Abstract
The innate immune response is, in the first place, elicited at the site of infection. Thus, the host response can be different among the infected cells and the cells surrounding them. Effector-triggered immunity (ETI), a form of innate immunity in plants, is triggered by specific recognition between pathogen effectors and their corresponding plant cytosolic immune receptors, resulting in rapid localized cell death known as hypersensitive response (HR). HR cell death is usually limited to a few cells at the infection site, and is surrounded by a few layers of cells massively expressing defense genes such as Pathogenesis-Related Gene 1 (PR1). This virtually concentric pattern of the cellular responses in ETI is proposed to be regulated by a concentration gradient of salicylic acid (SA), a phytohormone accumulated around the infection site. Recent studies demonstrated that jasmonic acid (JA), another phytohormone known to be mutually antagonistic to SA in many cases, is also accumulated in and required for ETI, suggesting that ETI is a unique case. However, the molecular basis for this uniqueness remained largely to be solved. Here, we found that, using intravital time-lapse imaging, the JA signaling pathway is activated in the cells surrounding the central SA-active cells around the infection sites in Arabidopsis thaliana. This distinct spatial organization explains how these two phythormone pathways in a mutually antagonistic relationship can be activated simultaneously during ETI. Our results re-emphasize that the spatial consideration is a key strategy to gain mechanistic insights into the apparently complex signaling cross-talk in immunity.
Keywords: Arabidopsis thaliana, Effector-triggered immunity, Jasmonic acid, Salicylic acid, Pseudomonas syringae pv. tomato DC3000 carrying AvrRpt2, Time-lapse imaging
Introduction
Innate immunity is activated upon perception of pathogen-derived molecules by the host cells. Effector-triggered immunity (ETI), a form of innate immunity in plants, is activated by specific recognition of pathogen effector activities by their corresponding plant cytosolic immune receptors (Dodds and Rathjen 2010). Activation of ETI leads to transcriptional up-regulation of defense-related genes and is often associated with rapid localized cell death at the site of infection, known as hypersensitive response (HR) (Dodds and Rathjen 2010). Thus, HR cell death lesion limited to a few cells in the vicinity of the infection site appears to be surrounded by a few layers of cells massively expressing a number of defense genes such as Pathogenesis-Related Gene 1 (PR1) (Schmelzer et al. 1989, Ohshima et al. 1990, Enyedi et al. 1992, Murray et al. 2002).
The plant hormone salicylic acid (SA), which plays a key role in plant immunity against biotrophic pathogens, has been implicated in the formation of such a virtually concentric pattern of the cellular responses observed in HR (Enyedi et al. 1992, Dorey et al. 1997, Fu et al. 2012, Yan and Dong 2014). A recent study using Arabidopsis thaliana also proposed that a concentration gradient of SA formed around the pathogen infection site elicits different defense responses, such as cell death and PR1 activation, in a dose-dependent manner (Fu et al. 2012). Thus, the French flag model, which explains various processes of development, might also be applicable to explain how plant cells acquire positional information upon a pathogen attack to produce the organized local immune responses around the infection site (Wolpert 1969, Enyedi et al. 1992, Dorey et al. 1997, Fu et al. 2012).
A number of studies have demonstrated that the SA pathway functions antagonistically against a signaling pathway controlled by jasmonic acid (JA), another plant hormone required for immunity against necrotrophic pathogens as well as defense responses upon physical wounding (Glazebrook 2005, Vlot et al. 2009). Due to the mutually inhibitory relationship between the SA and JA pathways, plants are considered to select either pathway depending on lifestyles of invading pathogens (Glazebrook 2005). Several studies, however, reported that JA is accumulated massively when ETI is evoked (Kenton et al. 1999, Spoel et al. 2003). Moreover, the JA pathway was shown to contribute positively to ETI (Tsuda et al. 2009, Liu et al. 2016). These findings suggest that the SA and JA pathways are activated simultaneously in the same plants only in the case of ETI. Accordingly, the spatially controlled trade-off between the SA and JA pathways within a single leaf was not observed exceptionally in the case of ETI (Spoel et al. 2007). These findings that are apparently contradictory to the well-established antagonistic relationship between the SA and JA pathways might be explained by the characteristic compensatory signaling network structure governing ETI and/or the proposed unique interplay between SA and JA only found during ETI (Tsuda et al. 2009, Liu et al. 2016). Alternatively, it could be explained by simply introducing another layer of complexity, e.g. a spatiotemporal aspect, into the study of plant immunity. Indeed, our knowledge about the spatiotemporal dynamics of the plant immune response remains largely limited (Murray et al. 2002).
