Abstract
RpoS and RpoN are two alternative sigma factors typically associated with general stress responses in bacteria. To date, there has been no experimental evidence that RpoS and RpoN can directly control the expression of one another. Herein, using a combined strategy of gene disruption and genetic complementation targeting rpoN and rpoS in Borrelia burgdorferi strain 297, we describe a regulatory network for B. burgdorferi. In this network, RpoN controls the expression of RpoS, which, in turn, governs the expression of two important membrane lipoproteins, outer surface protein C and decorin-binding protein A, and likely other proteins of B. burgdorferi. Our findings provide a foundation for elucidating further key regulatory networks that potentially impact many aspects of B. burgdorferi's parasitic strategy, host range, and virulence expression.
A remarkable feature of Borrelia burgdorferi, the agent of Lyme disease, is its ability to thrive in diverse arthropod (Ixodes ticks) and mammalian (rodent) hosts (1). Upon exposure to blood, B. burgdorferi migrates from the tick midgut to the salivary glands and then is injected into mammalian dermal tissue (2, 3). During these processes, dramatic adaptive changes occur that are reflected in altered protein profiles of the spirochete, such as the reciprocal down-regulation of outer surface (lipo)protein (Osp) A and the up-regulation of OspC (4, 5). OspC is a circular plasmid (cp26)-encoded (6), 22-kDa lipoprotein (7) that varies in sequence (8, 9) and may or may not be an immune target, depending on expression levels and the strain of B. burgdorferi (10, 11). Although intensively studied (12, 13), the function of OspC remains unknown, albeit its up-regulation during tick feeding and preponderance among spirochetes exposed to blood suggest that it facilitates B. burgdorferi migration to tick salivary glands and/or transmission into mammalian tissues (14–16). Another lipoprotein, decorin-binding protein A (DbpA), is purported to facilitate the adherence of B. burgdorferi to extracellular matrix as the spirochete invades mammalian tissue (17). Studies that address the regulation of these lipoproteins will assist in clarifying their roles in Lyme disease pathogenesis and perhaps in elucidating their physiological functions.
A number of environmental cues (e.g., temperature, pH, and spirochete cell density) have been implicated in influencing differential antigen expression in B. burgdorferi (4, 18–22). More recently, it has been shown that the in vitro cultivation of virulent B. burgdorferi strain 297 (297) at elevated temperature (37°C), reduced pH (pH 6.8), and increased spirochete cell density, parameters ostensibly that mimic conditions of tick engorgement, caused an up-regulation of OspC, DbpA, OspF, and Mlp-8 (group I proteins) with a concomitant down-regulation of OspA, P22, and Lp6.6 (group II proteins) (23). Conditions that induced the group I proteins also induced the synthesis of RpoS (σs; σ38), one of two alternative sigma factors predicted to be present in B. burgdorferi B31 (B31) (24). Although conventionally associated with general stress responses (25), a role(s) for RpoS in the life cycle of B. burgdorferi remains unknown. However, the simultaneous induction of rpoS and the group I genes prompted the hypothesis that group I-like genes in 297 may be controlled through RpoS (23).
RpoN (σN; σ54) is another important sigma subunit that (i) does not share obvious homologies with other sigma factors, (ii) directs the core enzyme to a class of distinct −24/−12 promoters, and (iii) mediates enhancer-dependent transcription (26). Most bacteria possess one copy of rpoN, which generally is constitutively expressed and not essential (26). Because rpoN-mediated transcription has been associated with bacterial pathogenicity (27–30), we examined the potential influence of rpoN, as well as rpoS, on patterns of adaptive gene expression in low-passage 297. Using a combined strategy of targeted gene disruption and genetic complementation, it was found that RpoN regulates the expression of RpoS, which, in turn, influences the expression of OspC and DbpA. These studies provide a foundation for elucidating further key regulatory networks that potentially impact many aspects of B. burgdorferi's parasitic strategy.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
A virulent population of 297 (31) was recovered from needle-inoculated mice; generally, once-passaged organisms were used. Unless otherwise indicated, borreliae were cultivated at 34°C in complete Barbour-Stoenner-Kelly medium (BSK) (Sigma) (32) under an atmosphere of 1% CO2. pBSK solid medium (33) supplemented with either erythromycin (Erm) (0.06 μg/ml) or kanamycin (Kan) (170 μg/ml) was used to select for transformants. All plasmids and recombinant constructs (Table 1) were propagated in Escherichia coli DH5α (GIBCO/Life Technologies, Grand Island, NY).
Table 1.
