Abstract
Oral premalignant lesions (OPLs) have malignant transformation potential, with no reliable markers available. This study aimed to assess molecular events to identify biomarkers that can reflect high-risk lesions as predictive factors to tailor clinical decision for patients on the basis of long noncoding RNAs (lncRNA) expression profiling by serial analysis of gene expression. The GSE31021 and GSE8127 datasets were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) and lncRNAs were identified using the LIMMA package in R language. The genes targeted by lncRNAs were predicted among screened DEGs using Pearson’s correlation. Gene ontology function and Kyoto Encyclopedia of Genes and Genomes pathway analyses were carried out for genes targeted by lncRNAs using the Database for Annotation, Visualization, and Integrated Discovery online tool. A total of 674 DEGs and differentially expressed lncRNAs were screened. Thirty-two interactions of 10 lncRNAs and 524 target genes were predicted. The lncRNA NEAT1 was among the top 10 lncRNAs. The coregulated target genes RP4-684O24, RP11-283I3, and RP11-350G8 were significantly enriched in the immune response and mannosyl-oligosaccharide mannosidase activity. The target genes coregulated by LINC00665 and MIR378D2 were significantly enriched in the ubiquitin-dependent protein catabolic process, ubiquitin-protein ligase activity, and neurotrophin signaling. The lncRNA NEAT1 may play an important role in high-risk lesions. The novel lncRNAs and DEGs identified in OPLs may mediate the immune response and neurotrophin signaling and show ubiquitin ligase activity. These results improve our understanding of the molecular pathogenesis of OPLs and identify some potential targets for early diagnosis of high risk OPLs.
Keywords: biomarker, differentially expressed genes, long noncoding RNA, oral premalignant lesions, prevention, serial analysis of gene expression
Introduction
Oral squamous cell carcinoma (OSCC), the most common head and neck cancer, is associated with high mortality (Fuller et al., 2007). More than 170 000 individuals are diagnosed with OSCC every year worldwide (Petti, 2003). OSCC usually shows oral premalignant lesions (OPLs), which frequently progress from mild-to-severe dysplasia, and ultimately to invasive cancer. Oral leukoplakia, considered the most common OPLs, often transforms into OSCC with a canceration rate ranging widely from 0.13 to 36.4% according to different definitions and follow-up durations (Arduino et al., 2013); in cases with severe dysplasia, a rate of up to 43% was reported (Lumerman et al., 1995). Therefore, early diagnosis and intervention for high-risk lesions can help prevent the occurrence of OSCC. Oral epithelial dysplasia (OED) is a warning sign for malignant transformation. However, there are no objective and noninvasive methods to typify OED. The disease progression involves an accumulation of genetic changes. Detecting aberrant genes is beneficial for the identification and elimination of OPLs, as well as the prevention of malignant transformation. Therefore, there is a need to explore the molecular pathogenesis of OPLs to detect biomarkers that can help identify lesions at high risk of developing OSCC as predictive factors to tailor clinical decisions for individual patients.
Loss of heterozygosity at the chromosome 9p21 region is frequently observed in both OPLs and head and neck squamous cell carcinoma, whereas inactivation of p16 (INK4a) was described in OPL biopsies (Papadimitrakopoulou et al., 1997). Agarwal et al. (1999) reported a significant association of MDM2/p53 coexpression with OPLs. Recently, the focus on noncoding RNA (ncRNA) expression has outweighed the emphasis on protein-coding genes. MicroRNAs (miRNA), a group of small noncoding RNA molecules measuring 21–23 nucleotides in length, regulate the expression of almost one-third of the human genome, and play important roles in tumorigenesis (Boyd, 2008). Clague et al. (2010) found that a pri-miRNA single nucleotide polymorphism in mir26a-1 significantly alters the risk of OPL. Long noncoding RNAs (lncRNAs) constitute a novel class of long noncoding RNA molecules measuring 200 nucleotide to more than 100 kb (Costa, 2010). They serve as host genes for miRNAs, mediators of mRNA decay, and regulators of chromatin remodeling (Eis et al., 2005; Gong and Maquat, 2011; Kanduri, 2011). Abnormal expression of lncRNAs has been reported in various cancers (Huarte and Rinn, 2010; Gibb et al., 2011a, 2011b; Tsai et al., 2011). HOX antisense intergenic RNA is highly expressed in breast cancer (Gupta et al., 2010). The long noncoding RNA metastasis associated in lung adenocarcinoma transcript 1, which is highly expressed in non-small-cell lung cancer tissue, is related to patient survival (Schmidt et al., 2011). Gibb et al. (2011a, 2011b) reported the aberrant expression of lncRNAs in OPLs compared with normal oral mucosa using serial analysis of gene expression (SAGE). However, the role of aberrant expression of lncRNAs in the molecular pathogenesis of OPLs remains largely unknown.