Here, we report the spatiotemporal dynamics of defense-related promoter activities during ETI in A. thaliana. We established a time-lapse imaging assay of defense-related promoter activities using a fluorescent reporter protein in non-detached plant leaves, which enabled us to capture spatiotemporal development of plant immune responses. Using the system, we detected the spatiotemporal dynamics of the promoter activities of PR1, a conventional marker of SA activity, as well as of Vegetative Storage Protein 1 (VSP1), a JA marker, for 40 h after hand-infiltration of Pseudomonas syringae pv. tomato DC3000 carrying AvrRpt2 (Pst_a2). Our imaging data indicate that the SA and JA pathways are activated in distinct concentric domains; inner SA and outer JA domains around the HR cell death area, which explains the previous observations on the SA–JA relationship without contradiction. Our results shed light on the importance of spatial consideration as a key strategy to gain mechanistic insights into the apparently complex signaling cross-talk in plant immunity.
Results
The use of a nuclear-localized fluorescent protein discriminates fluorescent-based promoter activity from autofluorescence in planta
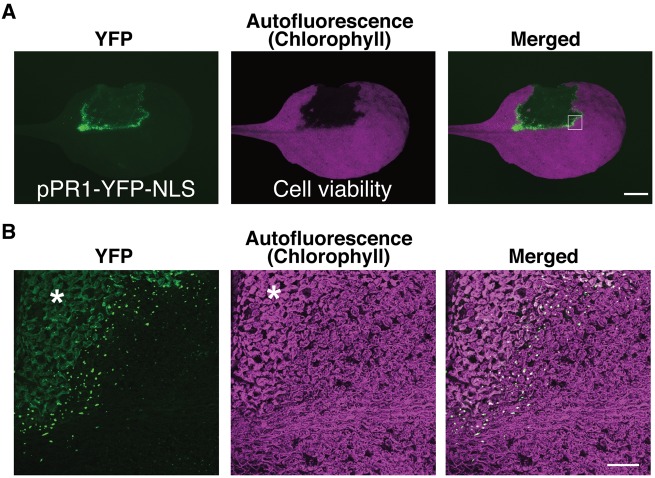
Plant immune response involves a dynamic transcriptional reprogramming regulated by multiple transcription factors as well as phytohormones (Tsuda and Somssich 2015). Accumulating data sets of a number of transcriptome profiling studies provide us with the opportunities to capture information on the plant immune system, e.g. the network structures of the signaling cascades as well as the temporal dynamics (Mine et al. 2014). However, our knowledge about the spatial dynamism of ETI still remains limited (Murray et al. 2002). In order to extend our understanding of the spatial aspect of the plant immune system, we generated transgenic Arabidopsis plants expressing yellow fluorescent protein (YFP) fused to the nuclear localization signal (YFP–NLS) under the control of the promoters of the defense marker genes (Kubo et al. 2005). First, we focused on the promoter activity of the PR1 gene, a well-established marker gene for immunity controlled by SA (Vlot et al. 2009). Hand-infiltration of A. thaliana (ecotype Col-0) leaves with a dense suspension of Pst_a2 results in Resistance to Pseudomonas syringae 2 (RPS2)-2- (RPS2) mediated ETI associated with HR cell death (Yu et al. 1993). Autofluorescence derived from plant chlorophylls (Chls) was utilized as a marker to detect living plant cells (Guadagno et al. 2017). Using a wide-field fluorescent stereomicroscope with our setting described in the Materials and Methods, we succeeded in visualizing the HR lesion by loss of Chl autofluorescence (Fig. 1A). The use of the characterized 4.5 kb upstream sequence of PR1 reproduces the characteristic pattern of PR1 promoter (pPR1) active cells surrounding the HR lesion 22 hours post-inoculation (h.p.i.) (Fig. 1A) (Lebel et al. 1998, Murray et al. 2002). Infiltration of the mock solution did not activate pPR1 (Supplementary Fig. S1). The pPR1 activity is relatively high around the mid-rib, as observed for ETI triggered by another Pseudomonas strain (Murray et al. 2002). Although pPR1 activity has been extensively studied and characterized by various means such as the luciferase reporter, the use of YFP–NLS enabled us to detect every single cell, in which pPR1 is activated, in the infiltrated leaves of the pPR1 reporter (hereafter, pPR1-YFP-NLS) plants, providing information on the promoter activation pattern with cellular resolution when combined with appropriate microscopes (Fig. 1B) (Murray et al. 2002). Furthermore, NLS-mediated accumulation of YFP in the nuclei allows us to distinguish the pPR1-driven YFP signal from plant autofluorescence accumulated during immune responses (Fig. 1B) (Bennett et al. 1996). In a magnified confocal image of the HR cell death border, only several layers of cells from the border exhibit massive YFP accumulation in the nuclei, indicating high pPR1 activity (Fig. 1B). The nuclear YFP signals are suddenly decreased outside of the cell layers showing the pPR1 maxima surrounding the HR lesion (Fig. 1B). This observation indicates that pPR1 activity is spatially regulated and strictly confined to the vicinity of the infection sites, which is in accordance with the published data of other defense-related genes activated in HR (Schmelzer et al. 1989, Ohshima et al. 1990). Thus, the promoter–YFP–NLS reporter transgenic plants are useful tools to give us a good spatial resolution in our understanding of ETI without any fixation or enzymatic reaction.