Recombinant plasmids
| Plasmid | Description | Source |
|---|---|---|
| pGEM-T | TA cloning vector, high copy number, Ampr | Promega |
| pCR2.1 | TA cloning vector, high copy number, Ampr, Kanr | Invitrogen |
| pUC4K | Aminoglycoside 3′-phosphotransferase gene from Tn903 (kan), Ampr, Kanr | Amersham Pharmacia |
| pJRS233 | Ermr | K. McIver (Univ. of Texas Southwestern) |
| pJRS525 | Specr, wide host range cloning vector | K. McIver (Univ. of Texas Southwestern) |
| pFAC | pGEM-T, for SP6 transcription of ospC- and flaB-specific competitor RNA | This study |
| pALH133 | pGEM-T, ermC from pJRS233, Ampr, Ermr | This study |
| pALH227 | pJRS525, Δ[spec, AlwNI-FspI(965 bp)] Ω[PflaB-kan, FspI–AlwNI (1,158 bp)], Kanr | This study |
| pALH251 | pALH227Ω[PflgB-rpoS, BamHI–NcoI (1,224 bp)], Kanr | This study |
| pALH293 | pALH227Ω[PrpoN-rpoN, BamHI–NcoI (1,628 bp)], Kanr | This study |
| pALH362 | pGEM-T, 4,631-bp fragment (analogous B31 chromosome 810459–815090), Ampr | This study |
| pALH364 | pGEM-T, 5,156-bp fragment (analogous B31 chromosome 468016–473172), Ampr | This study |
| pALH386 | pALH362, except rpoS∷ermC [BbsI (1 kb)], Ampr, Ermr | This study |
| pALH394 | pALH364, except rpoN∷ermC [AflII (1 kb)], Ampr, Ermr | This study |
| pALH400 | pALH394, except ermC∷PrpoN-rpoN-PflaB-kan [BclI (3 kb)], Ampr, Erms, Kanr | This study |
DNA Manipulations.
All recombinant DNA experiments and the use of antibiotic resistance markers in 297 were reviewed and approved by the University of Texas Southwestern Biological and Chemical Safety Advisory Committee. Plasmid DNA used for electroporation of 297 was purified by using the StrataPrep EF Plasmid Midiprep Kit (Stratagene). The design of oligonucleotide primers (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) to amplify 297 genes was based on published gene sequences for B31 (24); DNA amplifications were performed by using the Expand High Fidelity PCR System (Roche Diagnostics).
Construction of Suicide Plasmids Containing Disrupted Genes.
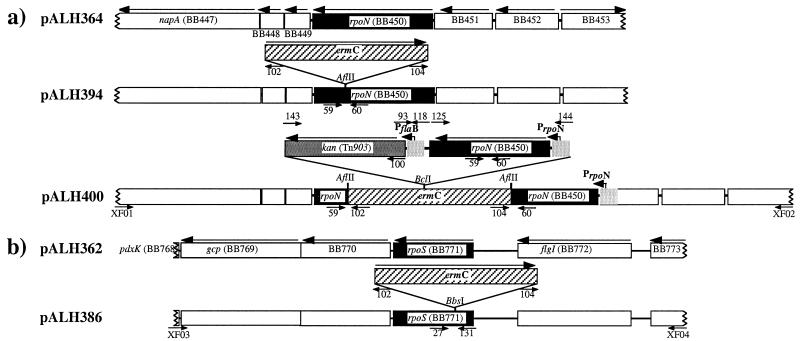
pGEM-T [encoding ampicillin (Amp) resistance] was the basis for all suicide plasmids; pALH394 and pALH386 were constructed with inactivated 297 gene homologs of rpoN (BB0450) and rpoS (BB0771), respectively (Fig. 1) (24). For rpoN, a 5.1-kb DNA fragment with flanking sequence was obtained by amplification (priXF01, priXF02) and cloned to yield pALH364 (Fig. 1a). For rpoS, pALH362 contained a 4.6-kb fragment that was obtained by using primers priXF03 and priXF04 (Fig. 1b). Unique restriction sites within each target structural gene were chosen as insertion sites for disruption.
Figure 1.
Strategy for gene inactivation and complementation of rpoN (a) and rpoS (b) in 297. rpoN and rpoS (black solid boxes) were first cloned in pGEM-T (pALH364 and pALH362, respectively). Only the relevant portions of the plasmids are shown (labeled at the left). rpoN and rpoS were insertionally disrupted with ermC (diagonal stripes) (pALH394 and pALH386, respectively). For complementation of rpoN, suicide plasmid pALH400 was constructed as described in Materials and Methods. Relevant restriction sites are shown, and relevant promoters are indicated by light gray boxes. Short arrows denote primers used for PCR (Table 2).
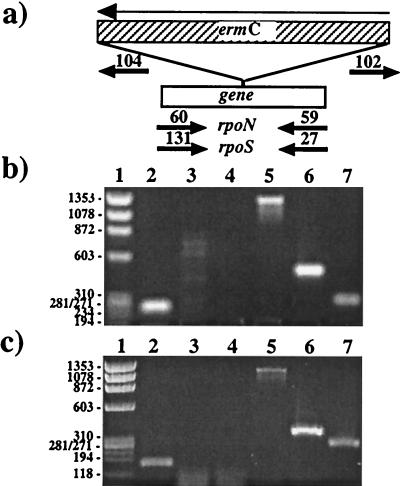
The Erm resistance gene, ermC, was chosen as a selectable marker for the genetic manipulation of 297 based on its prior use in pGK12 for B31 (34). The source of ermC was pJRS233, a derivative of the Staphylococcus aureus plasmid pE194 (34, 35). ErmC was PCR-amplified with its predicted promoter by using primers to introduce appropriate restriction sites for insertion into rpoN (AflII, priAH78 and priAH79) or rpoS (BbsI, priAH133 and priAH134), yielding pALH394 (Fig. 1b) and pALH386 (Fig. 1b), respectively. To minimize potential polar effects, ermC was inserted opposite in orientation to the gene being disrupted, which was confirmed by PCR using primers complementary to ermC (priAH102 and priAH104) and primers flanking the insertion site of the resistance marker (see Fig. 3a); the latter primers were priAH59 and priAH60 for rpoN (see Fig. 3b) and priAH131 and priAH27 for rpoS (see Fig. 3c).
