Summary
To enrich syntrophic acetate‐oxidizing bacteria (SAOB), duplicate chemostats were inoculated with sludge from syntrophic acetate oxidation (SAO)‐dominated systems and continuously supplied with acetate (0.4 or 7.5 g l−1) at high‐ammonia levels. The chemostats were operated under mesophilic (37°C) or thermophilic (52°C) temperature for about six hydraulic retention times (HRT 28 days) and were sampled over time. Irrespective of temperature, a methane content of 64–69% and effluent acetate level of 0.4–1.0 g l−1 were recorded in chemostats fed high acetate. Low methane production in the low‐acetate chemostats indicated that the substrate supply was below the threshold for methanization of acetate via SAO. Novel representatives within the family Clostridiales and genus Syntrophaceticus (class Clostridia) were identified to represent putative SAOB candidates in mesophilic and thermophilic conditions respectively. Known SAOB persisted at low relative abundance in all chemostats. The hydrogenotrophic methanogens Methanoculleus bourgensis (mesophilic) and Methanothermobacter thermautotrophicus (thermophilic) dominated archaeal communities in the high‐acetate chemostats. In line with the restricted methane production in the low‐acetate chemostats, methanogens persisted at considerably lower abundance in these chemostats. These findings strongly indicate involvement in SAO and tolerance to high ammonia levels of the species identified here, and have implications for understanding community function in stressed anaerobic processes.
Introduction
Syntrophic acetate‐oxidizing bacteria (SAOB) drive anaerobic conversion of acetate to methane in high‐ammonia biogas processes and thus play a major role in many commercial‐scale production systems (Karakashev et al., 2006; Sun et al., 2014; Frank et al., 2016; Mosbaek et al., 2016). SAOB work in close association with hydrogenotrophic methanogens, performing a two‐step reaction in which SAOB convert acetate to H2/format and CO2, which are then used by the methanogens for production of CH4 and CO2. Under conditions such as high ammonia, the syntrophic acetate oxidizers outnumber their competitors for acetate, the aceticlastic methanogens (Sun et al., 2014). This poses challenges regarding biogas digester operation, as low‐acetate conversion rates by the syntrophs may limit the overall efficiency and stability of the process. However, tailoring operation to underpin the syntrophic interactions, such as allowance of microbial adaptation during start‐up and changed operating conditions, long retention times and addition of trace elements has been shown to be effective in improving performance and mitigates the effect of ammonia toxicity in SAO processes (Westerholm et al., 2016).
To date, only a few SAOB have been isolated and characterized. These are the thermophilic Thermacetogenium phaeum (Hattori et al., 2000) and Pseudothermotoga lettingae (Balk et al., 2002; Bhandari and Gupta, 2014), the thermotolerant Tepidanaerobacter acetatoxydans (Westerholm et al., 2011a,b) and the mesophilic [Clostridium] ultunense (Schnürer et al., 1996) and Syntrophaceticus schinkii (Westerholm et al., 2010). Presence and abundance of these SAOB during changed operating conditions in anaerobic systems have been indicated using species‐specific 16S rRNA gene‐targeting approaches (Westerholm et al., 2011a,b, 2012a,b, 2015). However, known SAOB are often low in abundance in relation to the overall microbial community (Westerholm et al., 2016). Their relative low abundance obstructs their detection, but also imposes limitations for identification of new potential SAOB using high‐throughput sequencing of complex anaerobic digester communities. Nevertheless, altering process operation towards SAO‐inducing conditions (such as increased ammonia level, injection of H2 or elevated temperature) may affect the microbial community enough for detection of potential SAOB using sequencing approaches. Through such approaches, SAOB candidates have been suggested within the families Thermoanaerobacteraceae (which also includes the known SAOB T. phaeum and S. schinkii) and Thermodesulfobiaceae (Ho et al., 2014; Yamada et al., 2014; Bassani et al., 2015; Müller et al., 2016) and the phylum Spirochaetes (Lee et al., 2015). However, the inconceivable numbers of microbes and high microbial diversity in anaerobic digesters pose a major challenge when seeking to establish reliable links between abundant species and SAO‐function and further research is required to confirm species within these groups as SAOB.
Most known SAOB are affiliated to the physiological group of acetogens, a feature that has been used to reveal further information about potential SAOB by targeting the fhs gene, encoding a key enzyme of both acetogenic and SAO metabolism (Müller et al., 2016). Through this method, acetogenic groups unique to high‐ammonia biogas processes have been identified and are suggested to be involved in SAO (Müller et al., 2016). Results of other techniques to track down potential SAOB, such as stable isotope‐based functional probing and meta‐omics, suggest members of the orders Clostridiales and/or Thermoanaerobacterales (Zakrzewski et al., 2012; Lü et al., 2014; Müller et al., 2016), uncultured phylotypes affiliated with the Firmicutes (Frank et al., 2016), the Thermotogae (Zakrzewski et al., 2012; Nubo et al., 2015) and the phylum Synergistes (Ito et al., 2011) as candidates for SAO capacity. Taken together, the few SAOB isolates and the wide taxonomic diversity of the proposed SAOB currently pose an obstacle to predicting their function and behaviours in complex microbial communities. Identification of novel key players would thus be highly beneficial and would increase knowledge of SAO and help manage ammonia‐stressed anaerobic digesters and develop innovative operating guidance.
The aim of this study was to enrich acetate‐degrading microbial communities using a continuous cultivation approach to preserve and enrich core acetate‐utilizing communities occurring in high‐ammonia biogas systems. Continuous feeding with acetate for a long period was expected to enable microbial enrichment and facilitate identification of prominent SAOB, which due to their relatively low abundance are difficult to detect in more complex environments. The enrichments were initiated with inocula taken from anaerobic digesters previously demonstrated to be dominated by SAO. Differing factors between the enrichment chemostats included acetate influent concentration (0.4 or 7.5 g l−1), temperature (37°C or 52°C) and inoculum source, which were selected with the objective of enriching SAO populations, occupying different niches with regard to acetate concentration and optimal temperature conditions. Other operating parameters of the parallel acetate enrichments were set to mimic the continuous biogas system that was the source of the inoculum, that is high free ammonia level (0.6–0.9 g NH3 l−1) and ~30 day retention time. A combination of molecular methods, including Illumina sequencing of 16S rRNA genes, quantitative polymerase chain reaction (qPCR) and terminal restriction fragment length polymorphism (T‐RFLP) analysis, was used to identify microbial structure patterns over time and to quantify abundant species. This approach made it possible to focus on the metabolic group restricted to acetate degradation.
Results
Anaerobic chemostat performance
The mesophilic high‐acetate (7.5 g l−1) chemostats (MH) had a mean acetate effluent of 0.5 ± 0.1 g l−1 during hydraulic retention times (HRT) 2–6. During the corresponding period, the thermophilic high‐acetate chemostats (TH) had significantly higher values, averaging 1.2 ± 0.2 g acetate l−1 in TH1 and 0.7 ± 0.2 g acetate l−1 in TH2 (Fig. S1, Table S1). The high‐acetate chemostats produced gas with high methane content around 64–69% both at mesophilic and thermophilic conditions. All low‐acetate (0.4 g l−1) chemostats (ML, TL) had mean acetate effluent below the detection limit of 0.1 g l−1 and the methane level was as low as 0.3–9% throughout the trial. Ammonia concentration (g NH3 l−1) varied between 0.3–0.4 in ML, 0.7–0.8 in MH, 0.8–1.0 in TL and 1.5–1.8 in TH due to the different pH and temperature in the chemostats (Table S1).
As expected, propionate, butyrate, isobutyrate, valerate, isovalerate, capronate and isocapronate were not detected in any of the chemostats. Irrespective of operating temperature, pH remained stable over time around 7.7–7.8 and 8.1–8.2 in low‐acetate and high‐acetate chemostats respectively (Table S1).
Illumina sequencing and diversity indices
Illumina sequencing identified a total of 36 395 830 reads, of which 10 240 118 were of high quality. Rarefaction analysis is displayed in Fig. S2. The estimated coverage indicated that on average 79 ± 1% of the microbial community was covered in all samples. Alpha diversity of microbial communities showed significantly decreased richness and evenness (Sobs, Chao1 and Shannon diversity) throughout operation in all mesophilic chemostats and in the TH chemostats, whereas in TL there was no difference in alpha diversity values over time (Fig. S3, Tables S2 and S3). Richness and evenness indices in the high‐acetate chemostats were significantly higher in mesophilic than in thermophilic chemostats (Table S4).
Bacterial community structure and dynamics in mesophilic chemostats
Community OTU comparison by non‐metric multidimensional scaling (NMDS; OTU ≥ 97% identity) of each sample using the Bray–Curtis similarity metric revealed close grouping of MH chemostats, whereas the microbial communities in the low‐acetate chemostats drifted apart over time (Fig. S4A).