Fig. 1.
The use of YFP–NLS to visualize in planta promoter activity. (A) Fluorescent stereomicroscopic images of a pPR1-YFP-NLS leaf partially infiltrated with Pst_a2 (OD600 = 0.2) at 22 h.p.i. HR cell death was detected by the loss of Chl autofluorescence. Scale bar = 2.5 mm. (B) Confocal images of the region corresponding to the white square in (A). In our confocal setting described in the Materials and Methods, dead cells were visualized by whole-cell autofluorescence detected in both YFP and Chl autofluorescence images (in the area shown with an asterisk). They are distinguished from the nuclear- and chloroplast-localized signals. Scale bar = 200 μm. (See also Supplementary Fig. S1).
In toto live-imaging of a whole intact leaf undergoing ETI triggered by RPS2 revealed a dynamic spatiotemporal pattern of pPR1 activation
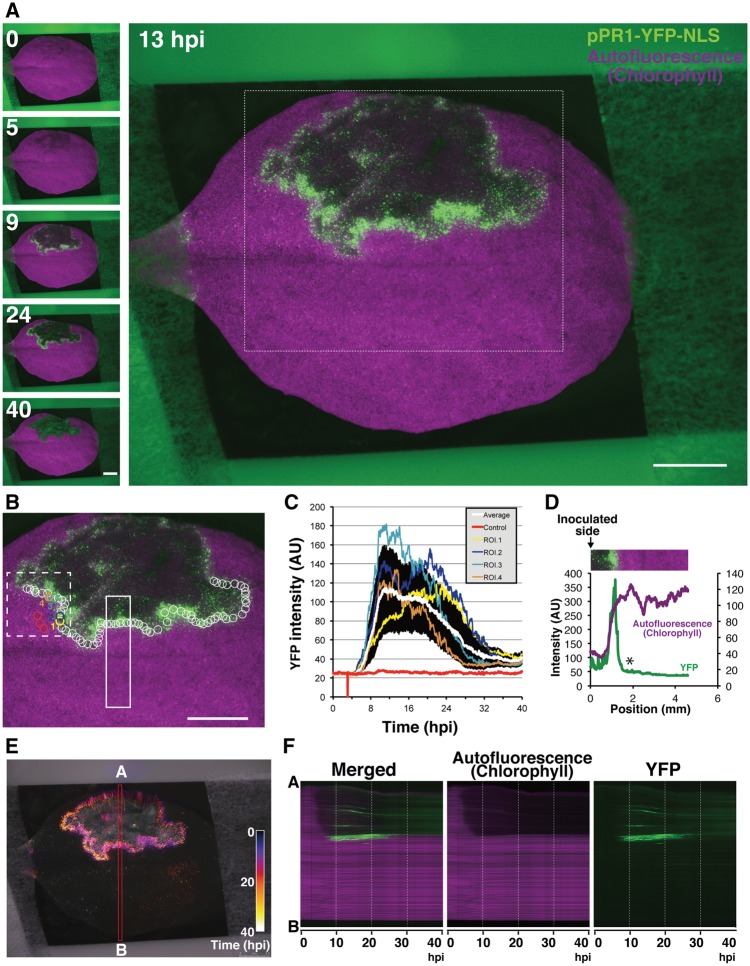
Biological events such as immunity and development progress over time. Thus, we combined the reporter plants with an automated fluorescent stereomicroscope, allowing us to perform intravital time-lapse imaging to capture the promoter dynamics in a soil-grown intact plant leaf over a few days. Since the light condition strikingly affected PR1 accumulation during ETI, we designed the time-lapse program in which YFP and autofluorescence images were taken at 3 min intervals programmed to expose the plant specimen to the light (Zeier et al. 2004). Time-lapse imaging of a pPR1-YFP-NLS leaf infiltrated with a dense suspension of Pst_a2 revealed that pPR1 is transiently activated around the HR cell death lesion (Fig. 2A; Supplementary Fig. S2A; Supplementary Movie S1). To analyze the temporal pPR1 activation profile, we set multiple regions of interest (ROIs) containing a few cells to cover almost all of the pPR1 active area at the margin of the HR lesion (white circles in Fig. 2B). The averaged data show an exponential increase of the YFP intensity, with a single peak around 12 h.p.i. followed by a gentle decrease by 40 h.p.i. (the white line in Fig. 2C). The YFP intensity profile after 24 h.p.i. could contain values derived from the background autofluorescence accumulated in the HR cell death area that cannot be distinguished from the YFP signal at this resolution (as in Fig. 1). Nevertheless, the averaged YFP intensity profile exhibits a transient activation of pPR1 during ETI. Focusing on the YFP profiles of the individual single ROIs, not the averaged data, however, showed notable variations in the temporal intensity profiles, even in comparisons of two ROIs located close to each other (i.e. ROI 1 vs. ROI 2 and ROI 3 vs. ROI 4 in Fig. 2B, C and Supplementary Movie S2). This is also recognized as large SDs of the YFP intensity profiles during the pPR1 active period (Fig. 2C). These tendencies of pPR1 activation around the HR lesion described above are also observed reproducibly (Supplementary Fig. S2B, C). These observations imply that the kinetics of pPR1 activation at the cellular level are not as simple as one might expect from the analyses of bulked tissues.
Fig. 2.
