Abstract
Cryptococcus neoformans is an important opportunistic fungal pathogen in humans. Recent studies have demonstrated that metals are critical factors for the regulation of fungal virulence in hosts. In this study, we systemically investigated the function of C. neoformans magnesium transporters in controlling the intracellular Mg balance and virulence‐associated factors. We identified three Mg transporters in C. neoformans: Mgt1, Mgt2, and Mgt3. While we could not detect a Mg2+‐related growth phenotype in mgt1 and mgt3 knockout strains, a GAL7p‐Mgt2 strain showed significant Mg‐dependent growth defects in the presence of glucose. Further analysis demonstrated that MGT2 is a homolog of MNR2 in Saccharomyces cerevisiae, which is localized to the vacuolar membrane and participates in intracellular Mg transport. Interestingly, a transcriptome analysis showed that Mgt2 influenced the expression of 19 genes, which were independent of Mg2+. We showed that melanin synthesis in C. neoformans required Mg2+ and Mgt2, and that capsule production was negatively regulated by Mg2+ and Mgt2. Repressing the expression of MGT2‐induced capsule, which resulted in an increased fungal burden in the lungs. Cumulatively, this study sets the stage for further evaluation of the important role of Mg homeostasis in the regulation of melanin and capsule in C. neoformans.
Keywords: Capsule, Cryptococcus neoformans, melanin, Mg homeostasis
1. INTRODUCTION
Cryptococcus neoformans is a major fungal pathogen in humans. It primarily afflicts immunodeficient and immunocompetent individuals, causing about 600,000 deaths per year globally (Idnurm et al., 2005; Kronstad et al., 2011; May, Stone, Wiesner, Bicanic, & Nielsen, 2016). Cryptococcus infections begin in the lung tissue. By unknown mechanisms, fungal cells disseminate into the central nervous system, resulting in lethal meningitis (Idnurm et al., 2005; Kronstad et al., 2011; May et al., 2016). Recent epidemiological studies have revealed that infections of Cryptococcus species are a significant health issue in the Pacific Northwest in Northern America, in addition to Europe, Africa, and China (Byrnes et al., 2010, 2011; Liu et al., 2017; May et al., 2016).
Two major fungal virulence factors have been extensively studied. These are the generation of a cell‐surface polysaccharide capsule structure and melanization using L‐DOPA as a substrate (Idnurm et al., 2005; Kronstad et al., 2011). The capsular structure is critical for shedding the fungal cells from the host immune cells and manipulating the responses of immune cells to C. neoformans invasion (May et al., 2016; Zaragoza et al., 2009). Melanin formation in C. neoformans protects fungal cells from many environmental stresses, such as UV radiation and free‐radical toxicity (Wang & Casadevall, 1994; Williamson, 1997). Numerous studies have demonstrated that metals, such as copper (Cu) and iron (Fe), regulate the production of the capsule structure and melanin pigment (Bairwa, Hee Jung, & Kronstad, 2017; Chun & Madhani, 2010; Ding, Festa, Sun, & Wang, 2014; Do et al., 2016; Jung, Sham, White, & Kronstad, 2006; Lian et al., 2005). It is known that the key melanin biogenesis protein (Lac1) utilizes Cu+ as a cofactor (Williamson, 1997). Disrupting key regulators of Cu homeostasis of C. neoformans influences melanin formation. For example, disrupting the Cu+ transporter genes CTR1 and CTR4, the Cu+ chaperone ATX1, or the P‐type ATPase CCC2 impairs the development of dark brown pigments in C. neoformans (Chun & Madhani, 2010; Ding et al., 2011; Walton, Idnurm, & Heitman, 2005; Waterman et al., 2007, 2012). Fe, the most well studied metal in C. neoformans, plays extremely important roles in modulating melanin and the capsule (Jung et al., 2006; Lian et al., 2005; Tangen, Jung, Sham, Lian, & Kronstad, 2007). A master Fe homeostasis regulator in C. neoformans, transcription factor Cir1 was demonstrated to inhibit melanin formation by repressing expression of LAC1 and LAC2. It also induces capsule biogenesis by activating capsule production‐related genes (Jung et al., 2006). Moreover, C. neoformans cells utilize melanin as a Fe reduction system (Nyhus, Wilborn, & Jacobson, 1997). Given the important functions of these metals in regulating fungal virulence factors, human fungal pathogens have evolved sophisticated acquisition and defense machineries to cope with metal deficiency and overload conditions at the pathogen‐host axis (Bairwa et al., 2017; Ding et al., 2014).
Of all‐important metals in biology, magnesium (Mg) is one of the most abundant metals in the cell and it plays critical roles as an enzymatic cofactor and structural component of biological molecules. Many mammalian disorders are associated with Mg2+ imbalance in the cell, such as depression, anxiety, agitation, and insulin resistance (Abdulatif, Ahmed, Mukhtar, & Badawy, 2013; Boyle, Lawton, & Dye, 2016, 2017; Serefko, Szopa, & Poleszak, 2016; Serefko et al., 2013). In fungal cells, supplementation of Mg2+ in the growth medium protects Saccharomyces cerevisiae cells from dehydration (Trofimova, Walker, & Rapoport, 2010). Mg2+ is also involved in mitotic spindle formation in Schizosaccharomyces pombe (Uz & Sarikaya, 2016). In S. cerevisiae, the Mg2+ transporters Alr1 and Alr2 were identified as plasma membrane‐bound Mg importers. The knockout strains for ALR1 and ALR2 display blocked Mg2+ uptake, which reduces intracellular Mg2+ concentrations and subsequently inhibits cell growth in normal growth condition (da Costa, Cornish, & Keasling, 2007). S. cerevisiae Mnr2 is a vacuolar Mg2+ transporter that pumps Mg2+ from the vascular lumen into the cytosolic space (Pisat, Pandey, & Macdiarmid, 2009). Mg2+ is required for mitochondrial function in fungi. Three mitochondrial Mg2+ transporters have been identified in S. cerevisiae. Lpe10 and Mrs2 are Mg2+ importers and Mme1 acts as a mitochondrial Mg2+ exporter. Inactivation of any mitochondrial Mg2+ transporters reduces Mg2+ accumulation in the mitochondria and causes defects in respiration (Cui et al., 2015; da Costa et al., 2007).
Although Mg2+ homeostasis plays vital functions in fungal biological processes, the role of Mg2+ in regulating virulence factors in human fungal pathogens remains largely unknown. In this study, we examined the function of three C. neoformans Mg2+ transporters with respect to the regulation of fungal virulence‐associated factors. We demonstrated that a vacuolar Mg2+ transporter (Mgt2) in C. neoformans positively regulated Mg2+ delivery and melanin formation, but negatively controlled capsule production. Using transcriptome analysis, we showed that knockdown of MGT2 differentially regulated the expression of 118 genes. Of these, 19 genes were influenced by MGT2 knockdown independently of exogenous Mg. In animal infection models, the mgt2 mutant displayed an elevated fungal burden in lung tissues as a result of an enlarged capsular structure.
2. MATERIAL AND METHODS
2.1. Fungal strains and growth conditions
The wild‐type strain H99 was used to generate Mg transporter mutants. Mutants were generated by homologous recombination and transformed using ballistic transformation as described previously (Toffaletti, Rude, Johnston, Durack, & Perfect, 1993). Fungal cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) at 30°C. To activate the C. neoformans GAL7 promoter, YPGal was used (1% yeast extract, 2% peptone, 2% galactose). After biolistic transformation, fungal cells were plated onto YPDSorb plates (1% yeast extract, 2% peptone, 2% galactose, 2% agar, 1 mol/L sorbitol). The transformant selection was performed using YPD agar supplemented with 200 μg/ml of G418. A final concentration of 50 mmol/L MgSO4 was used for GAL7p‐MGT2 cell growth.
2.2. Plasmid construction and mutant construction
The generation of C. neoformans strains in which Mg transporter genes were disrupted was performed as follows. All primers used for the generation of mutants are listed in Table S1. For the targeted disruption of the MGT1 or MGT3 open reading frame, the upstream sequence of MGT1 or MGT3 was amplified using primer pair CDp280/CDp281 or CDp288/CDp289, respectively. The PCR products were cloned between the ApaI and XmaI restriction sites in a NEO R plasmid. The downstream sequence of MGT1 or MGT3 was amplified using primer pair CDp282/CDp283 or CDp290/CDp291, respectively. The resulting PCR products were cloned between the BamHI and XbaI restriction sites of the NEO R plasmid. The resulting plasmids were sequenced, and the inserts were amplified using primer pair CDp23/CDp24 to generate the knockout integration cassette. The PCR products were purified, concentrated, and delivered using a PDS‐1000/He System (Bio‐rad). Transformants were picked and genomic DNA was isolated for PCR confirmation using primer pair CDp428/CDp131 for MGT1 and CDp962/CDp131 for MGT3.
The C. neoformans mgt2 mutant was constructed by replacing the MGT2 endogenous promoter with the GAL7 promoter. The upstream sequence from MGT2 was amplified using primers CDp284 and CDp285, and then this was cloned between the ApaI and XmaI restriction sites in a NEO R plasmid. The C. neoformans GAL7 promoter or MGT2 ORF (first ~1 kb of genomic DNA sequence) were amplified using primer pair CDp611/CDp612 or CDp613/CDp614, respectively. The resulting PCR fragments were purified and mixed, and overlapping PCRs were performed to join the GAL7 and MGT2 fragments using primers CDp611 and CDp614. The overlapping PCR product was purified and cloned between the SpeI and SacII restriction sites in the NEO R plasmid. The integration cassette was amplified using primers CDp23 and CDp24. The PCR fragment was transformed into C. neoformans by biolistic transformation. After 4 hr of recovery incubation, cells were transferred onto YPGal agar supplemented with G418. Transformants were confirmed by PCR using primers CDp632 and CDp131 and by Southern blotting.
For the construction of an overexpression strain of Mgt2, the C. neoformans TEF1 promoter was used to constitutively drive gene expression at the CMT2 locus. The upstream or downstream sequence of CMT2 was amplified from genomic DNA using primer pair CDp732/CDp733 or CDp738/CDp739, respectively. The upstream fragment was then cloned between the HindIII and EcoRI restriction sites in a NAT R plasmid, and the downstream fragment was cloned between the XbaI and SacII restriction sites in the same plasmid. The C. neoformans TEF1 promoter was amplified using primers CDp2204 and CDp2205, and the MGT2 ORF was amplified using primers CDp2206 and CDp2207. The fragments of the TEF1 promoter and the MGT2 ORF were joined by overlapping PCR using primers CDp2204 and CDp287. The resulting fragment was cloned at the XbaI site in the plasmid. The integration cassette was transformed using biolistic transformation. Successful integration at the CMT2 locus was confirmed using primers CDp861 and CDp131.