In this study, the GSE31021 dataset constructed by Gibb et al. (2011a, 2011b) and deposited in Gene Ontology Ominibus (GEO) was selected. Analysis was carried out using a series of bioinformatics methods. First, we identified differentially expressed lncRNAs and genes (DEGs), and predicted the target genes of lncRNAs. Then, gene ontology (GO) function and pathway enrichment analyses [Kyoto Encyclopedia of Genes and Genomes (KEGG)] were carried out to determine the target genes of lncRNAs. The resulting findings can help better understand OPLs and the underlying molecular mechanisms in lncRNA perspective, providing potential targets for further studies on strategies of lncRNA-based early diagnostics as well as justification of intervention timing and measures.
Materials and methods
SAGE datasets
This study was approved by the institutional review board of Beijing Stomatological Hospital Capital Medical University. To identify differentially expressed lncRNAs in OPLs, the SAGE dataset GSE31021 was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31021); it includes tissue samples of 10 OPLs (four severe, four moderate, and two mild dysplasia cases) with 10 SAGE libraries generated from oral biopsies. The expression profiling of the normal oral mucosa tissue was determined from the GSE8127 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE8127), which comprises six samples with three SAGE libraries generated from oral epithelial cells obtained from tongue brushes and three others obtained from oral biopsies. The two datasets were generated by the GPL4 platform (SAGE:10:NlaIII:Homo sapiens including 265 577 tag sequence) from nine individual patients with no histological evidence of invasion. Sixteen SAGE libraries deposited under GSE31021 and GSE8127 were constructed by the same research group according to the MicroSAGE protocol (http://www.sagenet.org/protocol/index.htm). The total tags sequenced per library ranged from 126 000 to 191 700, averaging 160 000 SAGE tags, excluding linker and duplicate ditags.
Data preprocessing
According to the information available in NCI (ftp://ftp1.nci.nih.gov/pub/SAGE/HUMAN/), tags specifically mapped to genes were screened out by commanding shell. On the basis of the lncRNA sequence information derived from GENCODE (ftp://ftp.sanger.ac.uk/pub/gencode/Gencode_human/release_18/gencode.v18.lncRNA_transcripts.fa.gz) (Harrow et al., 2012), unmapped tags were aligned with lncRNA sequences and the corresponding lncRNAs were screened out. When multiple tags mapped to the same lncRNA, tags were collapsed by summing up the tag counts to estimate the expression value of the lncRNA. Predicted mRNAs and lncRNAs were annotated on the basis of data available in the National Center of Biotechnology Information database.
Screening of differentially expressed lncRNAs and DEGs
Tag names were converted into genes or lncRNAs during the annotation. The remaining missing values were imputed using the k-nearest-neighbor algorithm, with the default k value set at 10 (Hastie et al., 2001). The expression levels of multiple tags corresponding to the same gene were averaged. The lncRNA expression matrices for the 16 samples were normalized using Robust Microarray Averaging. Differentially expressed lncRNAs and DEGs were screened with the LIMMA package in R (Smyth, 2005). The false discovery rate was determined by the Benjamini–Hochberg (BH) method using Multtest package (Pollard et al., 2008). False discovery rate less than 0.05 and |log2(fold change)|>0.585 were selected as criteria for lncRNA and DEG screening. The heat map of DEGs was obtained using the gplots package in R to assess their distribution in different samples.
Prediction of target genes of differentially expressed lncRNAs
To decipher the role of lncRNAs in the molecular pathogenesis of OPLs, the lncRNA-gene coexpression network was assessed. Coexpression levels of lncRNAs and genes were used to predict potential target genes for lncRNAs. The Pearson correlation coefficients of lncRNAs and genes were calculated using R, and interactions with Pearson’s correlation coefficient more than 0.95 and P value of less than 0.05 were selected to predict the target genes of lncRNAs.
Functional analysis of target genes regulated by lncRNAs
The target genes of lncRNAs were subjected to GO and KEGG enrichment analyses using Database for Annotation, Visualization, and Integrated Discovery (Dennis et al., 2003). The Enrichment Map Plugin in the Cytoscape was used to present enrichment results (Shannon et al., 2003; Merico et al., 2010).
Results
DEGs and differentially expressed lncRNAs
A total of 265 577 tags were identified in the GPL4 platform, with 43 455 mapped to 16 233 genes. After aligning the unmapped tags to the lncRNA sequences, 1651 tags were mapped to 1409 lncRNAs.
The box plots of expression data were distributed at similar intervals and densities after normalization, indicating successful adjustment (Fig. 1). The normalized data were used for further analysis. A total of 674 DEGs and differentially expressed lncRNAs showing statistical significance (P<0.05) were identified, including 648 downregulated DEGs, 16 upregulated DEGs, and 10 downregulated lncRNAs. No upregulated lncRNAs were found. The heat map (Fig. 2) shows a clear separation between OPLs and normal samples on the basis of the DEGs.