Visualization of pPR1 dynamics during RPS2-conditioned ETI. (A) Selected time-lapse images out of 800 images of the in vivo spatiotemporal dynamics of pPR1 activity for 40 h after Pst_a2 infiltration (OD600 = 0.2). Scale bars = 2.5 mm. (B) The region corresponding to the white box in (A) showing the positions of ROIs analyzed here. The red circles are used for background controls and the other circles are for calculating average intensity. The YFP intensity profiles in the numbered ROIs are individually shown in (C). The circles are all the same size. Scale bar = 2.5 mm. (C) YFP intensity plots in the ROIs shown in (B). Means � SD are plotted for the control (n = 3) and the others (n = 70). YFP profiles in four selected ROIs out of 70 ROIs are independently shown. The value 0 is due to the accidental loss of an image in the time-lapse system. (D) The intensity profiles of YFP and Chl autofluorescence in the white closed box shown in (B). A white asterisk indicates a sudden fall of YFP intensity outside the infection site. (E) Spatiotemporal dynamics of pPR1 activity are shown in Temporal-Color Code. All the YFP images corresponding to (A) are re-colored by a specific Temporal-Color Code shown in the picture. (F) Kymographs corresponding to the red box along the A–B axis (E) are generated. (See also Supplementary Fig. S2; Supplementary Movies S1, S2).
As previously reported, we also observed sharp borders between the HR cell death area and the surrounding pPR1 active area, with the remainder of the tissue showing no apparent response (Fig. 2D) (Schmelzer et al. 1989). Using 800 time-lapse images for 40 h after Pst_a2 inoculation, we generated a Temporal-Color-Coded image of the YFP signals as well as a kymograph, both enabling us to capture the spatiotemporal dynamics of pPR1 activation in RPS2-mediated ETI (Fig. 2E, F). The images reveal that the clear demarcation of the pPR1 active zone was strictly maintained over 40 h around the lesion, indicating the presence of an active regulatory mechanisms to limit the propagation of the pPR1 active area (Fig. 2E, F). In addition, we detected weak and transient pPR1 activation in the uninfiltrated side of the leaf after the local pPR1 activity declines (Fig. 2E; Supplementary Fig. S2D, E). Although the spatial pattern of this pPR1 active area differs in every experiment, the weak pPR1 activation in the uninfiltrated side of the inoculated leaves is reproducible (Supplementary Fig. S2F). Plants possess an ability to activate immunity systemically upon local pathogen challenges (Vlot et al. 2009). Our observation might indicate that a similar phenomenon happens even at the uninfected side of the inoculated leaf. Thus, our data obtained using intravital time-lapse imaging demonstrate that pPR1 activation in RPS2-conditioned ETI appears to possess two modes of action; the primary strong activation around the infection sites within the first 24 h followed by the secondary weak activation in the uninfiltrated area. Spatial resolution achieved by imaging techniques enables us to detect, for the first time, these two phases of pPR1 activation in spatially distinct area of leaves. Furthermore, it also reveals that individual cells exhibit temporal variation in pPR1 activation, although the leaf taken as a whole appears to activate pPR1 transiently with a single peak. Conventional molecular and biochemical techniques analyzing whole-leaf extracts might have overlooked such fluctuations in pPR1 activation at the single-cell level.