For the expression of S. cerevisiae ALR1 or MNR2 in the GAL7‐MGT2 strain, a C. neoformans GPD1 expression plasmid (pXL‐HYG) was used as described previously. The genomic DNA sequence from S. cerevisiae ALR1 or MNR2 was amplified using primer pair CDp1699/CDp1700 or CDp1954/CDp1955, respectively. The resulting PCR products were cloned into the PacI site in the pXL‐HYG plasmid. The plasmids with the correct gene orientation were transformed in the C. neoformans GAL7‐MGT2 strain.
2.3. Genomic DNA isolation and Southern blotting
Genomic DNA isolation and Southern blotting were performed as previously described (Ding & Butler, 2007). Briefly, 5 ml of cell culture (YPD medium or YPGal medium) was washed twice with water and then resuspended in lysis buffer (100 mM Tris‐HCl pH 8.0, 50 mmol/L EDTA, 1% SDS, 100 mM NaCl, 2% Triton X‐100). Cells were disrupted using a bead beater (Biospec). The supernatant was mixed with 7 mol/L ammonia acetate (pH 7.0). Genomic DNA was extracted using chloroform and precipitated with isopropanol. DNA pellets were then washed, dried, and resuspended in water. Next, 1 μg of DNA was used for PCR. Genomic DNA (20 μg) was treated with RNase A and digested with StuI and SacII overnight for Southern hybridization analysis. Southern blotting was performed using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche), and the experimental procedure was carried out according to the manufacturer's manual. The DIG‐labeled DNA probe was synthesized by PCR using primers CDp1045 and CDp612 (Table S1).
2.4. RNA isolation, real‐time PCR, and transcriptome analysis
RNA samples used for quantitative real‐time PCR and transcriptome library construction were isolated using TRIzol reagent (Thermo Fisher Scientific) followed by Turbo DNase I treatment (Thermo Fisher Scientific) to eliminate DNA contamination. Next, 1 μg of total RNA was reverse‐transcribed into cDNA using the Go Script Reverse Transcription System (Promega). Real‐time PCR was performed using a CFX Connect Thermocycler (Bio‐Rad). The data were analyzed using 2−ΔΔCt method. ACT1 was used as a loading control. Primers are listed in Table S1. For RNA‐seq analysis, 3 μg of total RNA were processed using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Next, mRNA was purified from total RNA using the polyT oligo‐attached magnetic beads. An Illumina proprietary fragmentation buffer was used for mRNA fragmentation. First‐strand cDNA was synthesized with random hexamer primers using SuperScript II followed by second‐strand cDNA synthesis using RNase H and DNA polymerase I. Overhang sequences were then blunted and adenylated at the 3′ end of the cDNA sequence. Subsequently, cDNA sequences 200 bp in length were selected using the AMPure XP system (Beckman Coulter) and enriched using Illumina PCR Primer Cocktail in a 15‐cycle PCR reaction. The PCR products were purified and their integrity was confirmed using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 (Agilent). The sequencing library was then sequenced on a Hiseq platform (Illumina) by Shanghai Personal Biotechnology Cp. Ltd. Sequenced reads were mapped using TopHat (http://ccb.jhu.edu/software/tophat/), and the transcript assembly was performed using Cufflinks (http://cufflinks.cbcb.umd.edu) (Trapnell et al., 2012). Genes with differential expression were analyzed using DESeq, as previously described (Chen et al., 2014). Genes of the mutant strains with fold changes greater or less than 1.5 in comparison to that of the wild‐type strain were considered to be upregulated or downregulated, respectively.
2.5. Microscopy
The capsule was observed using a Nikon Eclipse Ni‐E microscope with the 100× objective (Nikon). Lung tissue mucicarmine and hematoxylin/eosin staining were visualized using a Leica DM4000B LED microscope (Leica) with the 20× objective. Protein localization of GFP‐Mgt2 was performed by inducing cells in medium supplemented with galactose and 200 μM EDTA for 6 hr. The GFP signal was observed using a Leica TCS‐SP8 confocal microscope under the 65× oil objective (Leica).
2.6. Animal virulence assay, fungal burden, and histopathology
All animal experiments were reviewed and ethically approved by the Research Ethics Committees of the College of Life and Health Sciences of Northeastern University. The animal infection experiments were performed as previously described (Sun et al., 2014). Briefly, 6–8‐week‐old female C57BL/6 mice were used for survival rate and fungal burden analysis, and they were purchased from Changsheng Biotech (China). For the survival rate assay, mice were anesthetized and intranasally administered 105 fungal cells in 50 μl of PBS. Animals were monitored twice a day for morbidity. For the fungal burden analysis, infected mice at 14 days post infection were killed by exposure to CO2. Lung and brain tissues were collected, weighed, and homogenized in 5 ml of PBS. The suspensions were diluted with PBS, and serial dilutions were plated onto YPGal agar at 30°C for 3 days. For the histopathological assay, lung tissues were collected at 14 days post infection and fixed with paraformaldehyde. Fixed lung tissues were frozen and processed using a Crypstat Microtome Leica CM1850 (Leica). Tissue sections (10 μm in thickness) were stained with mucicarmine or hematoxylin/eosin (Sigma).
2.7. Melanin formation and capsule induction
Melanin was quantified using L‐DOPA agar medium. Overnight cell cultures were diluted to an A 600 of 1.0 and spotted onto L‐DOPA plates supplemented with various concentrations of MgSO4 (0 g/L, 0.25 g/L, 0.5 g/L, or 1 g/L). The plates were incubated at 37°C until black pigment was observed. Photographs were then taken. The capsule induction was performed using two induction conditions. Cells were either incubated in DMEM supplemented with 10% FCS at 37°C and 5% CO2 for 3 days, or in ACSF medium supplemented with 0.1% glucose at 37°C for 3 days (Rathore, Raman, & Ramakrishnan, 2016). The capsule was stained with India ink.
2.8. Atomic absorption spectroscopy
To quantify intracellular Mg concentrations, the wild‐type and Gal7p‐MGT2 cells were inoculated in YPGal medium overnight. The cultures were diluted to an A 600 of 0.2 in 10 ml of YPD medium and incubated on an orbital shaker at 30°C for 6 hr. Cells were then washed twice with 10 ml of 1 mmol/L EDTA and twice with deionized water to remove Mg ions from the medium and cell surface. The washed cells were collected in preweighed Eppendorf tubes, and the tubes were incubated at 80°C overnight to eliminate residual water. The dried cells were weighed. Next, 400 ml of 70% nitric acid was added to digest the cells at 100°C for 30 min. The resulting suspensions were diluted with 1% nitric acid. Mg concentrations were quantified using a JENA Atomic Absorption Spectroscopy ZEE nit 700P (JENA). The operation procedure was performed according to the manufacturer's manual. A standard curve was first constructed using an Mg atomic spectroscopy standard solution (Sigma). The Mg concentrations were then calculated using the standard curve.
3. RESULTS
3.1. Identification C. neoformans Mg transporters
Previous studies in baker's yeast identified five Mg2+ transporters, including two plasma membrane‐bound Mg transporters (Alr1 and Alr2), a vacuolar Mg2+ transporter (Mnr2), and two mitochondrial Mg2+ transporters (Lpe10 and Mrs2) (Figure 1a) (Bui, Gregan, Jarosch, Ragnini, & Schweyen, 1999; Cui et al., 2015; da Costa et al., 2007; Pisat et al., 2009). To identify Mg2+ transporters in C. neoformans, a protein phylogenetic analysis was employed. Three S. cerevisiae Mg2+ transporter homologs were found in a proteome database for C. neoformans, including CNAG_03901, CNAG_03502, and CNAG_00010. These were re‐named Mgt1, Mgt2, and Mgt3, respectively. The phylogenetic analysis showed that Mgt1 grouped with S. cerevisiae mitochondrial Mg2+ transporters, Mgt2 had a close evolutionary relationship with the vascular Mg2+ transporter, and Mgt3 was similar to plasma membrane Mg2+ transporters (Figure 1b). Because the gene expression of many metal transporters is regulated by environmental metal levels, real‐time PCRs were carried out to measure the endogenous expression levels of MGT1, MGT2, and MGT3 in response to elevated Mg2+ conditions (Figure 1c). Our data showed that the expression of C. neoformans Mg2+ transporter‐encoding genes was not regulated by environmental Mg levels.
Figure 1.