Firmicutes (37–53%) and Bacteroidetes (17–30%) were the most abundant phyla during initial operation of the four mesophilic chemostats (Fig. 1). The majority of the remaining bacterial sequences were not classified at phylum level (22–30% of total reads). In both MH chemostats, Firmicutes remained abundant at 35–71% of total sequences throughout operation, whereas Bacteroidetes gradually decreased to constitute 8–13% after 4 HRT. Proteobacteria became more abundant over time and represented 22–47% after 4 HRT (initial level ≤ 2%). Microbial dynamics in ML chemostats included increased relative abundances of Proteobacteria (to 18–22%) and Bacteroidetes (to 29–49%) and stable proportions of Firmicutes (23–42% after 4 HRT), whereas unclassified bacteria decreased to < 8% (Fig. 1).
Figure 1.
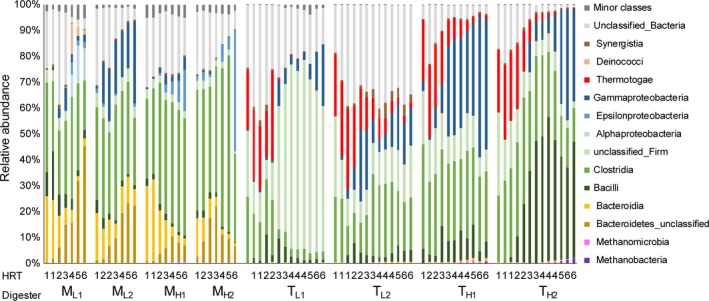
Percentage contribution of 16S rRNA gene sequences (grouped by class) during acetate enrichment under mesophilic (M) or thermophilic (T) temperature conditions, as determined using Illumina MiSeq sequencing. Chemostats designated MH/TH and ML/TL received medium containing 7.5 and 0.4 g acetate l−1 respectively. The corresponding hydraulic retention time (HRT) at the point of sampling is given on the x‐axis. Yellow, green, blue and red bars represent classes within the phyla Bacteriodetes, Firmicutes, Proteobacteria and Thermotogae respectively. Purple indicates methanogenic archaea. Classes representing < 1% were included in minor classes.
On a class taxonomic scale, Clostridia (phylum Firmicutes) dominated the microbial communities in MH and was foremost comprised of one OTU (designated OTU_MHC), belonging to the family Clostridiaceae and sharing 96% identity with Alkaliphilus oremlandii and Alkaliphilus halophilus. The relative abundance of OTU_MHC increased from 2% to 3% during initial operation to 12–23% in MH1 and 26–44% in MH2 at the end of the experimental period. In ML, the relative abundance of OTU_MHC remained below 3%.
Epsilonproteobacteria was the dominant class in Proteobacteria in MH chemostats, and the vast majority of sequences were from an OTU associated with the genus Sulfurospirillum (96% gene sequence identity to Sulfurospirillum alkalitolerans). During operation of the MH chemostats, this Sulfurospirillum sp. increased from < 3% in initial operation to 5–8% after 4 HRT and 16–47% in final HRT. Under lower acetate conditions, other classes dominated the phylum Proteobacteria, that is Alphaproteobacteria in ML1 and Gammaproteobacteria in ML2. In the class Gammaproteobacteria, the most dominant populations belonged to the family Pseudomonadaceae. These Pseudomonadaceae sp. were also found in thermophilic enrichment chemostats.
T‐RFLP analysis was performed to characterize the dynamics of the SAOB community by targeting the formyltetrahydrofolate synthetase (fhs) gene, a functional marker for SAOB and the acetogenic community in general. The majority of the fragments appearing in the high‐acetate chemostats were phylogenetically assigned by means of clone library analyses (yield of 35 partial fhs gene sequences in total). The T‐RFLP analysis showed some discrepancies between samples taken from initial operation of the duplicate MH chemostats (Fig. S5A). However, both replicate communities had a high relative abundance of a restriction fragment of 28 bp, representing 15–74% in the MH chemostats. A 58 bp restriction fragment constituted 6–27% in MH1 throughout operation, whereas in MH2 this fragment decreased from 8–17% to below detection limit at HRT 5–6. A fragment of 108 bp was detected in abundance of 4–15% at HRT 4–6 (except in MH1 at HRT 5). Throughout operation of MH, the T‐RFs 328 and 406 bp significantly decreased and increased respectively (Fig. S5A, Table S2). None of the T‐RFs that were highly abundant in later operation of MH were detected in the ML communities after 6 HRT. Clone sequences likely representing restriction fragments of 28, 58, 108 and 328 bp in the T‐RFLP had low nucleotide sequence identity (< 79%) to previously characterized species, but showed 97–99% identity to fhs gene sequences previously retrieved from high‐ammonia digesters (Westerholm et al., 2015; Moestedt et al., 2016; Müller et al., 2016) (Table S5). The T‐RF 406 bp had low sequence identity (< 79%) to previously recovered fhs genes but positioned close to the SAOB P. lettingae in the phylogenetic analyses (Fig. 2, Table S5).
Figure 2.
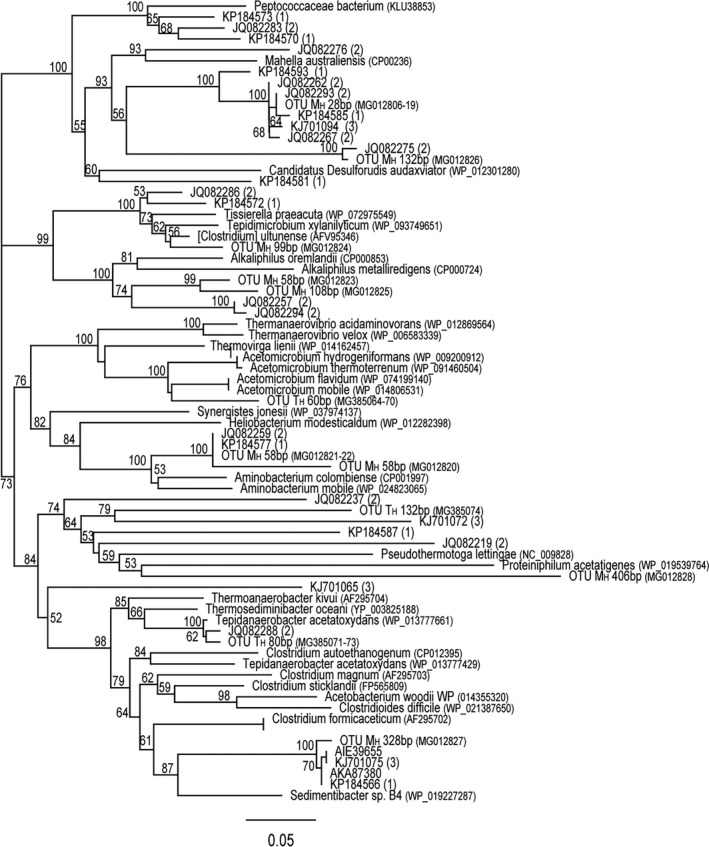
Phylogenetic placement of deduced FTHFS amino acid sequences of the partial fhs sequences retrieved from the high‐acetate mesophilic (MH) and thermophilic (TH) chemostats. Accession numbers are given for reference strains and for fhs gene sequences retrieved in this study (OTU MH/TH) and in previous studies of SAO‐dominated anaerobic digesters: (1) (Moestedt et al., 2016), (2) (Müller et al., 2016), (3) (Westerholm et al., 2015). The SAOB S. schinkii and T. phaeum and sulfate‐reducing bacteria formed a separate clade and were excluded from the alignment to reduce the size of the tree. The scale bar represents 50% sequence divergence. Values at the nodes are the percentage of 1000 bootstrap replicates; values below 50% are not shown.
Quantitative PCR analyses of known SAOB revealed stable abundance of C. ultunense (2.3 ± 0.5 × 106 gene copies μg−1 DNA) in all mesophilic chemostats, irrespective of acetate level. Over time, S. schinkii significantly increased in MH (from 105 to 107 gene copies μg−1 DNA) and decreased in ML chemostats (from 105 to 103–4 gene copies μg−1 DNA) and was thus significantly higher under high‐acetate than low‐acetate conditions after 2 HRT (Fig.ucode> 3, Table S2). T. acetatoxydans decreased in all mesophilic chemostats and was even below the limit of detection in ML1 after 2 HRT (Fig. 3, Table S2). T. phaeum was not detected in any mesophilic chemostat. Fhs gene abundances of potential SAOBs (previously identified in high‐ammonia processes; Müller et al., 2016) were stable at around 106 for fhsOTU8‐9 and at twofold higher values for fhsOTU3‐5 throughout operation of the MH chemostats. In the ML chemostats, these OTUs continually declined and became less abundant than in MH after 2 HRT. fhsOTU10 significantly decreased in all mesophilic chemostats (Fig. S6, Table S2). The total bacteria determined by qPCR analyses showed stable levels in all mesophilic chemostats relative to total extracted DNA (averaged 5.1 ± 0.5 × 108 gene copies μg−1 DNA). However, when analysed per volume chemostat sludge, the total bacteria decreased over time in ML (from 108 to 107 gene copies ml−1), whereas the levels remained relatively stable at around 108 gene copies ml−1 in the MH chemostats (Fig. S7A).