The promoter of VSP1, a JA marker gene, is activated in the periphery of the pPR1 active domain around the HR lesion
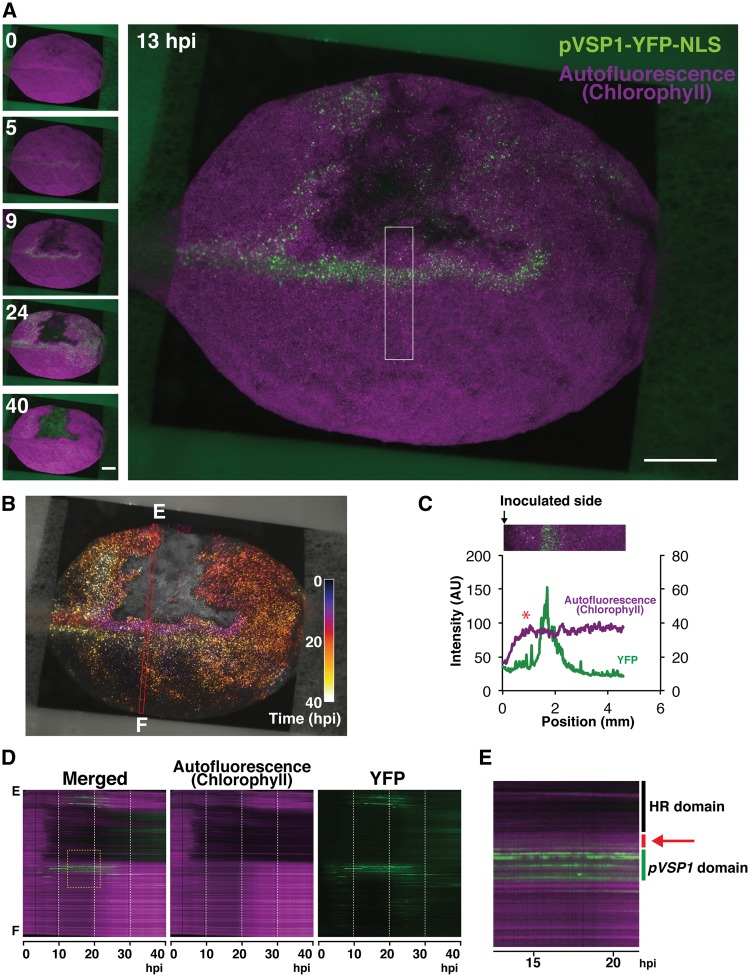
Recent studies showed a positive contribution of JA, in addition to SA, in invoking ETI conditioned by RPS2 (Tsuda et al. 2009, Liu et al. 2016). JA and SA are known to function usually in an antagonistic relationship, and thus simultaneous activation of SA and JA in ETI is considered to be a unique case (Vlot et al. 2009, Liu et al. 2016). However, those studies might have overlooked the spatial organization of two phytohormone signaling pathways. Therefore, we analyzed the activity of the promoter of VSP1 (pVSP1), a JA marker gene, in response to infiltration of Pst_a2 (Berger et al. 1995, Utsugi et al. 1998, Guerineau et al. 2003). Time-lapse imaging of a leaf of the transgenic plant carrying YFP–NLS under the control of pVSP1 (pVSP1-YFP-NLS) detected transient and strong activation of pVSP1 around the infection site (Fig. 3A; Supplementary Fig. S3A; Supplementary Movie S3). At the periphery of the infection site, pVSP1 activity is observed earlier than that of pPR1, but activation kinetics of pVSP1 show a variation to some extent (Supplementary Fig. S3B-F). Since a weak pVSP1 activity is often observed along the vasculature, the position of the infection site in relation to the vascular system might affect the pVSP1 activation kinetics during ETI (Supplementary Fig. S3A). After approximately 15 h.p.i., the pVSP1 active area is gradually propagated from the infection site to the uninfected area, especially at the side of the leaf tip (Fig. 3A, B; Supplementary Fig. S3, AG; Supplementary Movie S3).
