Abstract
The excessive production of type I IFNs is a hallmark and a main pathogenic mechanism of many autoimmune diseases, including systemic lupus erythematosus (SLE). In these pathologies, the sustained secretion of type I IFNs is dependent on the improper activation of plasmacytoid DCs (pDCs) by self–nucleic acids. However, the nature and origin of pDC-activating self–nucleic acids is still incompletely characterized. Here, we report that exosomes isolated from the plasma of SLE patients can activate the secretion of IFN-α by human blood pDCs in vitro. This activation requires endosomal acidification and is recapitulated by microRNAs isolated from exosomes, suggesting that exosome-delivered microRNAs act as self-ligands of innate single-stranded endosomal RNA sensors. By using synthetic microRNAs, we identified an IFN induction motif that is responsible for the TLR7-dependent activation, maturation, and survival of human pDCs. These findings identify exosome-delivered microRNAs as potentially novel TLR7 endogenous ligands able to induce pDC activation in SLE patients. Therefore, microRNAs may represent novel pathogenic mediators in the onset of autoimmune reactions and potential therapeutic targets in the treatment of type I IFN–mediated diseases.
Keywords: Autoimmunity, Immunology
Keywords: Cytokines, Dendritic cells, Innate immunity
MicroRNA-containing exosomes isolated from the plasma of SLE patients promote IFN-α secretion by human plasmacytoid DCs.
Introduction
Elevated levels of type I IFNs represent both a hallmark and a pathogenic mechanism of autoimmune diseases characterized by the so-called type I IFN signature, such as systemic lupus erythematosus (SLE) (1).
Plasmacytoid DCs (pDCs) are a rare subset of circulating leukocytes primarily responsible for the sustained production of type I IFNs in SLE patients (1, 2). pDCs express TLR7, a single-strand RNA (ssRNA) sensor, and TLR9, a CpG-rich DNA sensor (3). These 2 receptors are segregated into endosomes to allow prompt recognition of nucleic acids derived from engulfed microbes, while avoiding activation by extracellular self–nucleic acids (4). However, in SLE, the formation of complexes with autoantibodies or cationic peptides protect self-DNA from degradation and facilitate its transport to endosomes, where it activates TLR9 (5). A possible role for TLR7 activation by self-RNA has been postulated in autoimmune diseases, although it remains less characterized (6, 7).
MicroRNAs are short ssRNAs that exert posttranscriptional regulation of gene expression within the cell cytoplasm (8). However, extracellular microRNAs are also present in virtually all biological fluids associated with carrier proteins, such as Ago2, which makes them resistant to degradation (9). MicroRNAs can also be transferred from cell to cell through extracellular vesicles (10), suggesting that they may work as intercellular messengers (11). Indeed, vesicle-associated microRNAs were reported to elicit various physiological responses in target cells. However, the overall low concentration of extracellular microRNAs and the reported low copy number per vesicle (12) raised the question of whether extracellular microRNAs may reach the physiological limit of about 1,000 copies per cell estimated to be required to mediate significant paracrine or endocrine gene suppression in vivo (9). Recently, an alternative mechanism of cell regulation exerted by extracellular microRNAs has emerged, based on their ability to induce a broad, nonspecific stimulation of the innate immune responses via the induction of proinflammatory cytokines in murine models of cancer (13), neurodegeneration (14), and graft-versus-host disease (15).
The aim of this study was to investigate whether extracellular, exosome-delivered microRNAs may cause the activation of human pDCs in vitro and the production of type I IFNs in SLE.
Results
Exosomes from plasma of SLE patients activate the secretion of IFN-α by pDCs.
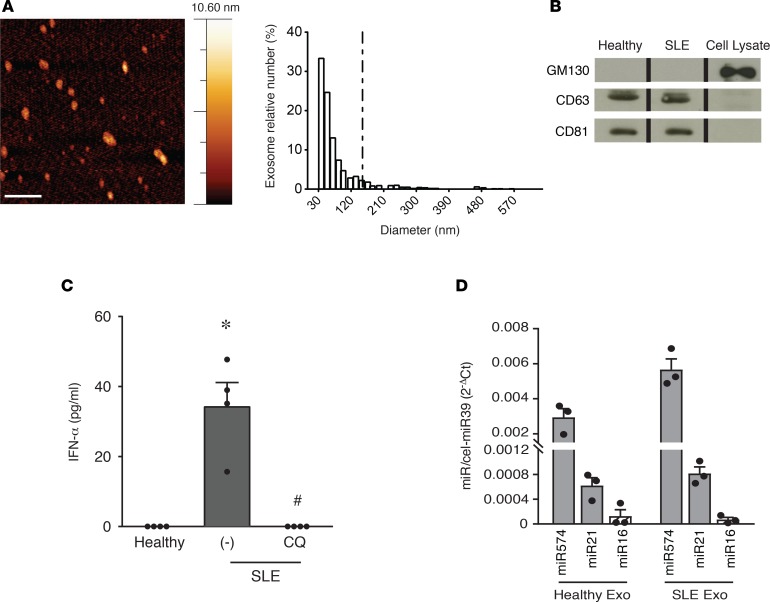
Small extracellular vesicles were separated by ultracentrifugation, followed by density gradient separation from a pool of plasma obtained from SLE patients (Table 1) or age-matched healthy controls. Extracellular vesicle preparations were then evaluated for purity and titrated by colorimetric nanoplasmonic (CONAN) assay (16), revealing equal amounts of vesicles (not shown). Extracellular vesicle morphology and size distribution was investigated by atomic force microscope (AFM) (17), proving that the population had typical vesicular morphology and exosome size ranging from 30–150 nm (Figure 1A) (18). The black background of the AFM images indicates the absence of any relevant nanoscale feature other than the EVs (with reference to the color code bar), confirming the absence of residual contaminants from the separation procedure and the purity of the samples (17). Finally, extracellular vesicles expressed specific exosomal markers (18) (Figure 1B). These results indicate that the used small extracellular vesicle population is prevalently composed by exosomes and so will be hereafter referred to as exosomes.
Table 1. Demographic and clinical characteristics of systemic lupus erythematosus (SLE) patients.
Figure 1. Exosomes from SLE plasma activate IFN-α secretion by pDCs.
(A) Representative AFM topography image of exosomes from systemic lupus erythematosus (SLE) plasma pools (scale bar: 500 nm; left). Particle size distribution obtained by image analysis of >1,200 round-shaped objects with a diameter ranging between 1 and 650 nm (5 μm × 5 μm fields, n = 6) demonstrating a prevailing exosome size (30–150 nm) (right). (B) Western blot of the exosome preparation showing the expression of exosome markers CD81 and CD63 and the lack of cis-Golgi matrix protein GM130 (negative control). One experiment out of 2 is represented; the lanes were run on the same gel but were noncontiguous. (C) pDCs from 4 donors were stimulated with exosomes purified from healthy or SLE plasma pools in the presence or absence of Chloroquine (CQ; 1 μM). The secretion of IFN-α was evaluated by ELISA after a 24-hour stimulation. Data are expressed as the mean ± SEM; *P < 0.05 versus (healthy) or #P < 0.05 versus (–) by paired Student’s t test. (D) Expression of selected microRNAs in exosomes obtained from plasma pools of healthy and SLE patients was evaluated by real-time PCR. Results were normalized over spiked-in cel-miR39. Data are expressed as the mean ± SEM of 3 replicates.