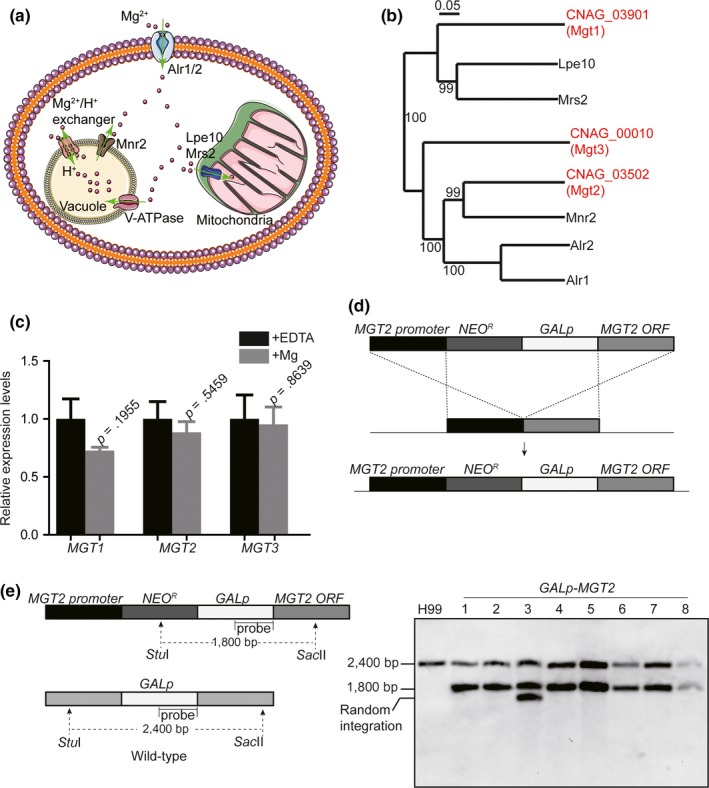
Identification of Mg transporters and generation of knockout mutants in C. neoformans. (a) Mg homeostasis in S. cerevisiae. The genome of baker's yeast encodes two Mg importers, Alr1 and Alr2 (Da Costa et al., 2007; Graschopf et al., 2001). Mnr2 is a vacuolar Mg transporter that pumps Mg into the cytosol (Pisat et al., 2009). A Mg2+/H+ exchanger also participates in Mg transport into the cytosol. Mg ions are delivered into the mitochondria via the Lpe10 and Mrs2 transporters (Gregan et al., 2001; Sponder et al., 2010). (b) Phylogenetic analysis of Mg transporters in S. cerevisiae and C. neoformans. A phylogenetic tree was generated as previously described (Ding et al., 2011). Numbers indicate the bootstrap resampling. Mg transporters in S. cerevisiae are labeled in black and those from C. neoformans are labeled in red. (c) Real‐time PCR quantifications of MGT1, MGT2, and MGT3. Overnight C. neoformans cell cultures were diluted to an A 600 of 0.2 in YPD medium, and the cultures were incubated on an orbital shaker at 30°C until an A 600 of 1.0. A final concentration of 50 mmol/L of MgSO4 or 200 μM EDTA were added to the cultures, and the incubations continued for 6 hr before RNA samples were isolated. Next, real‐time PCR was performed. Expression levels were normalized to ACT1. Four biological samples were used for the analysis. Standard deviations were determined, and the Student's t‐test was performed. (d). Scheme showing the construction of the GAL7p‐MGT2 strain. The construction of the integration cassette harboring a GAL7 promoter is described in the Material and Methods. Briefly, the promoter and ORF fragments from MGT2 were amplified using PCR and cloned into a NEO R ‐GAL7p plasmid. The resulting plasmid was introduced into the wild‐type strain using biolistic transformation. (e) Southern blot analysis of GAL7p‐MGT2 strains. The Southern blot analysis was performed as described in the Material and Methods. A probe for the GAL7p promoter was used. The restriction enzymes StuI and SacII were used for genomic DNA digestion. To verify the correct integration of the GAL7p‐MGT2 cassette, two GAL7p DNA species should be detected: one from the integrated cassette (resulting in a 1800‐bp band) and the other from the wild‐type GAL7p (resulting in a 2400‐bp band). For the wild‐type strain, only one band was detected at 2400 bp. Eight isolates (including two biological integrations) were used for the analysis. All isolates demonstrated correct integrations except isolate 3, which contained a random integration of the cassette in the genome
To determine the function of C. neoformans Mg2+ transporters in maintaining Mg2+ balance, Mg2+ transporter knockout strains were generated using the targeted homologous integration method, to obtain mgt1Δ, mgt3Δ, and mgt1Δ mgt3Δ. In our hands, we could not disrupt MGT2 after multiple attempts, implying an essential function for MGT2 under normal yeast growth conditions. We therefore substituted the MGT2 promoter with a C. neoformans galactose‐regulated promoter (GAL7, CNAG_06052: the gene coding for galactose‐1‐phosphate uridyl transferase), which was inserted before the start codon of the MGT2 ORF. This generated a GAL7p‐MGT2 strain in which the expression of MGT2 was activated by galactose and repressed by glucose (Figure 1d). A DNA probe from the GAL7 promoter was used for Southern blot analysis to evaluate the precise integration of the GAL7 promoter cassette in the genome (Figure 1e). For the wild‐type strain, a 2400‐bp band was detected, representing the wild‐type GAL7 promoter with the StuI‐SacII restriction‐digested fragment. The correct GAL7 cassette integrations yielded a smaller band of 1800 bp. The Southern blot analysis demonstrated successful construction of the GAL7p‐MGT2 strain in almost all transformation isolates (Figure 1e).
Using the spotting assay, the GAL7p‐MGT2 strain showed severe growth defects when glucose was added to suppress the activity of the galactose promoter in the medium. This observation was in agreement with the difficulties related to generating an mgt2Δ strain (Figure 2a). The mgt1Δ, mgt3Δ, and mgt1Δ mgt3Δ strains had similar cell growth rates compared to the wild‐type strain. The cell growth of the GAL7p‐MGT2 strain was rescued when Mg was supplemented into the medium, but not with other metals (Figure 2a and b). Cumulatively, our data demonstrated that Mgt2 functions as a specific Mg transporter in C. neoformans. We speculated that the impairment in cell growth was due to an intracellular Mg deficiency. Therefore, atomic absorption spectroscopy was used to measure cellular Mg levels. Our data showed a 50% reduction in the Mg2+ content in the GAL7p‐MGT2 strain (Figure 2c). These results strongly suggested that the Mgt2 protein is a critical C. neoformans Mg transporter involved in intracellular Mg accumulation.
Figure 2.
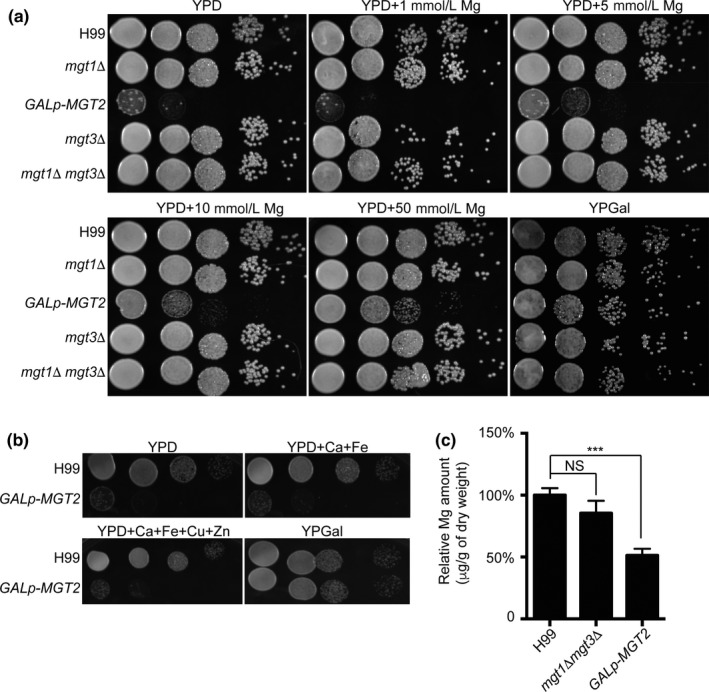
The growth phenotypes of Mg transporter mutants. (a) Cell growth assay on the Mg transporter mutants following Mg exposure. Overnight galactose liquid cell cultures of the wild‐type H99, mgt1Δ, GAL7p‐MGT2, mgt3Δ, and mgt1Δ mgt3Δ were diluted in PBS to an A 600 of 1.0. Ten‐fold serial cell dilutions were spotted onto YPGal agar, YPD agar, or YPD agar supplemented with 1 mmol/L, 5 mmol/L, 10 mmol/L, or 50 mmol/L MgSO4. Plates were incubated at 30°C for 3 days before photographs were taken. (b) Cell growth assay of Mg transporter mutants following exposure to other metals. The spotting assay was performed as described in Figure 2a, except YPD agar supplemented with 50 mmol/L Ca, 200 μmol/L Fe, 1 mmol/L Cu, and 200 μmol/L Zn was used. (C) Atomic absorption spectroscopy analysis of Mg transporter mutants. Overnight galactose liquid cell cultures of the wild‐type H99, GAL7p‐MGT2, and mgt1Δ mgt3Δ strains were diluted in YPD medium to an A 600 of 1.0, and the cultures were incubated for 6 hr. Cells were collected, washed, and placed in preweighed Eppendorf tubes. Tubes were then dried in an oven overnight. Atomic absorption spectroscopy was performed as described in the Material and Methods. Relative Mg concentrations were calculated. Four biological samples were used for the analysis. Standard deviations were determined, and the Student's t‐test was performed. Triple asterisks represent p < .001 and NS indicates no statistical significance
3.2. Mgt2 is an Mnr2 homolog
Our phylogenetic analysis demonstrated that Mgt2 is a putative Mg2+ transporter and shares significant sequence similarity to Mnr2 from S. cerevisiae (Figure 1b). To assess whether Mgt2 functionally resembles Mnr2, a GAL7p‐GFP‐MGT2 strain was generated. This strain expressed GFP‐Mgt2 chimeric protein under the regulation of a galactose‐inducible promoter (Figure 3a). The GAL7p‐GFP‐MGT2 strain generated similar growth phenotypes as that of the GAL7p‐MGT2 strain on YPD (Yeast Peptone Dextrose), and the growth phenotype was complemented in the presence of galactose (Figure 3a), demonstrating that this was a function specific to GFP‐Mgt2 protein. Confocal microscopy was used to show that GFP‐Mgt2 was localized to the vacuolar membrane (Figure 3b). We also observed GFP signal in unidentified intracellular spaces. Because S. cerevisiae Mg2+ transporters have been shown to localize to the plasma membrane, vacuolar membrane, and mitochondria (Bui et al., 1999; Cui et al., 2015; Pisat et al., 2009), we employed a mitochondrial staining method using MitoTracker to ascertain whether Mgt2 was a mitochondrial‐localized Mg2+ transporter in C. neoformans. However, no colocalization of Mgt2 and mitochondria was observed, indicating that Mgt2 is not a mitochondrial‐associated Mg2+ transporter (Figure 3b). A vacuolar specific dye, FM4‐64, was used to stain the GFP‐MGT2 strain. Fluorescent signals corresponding to FM4‐64 and GFP were colocalized on the vacuolar membrane, demonstrating that the Mgt2 is a vacuolar Mg2+ transporter (Figure 3c). An immunoblot assay was performed to measure the protein stability of Mgt2 in response to exogenous Mg2+ levels. Our data demonstrated stable protein expression of Mgt2 protein under high Mg2+ concentrations (Figure 3d). To further address Mgt2 function, S. cerevisiae MNR2 or ALR1 were cloned into a C. neoformans expression vector containing the constitutive promoter GPD1p. Vectors expressing S. cerevisiae MNR2 or ALR1 were transformed into the C. neoformans GAL7p‐MGT2 strain. Fungal cells were spotted onto YPD agar medium. This revealed that the S. cerevisiae MNR2 gene functionally complemented the growth phenotype of the GAL7p‐MGT2 strain on YPD, whereas ScALR1 did not (Figure 3e). These data strongly suggested that C. neoformans Mgt2 is a vacuolar Mg2+ transporter that functionally analogous to S. cerevisiae Mnr2.