Fig. 3.
Visualization of pVSP1 dynamics during RPS2-conditioned ETI. (A) Selected time-lapse images out of 800 images of in vivo spatiotemporal dynamics of pVSP1 activity for 40 h after Pst_a2 infiltration (OD600 = 0.2). Scale bars = 2.5 mm. (B) Spatiotemporal dynamics of pVSP1 activity are shown in Temporal-Color Code. All the YFP images corresponding to (A) are re-colored by a specific Temporal-Color Code shown in the picture. (C) The intensity profiles of YFP and Chl autofluorescence in the white closed box shown in (A). A red asterisk indicates a spatial gap between the HR cell death lesion and pVSP1 active domain. (D) Kymographs corresponding to the red box along the E–F axis in (B). (E) A magnified view of the yellow dashed box in (D). A spatial gap between the HR cell death lesion and pVSP1 active domain is indicated by a red bar and a red arrow. (See also Supplementary Fig. S3; Supplementary Movie S3).
In contrast to the pPR1 active domain observed just around the HR cell death border, a spatial gap was detected reproducibly between the pVSP1 active domain and the HR cell death area despite a variation of pVSP1 activation kinetics in ETI (Fig. 3C–E; Supplementary Fig. S3H). Mock treatment does not trigger such a pattern of pVSP1 activation (Supplementary Fig. S3I). In addition, Pst_a2-infiltrated leaves without time-lapse imaging also produced a similar pVSP1 activation pattern with a pVSP1 inactive gap around the HR lesion (Supplementary Fig. S3J). These data suggest that this characteristic pVSP1 activation pattern is not due to any stress from infiltration or continuous imaging. Since pPR1 is activated just at the margin of the HR cell death domain, pVSP1 activation is considered to occur mainly outside the pPR1 active domain around the infection sites (Figs. 2D, F, 3C–E). These data strongly indicate that this virtually concentric pattern of the inner pPR1 active and the outer pVSP1 active domains around the infection foci is prominent in RPS2-triggered immunity.
SA accumulates in the pPR1 active cells in early ETI
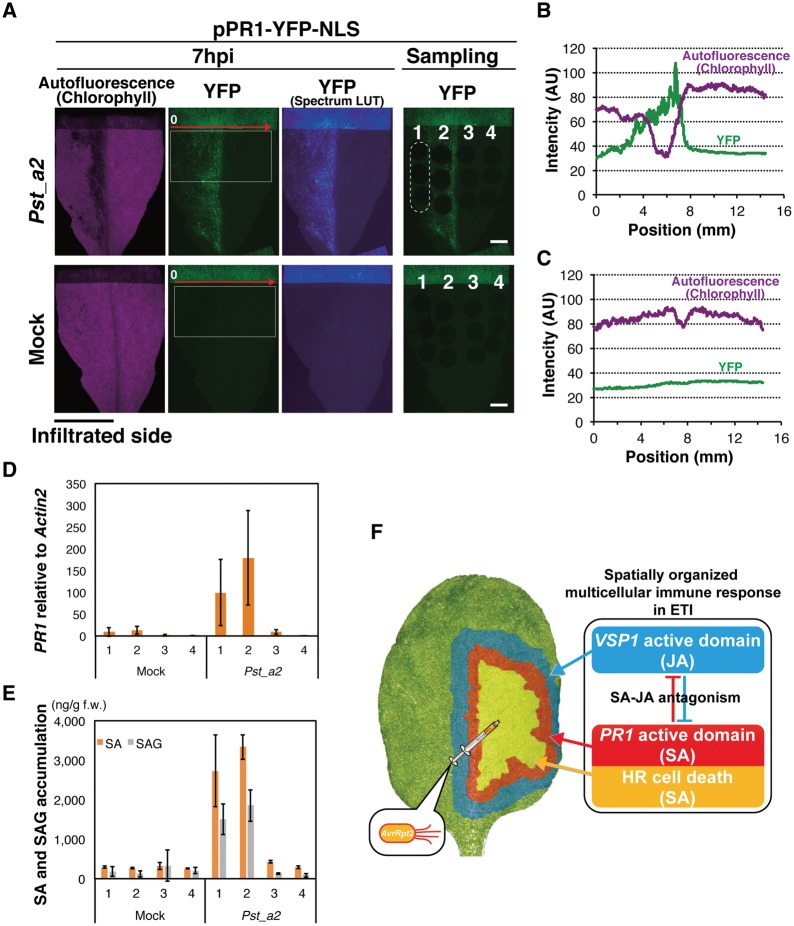
A recent study showed that, in ETI triggered by RPS2, PR1 accumulation was maintained through MPK3/6 activity in the salicylic acid induction-deficient 2 (sid2) mutants that fail to accumulate SA fully upon pathogen challenge, arguing that PR1 might no longer be an SA marker in ETI (Tsuda et al. 2013). Therefore, we analyzed the spatial distribution of SA in relation to pPR1 activity in leaves elicited by RPS2. Infiltration of one half of pPR1-YFP-NLS leaves with Pst_a2, not with the mock solution, resulted in YFP accumulation detectable only in the infiltrated side at 7 h.p.i. (Fig. 4A–C). Using these leaves, four different zones along the border between the infiltrated (zones 1 and 2) and uninfiltrated (zones 3 and 4) halves were analyzed for SA levels as well as endogenous PR1 transcript levels (Fig. 