Figure 3.
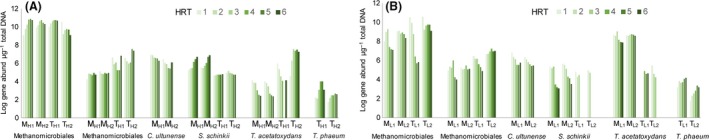
Log gene abundances of the methanogens Methanomicrobiales and Methanobacteriales and the syntrophic acetate‐oxidising bacteria (SAOB) C. ultunense, S. schinkii, T. acetatoxydans and T. phaeum in duplicate mesophilic (M) and thermophilic (T) chemostat fed 7.5 (A, H) and 0.4 (B, L) g acetate l−1. Each hydraulic retention time (HRT) represents 28 days of operation.
Methanogenic community structure and dynamics in mesophilic chemostats
Illumina sequencing revealed < 0.02% methanogenic reads during initial operation of the MH chemostats and throughout complete operation of the ML chemostats. The relative contribution of methanogens increased to 0.1–3.0% over time in MH (HRT 2‐6), primarily due to increased relative abundance of an OTU with 100% 16S rRNA gene sequence identity with Methanoculleus bourgensis. The mcrA (methyl coenzyme‐M reductase) genes retrieved from the duplicate MH chemostats after 6 HRT showed 100% identity to a mcrA gene previously recovered in the biogas digester that was the source of the inoculum for the enrichment chemostats (OTU15 in DTE37, accession number KJ701175; Westerholm et al., 2015). The qPCR analyses revealed significantly (P < 0.001) higher Methanomicrobiales abundance in MH (3.7 ± 0.7 × 1010 gene copies μg−1 DNA) than in ML (7.2 ± 2.0 × 108 gene copies μg−1 DNA) at HRT 2‐6 (Fig. 3, Table S2). In contrast, the differences in abundance of Methanobacteriales in high‐acetate (6.6 ± 0.7 × 104 gene copies μg−1 DNA) compared with low‐acetate chemostats (1.9 ± 0.6 × 105 gene copies μg−1 DNA) were not statistically (P > 0.05) significant. Methanosarcina were not detected in any chemostat.
Bacterial community structure and dynamics in thermophilic chemostats
Non‐metric multidimensional scaling visualization showed separated clustering by chemostats. The microbial community structure in all chemostats was highly dynamic over time, in particular in TL1 (Fig. S4B). The phyla Firmicutes (47–74%) and Thermotogae (16–29%) were the most abundant bacterial populations in the first and second HRT under thermophilic conditions at both acetate levels (Fig. 1). Thermotogae thereafter decreased significantly in all thermophilic chemostats and represented < 8% at HRT 4–6. Firmicutes remained relatively constant, whereas the relative representation of Proteobacteria increased from < 4% to 3–24% in TL, 32–54% in TH1 and 6–43% in TH2 after HRT 4. In TH, unclassified bacteria comprised < 3% during the final operating period, whereas in TL about 14–39% of the total reads were not classified at phylum level (Fig. 1).
Although the relative abundance of Firmicutes remained at a similar level throughout operation, the relative contribution of the family Thermoanaerobacteraceae within this phylum rapidly increased, from 2–3% at HRT 1 to 8–27% at HRT 2–6, in the TH chemostats. The majority of the Thermoanaerobacteraceae sequences clustered into one OTU (referred to as OTU_THS), with a representative sequence that exhibited 97% and 93% identity to the previously identified mesophilic SAOB S. schinkii and thermophilic SAOB T. phaeum respectively. In TL, the relative abundance of OTU_THS accounted for < 2% of total sequences at all sampling points. The genus Tepidimicrobium (phylum Firmicutes, class Clostridia) was initially present at 7–22% in all thermophilic samples, but decreased below detection after a few retention times. An OTU, of which the representative sequence showed an identical match to the previously characterized bacteria Bacillus infernus (phylum Firmicutes, class Bacillus), increased in relative abundance over time in TH2 and represented 34–53% between HRT 3–6. The relative abundance of this OTU was < 10% in T H1 and T L chemostats. Large proportions of the phylum Firmicutes were unclassified below phylum level in T L1, whereas in T L2 the majority of the sequences belonged to the class Clostridia (Fig. 1).
Gammaproteobacteria was the dominant class in Proteobacteria, mainly constituted by two OTUs classified into the family Pseudomonadaceae. These OTUs were closely related (96.8% gene sequence similarity) and together represented 3–22% and 6–53% at HRT 3‐6 in TL and TH chemostats respectively. These OTUs were also found to be highly abundant (17–32% of total sequences) in later operation of the low‐acetate mesophilic ML2 chemostat.
T‐RFLP analysis of the TH chemostats identified a restriction fragment of 536 bp that significantly increased from 10–11% to 32–35% during the course of the experiment (Fig. S5B). T‐RFs 60, 132 and 636 bp were other highly abundant restriction fragments in one or both duplicate chemostats during the operating trial. However, the majority of the abundant T‐RFs in the TH chemostats also represented 9–29% of the total fhs genes recovered in TL2 at HRT 6, which obstructed interpretation of their correlation to acetate degradation. Sequencing of fhs genes allowed phylogenetic assignment of T‐RFs 60, 80 and 132 bp in the T‐RFLP (in total 12 clones, Table S5). T‐RF 80 bp had close identity to T. acetatoxydans (96%) and fhs genes previously retrieved from a high‐ammonia SAO‐dominated digester (Müller et al., 2016). The 536 and 636 bp fragments appearing at high abundance in the T‐RFLP were not matched in the clone library. Furthermore, none of the fhs genes was closely related to S. schinkii and T. phaeum FTHFS, and they were thus not included as reference strains in the phylogenetic analyses (Fig. 2).
The 16S rRNA gene‐targeting qPCR analyses revealed constant levels of S. schinkii in TH (4.0–8.7 × 104 gene copies μg−1 DNA), whereas this species was not detected in TL after 3 HRT. T. phaeum increased from 101 to 103 gene copies μg−1 DNA during the experimental trial in all chemostats. T. acetatoxydans decreased to below detection in TH1, but in TH2, it increased from 105–6 to 107–8 gene copies μg−1 DNA after 3 HRT (Fig. 3). C. ultunense 16S rRNA genes and fhs genes previously linked to potential SAOB (Müller et al., 2016) were not detected in any of the thermophilic chemostats.
Total bacterial abundances per ml sludge declined significantly in TH chemostats and TL2 (Fig. S7B, Table S3). However, in TL1, the levels were qualitatively similar throughout operation (Fig. S7B), although, as in the mesophilic chemostats, the relative abundance observed over time showed similar levels in all chemostats when related to total DNA. This supports the results obtained in comparative analyses of the chemostats.
Methanogenic community structure and dynamics in thermophilic chemostats
Methanogenic relative abundances did not exceed 0.3% in the Illumina sequencing data for the TL chemostats. In the TH chemostats, the methanogenic contribution increased from < 0.1% to 5–10% of total sequences by the end of the trial. The Illumina sequencing indicated equal relative abundance of an OTU belonging to the genus Methanothermobacter (order Methanobacteriales, 100% similarity to M. thermophilus, M. wolfeii and M. defluvii) and an OTU with identical sequence to M. bourgensis (type strain MS2T, order Methanomicrobiales) in TH1. In TH2, Methanothermobacter was the dominant methanogen according to the Illumina sequencing. However, based on qPCR data, Methanobacteriales abundance was 3.4 ± 1.8 × 106 in TH1 and 3.3 ± 1.0 × 107 in TH2 at HRT 5–6, which was several orders of magnitude lower than Methanomicrobiales abundance (4.5 ± 0.8 × 1010 gene copies μg DNA−1 in both TH chemostats; Fig. 3, Table S3).