Exosomes purified from SLE, but not from healthy plasma, induced the release of IFN-α by freshly purified human circulating pDCs (Figure 1C). This action was reverted by chloroquine, indicating that endosomal acidification is required for pDC activation. This finding suggests that exosomes contain a putative ligand of endosomal innate immune receptors expressed by pDCs, namely TLR7 and/or TLR9 (4). Exosomes contain proteins, lipids, and short RNAs (19), among which microRNAs account for about 76% of total nucleic acids (20). Because pDCs do not bear endosomal receptors able to recognize proteins or lipids, but express the ssRNA receptor TLR7 (4, 13, 14, 21), our attention was focused on microRNAs. Equal amounts of total microRNAs were detected in exosome preparation from healthy and SLE individuals using the Quant-iT RiboGreen kit (not shown). Real-time PCR amplification revealed the presence of 3 microRNAs previously characterized for being expressed in SLE plasma (22, 23) (Figure 1D).
MicroRNAs delivered by exosomes activate the secretion of IFN-α in pDCs.
In order to further characterize the pDC-activating TLR ligand, amounts of exosomes were needed that could not be obtained from patient samples. Therefore, we decided to switch to exosomes obtained from cell lines. After testing different cell lines, we chose the keratinocyte cell line HaCaT both for the amounts of released exosomes and because of their biological significance, since the skin is a major site of pDC activation in SLE.
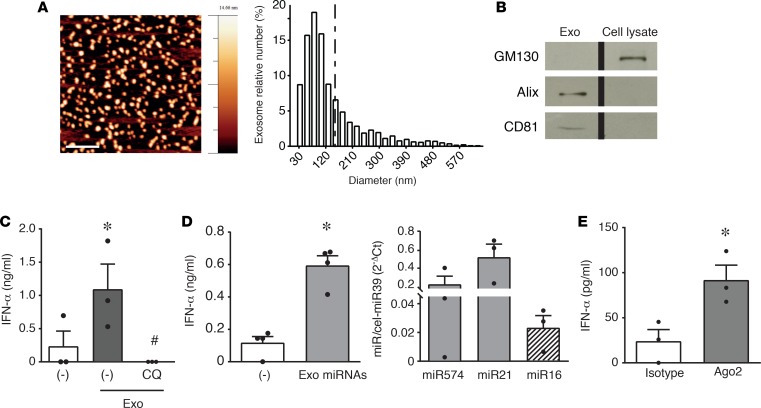
HaCaT-derived exosomes were characterized by AFM (Figure 2A) and for the expression of specific exosomal markers (Figure 2B) (18). These exosomes recapitulated the effects of SLE patient–derived exosomes (Figure 2C). MicroRNAs purified from HaCaT exosomes and complexed with a cationic liposomal vehicle (DOTAP) to protect them from degradation and to facilitate endocytosis (13, 21) could efficiently induce the secretion of IFN-α by pDCs (Figure 2D, left panel). In this preparation, real-time PCR revealed the presence of the 3 microRNAs previously detected in circulating exosomes (Figure 2D, right panel). To formally identify microRNAs as pDC activators and exclude other small RNAs (which may contaminate the microRNA fraction obtained with commercial purification kits used in Figure 2D), microRNAs were isolated, taking advantage of their constitutive association with the carrier protein Ago2. Figure 2E shows that microRNAs immunoprecipitated with an anti-Ago2 antibody, but not with an isotype control antibody, were indeed able to induce IFN-α secretion by human pDCs.
Figure 2. Exosomal microRNAs from HaCaT-conditioned media activate IFN-α secretion by pDCs.
(A) AFM topography image of HaCaT exosomes (scale bar: 500 nm) (left); size distribution obtained by image analysis of >1,400 round-shaped objects with a diameter ranging between 1 and 650 nm (5 μm × 5 μm fields, n = 3) demonstrating a prevailing exosome size (30–150 nm) (right). (B) Western blot of HaCaT exosomes showing the expression of exosome markers Alix and CD81 and the lack of cis-Golgi matrix protein GM130 (negative control). One experiment out of 2 is represented; the lanes were run on the same gel but were noncontiguous. (C) pDCs were stimulated with exosomes obtained from HaCaT-conditioned media. Where indicated, pDCs were pretreated with 1 μM CQ for 1 hour. Data are expressed as the mean ± SEM (n = 3); *P < 0.05 versus (–) or #P < 0.05 versus “(–)Exo” by paired Student’s t test. (D, left) MicroRNAs from HaCaT exosomes were purified and used to stimulate pDCs. Data are expressed as the mean ± SEM (n = 4); *P < 0.05 versus (–) by paired Student’s t test. (D, right) Expression of selected microRNAs in exosomes obtained from HaCaT-conditioned media was evaluated by real-time PCR. Results were normalized over spiked-in cel-miR39. Data are expressed as the mean ± SEM (n = 3). (E) Native, Ago2-bound microRNAs were immunoprecipitated with anti-Ago2 or isotype control mAb, purified with a commercial microRNA enrichment kit, and used to stimulate pDCs. Data are expressed as the mean ± SEM (n = 3); *P < 0.05 versus (isotype) by paired Student’s t test. In all the experiments, the secretion of IFN-α was evaluated by ELISA after a 24-hour stimulation.
Taken together, these results demonstrate that exosome-delivered microRNAs can activate the secretion of type I IFNs by pDCs.