Fig. 1.
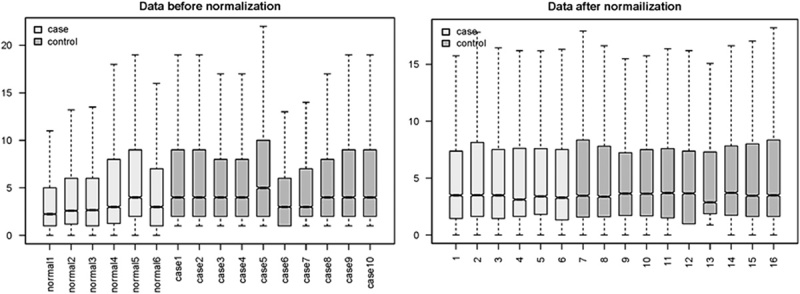
Box plots of expression data before and after normalization using Robust Microarray Averaging.
Fig. 2.
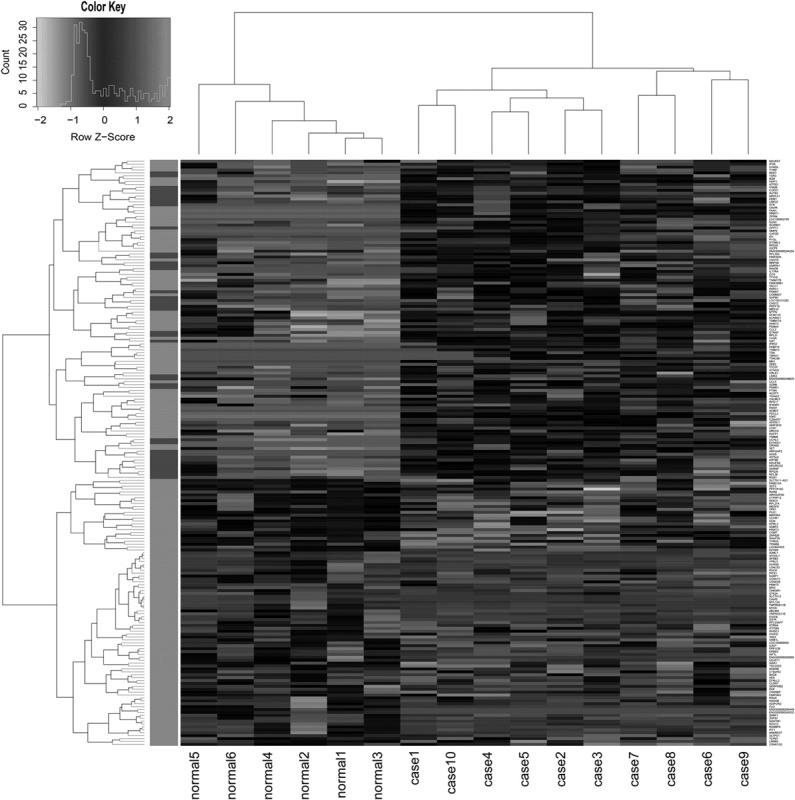
Heat map of differentially expressed genes. In the figure, ‘normal 1–3’ represent biopsies from control samples and ‘normal 4–6’ are brushes from control samples; ‘cases 4–5’ represent mild dysplasia; ‘cases 1, 2, 3, and 10’ represent severe dysplasia, and ‘cases 6–9’ represent moderate dysplasia. Gene expression ratios were displayed by applying progressively brighter shades of black (downregulated) or white (upregulated) to normalized values that increasingly deviate from zero.
Target genes of lncRNAs
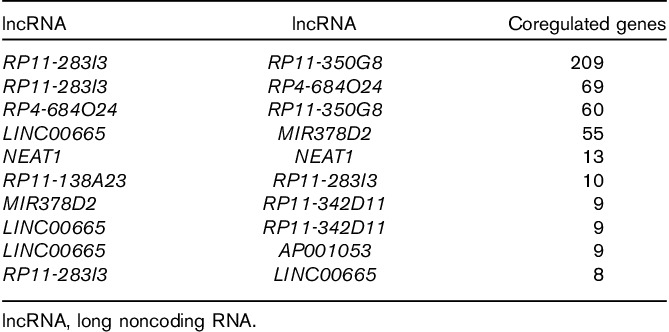
Interactions with Pearson’s correlation coefficient more than 0.95 and P value of less than 0.05 were determined. The coexpression network of lncRNAs and DEGs is presented in Fig. 3. Finally, we obtained 946 interactions for 10 downregulated lncRNAs and 531 downregulated DEGs. The top 10 interactions with the most target genes are listed in Fig. 4. RP4-684O24, RP11-283I3, and RP11-350G8 coregulated 60 DEGs, and LINC00665 and MIR378D2 coregulated 55 DEGs (Table 1). Excluding NEAT1, all the predicted lncRNAs coregulating target genes were new.