Discussion
Possible acetate dependencies of SAOB yet to be revealed
A continuous enrichment approach was employed in the present study to select for syntrophic acetate oxidizers, which due to their relatively low abundance are difficult to detect in more complex environments. The experimental set‐up was designed to distinguish mesophilic and thermophilic key populations during continuous feeding of two different levels of acetate. Acetate‐dependent growth rate of SAOB has been indicated in co‐cultivation and metagenomic studies (Oehler et al., 2012; Manzoor et al., 2015; Müller et al., 2015; Westerholm et al., 2016). Estimation of the degree to which the acetate level shapes syntrophic acetate‐degrading communities in biogas digesters is complex, particularly as high acetate levels often co‐occur with high levels of ammonia, which is a strong driver of development of the microbial community (Werner et al., 2014; De Vrieze et al., 2015; Müller et al., 2016). The set‐up of the present study was designed to shed more light on this subject. However, absence of methane formation in the mesophilic and thermophilic low‐acetate chemostats indicated that the influent acetate level was below the threshold for methane production via SAO. Aceticlastic methanogens are known to degrade acetate to concentrations below that level (Smith and Ingram‐Smith, 2007); however, in present chemostats, they were likely inhibited by the high ammonia level. Nevertheless, acetate was apparently consumed, judging by the lower acetate level in effluent than in influent. Consistent with this, Illumina sequencing analyses indicated presence of relatively diverse bacterial communities in ML and TL. These microorganisms might degrade acetate and/or remain in the chemostats by consuming compounds included in the medium for growth support or as reducing agents (i.e. yeast extract, cysteine). However, the microbial community profiles obtained for the low‐acetate chemostats were still useful as references to distinguish SAOB candidates prevalent in MH and TH. Given the declining trends in S. schinkii and T. acetatoxydans abundance in the ML and TL chemostats, the acetate level in the chemostats was below the threshold for SAO activity of these known SAOB. However, the acetate level did not affect the abundance of C. ultunense, indicating that this species was able to stay syntrophically active at low acetate levels or used cysteine supplied in the medium for its growth (Schnürer et al., 1996).
In particular at thermophilic temperature, the overall community sequencing and the T‐RFLP profiling revealed surprisingly high structural variations between duplicate chemostats (representing biological replicates). Quantification of known SAOB, however, revealed quite similar trends in the duplicate chemostats. The explanation for this and possible impact by microbial variations on community functions and performances are unclear at this point. Similar results have been shown in previous microbial studies of parallel digesters under high‐stress, showing diverse structures at genus and species level OTUs (Goux et al., 2015; De Vrieze et al., 2016). In line with present result, these studies report on similar and stable digester performances despite microbial divergences. This highlights the challenges in interpreting links between microbial dynamics, operating conditions and process performance and emphasizes the need for increased understanding within this area to search for potential sources of variability and significances for process performances.
Differences in richness and dynamics between mesophilic and thermophilic enrichment communities mimic trends seen in complex anaerobic communities
The initial strain richness and evenness were significantly lower in the thermophilic than in the mesophilic microbial communities, which are in line with other studies (Levén et al., 2007; Guo et al., 2014; Jang et al., 2016). We observed declining trends for these indices in all mesophilic and thermophilic high‐acetate chemostats throughout operation, which was expected as feeding a restricted number of substrates limits the potential metabolic pathways used for microbial growth and thus likely affects the phylogenetic distribution. Notably, the MH communities were still significantly higher in richness at the end of the operating trial, indicating that mesophilic temperatures support a more diverse community than thermophilic conditions during a restricted feeding strategy.
Alpha diversity of microbial communities displayed significantly decreased richness and evenness in the ML chemostats over time but, contradicting expectations, these indices remained similar at all TL sampling points. However, due to the higher initial level at mesophilic temperature, similar ranges were recorded at both temperatures by the end of the trial, indicating that the low content of yeast extract and/or cysteine included in the medium was enough to support growth of a relatively diverse microbiota at both temperatures.
Highly abundant OTUs suggest that mesophilic Clostridiaceae sp. and thermophilic Syntrophaceticus sp. are drivers of syntrophic acetate degradation
Despite the SAO‐selective conditions applied in the chemostats, previously known SAOB did not represent dominant populations. Thus, even though their abundances were higher in high‐acetate than in low‐acetate conditions (which suggests SAO activity), these SAOB were probably not the major acetate degraders in the systems investigated. However, the T‐RFLP profiling and partial fhs gene sequencing strongly indicated presence of bacteria previously found in high‐ammonia SAO‐dominated digesters (Westerholm et al., 2015; Moestedt et al., 2016; Müller et al., 2016). Unfortunately, it is currently not possible to phylogenetically position these bacteria based on the fhs gene. However, the Illumina results highlighted ubiquity and high relative abundance of a Clostridiaceae sp. (OTU_MHC) and a Syntrophaceticus sp. (OTU_THS), which strongly suggests them as novel SAOB candidates.
The closest relatives of Clostridiaceae sp. OTU_MHC (A. halophilus and A. oremlandii) are moderately halophilic (optimum growth at pH 8) and fermentative bacteria (Fisher et al., 2008; Wu et al., 2010). Some characteristics of these Alkaliphilus species resemble the features of SAOB when grown in pure culture, such as similar substrate patterns, formation of formate and acetate as main fermentation products (A. halophilus) and respiratory capabilities (Schnürer et al., 1996; Hattori et al., 2000; Fisher et al., 2008; Westerholm et al., 2010, 2011a,b; Wu et al., 2010). A. oremlandii also oxidizes acetate, with reduction of thiosulfate as the electron acceptor (Fisher et al., 2008), which is a capability it shares with the SAOB T. phaeum (Hattori et al., 2000). Alkaliphilus species have also been detected in high‐ammonia biogas digesters (> 0.3 g NH3‐N l−1/8–10 g NH+ 4‐N l−1, Kovács et al., 2013; Müller et al., 2016; Tsapekos et al., 2017; Ziganshina et al., 2017), suggesting that they play a critical role in biogas digesters operating under such conditions. Their presence has been attributed to the ability of certain members to encode crucial peptidases for proteolysis of proteins (Stolze et al., 2015). Moreover, the levels of these bacteria have been shown to correlate with an ammonia‐induced shift from aceticlastic methanogenesis to SAO (Müller et al., 2016). Phylogenetic analyses of the deduced FTHFS amino acid sequences positioned retrieved sequences (represented by the T‐RFs 58 and 108 bp) close to fhs genes of Alkaliphilus sp., which indicates Clostridiaceae sp. OTU_MHC. Other sequences assigned to the T‐RF 406 bp significantly increased in abundance during operation of both MH chemostats, positioned close to the known SAOB P. lettingae in the phylogenetic analyses and are thus considered interesting SAOB candidates.
The taxonomic survey of the OTU_THS, which represented a significant proportion of the bacterial community in the thermophilic TH chemostats, revealed phylogenetic relatedness (97% identity) with the cultured representative of S. schinkii. The mesophilic S. schinkii oxidizes acetate in association with the hydrogenotrophic M. bourgensis, tolerates high ammonium levels and has a growth temperature ranging from 25 to 40°C (Westerholm et al., 2010). Despite the narrow temperature range for growth of this type strain, relatives to this species have been found in mesophilic biogas digesters (37–40°C; Westerholm et al., 2011a,b, 2012a,b, 2015; Karlsson et al., 2012; Moestedt et al., 2014; Sun et al., 2014; Müller et al., 2016) and in digesters operating at moderate (42–45°C; Moestedt et al., 2014; Westerholm et al., 2015) and thermophilic (49–60°C; Weiss et al., 2008; Sun et al., 2014; Lebuhn et al., 2015; Müller et al., 2016) temperatures. Together, these findings indicate that, apart from the wide‐ranging temperature span, relatives to S. schinkii are able to remain active under varying operating conditions in terms of ammonia concentration, HRT, substrate feed and digester configuration. Despite the lower phylogenetic relationship between OTU_THS and T. phaeum, the thermophilic growth condition was a shared capability of these species. Searches in the Blast database furthermore revealed that sequences with identical identity to OTU_THS have been discovered in syntrophic propionate‐degrading communities (Sugihara et al., 2007), indicating ability to degrade both propionate and acetate or involvement in acetate degradation in association with syntrophic propionate‐degrading bacteria. 16S rRNA gene sequences identical to OTU_THS have previously also been found in thermophilic (53°C) dry anaerobic digestion of waste paper‐based medium and suggested to be involved in SAO (OTU 1‐1B‐29 in Tang et al., 2011) The fhs gene sequencing indicated that the gene from OTU_THS was not targeted by the current primers. Hence, further development of the fhs‐primers targeting thermophilic species is needed for a more complete coverage of potential SAOB. However, the significantly higher relative abundance of OTU_THS in TH than in TL, the relatively close identity to the SAOB S. schinkii and T. phaeum and the detection of genes with high identity in systems with a high possibility of being SAO‐dominated indicate that OTU_THS may be a hitherto undescribed species in the genus Syntrophaceticus that is capable of SAO.