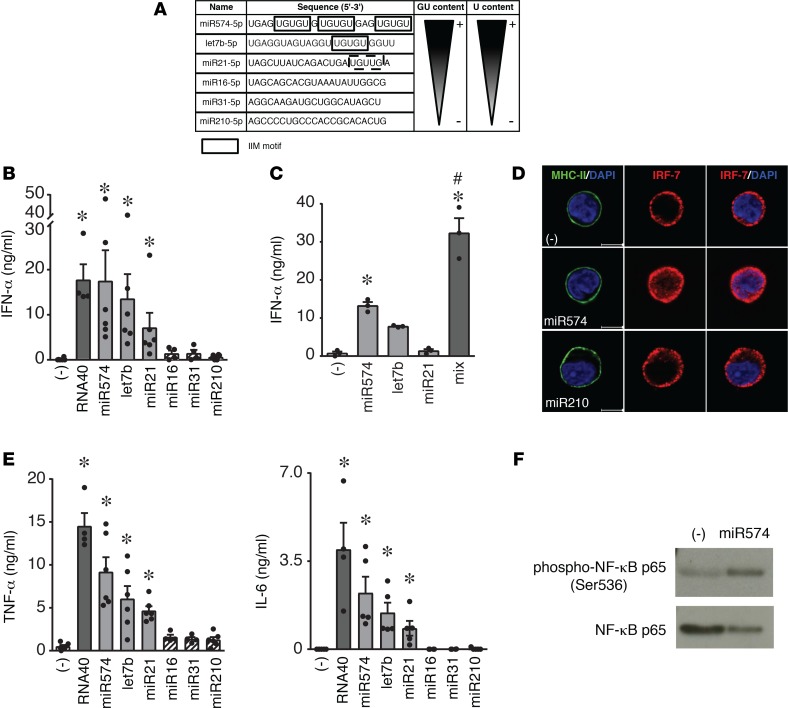
Synthetic microRNAs containing an IFN induction motif (IIM) activate cytokine secretion by pDCs and induce pDC maturation.
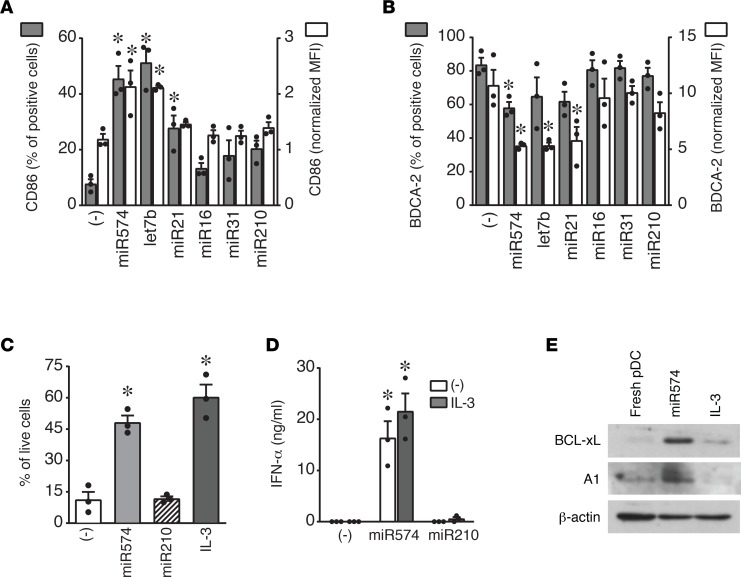
Previous work on small interfering RNAs identified an IIM (5′-UGUGU-3′) capable of activating the innate immune response (24). Thus, we selected microRNAs upregulated in SLE plasma (23) and bearing this or similar motives (IIM microRNAs: miR574, let7b, and miR21) to evaluate their ability to activate pDCs. MicroRNAs devoid of this motif were selected as negative controls (Figure 3A), while the viral RNA40 was used as a positive control for pDC activation (21). Figure 3B shows that IIM-bearing microRNAs activated the secretion of IFN-α by pDCs with a magnitude comparable with RNA40. This effect was observed when microRNAs were complexed with DOTAP but not when administered free into the culture media (not shown). In addition, IIM microRNAs administered at suboptimal concentrations cooperated in inducing a strong IFN-α secretion (Figure 3C). Consistent with this, IIM microRNAs induced the nuclear translocation of IRF-7, the master transcription factor for type I IFN production (Figure 3D). IIM microRNAs also induced the secretion of the proinflammatory cytokines TNF-α and IL-6 (Figure 3E) and the phosphorylation of the p65 subunits of NF-κB (Figure 3F). In addition, IIM microRNAs induced the phenotypic maturation of pDCs, as shown by the increased expression of CD86 (Figure 4A) and the decrease of the BDCA-2 levels (Figure 4B). Finally, IIM microRNAs rescued pDC survival when cultured in the absence of IL-3, the main growth factor for pDCs (3) (Figure 4C), whereas IFN-α production was similarly induced regardless of the presence of IL-3 (Figure 4D). Consistent with increased survival, IIM microRNAs induced the expression of the antiapoptotic proteins BCL-xL and A1 (Figure 4E) (25). None of these effects were observed using control, IIM-negative microRNAs (e.g., miR16, miR31, and miR210) (Figures 3 and 4, and data not shown).
Figure 3. Synthetic IIM microRNAs activate human pDCs to secrete IFN-α and proinflammatory cytokines.
(A) Sequence of synthetic microRNAs. The IFN induction motif (IIM) is boxed, while a similar motif is dashed. The relative content of GU and U are also indicated. (B) pDCs were stimulated with viral RNA40 or with the indicated microRNAs (10 μg/ml) or with vehicle alone (–) for 24 hours. The production of IFN-α was evaluated by ELISA in cell-free supernatants. Data are expressed as mean ± SEM (n = 4–6); *P < 0.05 versus (–) by 1-way ANOVA with Dunnett’s post-hoc test. (C) pDCs were stimulated with a suboptimal concentration of active microRNAs (2.5 μg/ml) alone or in combination. IFN-α production was evaluated by ELISA. Data are expressed as mean ± SEM (n = 3); *P < 0.05 versus (–) by 1-way ANOVA with Dunnett’s post-hoc test or #P < 0.05 by Tukey’s multiple comparison test. (D) pDCs were stimulated with 10 μg/ml microRNAs for 4 hours. Cell membrane was identified by an anti–MHC class II–Alexa 488 (green) mAb and the nucleus by DAPI staining (blue). IRF-7 nuclear translocation was visualized by a specific anti–IRF-7 Ab (red). The figure shows 1 representative donor out of 3; scale bars: 5 μm. (E) pDCs were stimulated as in B, and the production of TNF-α and IL-6 was evaluated by ELISA in cell-free supernatants. Data are expressed as mean ± SEM (n = 4–6); *P < 0.05 versus (–) by 1-way ANOVA by Dunnett’s post-hoc test. (F) pDCs were stimulated with miR574 for 90 minutes, lysed, and analyzed by Western blot using an anti–phosphorylated NF-κB p65 mAb. Image depicts 1 representative fluorogram out of 3 performed.
Figure 4. Synthetic IIM microRNAs induce pDC maturation and promote pDC survival.
(A and B) pDCs were stimulated with the indicated microRNAs (10 μg/ml) or with vehicle alone (–) for 24 hours. The surface expression of CD86 (A) and BDCA-2 (B) was evaluated by FACS analysis. Data are expressed as the mean ± SEM of the percentage of positive cells (left y axis), as well as the mean ± SEM of the median fluorescence intensity (MFI) of positive cells (right y axis) normalized over unstained controls (n = 3); *P < 0.05 versus (–) by 1-way ANOVA with Dunnett’s post-hoc test. (C) pDCs were stimulated as indicated in culture medium devoid of IL-3 for 24 hours. Cell viability was evaluated by propidium iodide staining (mean SEM; n = 3); *P < 0.05 versus (–) by paired Student’s t test. (D) pDCs were stimulated with the indicated microRNAs (10 μg/ml) in the presence or absence of IL-3 for 24 hours. IFN-α production was evaluated by ELISA. Data are expressed as mean ± SEM (n = 3); *P < 0.05 versus (–) by paired Student’s t test. (E) pDCs were stimulated as indicated or lysed immediately after purification (fresh). The expression of BCL-xL, A1, and β-actin were determined by Western blot. One representative fluorogram out of 3 performed is shown.