Fig. 3.
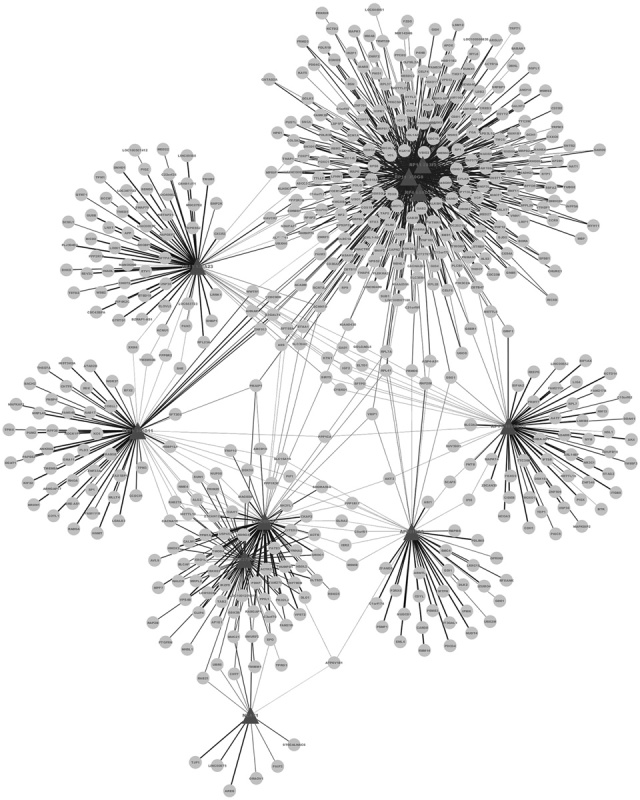
Coexpression network of lncRNAs and differentially expressed genes (DEGs). Nodes represent downregulated DEGs. Angles represent the lncRNAs coregulating DEGs.
Fig. 4.
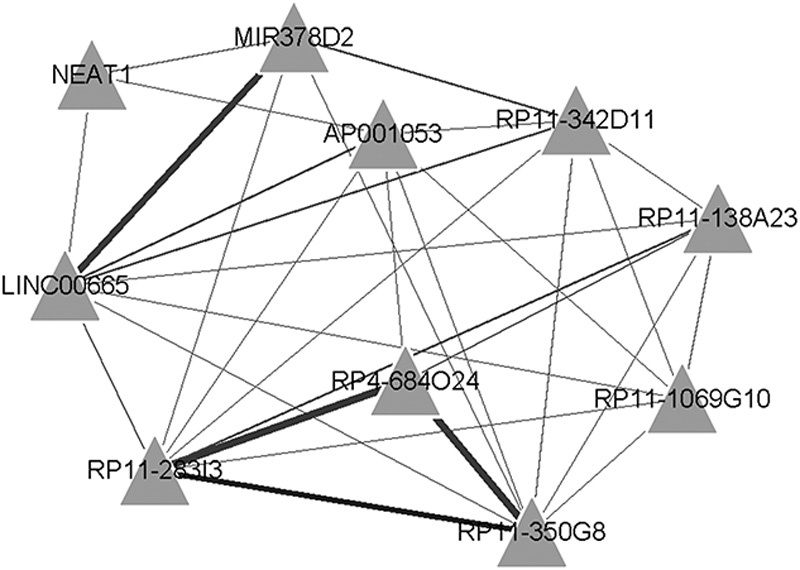
Interaction network of long noncoding RNAs (lncRNAs) and their target genes (top 10 interactions with the most target genes). Angles represent lncRNAs. Line thickness reflects the number of target genes.
Table 1.
List of coregulated lncRNAs and the top 10 target genes
GO function and KEGG enrichment findings
Cytoscape was used to construct the functional network. The target genes coregulated by RP4-684O24, RP11-283I3, and RP11-350G8 mediated the immune response, protein complex biogenesis, and assembly. They were mainly located in the membrane and insoluble cellular fractions, and manifested mannosyl-oligosaccharide mannosidase, anion binding, and mannosidase activities (Table 2 and Fig. 5). The target genes coregulated by LINC00665 and MIR378D2 mainly regulated ubiquitin-dependent protein catabolism and response to calcium ion. They are mainly located in the cytosol, axons and dendrites, and synapses; they were predicted to bind protein C. They showed ubiquitin-protein ligase and small conjugating protein ligase activity, and participated in neurotrophin signaling, Vibrio cholera infection, and the insulin signaling pathway (Table 3 and Fig. 6).
Table 2.