The strict hydrogenotrophs M. bourgensis and Methanothermobacter are likely SAO partners, but may compete with formate‐utilizing bacteria
The dominance of the strictly hydrogenotrophic M. bourgensis and Methanothermobacter in the high‐acetate chemostats strongly suggests SAOB partnerships. The mcrA gene analysis for MH also suggested dominance of a M. bourgensis strain previously found in a biogas digester supplied with trace elements (OTU15 in Westerholm et al., 2015) that was the inoculum source for our mesophilic enrichment chemostats. This parental biogas digester had relatively high richness of mcrA genes (~14 dominant OTUs with relatively even distribution, Westerholm et al., 2015), and the dominance of one particular strain in the MH chemostats was thus somewhat surprising. A relevant finding in the previous study is that OTU15 was not detected in a digester operating under corresponding conditions, which did not receive trace elements (Westerholm et al., 2015). This indicates that continuous feeding of the trace element‐rich medium in the MH chemostats may have been particularly advantageous for this M. bourgensis strain. Relatively low partial hydrogen partial pressure was another differing operating parameter between the parental digester and other digesters in the previous study (Westerholm et al., 2015). While the influence of this particular strain on hydrogen removal remains to be investigated, the ability for hydrogen removal to low levels (due to its high affinity for hydrogen) and the tolerance to high ammonia levels have been suggested as key drivers for M. bourgensis suitability as a SAO methanogenic partner (Westerholm et al., 2015; Neubeck et al., 2016). This is also stressed by the frequent detection of M. bourgensis in SAO‐dominated biogas digesters (reviewed in Westerholm et al., 2016).
In thermophilic SAO digesters, Methanomicrobiales and Methanobacteriales (Methanothermobacter) are frequently reported as the dominant hydrogenotrophic methanogens (reviewed in Westerholm et al., 2016). Similarly, the Illumina results in the present study demonstrated high relative abundance of Methanothermobacter. Methanobacteriales was also detected by qPCR analyses of the TH chemostats, but corresponding analyses for Methanomicrobiales indicated that it was present in considerably higher abundance in these chemostats. These conflicting findings obtained in Illumina sequencing and specific qPCR analyses of ratios between Methanomicrobiales and Methanobacteriales can be related to differences in specificity in the primers used for these analyses. Due to the need for high sensitivity, encompassing as many 16S rRNA variants as possible, the Illumina primers will likely not have the same specificity for archaeal methanogens as the qPCR primers.
The relatively high abundances of Sulfurospirillum sp. in MH and Bacillus sp. in the TH chemostats were puzzling, as SAOB candidates have not previously been suggested to belong to these genera. However, negligible detection of these OTUs in ML and TL indicates involvement in acetate degradation. The strict anaerobic requirements and temperature ranges for growth reported for the isolated type strains of the closest relatives, that is S. alkalitolerans and B. infernus, are in agreement with the conditions in MH and TH2. Furthermore, the substrate patterns of these bacteria resemble what has been observed in pure cultures of known SAOB (e.g. lactate, pyruvate, fumarate or glucose, Boone et al., 1995; Schnürer et al., 1996; Hattori et al., 2000; Balk et al., 2002; Westerholm et al., 2010, 2011a,b; Sorokin et al., 2013). However, S. alkalitolerans is also capable of utilizing formate and H2 (with acetate as carbon source, Sorokin et al., 2013), and all components for growth of this bacterium would thus be provided in the present acetate‐enriched chemostats, with formate or H2 produced by SAOB (Müller et al., 2015) and acetate present in the medium. B. infernus can also use formate with Fe(III), MnO2, trimethylamine oxide or nitrate as an electron acceptor, but cannot grow on acetate (Boone et al., 1995). Hence, these data indicate that growth of B. infernus and Sulfurospirillum sp. may rely on supply of formate and/or H2 from acetate oxidation by SAOB. This would thus lead to competition between B. infernus and Sulfurospirillum sp. and the hydrogenotrophic methanogens for substrate, although further investigation is required to establish such a scenario. However, despite the uncertain metabolic roles of Sulfurospirillum OTU_MHEp and B. infernus in our enrichment chemostats, their detection confirms them to be ammonia‐tolerant.
To conclude, the continuous enrichment approach applied in the present study permitted analysis of a community specialized in acetate conversion, with a limited confounding effect of other metabolic groups. As anticipated, the populations identified as dominant SAOB were present at very low abundances or below detection in the initial operating period. The continuous acetate feeding then strongly restricted the microbial community structure and decreased phylotype richness, which enabled detection of potential SAOB after a few HRT. Based on our results, we posit SAO capability of the highly abundant OTU_MHC (GenBank accession number MG356789) and OTU_THS (GenBank accession number MG356790) in mesophilic and thermophilic conditions respectively. However, this requires further investigation. Considering the importance of acetate removal for the overall biogas production system, identifying acetate‐degrading strains capable of remaining active in the presence of high ammonia concentrations is of the utmost importance. Whether bioaugmentation of key SAO populations or/and altered operating conditions to support their activity is a suitable approach to improve biogas yield remains to be determined. Nonetheless, this is an area in which increased insights into how to predict behaviours of key microbial populations and support their activity shows great promise for maintaining robust performance under stressed conditions.
Experimental procedures
Anaerobic chemostat set‐up and operation
Four identical laboratory‐scale continuously stirred (80 rpm) tank chemostats (Belach Bioteknik, Stockholm, Sweden) with working volume 1.1 l were operated in parallel in continuous mode under two temperature regimes; mesophilic (M, 37°C) and thermophilic (T, 52°C). Anoxic and sterile bicarbonate‐buffered basal medium (BM, Westerholm et al., 2010) supplemented with 16.1 g NH4Cl l−1 and 0.4 g sodium acetate l−1 (mesophilic: ML1, ML2 thermophilic: TL1, TL2) or 7.5 g sodium acetate l−1 (mesophilic: MH1, MH2; thermophilic: TH1, TH2) was fed to the chemostats using four separate peristaltic pumps (Belach Bioteknik) at a speed of 26 μl/min. The chemostat compartments were placed on balance, which activated the outflow pumping when required, to maintain a hydraulic retention time (HRT) of 28 days. Each operating set was maintained for about 160 days (> 6 retention times).
The chemostats were inoculated with 1 l sterile BM and 0.1 l sludge during flushing with N2. The MH and ML chemostats were inoculated with sludge from a mesophilic high‐ammonia (5.4 g NH4 +‐N l−1, 0.6–0.9 g NH3 l−1, Table S6) digester degrading food waste and albumin, and TH and TL were started with sludge from a thermophilic (52°C) commercial‐scale biogas plant degrading industrial food waste and manure (Kungsängens gård, Uppsala, Sweden, 3.2 g NH4 +‐N l−1, 0.6 g NH3 l−1, Table S6). Both these digesters have been shown in previous studies to have SAO as the dominant acetate‐degrading pathway and to include high abundance of known SAOB (digesters DTE37 and M; (Sun et al., 2014; Westerholm et al., 2015; Müller et al., 2016).
Analytical investigations
Volatile fatty acid (VFA) concentrations (acetate, propionate, butyrate, isobutyrate, valerate, isovalerate, capronate and isocapronate) were analysed continuously using high‐performance liquid chromatography (HPLC), and methane content in the gas was determined by gas chromatography (GC) as described previously (Westerholm et al., 2012a,b).
DNA extraction, quantitative PCR, sequencing of the mcrA gene, terminal restriction fragment length polymorphism (T‐RFLP) analyses, cloning and phylogenetic analysis of partial fhs gene
Samples for molecular analyses were collected from each chemostat on a weekly basis and stored at ‐20°C until further use. DNA was extracted using the FastDNA soil kit (MP Biomedicals, France). Three DNA extracts from each time point and chemostat were included in the molecular analyses, to obtain three technical replicates per biological replicate (the two parallel chemostats).
To obtain information on absolute abundances of known SAOB (C. ultunense, T. phaeum, S. schinkii and T. acetatoxydans), 16S rRNA gene‐targeting qPCR assays were conducted as described previously (Westerholm et al., 2011a,b; Sun et al., 2014). The primer sets MBT and MMB (Yu et al., 2005) were used for targeting species of Methanobacteriales and Methanomicrobiales respectively. Descriptions of primer pair fhsOTU3‐fhsOTU10, targeting the fhs gene (encoding a key enzyme in acetogenic and SAO metabolism) of potential SAOB, and the qPCR protocol can be found in Müller et al. (2016).
To further investigate the methanogenic communities, PCR amplification with primers mlas and mcrA‐rev was used in recommended reaction conditions (Steinberg and Regan, 2008). The amplicons were then sequenced with mcrA‐rev primer (Macrogen, Seoul, Korea). Quality check, editing and sequence assembly were performed with Geneious v10.1.3 (Kearse et al., 2012).