These results confirm the capability of encapsulated extracellular microRNAs to induce full pDC activation and maturation.
pDC activation by synthetic IIM microRNAs is mediated by an endosomal ssRNA receptor.
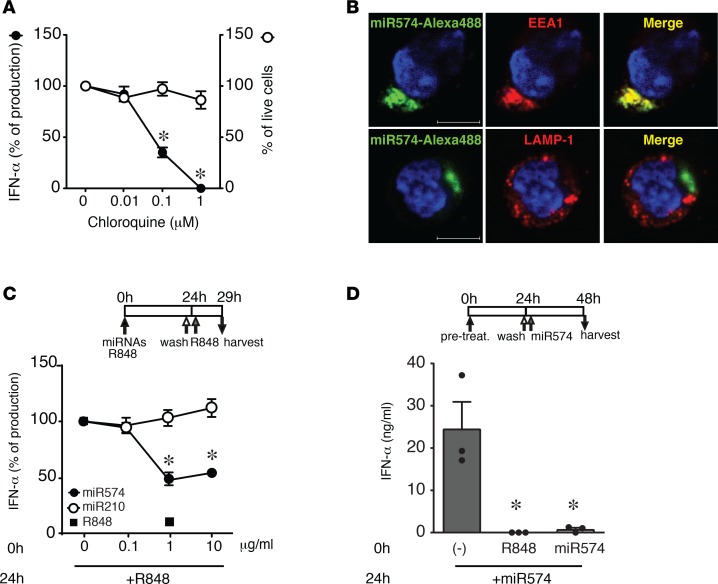
As previously observed with pDC activation by exosomes (Figure 1C and Figure 2C), chloroquine inhibited — in a dose-dependent manner — the production of IFN-α induced by synthetic miR574 (chosen for further experiments as prototypic IIM microRNA), in the absence of any toxicity as measured by pDC viability (Figure 5A). This result further demonstrates that endosomal acidification is functional for IFN-α secretion by microRNAs. In addition, fluorescent miR574 colocalized with the early endosome marker EEA1, but not with the late endosome marker LAMP-1 (Figure 5B), confirming the endosomal localization and suggesting an interaction with an endosomal receptor. These findings strongly suggest TLR7, the only endosomal ssRNA receptor present in pDCs, as the potential receptor involved.
Figure 5. Synthetic IIM microRNAs localize within early endosomes and antagonize the TLR7 ligand R848.
(A) pDCs were pretreated with increasing concentrations of CQ for 1 hour and then stimulated with miR574 (10 μg/ml) for 24 hours. The secretion of IFN-α was evaluated by ELISA (left y axis) and cell viability by propidium iodide staining (right y axis). Data are expressed as mean ± SEM of the percentage of IFN-α production or the percentage of live cells (n = 3); *P < 0.05 versus (0 µm CQ) by 1-way ANOVA with Dunnett’s post-hoc test. (B) pDCs were stimulated with 10 μg/ml miR574–Alexa 488 for 90 minutes. After fixation and permeabilization, pDCs were stained with an anti–early endosome antigen 1 (EEA1) mAb (red) or an anti–LAMP-1 Ab (red) and DAPI (blue). The image depicts results obtained with 1 representative donor out of 3; scale bars: 5 μm. (C) pDCs were pretreated (0h) with the indicated concentration of microRNAs or 1 μg/ml R848 for 24 hours, washed, and restimulated with 1 μg/ml R848 for an additional 5 hours. The production of IFN-α was evaluated by ELISA in cell-free supernatants. Data are expressed as the mean ± SEM of the percentage of IFN-α production (n = 4); *P < 0.05 versus (0 µm CQ) by 1-way ANOVA with Dunnett’s post-hoc test. (D) pDCs were pretreated (0h) with R848 (1 μg/ml) or miR574 (10 μg/ml) or left untreated for 24 hours, washed, and restimulated with miR574 for additional 24 hours. IFN-α was measured by ELISA in cell-free supernatants. Data are expressed as the mean ± SEM (n = 3); *P < 0.05 versus (–) by 1-way ANOVA with Dunnett’s post-hoc test.
TLR7, similarly to other TLRs, undergoes desensitization upon repeated stimulation (7). Accordingly, a 24-hour prestimulation with R848 completely blocked IFN-α production by a second R848 stimulation (Figure 5C). The release of IFN-α by R848 was also dose-dependently inhibited by the pretreatment with miR574, but not with miR210, suggesting that miR574 and R848 engage the same receptor (Figure 5C). This hypothesis was further supported by the ability of R848 to desensitize the action of miR574 (Figure 5D). As expected, miR574 pretreatment blocked IFN-α production by miR574 itself (Figure 5D)
Human TLR7 is a microRNA sensor in human pDCs.