Figure 3.
PCR analysis of rpoN and rpoS mutants. (a) Schematic of PCR primer pairs (short arrows). (b and c) Agarose gel patterns of amplicons for wild type (lanes 2–4), (b) a rpoN mutant, or (c) a rpoS mutant (lanes 5–7). Lanes 1 contain DNA markers of ΦX174/HaeIII. Gene disruption by ermC results in an increased size of the amplicons (compare lanes 2 and 5). A combination of ermC-specific and flanking primers yielded only products for the mutants (lanes 6 and 7), but not for 297 (lanes 3 and 4). Lanes 3 and 6: priAH102 (arrow 102) in combination with (b) priAH59 (arrow 59) or (c) priAH27 (arrow 27). Lanes 4 and 7: priAH104 (arrow 104) in combination with (b) priAH60 (arrow 60) or (c) priAH131 (arrow 131).
Electroporation of 297.
The 297 cells were made electrocompetent essentially as described (33) except that spirochetes were harvested from the midlogarithmic phase of growth (about 1 × 107 spirochetes per ml). Transformation (34) was carried out by using 15–20 μg of circular plasmid DNA to electroporate ≈1 × 109 bacteria (in 60 μl of electroporation buffer). After recovery, aliquots of 0.5, 1.0, and 1.5 ml (0.5–3.0 × 108 spirochetes) were added to ≈15 ml of pBSK (with antibiotic) and plated. Colonies appeared 12–20 days after plating. Transformation efficiencies tended to be about 2 × 10−8, determined from the ratio of antibiotic-resistant colony-forming units (cfu) relative to the total number of cfu per transformed culture, in the context of an overall plating efficiency of about 70%.
PCR Confirmation of Gene Disruptions.
To confirm marker exchange, Ermr colonies were picked and propagated in 1.5 ml of BSK-H containing Erm. Spirochetes (200 μl of culture) were collected by centrifugation. The cell pellet was suspended in 20 μl of sterile water and 3-μl aliquots were microwaved in 200-μl thin-wall PCR tubes and subjected to PCR. The PCR strategy (see Fig. 3c) was as described for confirmation of plasmids pALH394 and pALH386 (Fig. 1). For all Ermr clones tested, only PCR products indicative of disrupted genes (from double crossovers) were obtained.
Construction of Suicide and Shuttle Plasmids for Genetic Complementation.
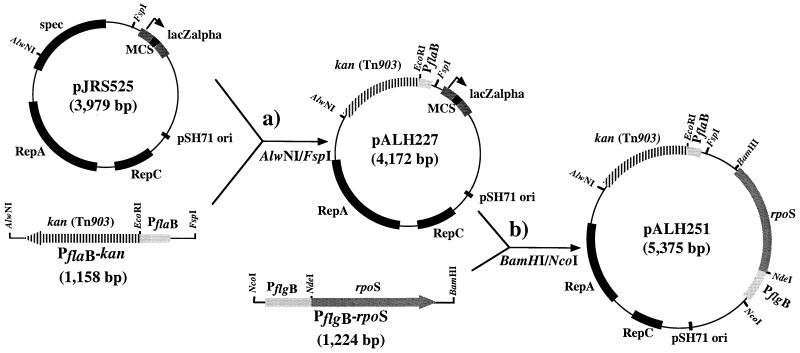
In each of two strategies for complementation, a Kan resistance gene (kan), derived from pUC4K containing the transposon Tn903 (Amersham Pharmacia, accession number X06404), was used as a selectable marker; its use for the genetic manipulation of B. burgdorferi has been described (36, 37). Promoterless kan was linked to the constitutively expressed 297 flaB promoter (PflaB) (36) via an EcoRI site; the kan gene and PflaB were PCR-amplified with primers that introduced an EcoRI site upstream of kan (priAH100, priAH119) and downstream of PflaB (priAH93, priAH118). The 224-bp PflaB region and the 934-bp kan fragments were cloned separately into pGEM-T (lacks an EcoRI site). Both plasmids were linearized with EcoRI and combined in a ligation reaction. The ligation products were subjected to PCR by using the forward primer for PflaB (priAH118) and the reverse primer for kan (priAH119), thereby selectively amplifying the desired product [1,158 bp with a junction of CATGGAGGAATTCGTTATGkan (EcoRI site underlined, RBSflaB bolded, and start codon of kan italicized)]. This amplicon was then inserted into FspI- and AlwNI-digested pJRS525 (38), which contains the same origin of replication as the recently described B. burgdorferi shuttle vector pGK12 (34). The resulting plasmid, pALH227 (Fig. 2a), confers Kan resistance in both E. coli and B. burgdorferi.