Terminal restriction fragment length polymorphism (T‐RFLP) was used to characterize acetogenic communities in the enrichment chemostats using the degenerated primer pair designed by Müller et al. (2012), which was slightly modified: 3‐SAOfhs‐fw (CCNACNCCNNNNGGNGANGGNAA) and 3‐SAO‐rev (ATITTIGCIAAIGGNCCNSCNTG). The fhs gene T‐RFLP data sets were collected as triplicates from one sampling point in each HRT in the high‐acetate chemostats and on two occasions in the low‐acetate chemostats. The protocol described in Müller et al.(2012) was used for the analyses, with the exception that AluI (New England BioLabs, Ipswich, MA, USA) was used for digestion of PCR products. The sample from chemostat TL1 HRT 6 did not fulfil the filtering criteria and was thus excluded from the analysis. The procedures in Müller et al.(2016) were followed to construct fhs clone libraries (from MH and TH chemostats at HRT 6) and to analyse and edit retrieved sequences. Multiple sequences alignment of the deduced amino acid fhs sequences and references sequences from NCBI (indicated accession numbers in Fig. 2) was executed with MAFFT v7.245 (Katoh and Standley, 2013) command line environment in 1000 iteration. The Maximum likelihood phylogenetic tree with 1000 bootstrap was constructed in the FastTree Version 2.1.8 SSE3 (Price et al., 2010) command line using the Whelan‐And‐Goldman 2001 model on the resulted MAFFT protein alignment. MAFFT and FastTree modules are available on the SLU Global Bioinformatics Servers described elsewhere in this article. All fhs sequence data reported in this study were deposited in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MG012806–MG012840 and MG385064–MG385074 for MH and TH chemostats respectively.
16S rRNA gene Illumina sequencing
Construction of 16S amplicon libraries using primers 515′F (GTGBCAGCMGCCGCGGTAA, Hugerth et al., 2014) and 805R (GACTACHVGGGTATCTAATCC, Herlemann et al., 2011) and Illumina sequencing of 16S rRNA genes was carried out in triplicate on DNA extracts from each time point and chemostat as described by Müller et al. (2016). Paired‐end sequencing was performed on an Illumina MiSeq instrument at SciLifeLab Stockholm, Sweden.
The resulting sequence analysis was performed on the Global Bioinformatics Servers at SLU, Uppsala, Sweden, running CentOS Linux release 7.1.1503, with module handling by Modules based on Lua: Version 6.0.1. The modules loaded were Cutadapt 1.13, Sickle 1.0 and Mothur 1.39.5. Raw data were trimmed using Cutadapt followed by removal of singletons and low quality reads by Sickle (Joshi and Fass, 2011; Martin, 2011; Schloss et al., 2012). Data were then processed using Mothur, following the guidelines of the Mothur MiSeq SOP with slight modifications (Kozich et al., 2013); pcr.seqs was performed using the custom primers utilized in amplification of the 16S rRNA gene, while the database used was the Mothur‐formatted SILVA. Full‐length sequences and taxonomy references were taken from the database release 128. Classification was performed using classify.seqs and the 16S rRNA reference (RDP) database, version 16. As no mock samples were processed in parallel with the other samples, no assessment of error rates was performed. The full history from the mother run and scripts can be found in Appendix S1. For sequence data and statistics of analysis, see Table S7.
Changes in phylogenetic diversity over time were tested using regression analyses in R. The MiSeq data are available at the National Center for Biotechnology Information Sequence Read Archive (accession PRJEB21746). Representative sequences of potential SAOB candidates were compared with publicly available sequences using the Basic Local Alignment Search Tool (BLAST) algorithm (Altschul et al., 1990) provided by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov). The representative partial 16S rRNA gene sequences for OTU_MHC and OTU_THS were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers MG356789 and MG356790 respectively.
Statistical analysis
The statistical significance of differences between low‐ and high‐acetate chemostats, and over time in the chemostats, was determined using analysis of variance (ANOVA) in R Studio version software (http://www.r-project.org, Team R, 2016). NMDS analysis was carried out on the sample‐OTU matrix using the Bray–Curtis distances (R package Vegan; Oksanen et al., 2015).
Conflict of interest
None declared.
Supporting information
Fig. S1. Acetate concentration in the high‐acetate chemostats (fed medium containing 7.5 g acetate l−1) operating at 37°C (A) and 52°C (B).
Fig. S2. Rarefaction curves generated from OTUs at 3% sequence dissimilarity occuring in the four mesophilic and the four thermophilic chemostats operated in the present study.
Fig. S3. Average richness (number of OTUs, Chao) and Shannon Diversity (H') of microbial communities in (A) mesophilic and (B) thermophilic temperature conditions.
Fig. S4. Non‐metric multidimensional scaling (NMDS) analysis of the Bray‐Curtis dissimilarity index of the microbial community OTUs (≥ 97% identity) based on Illumina sequencing of 16S rRNA genes in mesophilic (A) and thermophilic (B) chemostats fed 7.5 (MH/TH) or 0.4 g acetate l−1 (ML/TL).
Fig. S5. fhs (formyltetrahydrofolate synthetase) gene profiling by means of T‐RFLP in mesophilic (A) and thermophilic (B) chemostats fed 7.5 (MH/TH) and 0.4 (ML/TL) g acetate l−1.
Fig. S6. Average fhs gene copies obtained in quantitative PCR (qPCR) analyses targeting the fhs gene of potential SAOB in mesophilic chemostats fed high (MH, 7.5 g l−1) and low (ML, 0.4 g l−1) acetate.
Fig. S7. Total bacteria abundance per mL sludge in (A) mesophilic and (B) thermophilic chemostats.
Table S1. Summary of operating conditions of duplicate mesophilic (M) and thermophilic (T) chemostats receiving high‐acetate (H) or low‐acetate feed (L). The values are mean of > 3 analyses and standard error of the mean (SEM).
Table S2. Significant differences over time and between mesophilic high‐acetate (MH, 7.5 g l−1) and low‐acetate (ML, 0.4 g l‐1) chemostats in quantitative PCR (qPCR) data and diversity indices.
Table S3. Significant differences in quantitative PCR (qPCR) data and diversity indices between thermophilic (52°C) chemostats fed high (TH, 7.5 g l−1) and low (TL, 0.4 g l−1) acetate and over time.
Table S4. Significant differences in acetate levels, richness and evenness indices between mesophilic (M) and thermophilic (T) high‐acetate (H, 7.5 g l−1) chemostats.
Table S5. Accession numbers of fhs sequences retrived in clone libraries from the chemostats MH1 MH2, TH1 and TH2 after 6 hydraulic retention times of operation (HRT 28 days).
Table S6. Operating conditions for the two biogas digesters that were the inoculum source for the mesophilic (37°C) and thermophilic (52°C) chemostats in the present study.
Table S7. Sample name in Illumina sequencing data, sequencing results, source chemostat and the hydraulic retention time (HRT) for sampling.
Appendix S1. Scripts used for processing sequence reads in 16S rRNA analysis.
Acknowledgements
The authors wish to thank Simon Isaksson for assistance with chemostat operation. This research was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) under award number 2012‐808, The Swedish Energy Agency (2014‐000725) and Västra Götaland Regionen (VGR 93‐2014). BM was supported by Formas, project number 942‐2015‐1008.
Microbial Biotechnology (2018) 11(4), 680–693
Funding Information
This research was supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) under award number 2012‐808, The Swedish Energy Agency (2014‐000725) and Västra Götaland Regionen (VGR 93‐2014). BM was supported by Formas, project number 942‐2015‐1008.