Enrichment of target genes coregulated by RP4-684O24, RP11-283I3, and RP11-350G8
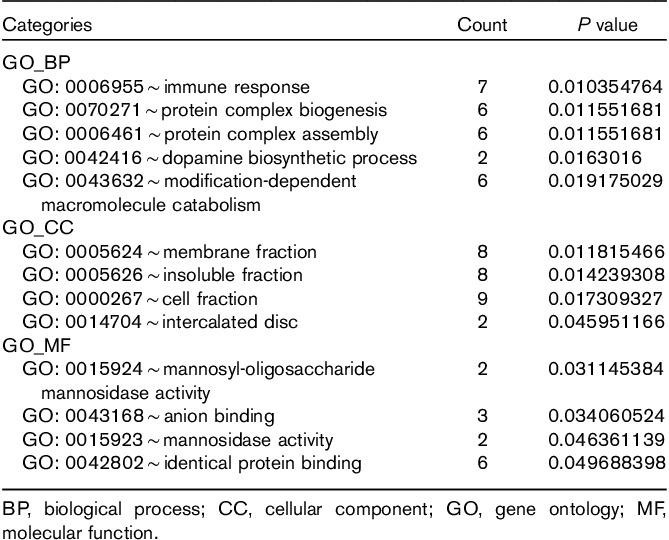
Fig. 5.
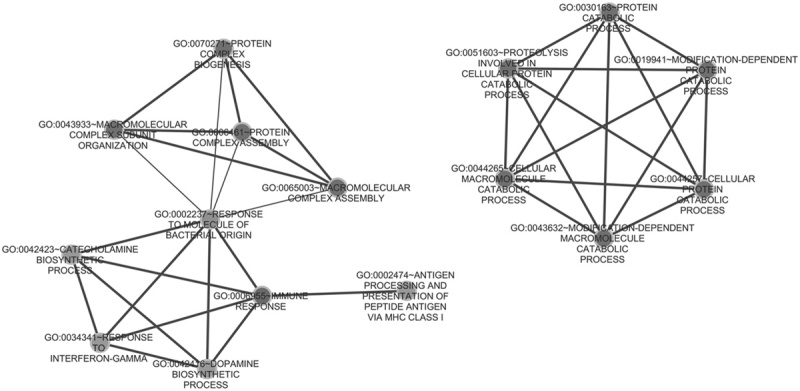
Gene ontology (GO) enrichment function modules of target genes coregulated by RP4-684O24, RP11-283I3, and RP11-350G8.
Table 3.
Enrichment of target genes coregulated by LINC00665 and MIR378D2
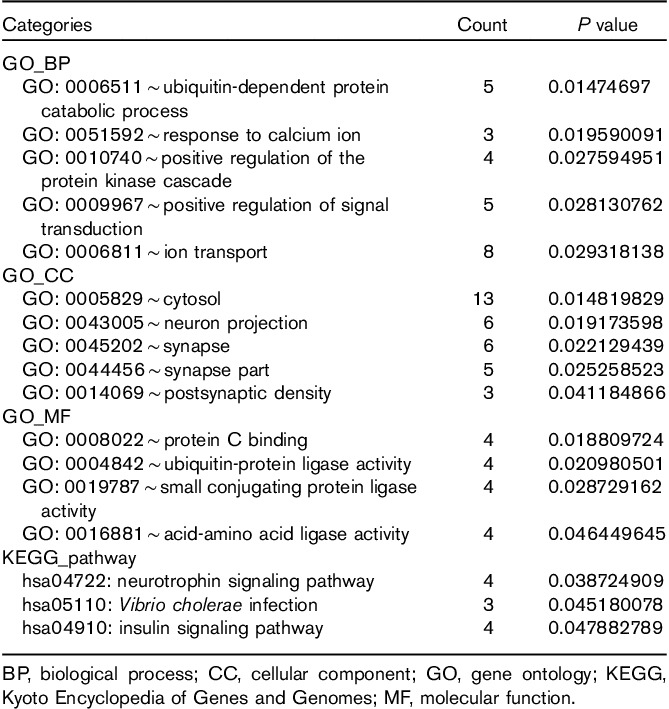
Fig. 6.
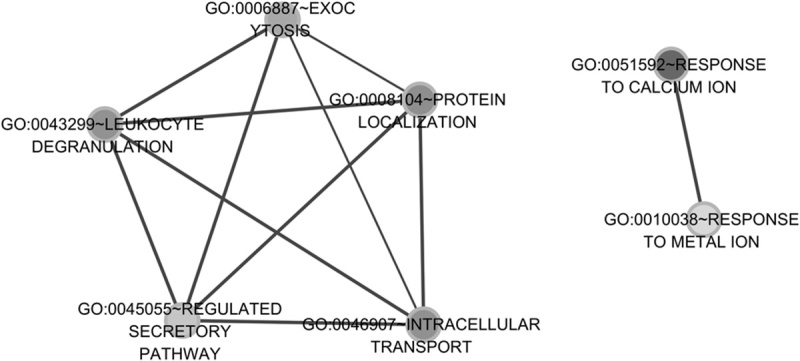
Gene ontology (GO) enrichment function modules of target genes coregulated by LINC00665 and MIR378D2.
