , Hochgräfe, F. , Kusch, H. , Albrecht, D. , Hecker, M. , and Engelmann, S. (2008) Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics 8: 3139–3153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Microorganisms, target vectors and oligonucleotides, which were applied in this study.
Fig. S1. Cluster of coxgenes in Oligotropha carboxidovorans OM5.
Fig. S2. Cultivation of strains of R. eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐PL, pBBR1MCS‐3::coxMSLDEFG Oc or pBBR1MCS‐3‐PL::coxMSLDEFG Oc in 1L Duran flasks filled with 50 ml mineral salts medium (Schlegel et al., 1961) at 30 °C and 130 r.p.m.
Fig. S3. Protein patterns of the soluble supernatant of lysed cells of R. eutropha, harbouring pBBR1MCS‐3::coxMSLDEFG Oc (A), pBBR1MCS‐3‐PL::coxMSLDEFG Oc (B); or pBBR1MCS‐3 (empty vector; C).
Fig. S4. Cultivation of strains of R. eutropha H16, harbouring pBBR1MCS‐3 (empty vector), pBBR1MCS‐3‐PL, pBBR1MCS‐3::coxMSLDEFG Oc, or pBBR1MCS‐3‐PL::coxMSLDEFG Oc, in 1L Duran flasks at 30 °C and an agitation of 130 r.p.m.
Fig. S5. Mass spectra of 3‐hydroxybutyrate methyl esters, extracted from cells of Ralstonia eutropha pBBR1MCS‐3‐PL::coxMSLDEFG Oc.