Figure 2.
Construction of pALH251 for complementation of inactivated rpoN with constitutively expressed rpoS. (a) A 965-bp fragment was excised from pJRS525 and a 1,158-bp fragment containing a PflaB-kan promoter fusion was inserted into the AlwNI and FspI sites, yielding pALH227. (b) pALH251 was obtained on ligating a PflgB-rpoS fusion construct (1,224 bp) into the BamHI and NcoI sites of the multiple cloning site of pALH227.
pALH227 served as a cloning vector for a wild-type copy of rpoN; a 1,628-bp fragment encompassing rpoN and its promoter (PrpoN) was amplified (priAH145, priAH125) and cloned into the NcoI and BamHI sites of pALH227, and the resulting plasmid (pALH293) served as a template for PCR amplification of the entire PrpoN-rpoN-(n)232-PflaB-kan region (priAH144, priAH143). The resulting 3-kb BclI fragment was ligated into the single BclI site within ermC of pALH394, yielding pALH400 (Fig. 1a).
pALH227 also was used to construct a shuttle vector (pALH251) (Fig. 2b) for RpoN-independent (constitutive) expression of rpoS in a rpoN mutant of 297. To effect this, the structural rpoS gene was fused to the flgB promoter (PflgB) by a strategy similar to the one described above. Briefly, the 410-bp PflgB region and the rpoS ORF (805 bp) were obtained by PCR, whereby an NdeI site was introduced at the 3′ end of PflgB (priAH122, priAH123) and at the 5′ end of rpoS (priAH126, priAH127). Both PCR products were individually cloned into pCR2.1 (lacks an NdeI site). The desired PflgB-rpoS fusion [junction region: GAGGGAGGTTTCCATATGrpoS (NdeI site underlined, RBSflgB bolded, start codon italicized)] was obtained by PCR (as described above) by using primers priAH122 (5′) and priAH127 (3′). This construct (1,224 bp) was ligated into the NcoI and BamH sites within the multiple cloning site of pALH227, yielding pALH251 (Fig. 2b), which was sufficiently stable to allow qualitative expression of RpoS (see Results).
Reverse Transcriptase–PCR (RT-PCR).
Total RNA from 297 or isogenic mutants was used in RT-PCRs (39). To detect transcripts for rpoS and confirm the disruption of rpoN, comparative RT-PCR was performed (rpoN, priAH59 and priAH60; rpoS, priAH131 and priAH132). To detect ospC and flaB transcripts, quantitative competitive RT-PCR was performed (39) by using primers ospC-5′, ospC-3′, flaB-5′, and flaB-3′ (39). Briefly, pFAC (Table 1) was constructed to allow SP6 RNA polymerase-dependent synthesis of an RNA with sequences for both flaB- and ospC-specific primers; in vitro-transcribed RNA served as a competitor for either natural transcript. About 1 μg of NcoI-linearized pFAC was used for in vitro transcription with SP6 RNA polymerase (SP6/T7 Transcription Kit; Roche Molecular Biochemicals). Competitor RNA was quantitated and 10-fold dilutions were used for RT-PCR in conjunction with constant quantities of test RNA (10 ng for ospC and 1 ng for flaB).
Results
Cloning and Disruption of 297 Genes Encoding Alternative Sigma Factors.
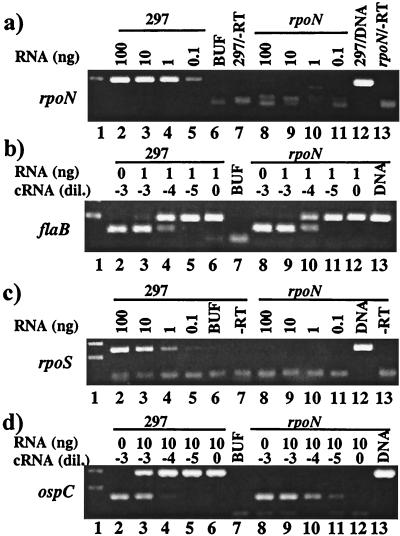
To assess the influence of RpoS and RpoN on differential antigen expression in 297, each theoretical 297 regulatory gene and its appropriate flanking regions were PCR-amplified (based on B31 sequence information) and inserted into pGEM-T, followed by insertional inactivation with the Erm resistance gene, ermC. The constructs (rpoN: pALH394, Fig. 1a) (rpoS: pALH386, Fig. 1b) were electroporated into 297. Generally, after plating on selective medium, several Erm-resistant (Ermr) colonies were obtained; typically, 12 were randomly chosen for PCR analysis. All clones yielded PCR amplification patterns consistent only with the desired genotypes (Fig. 3), with no evidence of a single crossover event and thus no evidence of plasmid (Amp resistance) incorporation. Also, no spontaneous Ermr mutants were observed in any experiment. RT-PCR performed on RNA from one of the rpoN∷ermC mutants confirmed that disrupted rpoN was not transcribed (Fig. 4a, lanes 8–11). The two mutants (rpoN and rpoS) displayed normal growth kinetics in BSK-H and appeared morphologically identical to 297 (not shown).