4A, D, E). As visualized by YFP, Pst_a2-infiltrated zones highly accumulate PR1 transcripts (zones 1 and 2). The PR1 level in zone 3 is only 5% of that in zone 2 in Pst_a2-treated leaves (Fig. 4D). A similar trend is observed for SA distribution. Zone 3 accumulates only 13% of the SA level in zone 2 in those leaves (Fig. 4D). Thus, in RPS2-conditioned ETI, massive SA accumulation largely coincides with high PR1 transcript levels. An earlier study has shown that the transgenic plants constitutively expressing NahG, a bacterial SA-catabolizing enzyme, were unable to accumulate PR1 transcripts in response to an incompatible pathogen (Delaney et al. 1994). Accordingly, the strong correlation between SA accumulation and the PR1 transcript levels cannot be rejected in ETI in the wild-type background, although MPK3/6-mediated compensation of PR1 activation might occur in the absence of SID2 activity in ETI. Thus, the spatiotemporal pattern of pPR1 activation observed in this study possesses a strong correlation with SA accumulation during ETI triggered by RPS2. Our data suggest formation of a steep SA concentration gradient around the infection site in early ETI (Fig. 4E), which is reflected as a sharp decrease of YFP intensity over the infiltrated/non-infiltrated border. Our data obtained in Arabidopsis agree with the previous findings in tobacco, suggesting conservation of a sharp SA gradient in ETI among plants(Enyedi et al. 1992).
Fig. 4.
Spatial regulation of SA accumulation leading to an organized multicellular response in ETI. (A) Site-specific sampling for SA and PR1 analyses at 7 h.p.i. A nearly complete half of a pPR1-YFP-NLS leaf was fully infiltrated with Pst_a2 (OD600 = 0.2, upper) or 10 mM MgCl2 (mock, lower). The leaf was divided into four areas along the mid-rib (numbered 1–4), and three leaf disks (2 mm in diameter) per area were sampled, as shown in a dashed ellipse for zone 1 in the upper right pictures. Representative sample pictures are shown. Scale bars = 2.5 mm. (B) Intensity profiles of YFP and autofluorescence in the white boxes in the Pst_a2-treated leaf in (A) along the red arrow. (C) Intensity profiles of YFP and autofluorescence in the white boxes in the mock-treated leaf in (A) along the red arrow. (D) The endogenous PR1 expression levels in the four zones were measured by qRT–PCR. Eighteen leaf disks, corresponding to one zone, from six leaves were pooled as one sample. Bars represent means � SD of three biological replicates. (E) The free SA and SA glycoside (SAG) levels in the four zones. Three disks from one zone from one leaf were pooled and analyzed. Bars represent means � SD of three leaves. Experiments were repeated twice with similar results. (F) A schematic summary of an organized concentric pattern of the inner SA and the outer JA active domains which appeared around the infection site of Pst_a2. (See also Supplementary Fig. S4).