Figure 3.
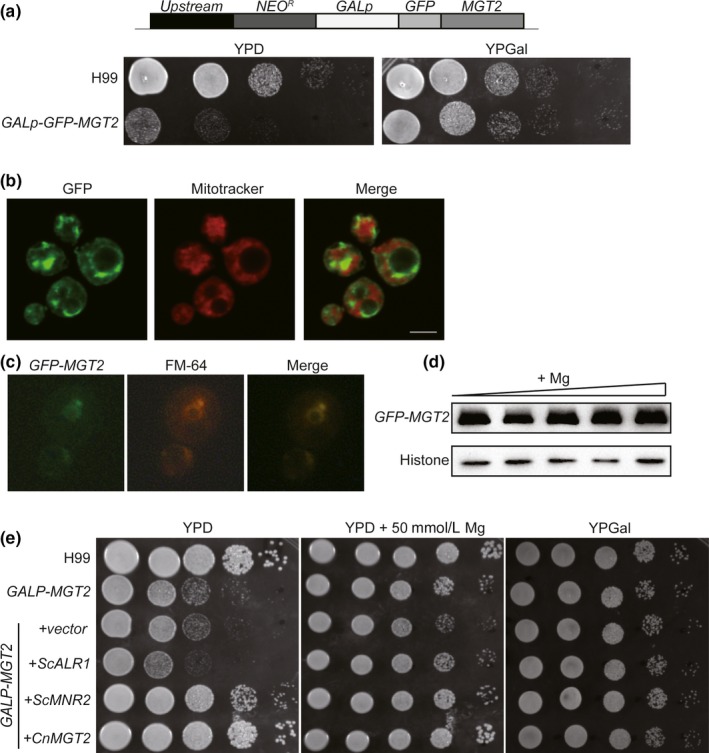
C. neoformans Mgt2 is a vacuolar Mg transporter. (a) Schematic for the construction of GAL7p‐GFP‐MGT2. A NEO R ‐GAL7p‐GFP cassette was integrated to replace the MGT2 endogenous promoter. The activation of GFP‐MGT2 expression is driven by galactose. A spotting assay was performed as described in Figure 2a to confirm the strain. (b) Subcellular localization analysis of Mgt2. Cells were grown in YPGal medium supplemented with 200 μmol/L EDTA. Cells were washed twice with PBS and then stained with Mitotracker. Visualization was performed using confocal microscopy (Leica TCS‐SP8). (c) Subcellular localization analysis of Mgt2. Cells were prepared as described above, except that cells were stained with FM4‐64. (d) Expression of Mgt2‐GFP protein in high Mg. Cells were grown in YPGal medium supplemented with 0 mmol/L, 1 mmol/L, 2 mmol/L, 5 mmol/L, 10 mmol/L of MgSO4. Protein samples were separated and immunoblot assays were performed using anti‐GFP and antihistone H3 antibodies. (e) Complementation of the GAL7p‐MGT2 strain phenotype by S. cerevisiae Mg transporter genes. C. neoformans expression vectors harboring S. cerevisiae ALR1, MNR2, or C. neoformans MGT2 were transformed into the GAL7p‐MGT2 strain. The resulting strains were cultured, diluted, and spotted onto YPD agar, YPD agar supplemented with 50 mmol/L MgSO4, or YPGal agar. The plates were incubated at 30°C for 3 days before the photographs were taken
3.3. Mgt2 modulates gene expression independent of Mg
The mgt2 knockout strain growth phenotype was complemented when exogenous Mg was supplemented in the growth medium. However, cell growth in the presence of Mg was only partially rescued, even at a very high level of Mg2+ (Figure 2a). This growth phenotype suggests that Mgt2 influences normal cell growth via a Mg2+‐independent pathway. To explore this hypothesis, we performed a high‐throughput transcriptome analysis using RNA sequencing (RNA‐seq). We compared the transcriptomes among the three conditions: wild‐type cells in YPD medium, GAL7p‐MGT2 cells in YPD medium, and GAL7p‐MGT2 cells grown in YPD medium supplemented with 50 mmol/L MgSO4 (Figure 4a). In our data, repressing the expression of MGT2 resulted in differential expression of 118 genes. Of these, the expression of 84 and 34 genes was down or upregulated in the GAL7p‐MGT2 cells, respectively (Table S2). Among all differentially expressed genes, expression of MGT2 was the most significantly downregulated in the GAL7p‐MGT2 cells, demonstrating the accuracy of our RNA‐seq data (Table 1, Table S2 and S3). We observed that genes encoding deacetylase, mitochondrial alternative oxidase, alternative cyclin Pcl12, Fe‐S cluster assembly factor, and proteins involved in sugar metabolism and transport were significantly downregulated. Genes encoding ferric‐chelate reductase, sulfite transporter, septin ring protein, and glucose‐methanol‐choline oxidoreductase were significantly upregulated (Table S2). On the other hand, supplementing Mg into the medium of GAL7p‐MGT2 cells only caused 27 genes to be differentially expressed. Of these, expression of 7 genes and 20 genes was up or downregulated, respectively (Table S3). We detected that genes encoding alcohol dehydrogenase, alternative cyclin pcl12, carnitine O‐acetyltransferase, and alternative oxidase were downregulated in the GAL7‐MGT2 strain when grown in high or low Mg conditions. Gene ontology analysis demonstrated that the addition of Mg into the medium of the GAL7p‐MGT2 strain caused dramatic changes in the GO enrichment (Figure 4b) compared to that of GAL7p‐MGT2 cells in YPD only. However, four GO terms remained unchanged in both conditions, regardless of Mg treatment, and these GO terms were significantly enriched in carbohydrate transport, single‐organism transport, single‐organism localization, and organic substance transport (Figure 4b). Two processes were differentially regulated upon the Mg2+ treatment, and these GO terms were enriched in monocarboxylic acid catabolic process and small molecule catabolic process. The expression of 19 genes remained constant in GAL7p‐MGT2 cells in YPD in the presence or absence of exogenous Mg (Figure 4b and Table 1), and this included MGT2 itself. Real‐time PCR was performed for five randomly selected putative target genes of the 19‐gene cohort to evaluate the RNA‐seq data, and expression regulation patterns resembled those obtained in the RNA‐seq analysis (Figure 4c).
Figure 4.
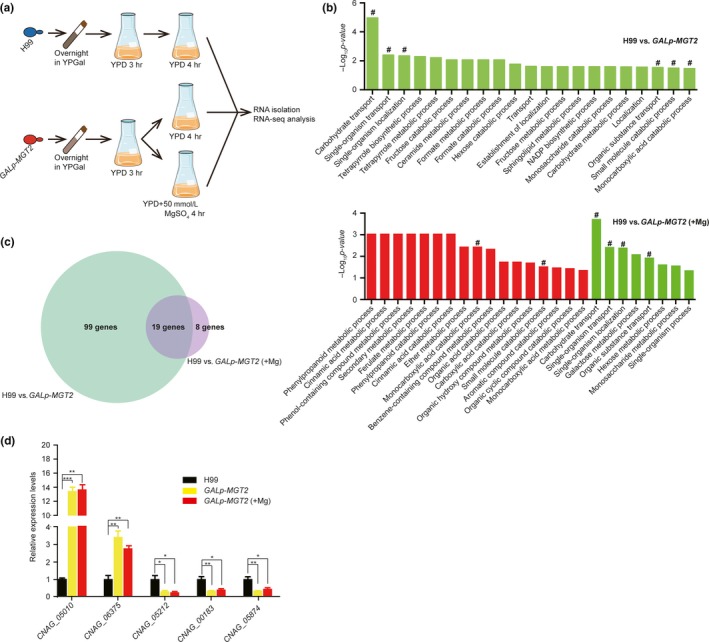
Transcriptomic analysis of the Gal7p‐MGT2 strain. (a) Strategy for performing the transcriptomic analysis on wild‐type and GAL7p‐MGT2 strains. Overnight YPGal cultures were diluted to an A 600 of 1.0 in YPD medium. The YPD cultures were incubated at 30°C for 3 hr to repress the activity of the GAL7 promoter in the GAL7p‐MGT2 strain. For the GAL7p‐MGT2 strain, the YPD culture was divided in two, and MgSO4 was added into one portion at a final concentration of 50 mmol/L. All cultures were incubated for additional 4 hr before RNA was isolated. Three biological replicates were carried out. B Gene ontology analysis. Gene ontology was performed using TopGO (Huber et al., 2015). GO terms were downloaded at http://www.bioconductor.org/packages/release/bioc/html/topGO.html. Significantly altered GO terms (classical fisher test, p < .05) are shown on the plot. Red bars indicate upregulation in the GAL7p‐MGT2 strain and green bars indicate downregulation in the GAL7p‐MGT2 strain. Hash signs show the unchanged GO terms after Mg treatment. (c) Venn diagram for transcriptomic analysis. Repressing MGT2 causes 118 genes to be differentially expressed compared to the wild‐type strain H99. Repressing MGT2 in the presence of exogenous Mg causes 27 genes to be differentially expressed compared to the wild‐type strain H99. Nineteen genes demonstrated similar expression dynamics in both growth conditions. (d) Real‐time PCR confirmation of transcriptomic data. Cells were grown as described in Figure 4a, and RNA was isolated, digested with DNase I, and reverse‐transcribed. Random genes were selected from 19 overlapping genes. Primers are listed in Table S1. Actin was used as a normalization control. Three biological replicates were performed. The Student's t‐test was performed to determine statistical significance (***p < .005, **p < .01, *p < .05).
Table 1.