Figure 4.
RT-PCR analysis of a rpoN mutant. Borreliae were cultivated in BSK-H at pH 6.8. (a and c) Comparative RT-PCR; 10-fold dilutions of RNA from the wild type (strain 297) or the mutant were tested for rpoN (a) and rpoS (c) transcripts. Buf, buffer only; −RT, lacking RT. (b and d) Quantitative competitive RT-PCR; competitor RNA (cRNA) was used at exponential dilution, as indicated above the panels. The expression of constitutive flaB was unaffected by the rpoN mutation (b), whereas the expression of ospC was abolished (d).
RpoN Controls the Expression of OspC, DbpA, and RpoS.
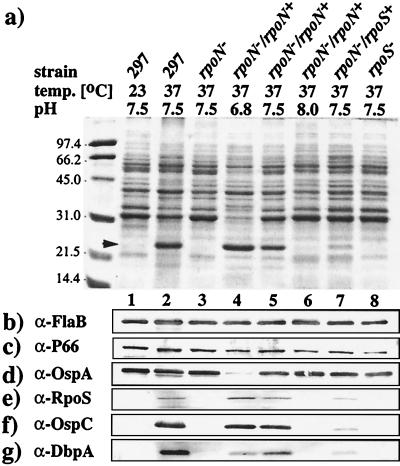
Parental 297 displayed its conventional temperature dependence for the expression of OspC (23) (Fig. 5a arrow, lanes 1 and 2). In contrast, OspC was notably absent in the rpoN mutant cultivated at 37°C (Fig. 5a, lane 3); this negative result was corroborated by immunoblotting (Fig. 5f, lane 3) and RT-PCR (Fig. 4d, lanes 8–12). Because OspC, DbpA, and RpoS have been hypothesized to be coregulated as group I proteins (23, 40), expression of DbpA and RpoS also was examined in the rpoN mutant. Neither DbpA (Fig. 5g, lane 3) nor RpoS (Fig. 5e, lane 3) was detectable in the rpoN mutant; the absence of mRNA for rpoS was confirmed by RT-PCR (Fig. 4c, lanes 8–11). In contrast to these group I proteins, the expression of OspA and Lp6.6, proteins down-regulated during 297 mammalian infection (group II proteins) (23), was not influenced by the rpoN mutation (Fig. 5d, lane 3; not shown for Lp6.6).
Figure 5.
SDS/PAGE (Coomassie blue stain) (a) and immunoblots (b–g) of whole cell lysates (39) from wild-type, mutant, and complemented strains of 297. Total protein from about 2 × 107 spirochetes (late-logarithmic phase of growth) was loaded per gel lane, except when probing for RpoS (10-fold more material). Monoclonal antibodies against OspA and FlaB and polyclonal antisera against all other proteins have been described (23, 51). Cultivation pH and temperature are indicated above each lane. Lanes 1 and 2, wild-type 297. Lane 3, rpoN mutant. Lanes 4–6, rpoN mutant complemented with wild-type rpoN. Lane 7, rpoN mutant complemented with pALH251 (rpoS). Lane 8, rpoS mutant.
To establish that the loss of expression of OspC, DbpA, and RpoS was the result of rpoN gene disruption, the mutant was complemented with a wild-type copy of rpoN (controlled by its native promoter) recombined into the chromosome by using pALH400 (Fig. 1a, line 3). A chromosomal location was selected to minimize gene dosage effects (i.e., copy number) and to maximize gene stability. As a result of complementation, not only was the expression of OspC, DbpA, and RpoS restored, but their expression patterns were pH-dependent (Fig. 5, lanes 4–6) as well as temperature-dependent (not shown), as has been reported (23).
RpoN Controls the Expression of OspC and DbpA Through RpoS.
Because of its role as a transcriptional activator, it remained possible that RpoS actually regulated OspC and DbpA expression. To test this, we examined the expression of OspC and DbpA in an rpoS mutant. In the rpoS mutant, the expression of OspC and DbpA was abolished (Fig. 5, lane 8), suggesting that RpoS works directly to regulate the expression of these two lipoproteins. As further evidence of the regulatory role of RpoS, the rpoN mutant was complemented with a wild-type copy of the rpoS by using the shuttle plasmid pALH251 (Fig. 2b). As a means of rendering rpoS expression independent of rpoN, rpoS was engineered to be driven by the constitutive B. burgdorferi flgB promoter (PflgB). In this rpoS diploid, OspC and DbpA were readily detectable (Fig. 5, lane 7), suggesting that constitutive production of RpoS could overcome the RpoN deficiency.