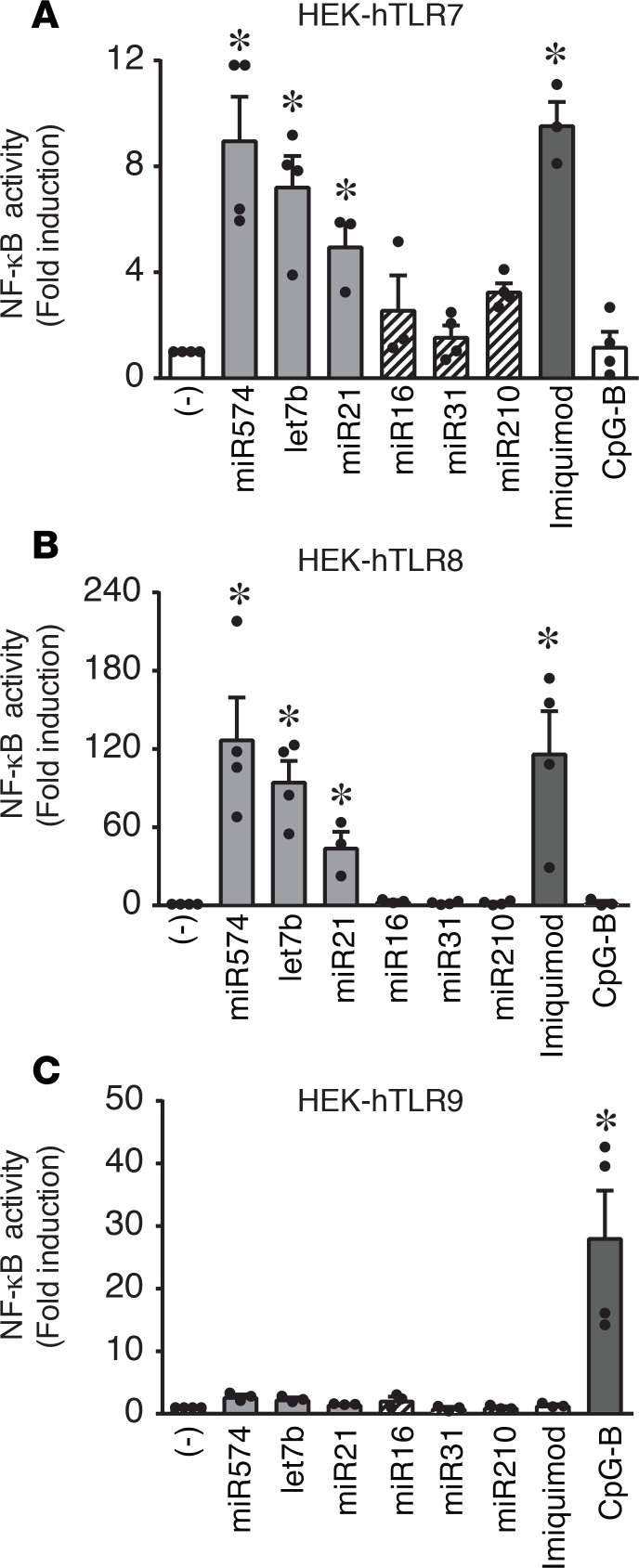
To formally demonstrate the ability of human TLR7 to sense IIM microRNAs, experiments were performed in HEK-293 cells stably transfected with human TLR7, TLR8, and TLR9 and containing a NF-κB reporter gene. Figure 6A shows that IIM microRNAs, similarly to imiquimod, stimulated TLR7-expressing cells. As expected (13), IIM microRNAs also potently activated NF-κB in transfectants expressing TLR8 (Figure 6B) but not cells expressing TLR9 (Figure 6C).
Figure 6. Synthetic IIM microRNAs engage human TLR7.
(A–C) HEK-293 cells stably transfected with human TLR7 (A), TLR8 (B), or TLR9 (C) were stimulated with 10 μg/ml of the indicated microRNAs or vehicle alone (–) for 24 hours. Imiquimod (1 μg/ml) or CpG-B (10 μg/ml) were used as TLR-specific positive controls. NF-κB activation was evaluated in terms of fold luciferase activity over unstimulated cells. Data are expressed as the mean ± SEM (n = 3–4); *P < 0.05 versus (–) by 1-way ANOVA with Dunnett’s post-hoc test.
Altogether, these results support the hypothesis that extracellular microRNAs activate human pDCs via TLR7 triggering.
Discussion
Our study reports several new findings that identify exosome-delivered microRNAs as new potential players in the pathogenesis of human autoimmune diseases depending on excessive type I IFN production.
First, we show that exosome-delivered microRNAs activate the secretion of type I IFNs by pDCs and that this effect is enhanced in SLE patients. SLE is the prototype of the growing family of autoimmune diseases known as type I IFN–dependent autoimmune diseases, in which undue pDC activation represents a major leading event (1). In addition to many genetic alterations clustering in the TLR and type I IFN pathways described in SLE individuals (26), the disease also recognizes environmental and inflammatory triggers (27). It is long known that, in the amplification phase of SLE, pDC activation is sustained by immunocomplexes of anti–nucleic acid antibodies (1). By contrast, self-ligands responsible for pDC activation at disease onset have only recently begun to emerge (1, 5). Our results now identify microRNAs as new pDC-activating molecules, setting the bases for future work to demonstrate a possible pathogenic role of microRNA-dependent pDC activation in murine models of type I IFN–dependent autoimmune diseases. MicroRNAs represent ideal candidates in virtue of the fact that they are released by virtually all cells in the body, are resistant to degradation, and were described to be deregulated in SLE and in other autoimmune diseases (22, 23, 28).
From a molecular point of view, our work demonstrates that extracellular microRNAs represent potentially novel endogenous ligands of human TLR7, the unique endosomal ssRNA receptor expressed by pDCs. The activation of endosomal TLRs by extracellular microRNAs was first reported by Fabbri and coworkers, who described the proinflammatory activation of murine macrophages by selected microRNAs aberrantly secreted by cancer cells, ultimately leading to tumor growth and metastasis (13). These authors identified murine TLR7 as the receptor involved in microRNA sensing, a finding independently confirmed by another group in a model of neuronal degeneration (14). Human TLRs involved in microRNA sensing were scarcely investigated. Our results now confirm that microRNAs activate human TLR8 transfectants, in line with previous evidence demonstrating the activation of TLR8-expressing conventional DCs in the presence of extracellular microRNAs (13, 15). In addition, using both pDCs as a physiological model of TLR7-expressing cells and TLR7 transfectants, we demonstrate that human TLR7 is a microRNA receptor. Our results also show that pDC-activating capability is not shared by all microRNAs. In this regard, we observed a strong correlation between IFN-α production and the presence of IIM motifs. Indeed, miR574, containing several IIMs, turned out to be the most potent stimulator, while all microRNAs devoid of IIM did not activate pDCs. In addition to IIM, other sequences, such as GU-rich elements or the number of U ribonucleotides, were described to be important for innate immune activation by ssRNAs (13, 14, 21, 29). In accordance, the IIM microRNAs used here are also rich in G and U ribonucleotides. According to a recently published analysis, G and U represent the most abundant ribonucleotides in human microRNAs, often present as GU dinucleotide, and at least 50 IIM microRNAs exist (30). Thus, it is conceivable to assume that, in vivo, many exosomal microRNAs possess the structural features required to stimulate pDCs. Such redundancy may overcome the theoretical limit to microRNA-dependent cell-to-cell regulation raised by the low levels of extracellular microRNAs, as also suggested by our experiments using combinations of activatory microRNAs at suboptimal concentrations. Of note, in our exosomal preparations, we could detect the expression of IIM microRNAs previously described as upregulated in SLE patients that possess the capability to activate pDCs in vitro.
The mechanisms underlying extracellular microRNA delivery and action on target cells remain poorly understood. Rettig and colleagues demonstrated that nanoparticles are selectively phagocytosed by pDCs (31), suggesting that exosome contents may be directly brought into contact with endosomal TLRs. Our results demonstrate that microRNAs protected by a liposomal vehicle effectively reach the endosome and recapitulate pDC activation observed using exosomes. In our hands, microRNAs administered to pDCs in the absence of liposomal vehicles did not induce IFN-α secretion. This is in line with the prevailing view that vesicle-associated microRNAs are responsible for cell-to-cell communications, while free microRNAs may represent cell byproducts devoid of biological function, although it was reported that free synthetic microRNAs may induce inflammatory neuronal activation (14). In addition, it was also shown that Ago2-bound microRNAs may enter the cells via the binding to neuropilin-1 (32). Of note, neuropilin-1 is expressed at very high levels by pDCs (1) and may, thus, allow the internalization of native, Ago2-bound microRNAs in our cell type. Further work is needed to investigate this possibility.