RNA‐seq analysis of differentially expressed genes in the GAL7‐MGT2 strain supplemented with or without MgSO4
| Gene ID | Log2(MGT2 ‐v‐ H99)a | Log2(MGT2 + Mg ‐v‐ H99)b | Description |
|---|---|---|---|
| CNAG_03502 | −4.28 | −3.44 | Magnesium transporter |
| CNAG_02489 | −1.20 | −2.37 | Alcohol dehydrogenase |
| CNAG_03772 | −0.92 | −2.28 | Glucose transporter |
| CNAG_07788 | −1.62 | −1.98 | Unknown |
| CNAG_05212 | −2.08 | −1.72 | Unknown |
| CNAG_07819 | −1.33 | −1.64 | Unknown |
| CNAG_05487 | −1.65 | −1.61 | Unknown |
| CNAG_07845 | −1.84 | −1.41 | Unknown |
| CNAG_05874 | −1.55 | −1.20 | Unknown |
| CNAG_00679 | −1.32 | −1.18 | Unknown |
| CNAG_00183 | −1.65 | −1.09 | Alternative cyclin Pcl12 |
| CNAG_01370 | −0.92 | −1.01 | Unknown |
| CNAG_06551 | −0.84 | −0.89 | Carnitine O‐acetyltransferase |
| CNAG_01065 | −0.78 | −0.72 | Unknown |
| CNAG_00162 | −0.82 | −0.64 | Alternative oxidase |
| CNAG_00664 | 0.91 | 1.16 | Unknown |
| CNAG_02392 | 0.98 | 1.33 | Unknown |
| CNAG_06375 | 2.14 | 2.25 | Unknown |
| CNAG_05010 | 2.80 | 3.23 | Unknown |
RNA‐seq analysis was performed and differential gene expression was compared between the wild‐type and GAL7‐MGT2 strains. Differential gene expression is represented by binary log of the fold change. Three biological replicates were performed. The full gene list is available in Table S2.
RNA‐seq analysis was performed and differential gene expression was compared between the wild‐type and GAL7‐MGT2 strains (supplemented with 50 mM MgSO4). Differential gene expression is represented by binary log of the fold change. Three biological replicates were performed. The full gene list is available in Table S3.
3.4. Mgt2 influences C. neoformans survival in the lungs
Because Mg2+ is an essential metal and the mgt2 mutant showed striking growth defects, we hypothesized that Mgt2 participates in regulating fungal virulence in mice. C57BL/J mice were infected with mgt1Δ mgt3Δ or GAL7p‐MGT2 strains (precultured in YPD medium), and the survival rates were recorded (Figure 5a). Mice infected with mgt1Δ mgt3Δ had similar death rates as the wild‐type. Surprisingly, mice infected with two independent GAL7p‐MGT2 strains had earlier death rates, but these data displayed poor reproducibility (Figure 5b). In the colony‐forming unit (CFU) analysis, we detected increased levels of fungal proliferation in two independent GAL7p‐MGT2 strains in the lungs (Figure 5c). This was subsequently recapitulated by the lung tissue mucicarmine stain in which more GAL7p‐MGT2 cells were observed in lung tissues compared to the lung tissue infected with the wild‐type strain (Figure 5f), with no significant changes observed for lung mass (data not shown). Furthermore, using the murine macrophage cell line RAW264.7, we showed that the mgt2 mutant inhibited the rate of phagocytisis, compared to the wild‐type cells (Figure 5e), suggesting that Mgt2 may participate in capsule formation (discussed below). A comparison of the CFUs from brain tissues between H99 and GAL7p‐MGT2 cells showed no significant difference (Figure 5d).
Figure 5.
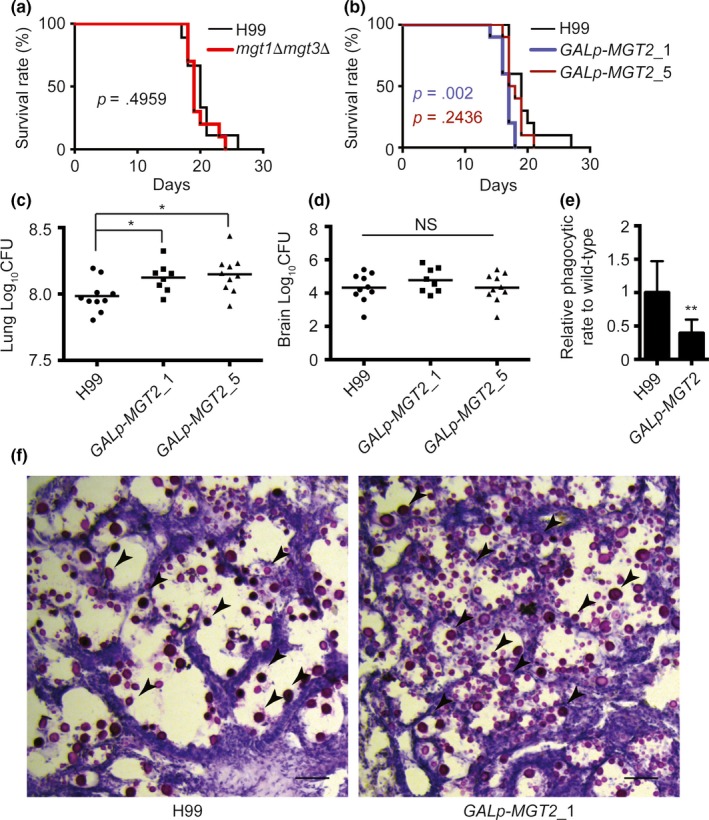
Analysis of C. neoformans Mg transporter mutants in mice. (a) Survival analysis of the wild‐type and mgt1Δ mgt3Δ strains. Twenty 4–6‐week‐old female C57BL/J mice were used for the analysis. Overnight fungal cell cultures were washed twice with PBS and diluted with PBS. A PBS suspension containing 105 cells of the wild‐type or mgt1Δ mgt3Δ strain were used for the infection of mice. Animal survival rates were recorded. (b) Survival analysis of the wild‐type and GAL7p‐MGT2 strains. Twenty 4–6‐week‐old female C57BL/J mice were used for the analysis. Overnight YPGal fungal cell cultures were washed twice with PBS and diluted in YPD medium. The YPD cultures were incubated for 6 hr to repress the GAL promoter. Cells were then washed and diluted. A PBS suspension containing 105 cells of the wild‐type or mgt1Δ mgt3Δ strain were used for infection in mice. Animal survival rates were recorded. (c) Fungal burden analysis in the lungs. Two independent GAL7p‐MGT2 strains were used to infect mice as described in Figure 5b. Fourteen days post infection, mice were killed, and CFUs from the lung tissues were analyzed. (d) Fungal burden analysis in the brain. The brain fungal burden tests were performed as described in Figure 5c. ANOVA was used for statistical analysis (*p < .05). (e) Phagocytic assay of mgt2 mutant. YPGal fungal cell cultures were washed twice with PBS, and then mixed with RAW264.7 cells in DMEM medium supplemented with 10% FCS. Extracellular fungal cells were washed with PBS buffer. Internalized and membrane‐bound fungal cells were counted. Relative phagocytic rates were calculated. The graph represents three biological replicates, and over 150 macrophages were counted for each replicate. The Student's t‐test was performed to determine statistical significance (**p < .01). (f) Histopathological analysis of C. neoformans‐infected lung tissues. Lung tissues from H99‐ and Gal7p‐MGT2‐infected mice were isolated, fixed, and stained with mucicarmine or H&E. Arrows indicates strained C. neoformans cells
To gain further insights into the mechanism by which Mgt2 influences fungal proliferation in the lung, we analyzed melanin and capsule formation in the GAL7p‐MGT2 strains. As shown in Figure 6a, melanin formation in the wild‐type cells required Mg2+. The GAL7p‐MGT2 strains demonstrated severe melanin production defects. The addition of Mg2+ only partially rescued the pigmentation defects. We also tested the mgt1Δ mgt3Δ strain, and it showed similar melanin production deficits when compared with the wild‐type strain (Figure 6a). Our data demonstrated that Mg2+ is a critical element for melanin formation in C. neoformans, and Mgt2 is required.
Figure 6.