Discussion
Significant advances have been made recently in the development of systems for the genetic manipulation of avirulent strains of B. burgdorferi (34, 36). Stewart et al. (37) in an initial attempt to transform virulent B. burgdorferi recently reported the construction of a shuttle plasmid that was stably maintained in strain N40. We chose to carry out our genetic studies in 297 because it is a human isolate (31) that is highly infectious (ID50 of about 50 organisms; unpublished data). Moreover, given that 297, unlike avirulent strain B31, expresses OspC very efficiently, it is particularly well suited to studies of the OspA/OspC regulatory paradigm. Although not a principal objective of this study, the use of 297 provided a chance for genetically manipulating virulent B. burgdorferi, a goal in borrelial genetics research that has been difficult to achieve. Nonetheless, herein we have extended the scope of B. burgdorferi genetics by performing genetic complementation in this highly intractable organism. Genetic complementation is an essential component of molecular Koch's postulates (41) and, as such, its application ultimately is vital for delineating borrelial gene function.
The use of Ermr as a selectable marker for genetic studies with virulent B. burgdorferi should not be controversial. Although the precise mechanism is unknown, Ermr occurs naturally among virulent B. burgdorferi (34, 42). Furthermore, antimicrobials other than Erm are preferred for the treatment of Lyme borreliosis (43). Finally, B. burgdorferi is not a free-living pathogen (requires a tick host), and thus inadvertent escape from the laboratory is virtually impossible. The presence of Amp resistance on pGEM-T, used as a convenient suicide plasmid for B. burgdorferi, also should be without measurable biohazardous risk because (i) pGEM-T is not stably maintained in B. burgdorferi, (ii) no attempt was made to select for Amp resistance in B. burgdorferi, and (iii) as stated earlier, it is highly implausible that a B. burgdorferi strain could escape from the laboratory.
The rpoN gene was selected for targeted disruption because it has been implicated in modulating the expression of bacterial virulence (27–30), and hence studies on rpoN could help to identify virulence-associated genes in B. burgdorferi. OspC was conspicuously absent in protein profiles of the rpoN mutant; this led to an initial hypothesis that RpoN directly regulated ospC, which was supported by the fact that ospC mRNA was undetectable by RT-PCR in the rpoN mutant. However, we also noted that under growth conditions that up-regulated OspC, DpbA, Mlp-8, OspF, and P35 (group I proteins) (23, 40) RpoS also was up-regulated (23). Moreover, analysis of the upstream promoter region of ospC predicted three putative −35/−10 promoters but did not highlight a −24/−12 consensus typical of an RpoN-dependent promoter (44). This prompted a revised hypothesis that perhaps RpoS, rather than RpoN, directly regulated OspC. Because the expression pattern for DbpA in all of the mutants mirrored that of OspC, it also was plausible that dbpA, and perhaps other group I genes, might be coordinately regulated along with ospC by RpoS.
RpoS is a stress-induced sigma factor of eubacteria (27), but its role in B. burgdorferi gene regulation and virulence expression has been obscure. Elias et al. (45) were the first to show that an rpoS mutant of avirulent B31 exhibited a growth phase-dependent sensitivity to 1 M NaCl. They also reported that a 454-bp intergenic region upstream of rpoS did not contain a sequence with more than 60% identity to a consensus σ70 promoter (45). Interestingly, computer analysis had predicted an excellent match for a RpoN-dependent −24/−12 promoter immediately (62 bp) upstream of the rpoS start codon in B. burgdorferi (46). We garnered experimental evidence for RpoN-dependent expression of rpoS in that both immunoblotting and RT-PCR failed to reveal any expression of rpoS in the rpoN mutant. Furthermore, a rpoS mutant did not express OspC or DbpA, and complementation of the rpoN mutant with constitutively expressed rpoS restored detectable levels of RpoS, OspC, and DbpA. Elias et al. (45) did not note an abrogation of ospC expression in a rpoS mutant of avirulent B31; however, expression of ospC in avirulent B31 tends to be low, thereby hampering studies of ospC expression. We thus conclude that RpoN controls the transcription of rpoS, which ultimately governs the expression of OspC, DbpA, and probably other group I-like proteins (23) of B. burgdorferi. Such a pathway of sigma factor interplay may represent an additional paradigm for bacterial gene regulation. It should be noted, however, that whereas our experiments imply that RpoN directly controls the expression of rpoS in B. burgdorferi, they do not entirely rule out the unlikely possibility that RpoN induces the expression of some other unknown regulator of rpoS; unfortunately, experiments to test this in B. burgdorferi currently are not feasible. We also have been unsuccessful thus far in isolating recombinant RpoN in a soluble form, a prerequisite for demonstrating in gel retardation assays RpoN binding specifically to the −12 region of PrpoS (47). However, the prediction of an RpoN-dependent −24/−12 promoter upstream of rpoS genes in B. burgdorferi as well as in Pseudomonas syringae and Enterobacter cloacae (46) provides circumstantial evidence that RpoN directly controls rpoS expression, and that this type of regulatory network may be relatively common among prokaryotes.