In summary, our work demonstrate that (a) exosomes purified from the plasma collected from SLE patients but not healthy donors, as well as microRNAs extracted from exosomes, induce the production of type I IFNs in human pDCs; (b) IIM microRNAs act as human pDC survival factors and activate the maturation and the production of IFN-α and proinflammatory cytokines by pDCs; and (c) IIM microRNAs represent potentially novel endogenous ligands of human TLR7, the unique endosomal ssRNA receptor expressed by pDCs. These results highlight the induction of type I IFN secretion by pDC as a new mechanism by which extracellular microRNAs modulate the inflammatory and adaptive immune response.
Methods
Patients.
Thirty-four premenopausal SLE patients were enrolled at the Department of Clinical and Experimental Sciences in Brescia. Patients met the 1997 American College of Rheumatology criteria for SLE (33), and their main clinical and demographic features are presented in Table 1. To be included in the study, patients had to be on stable disease inactivity, defined as absence of SLE flare and Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2K) score (34) below 6 in the year prior to enrollment.
Cell preparation and culture.
Peripheral blood mononuclear cells (PBMCs) were obtained from buffy coats (through the courtesy of Centro Trasfusionale, Spedali Civili, Brescia, Italy) by Ficoll gradient. Peripheral blood pDCs were obtained after a immunomagnetic separation with the Plasmacytoid Dendritic Cells Isolation kit II (Miltenyi Biotec). pDCs (1 × 106 cells/ml) were cultured in RPMI 1640 (Gibco, Thermo Fisher Scientific), supplemented with 10% heat-inactivated FBS, 2 mM L-Glutamine, penicillin and streptomycin (Gibco), and 20 ng/ml IL-3 (Miltenyi Biotec) (35). Human HaCaT cell line (CLS) was cultured in DMEM supplemented with 10% FBS.
Stimulation of peripheral blood pDCs.
Purified pDCs were stimulated with 10 μg/ml synthetic RNA oligonucleotide RNA40 or synthetic microRNAs (listed in Figure 3A; all bases depict a phosphorothioate linkage) synthesized by Integrated DNA Technologies (IDT). Complexation of RNA or microRNA with DOTAP Liposomal Transfection Reagent (Roche Diagnostics) was performed according to the manufacturer’s recommendation. Briefly, 5 μg RNA or microRNA in 50 μl HBS buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) was combined with 100 μl DOTAP solution (30 μl DOTAP plus 70 μl HBS buffer) and incubated for 15 minutes at 20°C. DOTAP/RNA (50 μl) or microRNA mixture were used for stimulation of a 96-well that contained pDCs in 100 μl complete medium. Where indicated pDCs were stimulated with TLR7 agonist R848 (1 μg/ml; InvivoGen) or pretreated with Chloroquine (InvivoGen) (36).
Cytokine measurement.
IFN-α in culture supernatants was detected using a specific Module Set ELISA kit (eBioscience, Affymetrix). IL-6 and TNF-α were measured using specific Duo-Set kits (R&D Systems).
Exosome isolation from plasma and HaCaT conditioned media.
To obtain conditioned media for exosome isolation, HaCaT cells were grown in complete DMEM until the reach of 70% confluence, and then the media were removed and renewed with DMEM without FBS for 48 hours. Afterward, conditioned media were collected and processed with a serial centrifugation protocol (37, 38). Briefly, conditioned media were centrifuged firstly at 800 g for 30 minutes to pellet and discard possibly contained apoptotic bodies and cellular debris. The supernatant was then centrifuged at 16,000 g for 45 minutes to pellet microvesicles and ultracentrifuged at 100,000 g for 2 hours to collect exosomes (Type 45 Ti rotor, Beckman Optima XPN 80, Beckman Coulter). Exosomes were washed with 1 ml of sterile PBS buffer to remove protein-based contaminants and pelleted at 100,000 g for 2 hours. Each centrifugation step was performed at 4°C.
To isolate exosomes from plasma samples, 8 ml of plasma derived from a pool of SLE patients or healthy donors were processed by serial centrifugations followed by gradient centrifugation. Briefly, exosome fraction was resuspended in 800 μl buffer (10 mM Tris–HCl 250mM sucrose, pH 7.4) and processed by a discontinuous sucrose gradient made by 6 fractions (15%, 20%, 25%, 30%, 40%, 60% sucrose in 10 mM Tris–HCl, pH 7.4) and centrifuged at 100,000 g for 16 hours (rotor MLS 50, Beckman Optima MAX). Twelve fractions with equal volumes (400 μl) were collected from the top of the gradient. The exosomes contained in fractions from 6–9 were pelleted by ultracentrifugation (100,000 g for 2 hours). Exosomes were evaluated for purity and titrated by CONAN assay as previously described (16). The total RNA content was calculated using the Quant-iT RiboGreen kit (Thermo Fisher Scientific) according to the manufacturer’s instructions.
AFM imaging of exosomes.
Exosomes isolated from HaCaT conditioned media or plasma pools were resuspended in 100 μl of PBS and diluted 1:100 with sterile Milli-Q water (MilliporeSigma). The diluted sample (5–10 μl) was then spotted onto freshly cleaved mica sheets (Grade V-1, thickness 0.15 mm, size 15 × 15 mm2). The samples were then air dried over a plate heated at 37°C–40°C for 20 minutes. They were finally imaged with a NaioAFM (Nanosurf AG) equipped with ContAl-G (Budget Sensors). Images were acquired in contact mode, with a scan size ranging from 2.5–25 μm and a scan speed from 0.8–1.2 seconds per scanning line. Size distribution analysis of the deposited exosomes was performed with the software WSxM 4.0 image (39). For the AFM size distribution analysis, topography images with an image scan size of 5 × 5 μm were used. A minimum of 1,200 round-shaped vesicles with a diameter ranging between 30 nm and 650 nm were analyzed. Obtained data were grouped in 50-nm classes, and off-scale and nonround-shaped objects were excluded from the analysis.
Immunofluorescence.
For the assay of the nuclear translocation of IRF-7, purified pDCs were stimulated for 4 hours with synthetic microRNAs (10 μg/ml). After incubation, cells were stained with an anti–human MHC class II-Alexa Fluor 488 (307619, BioLegend) and subsequently fixed overnight with 1% paraformaldehyde, seeded on glass slides by cytospin, and permeabilized with 100% ice-cold methanol (MilliporeSigma) for 10 minutes at –20°C. Samples were labeled with a rabbit polyclonal anti–human IRF-7 antibody (sc-9083, Santa Cruz Biotechnology Inc.). A conjugate Alexa Fluor 594 anti-rabbit (A-11072, Thermo Fisher Scientific) was used as a secondary antibody (40).