Discussion
The malignant transformation of OPLs may continue for a few years to decades. Such patients require long-term follow-up. Epithelial dysplasia is a key point in disease progression. Unfortunately, there are no reliable indicators of transforming OPLs for doctors to decide whether operation or other interventions should be carried out clinically. Meanwhile, carcinogenesis is a genetic process in which molecular changes occur earlier than histomorphological changes; indeed, carcinogenesis leads to changes in morphology and cellular behavior (Fatima et al., 2015). Therefore, the most efficient way to screen for high-risk lesions and prevent malignant transformation is to identify genetic alterations that might typify epithelial dysplasia.
In this study, we carried out gene expression profile analysis using high-throughput bioinformatics methods to identify DEGs in OPLs compared with normal controls. The main advantages of using this approach are as follows: (a) the samples used in the SAGE dataset were derived from patients with OED, which indicates high-risk lesions. We focused on lncRNA expression in OPLs and filtered crucial lncRNAs that could be used as biomarkers for the accurate detection of high-risk lesions. As with histopathological guidance, a reliable and objective molecular marker can help doctors make the right clinical decisions on OPLs without biopsies, which is more acceptable to patients. (b) Unlike microarrays, SAGE is a sequence-based transcriptome profiling technique that provides the absolute abundance values of transcripts according to sequence tag counts, thus enabling the identification of DEGs and novel lncRNAs by direct comparison of gene expression levels across SAGE data without disparity among housekeeping genes. (c) It is a direct and simple method to identify DEGs and lncRNAs by calculation fold changes and P-values, especially for the preliminary screening of target genes using SAGE datasets; however, the results require further validation with large-scale clinical samples.
Here, a total of 1409 lncRNAs were identified; meanwhile, 664 DEGs (including 648 downregulated and 16 upregulated genes) and 10 differentially expressed lncRNAs (including 10 downregulated and no upregulated lncRNAs) were screened from the expression profiling of OPLs compared with the normal oral mucosa. These results corroborated the heat map. LncRNAs are recognized widely to play important roles in many biological processes and to show aberrant expression in tumors (Hanahan and Weinberg, 2011). Therefore, considerable research has focused on lncRNAs as potential biomarkers for various tumor types, including OSCC (Mallardo et al., 2008; Guo et al., 2010; Schmidt et al., 2011; Tang et al., 2013; Zhao et al., 2014). Theoretically, the DEGs and lncRNAs identified can help clinicians differentiate the OPLs with dysplasia from those with low-risk malignant transformation. Nevertheless, this is far from clinical requirements and in-depth studies are warranted to translate potential biomarker candidates into clinically available diagnostic tools in the future.
The lncRNA NEAT1 was most highly expressed in OPLs in previous SAGE analyses (Gibb et al., 2011a, 2011b). NEAT1 plays an architectural role in the structure of paraspeckles (Clemson et al., 2009) and is involved in cancer metastasis (Gutschner and Diederichs, 2012). NEAT1 is overexpressed in lung cancer metastasis compared with matched primary tumors (Zhao et al., 2014), hepatocellular carcinoma (Mallardo et al., 2008), and cervical cancer (Guo et al., 2010). NEAT1 is highly expressed in saliva from patients with oral squamous cell carcinoma (Tang et al., 2013). Thus, NEAT1 may play an important role in OPLs and represent a target gene for the diagnosis of OPL.
The target genes of RP4-684O24, RP11-283I3, and RP11-350G8 were significantly enriched in immune response, protein complex biogenesis, and mannosyl-oligosaccharide mannosidase activity. The target genes of LINC00665 and MIR378D2 were significantly enriched in the ubiquitin-dependent protein catabolic process, ubiquitin-protein ligase activity, and the neurotrophin signaling pathway. Epidermal growth factor receptor was not only expressed in OPLs but also in lesions with mild dysplasia. MUC-1 and carcinoembryonic antigen were expressed in most patient samples. Further, autologous peripheral blood mononuclear leukocytes were stimulated to react with heterologous OPLs (Young et al., 2007). In addition, circulating immune complex levels are increased in oral precancer and cancer patients (Khanna and Karjodkar, 2006). Mannosyl-oligosaccharide mannosidase plays a central role in the N-glycosylation pathway. Abnormal glycosylation is associated with progression, metastasis, and poor clinical outcomes in breast, colon, and skin cancers (Goss et al., 1995; Dennis et al., 1999; Van den Elsen et al., 2001). Ubiquitin is an important signaling messenger in cell proliferation, cell cycle, and apoptosis (Hoeller et al., 2006). The ubiquitin ligase subunit S-phase kinase-associated protein shows oncogenic potential in breast epithelial cells and is highly expressed in breast carcinoma (Signoretti et al., 2002). Neurotrophins have been reported in cancers of the lung, bladder, and prostate (Okragly et al., 1999; Ricci et al., 2001; Satoh et al., 2001). They use Trk tyrosine kinase receptors and the p75 neurotrophin receptor (p75NTR) to regulate the growth, development, survival, and repair of the nervous system. Neurotrophin nerve growth factor enhances cell survival by two distinct signaling pathways mediated by p75NTR and nuclear factor-kB (Descamps et al., 2001). Activated fibroblasts increase the levels of the brain-derived neurotrophic factor, which interacts with neurotrophin receptor B in oral tumor progression (Dudás et al., 2011). However, the roles of mannosidase, ubiquitin, and neurotrophin in OPLs have never been reported. We conclude that they may be related to OPLs.