A caveat of our studies is that we could not conclude whether RpoN or RpoS is essential for virulence expression by B. burgdorferi. Although the rpoN mutant and the rpoN mutant complemented with wild-type rpoN replicated normally in dialysis membrane chambers implanted into the peritoneal cavities of rats (48) (implying that the rpoN mutant retained its ability to grow in a mammalian host-adapted state), thus far we have been unable to demonstrate that any of the 297 recombinants retained infectivity in the mouse model of Lyme borreliosis (49). No obvious differences in plasmid profiles between any of the recombinants and wild-type 297 have been observed. We thus have not discerned whether the inability of the rpoN- or rpoS-complemented strains to regain infectivity was caused by avirulent variants within the 297 population being preferentially transformed, or if repetitive in vitro manipulations somehow resulted in the loss of B. burgdorferi infectivity (50). Alternatively, if complete complementation was not achieved, the expression of other unknown proteins could have been adversely affected, resulting in loss of function(s) necessary for mouse infectivity. These uncertainties, however, do not detract from the demonstration that genetic complementation of the rpoN mutant with either rpoN or rpoS qualitatively restored the predicted protein expression of RpoS, OspC, and DbpA, which ultimately led to the discovery of a unique global regulatory pathway in B. burgdorferi. The genetic approaches described herein and the elucidation of the RpoN-RpoS regulatory pathway for key borrelial membrane lipoproteins have far-reaching implications for studying many aspects of B. burgdorferi infectivity, virulence, pathogenesis, and immune evasion.
The RpoN-RpoS regulatory pathway can be envisioned to serve the enzootic life cycle of B. burgdorferi in the following fashion. As a result of interdependent environmental signals (e.g., elevated temperature, reduced pH, and increased spirochete density) (23, 40) engendered during the tick's taking of a blood meal, activated RpoN activator protein likely binds to an enhancer region upstream of where RpoN is complexed with the RNA polymerase holoenzyme (−24/−12 region), leading to the synthesis of rpoS mRNA. RpoS then mediates the synthesis of OspC, DbpA, and ostensibly other group I-like proteins of B. burgdorferi. Other sensory information undoubtedly feeds into these processes, and thus details of this network likely will undergo refinements as new information emerges. The rpoN and rpoS mutants of 297 also provide systems in which to perform more global assessments of B. burgdorferi gene regulation.
Supplementary Material
Acknowledgments
We thank Kevin McIver for supplying pJRS233 and pJRS525 and Andrew Revel for assistance with implanting chambers in rats. This work was supported by Public Health Service Grants AI-45538 (to M.V.N.) and AI-43063 (to F.C.C.); A.H. was supported by National Institutes of Health Training Grant T32-AI07520 from the National Institute of Allergy and Infectious Diseases.
Abbreviations
- B31
Borrelia burgdorferi strain B31
- 297
B. burgdorferi strain 297
- BSK
Barbour-Stoenner-Kelly medium
- Dbp
decorin-binding protein
- Erm
erythromycin
- Kan
kanamycin
- Osp
outer surface protein
- Amp
ampicillin
- RT-PCR
reverse transcriptase–PCR
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Steere A C. Hosp Pract. 1993;28:37–44. doi: 10.1080/21548331.1993.11442776. [DOI] [PubMed] [Google Scholar]
- 2.de Silva A M, Fikrig E. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 3.Schwan T G. Infect Agents Dis. 1996;5:167–181. [PubMed] [Google Scholar]
- 4.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Silva A M, Telford S R, Brunet L R, Barthold S W, Fikrig E. J Exp Med. 1996;183:271–275. doi: 10.1084/jem.183.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadziene A, Wilske B, Ferdows M S, Barbour A G. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilske B, Preac-Mursic V, Jauris S, Hofmann A, Pradel I, Soutschek E, Schwab E, Will G, Wanner G. Infect Immun. 1993;61:2182–2191. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevenson B, Barthold S W. FEMS Microbiol Lett. 1994;124:367–372. doi: 10.1111/j.1574-6968.1994.tb07310.x. [DOI] [PubMed] [Google Scholar]
- 9.Theisen M, Borre M, Mathiesen M J, Mikkelsen B, Lebech A-M, Hansen K. J Bacteriol. 1995;177:3036–3044. doi: 10.1128/jb.177.11.3036-3044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bockenstedt L K, Hodzic E, Feng S, Bourrel K W, de Silva A, Montgomery R R, Fikrig E, Radolf J D, Barthold S W. Infect Immun. 1997;65:4661–4667. doi: 10.1128/iai.65.11.4661-4667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore R D, Kappel K J, Dolan M C, Burkot T R, Johnson B J B. Infect Immun. 1996;64:2234–2239. doi: 10.1128/iai.64.6.2234-2239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eicken C, Sharma V, Klabunde T, Owens R T, Pikas D S, Hook M, Sacchettini J C. J Biol Chem. 2001;276:10010–10015. doi: 10.1074/jbc.M010062200. [DOI] [PubMed] [Google Scholar]