For visualization of the trafficking of microRNA to endosome, pDCs were incubated with Alexa Fluor 488–tagged miR574 for 90 minutes. Cells were fixed overnight with 1% paraformaldehyde (MilliporeSigma), seeded on glass slides by cytospin, and permeabilized with 0.2% Triton X-100 (MilliporeSigma) for 10 minutes at RT. pDCs were labeled with a mouse monoclonal anti–human EEA1 antibody (610456, BD Biosciences) or with a rabbit polyclonal anti–human LAMP-1 antibody (ab24170, Abcam), and an Alexa Fluor 594 anti-mouse (A-21203, Thermo Fisher Scientific) or an Alexa Fluor 594 anti-rabbit were used as secondary antibodies, respectively.
Glass slides were mounted using Prolong antifade with DAPI (Thermo Fisher Scientific). Cell imaging was performed with a ZeissAxio Observer Z1 microscope equipped with Apotome system and a Plan-Apochromat 100×/1.4 NA oil objective.
FACS analysis.
pDCs were stained with Vioblue-conjugated anti-CD86 (clone FM95, Miltenyi Biotec) and APC-conjugated anti–BDCA-2 (clone AC144, Miltenyi Biotec) after 24 hours of the indicated treatments. Samples were read on a MACSQuant Analyzer (Miltenyi Biotec) and analyzed with FlowJo (Tree Star Inc.). To assess cell viability, pDCs were stained with Propidium Iodide (Thermo Fisher Scientific).
SDS-PAGE and Western blot.
Following the indicated stimulations, pDCs were lysed in NP-40/Triton lysis buffer (10 mM Tris-HCl, pH 7.9; 150 mM NaCl; 0.6% NP-40; and 0.5% Triton X-100) supplemented with inhibitors (1 mM Na3OV4, 2 mM DTT, 1 mM NAF, 1 mM PMSF, and protease inhibitor cocktail; all reagents were purchased from MilliporeSigma). Equal amounts of extracts were analyzed through SDS-PAGE followed by Western blotting with antibodies against BCL-xL (2764, Cell Signaling Tecnologies), A1 (14093, Cell Signaling Technology), pNF-κB p65 (Ser536, 3033, Cell Signaling Technology), NF-κB p65 (sc-372, Santa Cruz Biotechnology Inc.), and β-actin (sc-47778, Santa Cruz Biotechnology Inc.) (41).
Exosome preparations from plasma pools or HaCaT conditioned media were lysed in NP-40/Triton lysis buffer. Extracts (20–30 μg)were analyzed through SDS-PAGE, followed by Western blotting with antibodies against GM130 (610822, BD Biosciences), Alix (sc-53539, Santa Cruz Biotechnology Inc.), CD63 (CBL553, MilliporeSigma), and CD81 (sc-166029, Santa Cruz Biotechnology Inc.).
Protein bands were detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce).
NF-κB luciferase reporter assay.
TLR-specific activation assays were performed using human HEK-293 cells expressing luciferase under control of the NF-κB promoter and stably transfected with human TLR7, TLR8, and TLR9. HEK-293–transfected cells were maintained in DMEM supplemented with 10% FBS, and appropriate antibiotics were added. All the HEK-293–transfected cells were provided by U. D’Oro (GSK, Siena, Italy). For the NF-κB luciferase assay, 25,000 cells were seeded in complete DMEM without antibiotics in 96-well plates for 24 hours. Cells were stimulated with 10 μg/ml synthetic microRNAs, 1 μg/ml imiquimod (TLR7 agonist), 1 μg/ml R848 (TRL7-8 agonist), and 10 μg/ml CpG-B (TLR9 agonist) (all from Invivogen) for 24 hours. After stimulation, cells were lysed using ONE-Glo EX Luciferase Assay System (Promega) and assayed for luciferase activity using the EnSightMultimode Plate Reader (PerkinElmer).
Isolation of microRNA-enriched fraction and Ago2-associated microRNAs.
MicroRNAs contained in exosomes were purified with miRNeasy Mini Kit, together with RNeasy Mini Elute Cleanup Kit (Qiagen), according to the manufacturer’s instructions. Where indicated, 5 pg of synthetic miR39 from Caenorhabditis elegans (cel-miR39) were added as a spike-in control for purification efficiency. Alternatively, microRNAs were immunoprecipitated from protein lysates with 10 μg of anti-Ago2 antibody (clone 11A9, MilliporeSigma) or isotype control overnight at 4°C. To capture the immune complexes, anti–rat IgG agarose (ab7020, Abcam) was added to the antibody/lysate samples and incubated for 2 hours at 4°C. Immune complexes were washed 2 times with washing buffer (50 mM Tris-HCl; 150 mM NaCl; 1 mM PMSF), and the resin was dissolved in 700 μl of Qiazol reagent (Qiagen).
After purification, microRNA-enriched fraction and Ago2-associated microRNAs were complexed to DOTAP and used to stimulate pDCs.
MicroRNA expression analysis.
A fixed volume of 3 μl of microRNA fraction from 14-μl eluates obtained after microRNA isolation was used as an input into a reverse transcription reaction. Reverse transcription was performed using miScript II RT Kit (Qiagen). The levels of hsa-miR574, hsa-miR21, hsa-miR16, AND cel-miR39 were measured using specific miScript Primer assay in association with miScript SYBR Green PCR Kit (Qiagen) according to manufacturer’s instructions. Reactions were run in triplicate on a StepOne Plus Real-time PCR System (Applied Biosystems), and the generated products were analyzed by the StepOne Plus Software (Version 2.3, Applied Biosystem). Cel-miR39 was used for the normalization of microRNA levels.
Statistics.
Statistical significance among the experimental groups was determined using paired 2-tailed Student’s t test, 1-way ANOVA with Dunnett’s post-hoc test, or Tukey’s multicomparison test as appropriated (GraphPad Prism 7, GraphPad Software). P < 0.05 was considered significant.
Study approval.
The study was approved by the local institutional ethics committee in Brescia and conducted in accordance with the Declaration of Helsinki. Patients gave their written informed consent.
Author contributions
VS conceived and performed experiments with microRNAs and pDCs and analyzed data; VG performed experiments with microRNAs and pDCs, analyzed data, and prepared figures; SB performed EV purification and characterization and contributed to conception of exosome-related experiments; PB contributed to conception of and supervised exosome-related experiments; UD and AZ contributed to the design of experiments using HEK-293 transfected cells; LA and AT managed SLE sample selection and collection and study approval; SS conceived experiments and wrote the paper; DB conceived and supervised experiments, analyzed data, and wrote the paper. All authors critically reviewed and approved the final version of the manuscript and are accountable for all aspects of the work.