References
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. , and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Balk, M. , Weijma, J. , and Stams, A.J.M. (2002) Thermotoga lettingae sp. nov., a novel thermophilic, methanol‐degrading bacterium isolated from a themophilic anaerobic reactor. Int J Syst Evol Microbiol 52: 1361–1368. [DOI] [PubMed] [Google Scholar]
- Bassani, I. , Kougias, P.G. , Treu, L. , and Angelidaki, I. (2015) Biogas upgrading via hydrogenotrophic methanogenesis in two‐stage continuous stirred tank reactors at mesophilic and thermophilic conditions. Env Sci Technol 49: 12585–12593. [DOI] [PubMed] [Google Scholar]
- Bhandari, V. , and Gupta, R.S. (2014) Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Antonie Van Leeuwenhoek 105: 143–168. [DOI] [PubMed] [Google Scholar]
- Boone, D.R. , Liu, Y. , Zhao, Z. , Balkwill, D.L. , Drake, G.R. , Stevens, T.O. , and Aldrich, H.C. (1995) Bacillus infernus sp. nov., an Fe(III)‐ and Mn(IV)‐reducing anaerobe from the deep terrestrial subsurface. Int J Syst Bacteriol 45: 441–448. [DOI] [PubMed] [Google Scholar]
- De Vrieze, J. , Saunders, A.M. , He, Y. , Fang, J. , Nielsen, P.H. , Verstraete, W. , and Boon, N. (2015) Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Res 75: 312–323. [DOI] [PubMed] [Google Scholar]
- De Vrieze, J. , Regueiro, L. , Props, R. , Vilchez‐Vargas, R. , Jáuregui, R. , Pieper, D.H. , et al (2016) Presence does not imply activity: DNA and RNA patterns differ in response to salt perturbation in anaerobic digestion. Biotechnol Biofuel 9: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, E. , Dawson, A.M. , Polshyna, G. , Lisak, J. , Crable, B. , Perera, E. , et al (2008) Transformation of inorganic and organic arsenic by Alkaliphilus oremlandii sp. nov. strain OhILAs. Ann NY Acad Sci 1125: 230–241. [DOI] [PubMed] [Google Scholar]
- Frank, J.A. , Arntzen, M.Ø. , Sun, L. , Hagen, L.H. , McHardy, A.C. , Horn, S.J. , et al (2016) Novel syntrophic populations dominate an ammonia‐tolerant methanogenic microbiome. mSystems 1, e00092–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goux, X. , Calusinska, M. , Lemaigre, S. , Marynowska, M. , Klocke, M. , Udelhoven, T. , et al (2015) Microbial community dynamics in replicate anaerobic digesters exposed sequentially to increasing organic loading rate, acidosis, and process recovery. Biotechnol Biofuels 8: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Wang, C. , Sun, F. , Shu, W. , and Wu, W. (2014) A comparison of microbial characteristics between the thermophilic and mesophilic anaerobic digesters exposed to elevated food waste loadings. Bioresour Technol 152: 420–428. [DOI] [PubMed] [Google Scholar]
- Hattori, S. , Kamagata, Y. , Hanada, S. , and Shoun, H. (2000) Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate‐oxidizing bacterium. Int J Syst Evol Microbiol 50: 1601–1609. [DOI] [PubMed] [Google Scholar]
- Herlemann, D.P.R. , Labrenz, M. , Jürgens, K. , Bertilsson, S. , Waniek, J.J. , and Andersson, A.F. (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5: 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, D. , Jensen, P. , and Batstone, D. (2014) Effects of temperature and hydraulic retention time on acetotrophic pathways and performance in high‐rate sludge digestion. Environ Sci Technol 48: 6468–6476. [DOI] [PubMed] [Google Scholar]
- Hugerth, L.W. , Wefer, H.A. , Lundin, S. , Jakobsson, H.E. , Lindberg, M. , Rodin, S. , et al (2014) DegePrime, a program for degenerate primer design for broad‐taxonomic range PCR in microbial ecology studies. Appl Environ Microbiol 80: 5116–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T. , Yoshiguchi, K. , Ariesyady, H.D. , and Okabe, S. (2011) Identification of a novel acetate‐utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5: 1844–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, H.M. , Ha, J.H. , Kim, M. , Kim, J. , Kim, Y.M. , and Park, J.M. (2016) Effect of increased load of high‐strength food wastewater in thermophilic and mesophilic anaerobic co‐digestion of waste activated sludge on bacterial community structure. Water Res 99: 140–148. [DOI] [PubMed] [Google Scholar]
- Joshi, N. and Fass, J. (2011). Sickle: A sliding‐window, adaptive, quality‐based trimming tool for fastq files (Version 1.33) Retrieved from https://github.com/najoshi/sickle.
- Karakashev, D. , Batstone, D.J. , Trably, E. , and Angelidaki, I. (2006) Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae . Appl Environ Microbiol 72: 5138–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, A. , Einarsson, P. , Schnürer, A. , Eljertsson, J. , and Svensson, B.H. (2012) Impact of trace element addition on degradation efficiency of volatile fatty acids, oleic acid and phenyl acetate and on microbial populations in a biogas digester. J Biosci Bioeng 114: 446–452. [DOI] [PubMed] [Google Scholar]
- Katoh, K. , and Standley, D.M. (2013) MAFFT Multiple Sequence Alignment Software Version 7: improvments in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács, E. , Wirth, R. , Mároti, G. , Bagi, Z. , Rákhely, G. , and Kovács, K.L. (2013) Biogas production from protein‐rich biomass: fed‐batch anaerobic fermentation of casein and of pig blood and associated changes in microbial community composition. PLoS ONE 8: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich, J.J. , Westcott, S.L. , Baxter, N.T. , Highlander, S.K. , and Schloss, P.D. (2013) Development of a dual‐index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebuhn, M. , Weiss, S. , Munk, B. and Guebitz, G.M. (2015) Microbiology and molecular biology tools for biogas process analysis, diagnosis and control. Biogas Sci Technol 151, 1–40. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Park, J. , Kim, S.H. , Yu, B.J. , Yoon, J. , and Park, H. (2015) Evidence off syntrophic acetate oxidation by Spirochaetes during anaerobic methane production. Bioresour Technol 190: 543–549. [DOI] [PubMed] [Google Scholar]
- Levén, L. , Eriksson, A. , and Schnürer, A. (2007) Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol Ecol 59: 683–693. [DOI] [PubMed] [Google Scholar]
- Lü, F. , Bize, A. , Guillot, A. , Monnet, V. , Madigou, C. , Chapleur, O. , et al (2014) Metaproteomics of cellulose methanisation under thermophilic conditions reveals a surprisingly high proteolytic activity. ISME J 8: 88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor, S. , Bongcam‐Rudloff, E. , Schnürer, A. , and Müller, B. (2015) Genome‐guided analysis and whole transcriptome profiling of the mesophilic syntrophic acetate oxidising bacterium Syntrophaceticus schinkii . PLoS ONE 11: e0166520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Moestedt, J. , Nordell, E. , and Schnürer, A. (2014) Comparison of operational strategies for increased biogas production from thin stillage. J Biotechnol 175: 22–30. [DOI] [PubMed] [Google Scholar]
- Moestedt, J. , Müller, B. , Westerholm, M. , and Schnürer, A. (2016) Ammonia threshold for inhibition of anaerobic digestion of thin stillage and the importance of organic loading rate. Microbial Biotechnol 9: 180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbaek, F. , H., K. , Mulat, D.G. , Albertsen, M. , Ward, A.J. , Feilberg, A. and Nielsen, J.L. (2016) Identification of syntrophic acetate‐oxidizing bacteria in anaerobic digesters by combined protein‐based stable isotope probing and metagenomics. ISME J: 10, 2405–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B. , Sun, L. , and Schnürer, A. (2012) First insights into the syntrophic acetate‐oxidizing bacteria – a genetic study. MicrobiologyOpen 2: 35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B. , Manzoor, S. , Niazi, A. , Bongcam‐Rudloff, E. , and Schnürer, A. (2015) Genome‐guided analysis of physiological capacities of Tepidanaerobacter acetatoxydans provides insights into environmental adaptations and syntrophic acetate oxidation. PLoS ONE 10: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B. , Sun, L. , Westerholm, M. , and Schnürer, A. (2016) Bacterial community composition and fhs profiles of low‐ and high‐ammonia biogas digesters reveal novel syntrophic acetate‐oxidising bacteria. Biotechnol Biofuel 9: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubeck, A. , Sjöberg, S. , Price, A. , Callac, N. , and Schnürer, A. (2016) Effect of nickel levels on hydrogen partial pressure and methane production in methanogens. PLoS ONE 11: e0168357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nubo, K.M. , Narihiro, T. , Rinke, C. , Kamagata, Y. , Tringe, S.G. , Woyke, T. , and Liu, W.T. (2015) Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9: 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler, D. , Poehlein, A. , Leimbach, A. , Müller, N. , Daniel, R. , Gottschalk, G. , and Schink, B. (2012) Genome‐guided analysis of physiological and morphological traits of the fermentative acetate oxidizer Thermacetogenium phaeum . BMC Genom 13: 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, G. , Kindt, R. , Legendre, P. , Minchin, P.R. , O'Hara, R.B. , et al (2015) Community ecology package.