- 13.Kumaran D, Eswaramoorthy S, Luft B J, Koide S, Dunn J J, Lawson C L, Swaminathan S. EMBO J. 2001;20:971–978. doi: 10.1093/emboj/20.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore R D, Piesman J. Infect Immun. 2000;68:411–414. doi: 10.1128/iai.68.1.411-414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwan T G, Piesman J. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohnishi J, Piesman J, de Silva A M. Proc Natl Acad Sci USA. 2001;98:670–675. doi: 10.1073/pnas.98.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo B P, Brown E L, Dorward D W, Rosenberg L C, Hook M. Mol Microbiol. 1998;30:711–723. doi: 10.1046/j.1365-2958.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson B, Schwan T G, Rosa P A. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Indest K J, Ramamoorthy R, Sole M, Gilmore R D, Johnson B J B, Philipp M T. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramamoorthy R, Philipp M T. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll J A, Garon C F, Schwan T G. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Goldberg M S, Popova T G, Schoeler G B, Wikel S K, Hagman K E, Norgard M V. Mol Microbiol. 2000;37:1470–1479. doi: 10.1046/j.1365-2958.2000.02104.x. [DOI] [PubMed] [Google Scholar]
- 24.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 25.Hengge-Aronis R. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 26.Buck M, Gallegos M-T, Studholme D J, Guo Y, Gralla J D. J Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wosten M M S M. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]
- 28.Boucher J C, Schurr M J, Deretic V. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H, Li X, Johnson D E, Mobley H L. Microbiology. 1999;145:185–195. doi: 10.1099/13500872-145-1-185. [DOI] [PubMed] [Google Scholar]
- 30.Klose K E, Mekalanos J J. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 31.Norton Hughes C A, Kodner C B, Johnson R C. J Clin Microbiol. 1992;30:698–703. doi: 10.1128/jcm.30.3.698-703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollack R J, Telford S R, Spielman A. J Clin Microbiol. 1993;31:1251–1255. doi: 10.1128/jcm.31.5.1251-1255.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuels D S. In: Methods in Molecular Biology. Nickoloff J A, editor. Totowa, NJ: Humana; 1995. pp. 253–259. [Google Scholar]
- 34.Sartakova M, Dobrikova E, Cabello F C. Proc Natl Acad Sci USA. 2000;97:4850–4855. doi: 10.1073/pnas.080068797. . (First Published April 18, 2000; 10.1073/pnas.080068797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Casal J, Price J A, Maguin E, Scott J R. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 36.Bono J L, Elias A F, Kupko J J, Stevenson B, Tilly K, Rosa P. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart P E, Thalken R, Bono J L, Rosa P. Mol Microbiol. 2001;39:714–721. doi: 10.1046/j.1365-2958.2001.02256.x. [DOI] [PubMed] [Google Scholar]
- 38.McIver K S, Scott J R. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G B, Caimano M J, Radolf J D, Norgard M V. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramamoorthy R, Scholl-Meeker D. Infect Immun. 2001;69:2739–2742. doi: 10.1128/IAI.69.4.2739-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falkow S. Rev Infect Dis. 1998;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 42.Hardham J M, Rosey E L. J Mol Microbiol Biotechnol. 2000;2:425–432. [PubMed] [Google Scholar]
- 43.Wormser G P, Nadelman R B, Dattwyler R J, Dennis D T, Shapiro E D, Steere A C, Rush T J, Rahn D W, Coyle P K, Persing D H, et al. Clin Infect Dis. 2000;31:S1–S14. doi: 10.1086/314053. [DOI] [PubMed] [Google Scholar]
- 44.Marconi R T, Samuels D S, Garon C F. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elias A F, Bono J L, Carroll J A, Stewart P, Tilly K, Rosa P. J Bacteriol. 2000;182:2909–2918. doi: 10.1128/jb.182.10.2909-2918.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studholme D J, Buck M. Microbiology. 2000;146:4–5. doi: 10.1099/00221287-146-1-4. [DOI] [PubMed] [Google Scholar]
- 47.Wigneshweraraj S R, Chaney M K, Ishihama A, Buck M. J Mol Biol. 2001;306:681–701. doi: 10.1006/jmbi.2000.4393. [DOI] [PubMed] [Google Scholar]
- 48.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. J Clin Invest. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barthold S W, Beck D S, Hansen G M, Terwilliger A A, Moody K D. J Infect Dis. 1990;162:133–138. doi: 10.1093/infdis/162.1.133. [DOI] [PubMed] [Google Scholar]
- 50.Siebers A, Zhong W, Wallich R, Simon M M. Med Microbiol Immunol. 1999;188:125–130. doi: 10.1007/s004300050114. [DOI] [PubMed] [Google Scholar]
- 51.Hagman K E, Lahdenne P, Popova T G, Porcella S F, Akins D R, Radolf J D, Norgard M V. Infect Immun. 1998;66:2674–2683. doi: 10.1128/iai.66.6.2674-2683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.