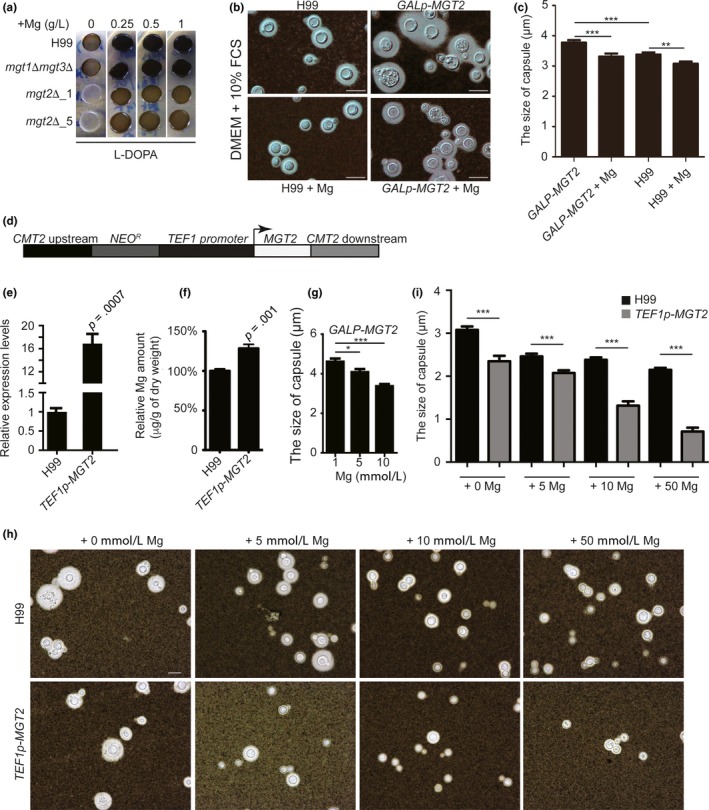
C. neoformans Mgt2 modulates melanin production and capsule structure. (a) Melanin formation in C. neoformans Mg transporter mutants. YPGal overnight cell cultures of H99, mgt1Δ mgt3Δ, and two independent Gal7p‐MGT2 strains were diluted and subcultured in YPD. Cells were then washed, diluted, and spotted onto L‐DOPA plates supplemented with various concentration of MgSO4. The plates were incubated at 37°C until black pigment developed. Photographs were taken at day 3 of incubation. (b) Capsule formation in the GAL7p‐MGT2 strain. YPGal overnight cell cultures of H99 and GAL7p‐MGT2 strains were diluted in DMEM supplemented with 10% FCS, with or without exogenous Mg. Cultures were incubated at 37°C (5% CO2) for 2 days. Pictures were taken using a Nikon Eclipse Ni‐E microscope. (c) Capsule structure quantifications in the GAL7p‐MGT2 strain in DMEM medium. The thickness of capsule structure was measured and quantified. At least 50 fungal cells from Figure 6b were measured. The Student's t‐test was performed (***p < .005, **p < .01). (d) Model for the overexpression of Mgt2. A linearized plasmid containing a NEO‐resistant gene, a constitutive promoter from C. neoformans (TEF1), CMT2 flanking sequences, and the MGT2 ORF were introduced into the H99 strain. The integration of the overexpression cassette occurred at the CMT2 locus. MGT2 expression was driven by the TEF1 promoter. (e) Real‐time PCR quantification of MGT2 in the TEF1p‐MGT2 strain. RNA samples were isolated from the H99 and GAL7p‐MGT2 strains and reversed‐transcribed into cDNA. Real‐time PCR on MGT2 cDNA was performed. Four biological replicates were performed. (f) Atomic absorption spectroscopy analysis in the TEF1p‐MGT2 strain. The assay was performed as described in Figure 2c. (g) Capsule structure quantifications in the GAL7p‐MGT2 strain in ACSF medium. At least 50 fungal cells were measured. The Student's t‐test was performed (***p < .005, *p < .05). (h) Capsule structure quantification in the TEF1p‐MGT2 strain. Overnight YPD cell cultures were diluted in ACSF medium and supplemented with various concentrations of MgSO4. Cultures were incubated at 37°C for 3 days. Pictures were taken using a Nikon Eclipse Ni‐E microscope. (i) Capsule structure quantifications in the TEF1‐MGT2 strain. The thickness of the capsule structure from Figure 6f was measured and quantified. At least 50 fungal cells from Figure 6f were measured. The Student's t‐test was performed (***p < .005)
Next, we examined capsule formation in Mg2+ transporter mutants in DMEM (Dulbecco's Modified Eagle Medium, high glucose) supplemented with 10% fetal calf serum (FCS) (Figure 6b). We observed a modest increase in capsule thickness in the GAL7p‐MGT2 strain compared to the wild‐type strain (Figure 6c). The addition of Mg2+ reduced the size of the capsule structure. To extend these observations, we constructed an Mgt2 overexpression strain in which MGT2 expression is under the control of the constitutive promoter, TEF1 (Figure 6d). To avoid random integration in the genome, the construct was designed to target the metallothionein 2 locus (CMT2) because previous studies demonstrated that the disruption of CMT2 had no effect on the expression of virulence factors (Ding et al., 2013). The expression of MGT2 was measured using real‐time PCR, which demonstrated a 15‐fold increase in gene expression compared to that of the wild‐type strain (Figure 6e). A significant increased intracellular Mg2+ concentration was detected in the TEF1‐MGT2 strain (Figure 6f). Since previous studies showed that different capsule induction environments result in the variations in capsule thickness (Rathore et al., 2016), we employed ACSF (artificial cerebrospinal fluid) medium to evaluate the capsule structure in the GAL7p‐MGT2 and TEF1p‐MGT2 strains. Significant reductions in the size of the capsule structure were observed in the GAL7p‐MGT2 strain in ACSF medium supplemented with a high concentration of Mg2+ (Figure 6g). These effects were similar to those of DMEM and ACSF media on capsule formation in the GAL7p‐MGT2 mutant. In contrast to the GAL7p‐MGT2 strain, the TEF1p‐MGT2 strain displayed a reduction in capsule thickness, and a greater reduction was observed in the presence of exogenous Mg2+ (Figure 6h and i). These results clearly demonstrated that expression of MGT2 has a negative effect on capsule production through the regulation of intracellular Mg2+ contents, increasing the cell survival of the GAL7‐MGT2 mutant strain in the lung tissue.
4. DISCUSSION
C. neoformans is a deadly human fungal pathogen, yet clinical antifungal therapies are ineffective. Many studies have demonstrated the important function of metal homeostasis in modulating C. neoformans virulence and host defenses (Ding et al., 2014; Samanovic, Ding, Thiele, & Darwin, 2012). Fe has been widely accepted as a key regulator of pathogenicity in major human fungal pathogens (Bairwa et al., 2017; Ding et al., 2014; Jung et al., 2006; Lian et al., 2005; Nyhus et al., 1997; Saikia, Oliveira, Hu, & Kronstad, 2014). In C. neoformans, mutations in genes associated with Fe acquisition often lead to dramatic attenuations in fungal virulence. A C. neoformans GATA‐type transcription factor, Cir1, is required for the regulation of expression of genes that are essential for Fe and Cu homeostasis. Moreover, important virulence factor genes, such as genes involved in the capsule and melanin biosynthesis processes, are also controlled by Cir1 (Jung et al., 2006). Using metabolomic analysis, it was demonstrated that Cir1 modulates the glycolytic pathway, ergosterol processes, and inositol metabolism. Other metal regulators, including HapX and Leu1, also contribute to metal homeostasis and regulate virulence in C. neoformans (Choi, Kim, Kim, Jung, & Lee, 2012; Do et al., 2015, 2016; Jung et al., 2010).
Numerous studies suggest that metal acquisition machinery has an essential function in C. neoformans virulence. Indeed, disrupting the high‐affinity Fe2+ permease Cft1 in C. neoformans significantly attenuates fungal virulence and fungal burden in animal models as a result of the poor adaptation of mutant cells during Fe limitation in the host. However, the disruption of Cft2 has no effect on fungal proliferation in the host (Han, Do, & Jung, 2012; Kim et al., 2012). Cu has been suggested to be an essential metal for C. neoformans because of its function in melanin formation, capsule production, respiration, and as an antioxidant agent cofactor (Chun & Madhani, 2010; Ding et al., 2013; Jiang et al., 2009; Mauch, Cunha, & Dias, 2013; Walton et al., 2005; Waterman et al., 2007, 2012; Zhang et al., 2016). Recent studies of Cu homeostasis focused on two Cu+ transporters, CTR1 and CTR4. It has been shown using two versions of a gene knockout system that a ctr4 mutant demonstrates high fungal burden in the lungs (Sun et al., 2014; Zhang et al., 2016). Meanwhile, the ctr1 and ctr4 double mutant has melanin defects, and this strain shows an increased fungal burden and brain dissemination (Sun et al., 2014).
Mg2+ is one of the most abundant divalent metal elements required for life, but its potential function in regulating fungal pathogenicity remains uninvestigated. Previous in vitro studies demonstrated that Mg2+ is an important element in the regulation of biofilm formation and capsule production in C. neoformans (Robertson, Wolf, & Casadevall, 2012). The addition of ethylenediaminetetraacetic acid (EDTA) reduces biofilm growth and vesicular secretion from the cell. However, these inhibitory effects are the result of divalent cation deficiency, rather than a Mg2+‐specific factor. This is because the supplementation of either Mg2+ or Ca2+ could rescue the inhibitory effects of EDTA. Interestingly, recent studies showed that Mg2+ has a potential influence on capsule structure in clinical C. neoformans isolates, but the molecular mechanism remains unknown (Rathore et al., 2016).
In our study, three Mg2+ transporter gene orthologs are identified in the genome of C. neoformans, based on BLAST searches using S. cerevisiae Mg transporters. Many key players involved in metal homeostasis are regulated transcriptionally by metal concentrations in the growth medium. For example, gene expression of CTR1 and CTR4 from C. neoformans decreases upon treatment with Cu+ (Ding et al., 2011). Unlike the response to other metals, the gene expression and protein levels of Mg transporters in C. neoformans are highly stable after supplementation with Mg2+ ions, demonstrating significantly different regulation patterns at the transcriptional and posttranslational level for Mg homeostasis. S. cerevisiae Alr1 and Alr2 are two Mg importers localized to the plasma membrane (Graschopf et al., 2001). Knocking out both genes impairs S. cerevisiae cell growth even at relatively high Mg concentrations (4 mmol/L Mg2+ supplementation in the growth medium), indicating the essential function of these Mg2+ importers in fungal biology (Graschopf et al., 2001; Pisat et al., 2009). Interestingly, the deletion of C. neoformans MGT3 (an ortholog of plasma membrane Mg2+ transporters in S. cerevisiae) causes no Mg‐dependent growth phenotypic difference compared to the wild‐type strain. Normal growth rates are observed in the mgt1 mutant, which is an ortholog of S. cerevisiae mitochondrial Mg transporters. A double mutant for MGT1 and MGT3 also has a comparable phenotype as the wild‐type C. neoformans strain, and no defects in cell growth, melanin, capsule formation, or virulence are observed in the animal model. These results imply that the C. neoformans genome encodes other Mg2+ transport factors that facilitate Mg2+ import into the cytosol and mitochondria.
A C. neoformans MGT2‐repressing mutant has a reduced intracellular Mg2+ content and cell growth defects under normal growth conditions, resembling the phenotype of S. cerevisiae (Pisat et al., 2009). Moreover, S. cerevisiae Mnr2 functionally complements C. neoformans Mgt2, and the C. neoformans Mgt2 has vacuolar protein localization. Taken together, our work demonstrates the conserved function of Mnr2 in C. neoformans and S. cerevisiae. Because of the striking cell growth defects in the mgt2 mutant, we speculate that the mgt2 mutant strain would have an impaired virulence and fungal burden in lung tissue. Surprisingly, our animal infection studies showed minor increases in mortality in the animals infected with the mgt2 mutants, in addition to significant inductions in the fungal burden in the lung tissue. Further analysis indicated that the increased fungal proliferation in the host results from an enlarged capsule structure when expression of Mgt2 was repressed, leading to the inhibition of macrophage phagocytosis. In agreement with the observation of capsule enlargement in the mgt2 mutant, the overexpression of Mgt2 resulted in less capsule production. Furthermore, the reduction in the capsule is inversely correlated with the addition of exogenous Mg2+ ions, demonstrating a direct connection between Mg2+ homeostasis and capsule production in C. neoformans. This highlights the important function of Mgt2 in controlling the capsule structure. Despite our data showing melanin formation defects in the Mgt2‐repressing strain, pigment development is not fully complemented when exogenous Mg is supplemented, indicating that both Mg2+ and Mgt2 are required for proper melanin production in C. neoformans.