Acknowledgments
This work was supported by the Italian Association for Cancer Research (AIRC) and Fondazione Berlucchi. VS was the recipient of a fellowship from Fondazione Italiana Ricerca sul Cancro (FIRC). VG was supported by the Interuniversity Attraction Poles (IAP) 7-40 program.
Version 1. 05/17/2018
Electronic publication
Footnotes
Conflict of interest: UD is to be listed as an inventor on patents owned by the GSK group of companies and has ownership of restricted GSK shares.
Reference information: JCI Insight. 2018;3(10):e98204. https://doi.org/10.1172/jci.insight.98204.
Contributor Information
Valentina Salvi, Email: valentina.salvi@unibs.it.
Veronica Gianello, Email: v.gianello@unibs.it.
Sara Busatto, Email: s.busatto@unibs.it.
Paolo Bergese, Email: paolo.bergese@unibs.it.
Laura Andreoli, Email: laura.andreoli@unibs.it.
Alessandra Zingoni, Email: Alessandra.Zingoni@uniroma1.it.
Angela Tincani, Email: angela.tincani@unibs.it.
Silvano Sozzani, Email: sozzani@med.unibs.it.
Daniela Bosisio, Email: daniela.bosisio@unibs.it.
References
- 1.Sozzani S, Del Prete A, Bosisio D. Dendritic cell recruitment and activation in autoimmunity. J Autoimmun. 2017;85:126–140. doi: 10.1016/j.jaut.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Sozzani S, Vermi W, Del Prete A, Facchetti F. Trafficking properties of plasmacytoid dendritic cells in health and disease. Trends Immunol. 2010;31(7):270–277. doi: 10.1016/j.it.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15(8):471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32(3):305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8(8):594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 6.Ganguly D, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206(9):1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayashi T, et al. Prevention of autoimmune disease by induction of tolerance to Toll-like receptor 7. Proc Natl Acad Sci USA. 2009;106(8):2764–2769. doi: 10.1073/pnas.0813037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: A Collision of Two Paradigms. Trends Biochem Sci. 2016;41(10):883–892. doi: 10.1016/j.tibs.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22(3):125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA. 2014;111(41):14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109(31):E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15(6):827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 15.Ranganathan P, et al. Serum miR-29a Is Upregulated in Acute Graft-versus-Host Disease and Activates Dendritic Cells through TLR Binding. J Immunol. 2017;198(6):2500–2512. doi: 10.4049/jimmunol.1601778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiolo D, et al. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87(8):4168–4176. doi: 10.1021/ac504861d. [DOI] [PubMed] [Google Scholar]
- 17.Paolini L, et al. Residual matrix from different separation techniques impacts exosome biological activity. Sci Rep. 2016;6:23550. doi: 10.1038/srep23550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tkach M, Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164(6):1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 20.Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 22.Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016;15(12):1171–1180. doi: 10.1016/j.autrev.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Peng W, Ouyang X, Li W, Dai Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res. 2012;160(3):198–206. doi: 10.1016/j.trsl.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Robbins M, Judge A, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19(2):89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- 25.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 26.Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus: A comprehensive review. J Autoimmun. 2015;64:125–136. doi: 10.1016/j.jaut.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Davidson A. Taming lupus-a new understanding of pathogenesis is leading to clinical advances. Nat Med. 2012;18(6):871–882. doi: 10.1038/nm.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang MJ, et al. Role of an imbalanced miRNAs axis in pathogenesis of psoriasis: novel perspectives based on review of the literature. Oncotarget. 2017;8(3):5498–5507. doi: 10.18632/oncotarget.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C. Nucleic acid agonists for Toll-like receptor 7 are defined by the presence of uridine ribonucleotides. Eur J Immunol. 2006;36(12):3256–3267. doi: 10.1002/eji.200636617. [DOI] [PubMed] [Google Scholar]
- 30.Rolle K, et al. The Sequence and Structure Determine the Function of Mature Human miRNAs. PLoS ONE. 2016;11(3):e0151246. doi: 10.1371/journal.pone.0151246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rettig L, et al. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010;115(22):4533–4541. doi: 10.1182/blood-2009-11-247817. [DOI] [PubMed] [Google Scholar]
- 32.Prud’homme GJ, Glinka Y, Lichner Z, Yousef GM. Neuropilin-1 is a receptor for extracellular miRNA and AGO2/miRNA complexes and mediates the internalization of miRNAs that modulate cell function. Oncotarget. 2016;7(42):68057–68071. doi: 10.18632/oncotarget.10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 34.Romero-Diaz J, Isenberg D, Ramsey-Goldman R. Measures of adult systemic lupus erythematosus: updated version of British Isles Lupus Assessment Group (BILAG 2004), European Consensus Lupus Activity Measurements (ECLAM), Systemic Lupus Activity Measure, Revised (SLAM-R), Systemic Lupus Activity Questionnaire for Population Studies (SLAQ), Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI-2K), and Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index (SDI) Arthritis Care Res (Hoboken) 2011;63 Suppl 11:S37–S46. doi: 10.1002/acr.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvi V, et al. IL-21 May Promote Granzyme B-Dependent NK/Plasmacytoid Dendritic Cell Functional Interaction in Cutaneous Lupus Erythematosus. J Invest Dermatol. 2017;137(7):1493–1500. doi: 10.1016/j.jid.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 36.Salvi V, et al. Dual regulation of osteopontin production by TLR stimulation in dendritic cells. J Leukoc Biol. 2013;94(1):147–158. doi: 10.1189/jlb.0412194. [DOI] [PubMed] [Google Scholar]
- 37.Montis C, et al. Size distribution of extracellular vesicles by optical correlation techniques. Colloids Surf B Biointerfaces. 2017;158:331–338. doi: 10.1016/j.colsurfb.2017.06.047. [DOI] [PubMed] [Google Scholar]
- 38.Radeghieri A, et al. Cultured human amniocytes express hTERT, which is distributed between nucleus and cytoplasm and is secreted in extracellular vesicles. Biochem Biophys Res Commun. 2017;483(1):706–711. doi: 10.1016/j.bbrc.2016.12.077. [DOI] [PubMed] [Google Scholar]
- 39.Horcas I, Fernández R, Gómez-Rodríguez JM, Colchero J, Gómez-Herrero J, Baro AM. WSXM: a software for scanning probe microscopy and a tool for nanotechnology. Rev Sci Instrum. 2007;78(1):013705. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 40.Salvi V, Bosisio D, Mitola S, Andreoli L, Tincani A, Sozzani S. Trichostatin A blocks type I interferon production by activated plasmacytoid dendritic cells. Immunobiology. 2010;215(9-10):756–761. doi: 10.1016/j.imbio.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Salvi V, et al. TLR Signalling Pathways Diverge in Their Ability to Induce PGE2. Mediators Inflamm. 2016;2016:5678046. doi: 10.1155/2016/5678046. [DOI] [PMC free article] [PubMed] [Google Scholar]