- Price, M.N. , Dehal, P.S. , and Arkin, A.P. (2010) FastTree 2 – approximately maximum‐likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryanbin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , et al (2012) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnürer, A. , Schink, B. , and Svensson, B.H. (1996) Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol 46: 1145–1152. [DOI] [PubMed] [Google Scholar]
- Smith, K.S. , and Ingram‐Smith, C. (2007) Methanosaeta, the forgotten methanogen? Trends Microbiol 15: 150–155. [DOI] [PubMed] [Google Scholar]
- Sorokin, D.Y. , Tourova, T.P. , and Muyzer, G. (2013) Isolation and characterization of two novel alkalitolerant sulfidogens from a Thiopaq bioreactor, Desulfonatronum alkalitolerans sp. nov., and Sulfurospirillum alkalitolerans sp. nov. Extremophiles 17: 535–543. [DOI] [PubMed] [Google Scholar]
- Steinberg, L.M. , and Regan, J.M. (2008) Phylogenetic comparision of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl Environ Microbiol 74: 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolze, Y. , Zakrzewski, M. , Maus, I. , Eikmeyer, F. , Jaenicke, S. , Rottmann, N. , et al (2015) Comparative metagenomics of biogas‐producing microbial communities from production‐scale biogas plants operating under wet or dry fermentation conditions. Biotechnol Biofuel 8: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara, T. , Shiratori, H. , Domoto, R. , Miyake, T. , Yaegashi, M. , Ayame, S. , et al (2007) Unique diversity of a thermophilic anaerobic microbial consortium that utilizes propionate in a synthetic medium. J Gen Appl Microbiol 53: 363–369. [DOI] [PubMed] [Google Scholar]
- Sun, L. , Müller, B. , Westerholm, M. , and Schnürer, A. (2014) Syntrophic acetate oxidation in industrial CSTR biogas digesters. J Biotechnol 171: 39–44. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Ji, P. , Hayashi, J. , Koike, Y. , Wu, X. , and Kida, K. (2011) Characteristic microbial community of a dry thermophilic methanogenic digester: its long‐term stability and change with feeding. Appl Microbiol Biotechnol 91: 1447–1461. [DOI] [PubMed] [Google Scholar]
- Team, R. RStudio (2016). Integrated Development for R. RStudio, Boston, MA: Retrieved from http://www.rstudio.com/. [Google Scholar]
- Tsapekos, P. , Kougias, P.G. , Treu, L. , Campanaro, S. , and Angelidaki, I. (2017) Process performance and comparative metagenomic analysis during co‐digestion of manure and lignocellulosic biomass for biogas production. Appl Energ 185: 126–135. [Google Scholar]
- Weiss, A. , Jerome, V. , Freitag, R. , and Mayer, H.K. (2008) Diversity of the resident microbiota in a thermophilic municipal biogas plant. Appl Microbiol Biotechnol 81: 163–173. [DOI] [PubMed] [Google Scholar]
- Werner, J.J. , Garcia, M.L. , Perkins, S.D. , Yarasheski, K.E. , Smith, S.R. , Muegge, B. , et al (2014) Microbial community dynamics and stability during an ammonia‐induced shift to syntrophic acetate oxidation. Appl Environ Microbiol 80: 3375–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerholm, M. , Roos, S. , and Schnürer, A. (2010) Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate‐oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol Lett 309: 100–104. [DOI] [PubMed] [Google Scholar]
- Westerholm, M. , Dolfing, J. , Sherry, A. , Gray, N.D. , Head, I.M. , and Schnürer, A. (2011a) Quantification of syntrophic acetate‐oxidizing microbial communities in biogas processes. Environ Microbiol Reports 3: 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerholm, M. , Roos, S. , and Schnürer, A. (2011b) Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate‐oxidizing bacterium isolated from two ammonium‐enriched mesophilic methanogenic processes. Syst Appl Microbiol 34: 260–266. [DOI] [PubMed] [Google Scholar]
- Westerholm, M. , Hansson, M. , and Schnürer, A. (2012a) Improved biogas production from whole stillage by co‐digestion with cattle manure. Bioresour Technol 114: 314–319. [DOI] [PubMed] [Google Scholar]
- Westerholm, M. , Levén, L. , and Schnürer, A. (2012b) Bioaugmentation of syntrophic acetate‐oxidising culture in biogas reactors exposed to increasing levels of ammonia. Appl Environ Microbiol 78: 7619–7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerholm, M. , Müller, B. , Isaksson, S. , and Schnürer, A. (2015) Trace element and temperature effects on microbial communities and links to biogas digester performance at high ammonia levels. Biotechnol Biofuel 8: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerholm, M. , Moestedt, J. , and Schnürer, A. (2016) Biogas production through syntrophic acetate oxidation and deliberate operating strategies for improved digester performance. Appl Energ 179: 124–135. [Google Scholar]
- Wu, X. , Shi, K. , Xu, X. , Wu, M. , Oren, A. , and Zhu, X. (2010) Alkaliphilus halophilus sp. nov., a strictly anaerobic and halophilic bacterium isolated from a saline lake, and emended description of the genus Alkaliphilus . Int J Syst Evol Microbiol 60: 2898–2902. [DOI] [PubMed] [Google Scholar]
- Yamada, C. , Kato, S. , Ueno, Y. , Ishii, M. and Igarashi, Y. (2014) Conductive iron oxides accelerate thermophilic methnogenesis from acetate and propionate. J Biosci Bioeng 119, 678–682. [DOI] [PubMed] [Google Scholar]
- Yu, Y. , Lee, C. , Kim, J. , and Hwang, S. (2005) Group specific primer and probe sets to detect methanogenic communities using quantitative real‐time polymerase chain reaction. Biotechnol Bioeng 89: 670–679. [DOI] [PubMed] [Google Scholar]
- Zakrzewski, M. , Goesmann, A. , Jaenicke, S. , Jünemann, S. , Eikmeyer, F. , Szczepanowski, R. , et al (2012) Profiling of the metabolically active community from a production‐scale biogas plant by means of high‐throughput metatranscriptome sequencing. J Biotechnol 158: 248–258. [DOI] [PubMed] [Google Scholar]
- Ziganshina, E.E. , Ibragimov, E.M. , Vankov, P.Y. , Miluykov, V.A. , and Ziganshin, A.M. (2017) Comparison of anaerobic digestion strategies of nitrogen‐rich substrates: performance of anaerobic reactors and microbial community diversity. Waste Manage 59: 160–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Acetate concentration in the high‐acetate chemostats (fed medium containing 7.5 g acetate l−1) operating at 37°C (A) and 52°C (B).
Fig. S2. Rarefaction curves generated from OTUs at 3% sequence dissimilarity occuring in the four mesophilic and the four thermophilic chemostats operated in the present study.
Fig. S3. Average richness (number of OTUs, Chao) and Shannon Diversity (H') of microbial communities in (A) mesophilic and (B) thermophilic temperature conditions.
Fig. S4. Non‐metric multidimensional scaling (NMDS) analysis of the Bray‐Curtis dissimilarity index of the microbial community OTUs (≥ 97% identity) based on Illumina sequencing of 16S rRNA genes in mesophilic (A) and thermophilic (B) chemostats fed 7.5 (MH/TH) or 0.4 g acetate l−1 (ML/TL).
Fig. S5. fhs (formyltetrahydrofolate synthetase) gene profiling by means of T‐RFLP in mesophilic (A) and thermophilic (B) chemostats fed 7.5 (MH/TH) and 0.4 (ML/TL) g acetate l−1.
Fig. S6. Average fhs gene copies obtained in quantitative PCR (qPCR) analyses targeting the fhs gene of potential SAOB in mesophilic chemostats fed high (MH, 7.5 g l−1) and low (ML, 0.4 g l−1) acetate.
Fig. S7. Total bacteria abundance per mL sludge in (A) mesophilic and (B) thermophilic chemostats.
Table S1. Summary of operating conditions of duplicate mesophilic (M) and thermophilic (T) chemostats receiving high‐acetate (H) or low‐acetate feed (L). The values are mean of > 3 analyses and standard error of the mean (SEM).
Table S2. Significant differences over time and between mesophilic high‐acetate (MH, 7.5 g l−1) and low‐acetate (ML, 0.4 g l‐1) chemostats in quantitative PCR (qPCR) data and diversity indices.
Table S3. Significant differences in quantitative PCR (qPCR) data and diversity indices between thermophilic (52°C) chemostats fed high (TH, 7.5 g l−1) and low (TL, 0.4 g l−1) acetate and over time.
Table S4. Significant differences in acetate levels, richness and evenness indices between mesophilic (M) and thermophilic (T) high‐acetate (H, 7.5 g l−1) chemostats.
Table S5. Accession numbers of fhs sequences retrived in clone libraries from the chemostats MH1 MH2, TH1 and TH2 after 6 hydraulic retention times of operation (HRT 28 days).
Table S6. Operating conditions for the two biogas digesters that were the inoculum source for the mesophilic (37°C) and thermophilic (52°C) chemostats in the present study.
Table S7. Sample name in Illumina sequencing data, sequencing results, source chemostat and the hydraulic retention time (HRT) for sampling.
Appendix S1. Scripts used for processing sequence reads in 16S rRNA analysis.