It is interesting that Mgt2 has a positive effect on the cell growth, which is independent of intracellular Mg level. This result indicates the potential novel functions of Mg2+ transporters in C. neoformans. In fact, Mg2+ transporters have been demonstrated to be involved in the RNA group II splicing process in S. cerevisiae (Gregan et al., 2001; Sponder et al., 2010). While it is still unclear what the regulation mechanism is in, it is possible that the Mgt2‐repressing strain dramatically dampens the intracellular Mg levels, and thus impairs the RNA splicing process in C. neoformans, thus blocks the gene expression. Using transcriptomic analysis, we show here that C. neoformans Mgt2 differentially regulates the expression of 19 genes, regardless of Mg concentration. We observed genes encoding alternative cyclin Pcl12 and carnitine O‐acetyltransferase were decreased in expression when MGT2 was repressed. Previous studies have demonstrated the critical functions of Pcl12 and carnitine O‐acetyltransferase in the regulation of cell cycle progression and cell morphology (Brunner, Kramar, Denhardt, & Hofbauer, 1997; Flor‐Parra, Castillo‐Lluva, & Perez‐Martin, 2007; Perez‐Martin & Castillo‐Lluva, 2008). Therefore, it is most likely that the growth defects of the GAL7‐MGT2 strain after Mg treatment is due to the inhibition of gene expression of cell proliferation regulators.
Despite a previous study using semiquantitative RT‐PCR showing that Mg2+ influences the expression of genes involved in capsule production, we do not detect capsule‐related RNA expression differences in our RNA‐seq analysis. This is probably because different strain isolates were used in each study, causing different responses to metal ions (Sun et al., 2014; Waterman et al., 2012; Zhang et al., 2016). In fact, C. neoformans is a fast‐evolving human fungal pathogen. Comparative genomic analyses have demonstrated inconsistent phenotypic variations in stress responses, mating and virulence factor expression among H99 lineages because of these microevolutionary processes (Bahn, Hicks, Giles, Cox, & Heitman, 2004; Geddes et al., 2016; Gong, Grodsky, Zhang, & Wang, 2014; Janbon et al., 2014; Maeng et al., 2010; Pukkila‐Worley & Alspaugh, 2004; Shen, Wang, Whittington, Li, & Wang, 2008). Most importantly, the enlargement of capsule structure and increased fungal lung proliferation in MGT2‐repressed cells may also be determined by other factors, because C. neoformans Mgt2 differentially regulates the expression of 19 different genes. Previous studies demonstrated that C. neoformans capsule formation is induced by low glucose conditions, via the activation of the cAMP‐PKA pathway, which is also an extremely important regulator of C. neoformans virulence. Interestingly, the expression of the glucose transporter (CNAG_03772) is repressed when Mgt2 expression is inhibited. It is likely that the mgt2 mutant activates the cAMP‐PKA pathway via intracellular glucose deficiency, which in turns, causes capsule structure induction and increases lung fungal burden. Therefore, we suggest that multiple inputs are needed for the capsule development and the induction of fungal virulence in the mgt2 mutant. Moreover, we also detected that Mgt2 regulates 13 hypothetical genes that may play important roles in C. neoformans capsule formation.
In conclusion, we have demonstrated an evolutionarily conserved role for a vacuolar Mg transporting protein in C. neoformans, and highlighted the important function of Mg2+ in modulating C. neoformans virulence factors.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
We thank Mr. Yang Liu and Personalbio, Inc for assistance with the RNA sequencing experiments. W.D. acknowledges training received from the Medical Fungi Summit Forum and Advanced Technology Training Course. This research was supported by the funding support from the National Natural Science Foundation of China (31300974 to C.D. and 81501784 to S‐X.S.), the Fundamental Research Funds for Central Universities of China (N142005001 to C.D.), and the Thousand Youth Talents Plan Project of China. The funding agents had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Suo C‐H, Ma L‐J, Li H‐L, et al. Investigation of Cryptococcus neoformans magnesium transporters reveals important role of vacuolar magnesium transporter in regulating fungal virulence factors. MicrobiologyOpen. 2018;7:e564 10.1002/mbo3.564
REFERENCES
- Abdulatif, M. , Ahmed, A. , Mukhtar, A. , & Badawy, S. (2013). The effect of magnesium sulphate infusion on the incidence and severity of emergence agitation in children undergoing adenotonsillectomy using sevoflurane anaesthesia. Anaesthesia, 68, 1045–1052. 10.1111/anae.12380 [DOI] [PubMed] [Google Scholar]
- Bahn, Y. S. , Hicks, J. K. , Giles, S. S. , Cox, G. M. , & Heitman, J. (2004). Adenylyl cyclase‐associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP‐protein kinase A cascade. Eukaryotic Cell, 3, 1476–1491. 10.1128/EC.3.6.1476-1491.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa, G. , Hee Jung, W. , & Kronstad, J. W. (2017). Iron acquisition in fungal pathogens of humans. Metallomics, 9, 215–227. 10.1039/C6MT00301J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, N. B. , Lawton, C. L. , & Dye, L. (2016). The effects of magnesium supplementation on subjective anxiety. Magnesium Research, 29, 120–125. [DOI] [PubMed] [Google Scholar]
- Boyle, N. B. , Lawton, C. , & Dye, L. (2017). The effects of magnesium supplementation on subjective anxiety and stress‐a systematic review. Nutrients, 9, E429 10.3390/nu9050429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S. , Kramar, K. , Denhardt, D. T. , & Hofbauer, R. (1997). Cloning and characterization of murine carnitine acetyltransferase: Evidence for a requirement during cell cycle progression. The Biochemical Journal, 322(Pt 2), 403–410. 10.1042/bj3220403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, D. M. , Gregan, J. , Jarosch, E. , Ragnini, A. , & Schweyen, R. J. (1999). The bacterial magnesium transporter CorA can functionally substitute for its putative homologue Mrs2p in the yeast inner mitochondrial membrane. Journal of Biological Chemistry, 274, 20438–20443. 10.1074/jbc.274.29.20438 [DOI] [PubMed] [Google Scholar]
- Byrnes, E. J. III , Li, W. , Lewit, Y. , Ma, H. , Voelz, K. , Ren, P. , … Heitman, J. (2010). Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathogens, 6, e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes, E. J. III , Li, W. , Ren, P. , Lewit, Y. , Voelz, K. , Fraser, J. A. , … Heitman, J. (2011). A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathogens, 7, e1002205 10.1371/journal.ppat.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Toffaletti, D. L. , Tenor, J. L. , Litvintseva, A. P. , Fang, C. , Mitchell, T. G. , … Perfect, J. R. (2014). The Cryptococcus neoformans transcriptome at the site of human meningitis. MBio, 5, e01087–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. N. , Kim, J. , Kim, J. , Jung, W. H. , & Lee, C. H. (2012). Influence of iron regulation on the metabolome of Cryptococcus neoformans . PLoS ONE, 7, e41654 10.1371/journal.pone.0041654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, C. D. , & Madhani, H. D. (2010). Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans . PLoS ONE, 5, E12503 10.1371/journal.pone.0012503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, Y. , Zhao, S. , Wang, J. , Wang, X. , Gao, B. , Fan, Q. , … Zhou, B. (2015). A novel mitochondrial carrier protein Mme1 acts as a yeast mitochondrial magnesium exporter. Biochimica et Biophysica Acta, 1853, 724–732. 10.1016/j.bbamcr.2014.12.029 [DOI] [PubMed] [Google Scholar]
- da Costa, B. M. , Cornish, K. , & Keasling, J. D. (2007). Manipulation of intracellular magnesium levels in Saccharomyces cerevisiae with deletion of magnesium transporters. Applied Microbiology and Biotechnology, 77, 411–425. 10.1007/s00253-007-1177-4 [DOI] [PubMed] [Google Scholar]
- Ding, C. , & Butler, G. (2007). Development of a gene knockout system in Candida parapsilosis reveals a conserved role for BCR1 in biofilm formation. Eukaryotic Cell, 6, 1310–1319. 10.1128/EC.00136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, C. , Festa, R. A. , Chen, Y. L. , Espart, A. , Palacios, O. , Espin, J. , … Thiele, D. J. (2013). Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host & Microbe, 13, 265–276. 10.1016/j.chom.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, C. , Festa, R. A. , Sun, T. S. , & Wang, Z. Y. (2014). Iron and copper as virulence modulators in human fungal pathogens. Molecular Microbiology, 93, 10–23. 10.1111/mmi.12653 [DOI] [PubMed] [Google Scholar]
- Ding, C. , Yin, J. , Tovar, E. M. , Fitzpatrick, D. A. , Higgins, D. G. , & Thiele, D. J. (2011). The copper regulon of the human fungal pathogen Cryptococcus neoformans H99. Molecular Microbiology, 81, 1560–1576. 10.1111/j.1365-2958.2011.07794.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, E. , Hu, G. , Caza, M. , Oliveira, D. , Kronstad, J. W. , & Jung, W. H. (2015). Leu1 plays a role in iron metabolism and is required for virulence in Cryptococcus neoformans . Fungal Genetics and Biology, 75, 11–19. 10.1016/j.fgb.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, E. , Park, M. , Hu, G. , Caza, M. , Kronstad, J. W. , & Jung, W. H. (2016). The lysine biosynthetic enzyme Lys4 influences iron metabolism, mitochondrial function and virulence in Cryptococcus neoformans . Biochemical and Biophysical Research Communications, 477, 706–711. 10.1016/j.bbrc.2016.06.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor‐Parra, I. , Castillo‐Lluva, S. , & Perez‐Martin, J. (2007). Polar growth in the infectious hyphae of the phytopathogen ustilago maydis depends on a virulence‐specific cyclin. Plant Cell, 19, 3280–3296. 10.1105/tpc.107.052738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes, J. M. , Caza, M. , Croll, D. , Stoynov, N. , Foster, L. J. , & Kronstad, J. W. (2016). Analysis of the protein kinase a‐regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin‐proteasome pathway in capsule formation. MBio, 7, e01862–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, J. , Grodsky, J. D. , Zhang, Z. , & Wang, P. (2014). A Ric8/synembryn homolog promotes Gpa1 and Gpa2 activation to respectively regulate cyclic AMP and pheromone signaling in Cryptococcus neoformans . Eukaryotic Cell, 13, 1290–1299. 10.1128/EC.00109-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graschopf, A. , Stadler, J. A. , Hoellerer, M. K. , Eder, S. , Sieghardt, M. , Kohlwein, S. D. , & Schweyen, R. J. (2001). The yeast plasma membrane protein Alr1 controls Mg2 + homeostasis and is subject to Mg2 + ‐dependent control of its synthesis and degradation. Journal of Biological Chemistry, 276, 16216–16222. 10.1074/jbc.M101504200 [DOI] [PubMed] [Google Scholar]