In brief, lncRNAs and DEGs mediate neurotrophin signaling in the pathogenesis of OPLs. In addition, mannosidase and ubiquitin may play important roles in OPLs. On the basis of the current understanding of molecular events, it may be possible to prevent the occurrence of OSCC by reversing the lesions by means of gain or loss of function of specific lncRNAs in epithelial dysplasia.
In this study, the two SAGE libraries used were constructed by the same research group using the same patient cohort, which provides a homogeneous and comparable basis for data analysis. Moreover, after normalization by tags per million value, the quality of raw data was confirmed from a consistent tag abundance distribution, a good match degree of every tag between the two samples, and the high quality of reads. However, methodological deficiencies of our approach still exist. Because of the complex diversity of gene networks, a huge volume of data, and sequencing errors, the strategy of screening DEGs according to fold change and linking them to biology activity of OPLs cannot provide a factual mechanism of how DEGs and lncRNAs are involved in OPLs. Clinical validation and functional experiments should be conducted to correct screening errors and investigate the regulatory mechanisms of coexpressed lncRNAs and DEGs.
Nevertheless, our findings indicated that some lncRNAs related to dysplasia were identified in OPLs using SAGE profiling analysis. The lncRNA NEAT1 may play an important role in OPLs. However, most of the lncRNAs found are new, with unknown functions. Although the functions of target genes of lncRNAs were analyzed in GO and the KEGG pathway, additional studies are needed to confirm the findings and elucidate their actual mechanisms in the pathogenesis of OPLs, laying the foundation for developing OSCC prevention strategies on the basis of lncRNAs.
Acknowledgements
This work was supported by grants from the Beijing Municipal Administration of Hospitals Key Medical Project (ZYLX201407) and the National Natural Science Foundation of China (81372897).
Conflicts of interest
Zheng Sun is currently receiving a grant (ZYLX201407) from the Beijing Municipal Administration of Hospitals Key Medical Project and a grant (81372897) from the National Natural Science Foundation of China. For the remaining authors, there are no conflicts of interest.
Footnotes
Hongcheng Jia and Xuan Wang contributed equally to the writing of this article.
References
- Agarwal S, Mathur M, Srivastava A, Ralhan R. (1999). MDM2/p53 co-expression in oral premalignant and malignant lesions: potential prognostic implications. Oral Oncol 35:209–216. [DOI] [PubMed] [Google Scholar]
- Arduino PG, Bagan J, El-Naggar AK, Carrozzo M. (2013). Urban legends series: oral leukoplakia. Oral Dis 19:642–659. [DOI] [PubMed] [Google Scholar]
- Boyd SD. (2008). Everything you wanted to know about small RNA but were afraid to ask. Lab Invest 88:569–578. [DOI] [PubMed] [Google Scholar]
- Clague J, Lippman SM, Yang H, Hildebrandt MA, Ye Y, Lee JJ, et al. (2010). Genetic variation in microRNA genes and risk of oral premalignant lesions. Mol Carcinog 49:183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. (2010). Non-coding RNAs: meet thy masters. Bioessays 32:599–608. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Granovsky M, Warren CE. (1999). Protein glycosylation in development and disease. Bioessays 21:412–421. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3. [PubMed] [Google Scholar]
- Descamps S, Toillon RA, Adriaenssens E, Pawlowski V, Cool SM, Nurcombe V, et al. (2001). Nerve growth factor stimulates proliferation and survival of human breast cancer cells through two distinct signaling pathways. J Biol Chem 276:17864–17870. [DOI] [PubMed] [Google Scholar]
- Dudás J, Bitsche M, Schartinger V, Falkeis C, Sprinzl GM, Riechelmann H. (2011). Fibroblasts produce brain-derived neurotrophic factor and induce mesenchymal transition of oral tumor cells. Oral Oncol 47:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, et al. (2005). Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102:3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima R, Akhade VS, Pal D, Rao SM. (2015). Long noncoding RNAs in development and cancer:potential biomarkers and therapeutic targets. Mol Cell Ther 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller CD, Wang SJ, Thomas CR, Jr., Hoffman HT, Weber RS. (2007). Conditional survival in head and neck squamous cell carcinoma: results from the SEER dataset 1973–1998. Cancer 109:1331–1343. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Enfield KS, Stewart GL, Lonergan KM, Chari R, Ng RT, et al. (2011a). Long non-coding RNAs are expressed in oral mucosa and altered in oral premalignant lesions. Oral Oncol 47:1055–1061. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Brown CJ, Lam WL. (2011b). The functional role of long non-coding RNA in human carcinomas. Mol cancer 10:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470:284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss PE, Baker MA, Carver JP, Dennis JW. (1995). Inhibitors of carbohydrate processing: a new class of anticancer agents. Clin Cancer Res 1:935–944. [PubMed] [Google Scholar]