- Gregan, J. , Bui, D. M. , Pillich, R. , Fink, M. , Zsurka, G. , & Schweyen, R. J. (2001). The mitochondrial inner membrane protein Lpe10p, a homologue of Mrs2p, is essential for magnesium homeostasis and group II intron splicing in yeast. Molecular and General Genetics, 264, 773–781. 10.1007/s004380000366 [DOI] [PubMed] [Google Scholar]
- Han, K. , Do, E. , & Jung, W. H. (2012). A human fungal pathogen Cryptococcus neoformans expresses three distinct iron permease homologs. Journal of Microbiology and Biotechnology, 22, 1644–1652. 10.4014/jmb [DOI] [PubMed] [Google Scholar]
- Huber, W. , Carey, V. J. , Gentleman, R. , Anders, S. , Carlson, M. , Carvalho, B. S. , … Morgan, M. (2015). Orchestrating high‐throughput genomic analysis with bioconductor. Nature Methods, 12, 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm, A. , Bahn, Y. S. , Nielsen, K. , Lin, X. , Fraser, J. A. , & Heitman, J. (2005). Deciphering the model pathogenic fungus Cryptococcus neoformans . Nature Reviews Microbiology, 3, 753–764. 10.1038/nrmicro1245 [DOI] [PubMed] [Google Scholar]
- Janbon, G. , Ormerod, K. L. , Paulet, D. , Byrnes, E. J. III , Yadav, V. , Chatterjee, G. , … Dietrich, F. S. (2014). Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genetics, 10, e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. , Sun, N. , Xiao, D. , Pan, J. , Wang, Y. , & Zhu, X. (2009). A copper‐responsive factor gene CUF1 is required for copper induction of laccase in Cryptococcus neoformans . FEMS Microbiology Letters, 296, 84–90. 10.1111/j.1574-6968.2009.01619.x [DOI] [PubMed] [Google Scholar]
- Jung, W. H. , Saikia, S. , Hu, G. , Wang, J. , Fung, C. K. , D'Souza, C. , … Kronstad, J. W. (2010). HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans . PLoS Pathogens, 6, e1001209 10.1371/journal.ppat.1001209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, W. H. , Sham, A. , White, R. , & Kronstad, J. W. (2006). Iron regulation of the major virulence factors in the AIDS‐associated pathogen Cryptococcus neoformans . PLoS Biology, 4, e410 10.1371/journal.pbio.0040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Cho, Y. J. , Do, E. , Choi, J. , Hu, G. , Cadieux, B. , … Jung, W. H. (2012). A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genetics and Biology, 49, 955–966. 10.1016/j.fgb.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad, J. W. , Attarian, R. , Cadieux, B. , Choi, J. , D'Souza, C. A. , Griffiths, E. J. , … Wang, J. (2011). Expanding fungal pathogenesis: cryptococcus breaks out of the opportunistic box. Nature Reviews Microbiology, 9, 193–203. 10.1038/nrmicro2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, T. , Simmer, M. I. , D'Souza, C. A. , Steen, B. R. , Zuyderduyn, S. D. , Jones, S. J. , … Kronstad, J. W. (2005). Iron‐regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans . Molecular Microbiology, 55, 1452–1472. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Kang, M. , Wu, S. Y. , Ma, Y. , Chen, Z. X. , Xie, Y. , & Tang, J. T. (2017). Different characteristics of cryptococcal meningitis between HIV‐infected and HIV‐uninfected patients in the Southwest of China. Medical Mycology, 55, 255–261. [DOI] [PubMed] [Google Scholar]
- Maeng, S. , Ko, Y. J. , Kim, G. B. , Jung, K. W. , Floyd, A. , Heitman, J. , & Bahn, Y. S. (2010). Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans . Eukaryotic Cell, 9, 360–378. 10.1128/EC.00309-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch, R. M. , Cunha, V. D. E. O. , & Dias, A. L. (2013). The copper interference with the melanogenesis OF Cryptococcus neoformans . Revista do Instituto de Medicina Tropical de Sao Paulo, 55, 117–120. 10.1590/S0036-46652013000200009 [DOI] [PubMed] [Google Scholar]
- May, R. C. , Stone, N. R. , Wiesner, D. L. , Bicanic, T. , & Nielsen, K. (2016). Cryptococcus: From environmental saprophyte to global pathogen. Nature Reviews Microbiology, 14, 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhus, K. J. , Wilborn, A. T. , & Jacobson, E. S. (1997). Ferric iron reduction by Cryptococcus neoformans . Infection and Immunity, 65, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Martin, J. , & Castillo‐Lluva, S. (2008). Connections between polar growth and cell cycle arrest during the induction of the virulence program in the phytopathogenic fungus Ustilago maydis. Plant Signal and Behavior, 3, 480–481. 10.4161/psb.3.7.5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisat, N. P. , Pandey, A. , & Macdiarmid, C. W. (2009). MNR2 regulates intracellular magnesium storage in Saccharomyces cerevisiae. Genetics, 183, 873–884. 10.1534/genetics.109.106419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila‐Worley, R. , & Alspaugh, J. A. (2004). Cyclic AMP signaling in Cryptococcus neoformans . FEMS Yeast Research, 4, 361–367. 10.1016/S1567-1356(03)00241-1 [DOI] [PubMed] [Google Scholar]
- Rathore, S. S. , Raman, T. , & Ramakrishnan, J. (2016). Magnesium ion acts as a signal for capsule induction in Cryptococcus neoformans . Frontiers in Microbiology, 7, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, E. J. , Wolf, J. M. , & Casadevall, A. (2012). EDTA inhibits biofilm formation, extracellular vesicular secretion, and shedding of the capsular polysaccharide glucuronoxylomannan by Cryptococcus neoformans . Applied and Environment Microbiology, 78, 7977–7984. 10.1128/AEM.01953-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia, S. , Oliveira, D. , Hu, G. , & Kronstad, J. (2014). Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans . Infection and Immunity, 82, 839–850. 10.1128/IAI.01357-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic, M. I. , Ding, C. , Thiele, D. J. , & Darwin, K. H. (2012). Copper in microbial pathogenesis: Meddling with the metal. Cell Host & Microbe, 11, 106–115. 10.1016/j.chom.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serefko, A. , Szopa, A. , & Poleszak, E. (2016). Magnesium and depression. Magnesium Research, 29, 112–119. [DOI] [PubMed] [Google Scholar]
- Serefko, A. , Szopa, A. , Wlaz, P. , Nowak, G. , Radziwon‐Zaleska, M. , Skalski, M. , & Poleszak, E. (2013). Magnesium in depression. Pharmacological Reports, 65, 547–554. 10.1016/S1734-1140(13)71032-6 [DOI] [PubMed] [Google Scholar]
- Shen, G. , Wang, Y. L. , Whittington, A. , Li, L. , & Wang, P. (2008). The RGS protein Crg2 regulates pheromone and cyclic AMP signaling in Cryptococcus neoformans . Eukaryotic Cell, 7, 1540–1548. 10.1128/EC.00154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponder, G. , Svidova, S. , Schindl, R. , Wieser, S. , Schweyen, R. J. , Romanin, C. , … Weghuber, J. (2010). Lpe10p modulates the activity of the Mrs2p‐based yeast mitochondrial Mg2 + channel. FEBS Journal, 277, 3514–3525. 10.1111/j.1742-4658.2010.07761.x [DOI] [PubMed] [Google Scholar]
- Sun, T. S. , Ju, X. , Gao, H. L. , Wang, T. , Thiele, D. J. , Li, J. Y. , … Ding, C. (2014). Reciprocal functions of Cryptococcus neoformans copper homeostasis machinery during pulmonary infection and meningoencephalitis. Nature Communications, 5, 5550 10.1038/ncomms6550 [DOI] [PubMed] [Google Scholar]
- Tangen, K. L. , Jung, W. H. , Sham, A. P. , Lian, T. , & Kronstad, J. W. (2007). The iron‐ and cAMP‐regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans . Microbiology, 153, 29–41. 10.1099/mic.0.2006/000927-0 [DOI] [PubMed] [Google Scholar]
- Toffaletti, D. L. , Rude, T. H. , Johnston, S. A. , Durack, D. T. , & Perfect, J. R. (1993). Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. Journal of Bacteriology, 175, 1405–1411. 10.1128/jb.175.5.1405-1411.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D. R. , … Pachter, L. (2012). Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofimova, Y. , Walker, G. , & Rapoport, A. (2010). Anhydrobiosis in yeast: Influence of calcium and magnesium ions on yeast resistance to dehydration‐rehydration. FEMS Microbiology Letters, 308, 55–61. 10.1111/j.1574-6968.2010.01989.x [DOI] [PubMed] [Google Scholar]
- Uz, G. , & Sarikaya, A. T. (2016). The effect of magnesium on mitotic spindle formation in Schizosaccharomyces pombe. Genetics and Molecular Biology, 39, 459–464. 10.1590/1678-4685-GMB-2015-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, F. J. , Idnurm, A. , & Heitman, J. (2005). Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans . Molecular Microbiology, 57, 1381–1396. 10.1111/j.1365-2958.2005.04779.x [DOI] [PubMed] [Google Scholar]
- Wang, Y. , & Casadevall, A. (1994). Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Applied and Environment Microbiology, 60, 3864–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman, S. R. , Hacham, M. , Hu, G. , Zhu, X. , Park, Y. D. , Shin, S. , … Williamson, P. R. (2007). Role of a CUF1/CTR4 copper regulatory axis in the virulence of Cryptococcus neoformans . Journal of Clinical Investigation, 117, 794–802. 10.1172/JCI30006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman, S. R. , Park, Y. D. , Raja, M. , Qiu, J. , Hammoud, D. A. , O'Halloran, T. V. , & Williamson, P. R. (2012). Role of CTR4 in the Virulence of Cryptococcus neoformans . MBio, 3, e00285–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, P. R. (1997). Laccase and melanin in the pathogenesis of Cryptococcus neoformans . Frontiers in Bioscience, 2, e99–e107. 10.2741/A231 [DOI] [PubMed] [Google Scholar]
- Zaragoza, O. , Rodrigues, M. L. , de Jesus, M. , Frases, S. , Dadachova, E. , & Casadevall, A. (2009). The capsule of the fungal pathogen Cryptococcus neoformans . Advances in Applied Microbiology, 68, 133–216. 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Zhang, D. , Zhao, X. , Wei, D. , Wang, Y. , & Zhu, X. (2016). Effects of CTR4 deletion on virulence and stress response in Cryptococcus neoformans . Antonie van Leeuwenhoek, 109, 1081–1090. 10.1007/s10482-016-0709-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