- Guo F, Li Y, Liu Y, Wang J, Li G. (2010). Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 42:224–229. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T, Diederichs S. (2012). The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9:703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011). Hallmarks of cancer: the next generation. Cell 144:646–674. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. (2012). GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Narasimhan B, Chu G. (2001). Impute: imputation for microarray data. Bioinformatics 17:520–525.11395428 [Google Scholar]
- Hoeller D, Hecker CM, Dikic I. (2006). Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer 6:776–788. [DOI] [PubMed] [Google Scholar]
- Huarte M, Rinn JL. (2010). Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C. (2011). Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol 22:343–350. [DOI] [PubMed] [Google Scholar]
- Khanna SS, Karjodkar FR. (2006). Circulating Immune Complexes and trace elements (Copper, Iron and Selenium) as markers in oral precancer and cancer: a randomised, controlled clinical trial. Head Face Med 2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumerman H, Freedman P, Kerpel S. (1995). Oral epithelial dysplasia and the development of invasive squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 79:321–329. [DOI] [PubMed] [Google Scholar]
- Mallardo M, Poltronieri P, D’Urso OF. (2008). Non-protein coding RNA biomarkers and differential expression in cancers: a review. J Exp Clin Cancer Res 27:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD. (2010). Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okragly AJ, Niles AL, Saban R, Schmidt D, Hoffman RL, Warner TF, et al. (1999). Elevated tryptase, nerve growth factor, neurotrophin-3 and glial cell line-derived neurotrophic factor levels in the urine of interstitial cystitis and bladder cancer patients. J Urol 161:438–442. [PubMed] [Google Scholar]
- Papadimitrakopoulou V, Izzo J, Lippman SM, Lee JS, Fan YH, Clayman G, et al. (1997). Frequent inactivation of p16INK4a in oral premalignant lesions. Oncogene 14:1799–1803. [DOI] [PubMed] [Google Scholar]
- Petti S. (2003). Pooled estimate of world leukoplakia prevalence: a systematic review. Oral Oncol 39:770–780. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Ge Y, Dudoit S. (2008). multtest: Resampling-based multiple hypothesis testing R package version 1. New York: Springer. [Google Scholar]
- Ricci A, Greco S, Mariotta S, Felici L, Bronzetti E, Cavazzana A, et al. (2001). Neurotrophins and neurotrophin receptors in human lung cancer. Am J Respir Cell Mol Biol 25:439–446. [DOI] [PubMed] [Google Scholar]
- Satoh F, Mimata H, Nomura T, Fujita Y, Shin T, Sakamoto S, et al. (2001). Autocrine expression of neurotrophins and their receptors in prostate cancer. Int J Urol 8:S28–S34. [DOI] [PubMed] [Google Scholar]
- Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, et al. (2011). The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small-cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6:1984–1992. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoretti S, Di Marcotullio L, Richardson A, Ramaswamy S, Isaac B, Rue M, et al. (2002). Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J Clin Invest 110:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smyth GK.Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. (2005). Limma: linear models for microarray data. Bioinformaticsand computational biology solutions using R and Bioconductor. New York: Springer; 397–420. [Google Scholar]
- Tang H, Wu Z, Zhang J, Su B. (2013). Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep 7:761–766. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Spitale RC, Chang HY. (2011). Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res 71:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Elsen JM, Kuntz DA, Rose DR. (2001). Structure of Golgi α‐mannosidase II: a target for inhibition of growth and metastasis of cancer cells. EMBO J 20:3008–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MRI, Neville BW, Chi AC, Lathers DM, Gillespie MB, Day TA. (2007). Oral premalignant lesions induce immune reactivity to both premalignant oral lesions and head and neck squamous cell carcinoma. Cancer Immunol Immunother 56:1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, An Y, Liang Y, Xie X. (2014). Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci 18:1930–1936. [PubMed] [Google Scholar]