As for JA, we detected higher JA accumulation from extracts of whole leaves challenged by Pst_a2 at 24 h.p.i., compared with mock treatment (Supplementary Fig. S4A). Accordingly, the VSP1 transcript levels are also found to be accumulated in the pathogen-challenged whole-leaf samples at 24 h.p.i. (Supplementary Fig. S4B). Using the same site-specific samples as used for PR1 analysis, we examined the spatial pattern of VSP1 accumulation (Supplementary Fig. S4C). Although the variance in the VSP1 transcript levels in the respective samples is very high, as expected from live-imaging data, there appears to be a trend that the uninfiltrated area (zones 3 and 4) showed higher VSP1 accumulation than the infiltrated area (zones 1 and 2) in the pathogen-treated leaves, supporting our finding that VSP1 is activated outside the infected area where PR1 is activated (Supplementary Fig. S4C). Thus, spatial separation of the pPR1 and pVSP1 active domains are confirmed, at least at the level of mRNA accumulation in the early RPS2-mediated immunity.
Discussion
Our data revealed that PR1 and VSP1 genes are activated in different domains, namely the local PR1 and the peripheral VSP1 domains, around the infection site where RPS2-conditined immunity is triggered. PR1 has been considered to be a conventional marker gene for SA in the case of the wild-type background (Vlot et al. 2009) and PR1 transcript accumulation spatially correlates with SA accumulation in early RPS2-mediated immunity (Fig. 4A, D, E). VSP1 is a well-established marker for a branch of the JA signaling pathway (Kazan and Manners 2013). Collectively, our finding indicates that the SA and JA pathways are spatially separated domains as exemplified by two respective marker gene promoter activities in early RPS2-trrigered immunity. Since the loss of SA accumulation by constitutive expression of NahG compromises not only PR1 activation but also HR cell death, these local events are mainly considered to be under the control of SA accumulated around the infection foci (Delaney et al. 1994). The SA concentration gradient formed around the infection center appears to be rather steep not only in RPS2-conditioned ETI (Fig. 4E), but also in tobacco ETI against a viral pathogen (Enyedi et al. 1992), indicating the presence of strict spatial regulation of the SA activation conserved in ETI. Considering the well-characterized mutually antagonistic relationship between SA and JA across multiple plant species (Spoel et al. 2007, Vlot et al. 2009), the JA active domain outside the SA active infection foci might contribute to limit overactivation of the SA pathway spatially around the infection site. This hypothesis should be further tested carefully by means of genetics and cell biology. In addition, the outer JA active domain may constitute another layer of ETI outside the SA domain, since the JA pathway also contributes positively to ETI conditioned by RPS2 (Tsuda et al. 2009, Liu et al. 2016). The SA active cells undergoing programmed cell death during HR could be a target of necrotrophic pathogens (Spoel et al. 2007, Liu et al. 2016). The JA active domain surrounding the central SA active domain may have a function to protect living plant cells around the necrotic HR lesion from such secondary infections of necrotrophs. Taken together, this virtually concentric pattern of the inner SA and the outer JA active domains found in this study may constitute a field of cells expressing orchestrated and comprehensive defense responses around the infection site during ETI. The biological relevance of this spatially organized ETI active field formation around the infection site now needs to be dissected further in detail. Another open question is whether or not this SA–JA concentric pattern in the ETI active field, which emerged around the infection site, is only specific to ETI conditioned by RPS2. It will be worth investigating other pathosystems including non-ETI-causing pathogens using these promoter–reporters.
Our imaging-based analysis shed light on the importance of spatial aspects in understanding the complex plant immune signaling. Liu et al. (2016) demonstrated that the early activation of the JA pathway requires SA through SA receptors, instead of the conventional JA pathway (Liu et al. 2016). However, our data indicated that VSP1 activation precedes PR1. More detailed genetic, biochemical and imaging-based studies, including other ETI responses, are required to understand how the concentric SA and JA active domains are formed from the spatiotemporal point of view. In addition, our data suggested, for the first time, that an apparent transient PR1 activation is achieved through highly variable PR1 activation in individual cells. Further detailed analysis at the single-cell level would answer the question of how plant tissues organize variable individual cellular activities into such collective behavior to form the concentric SA and JA active domains around the infection center. The complex activation of PR1 and VSP1 in the uninfiltrated side of the leaves could also be further studied by single-cell level analyses. Thus, our imaging-based study stimulates further research to explore plant immunity spatiotemporally.
Materials and Methods
Plant materials and growth conditions
The A. thaliana wild type used in this study was Col-0. Water-soaked seeds were sown on soil and grown in a growth room at 23 �C under continuous white light (20–50 mmol m−2 s−1). Two- to three-week-old plants were used for all the experiments in this study.
Construction of transgenic promoter reporter plants
The 4.5 kb promoter of the PR1 gene and the 3.0 kb promoter of the VSP1 gene, both of which covered the previously analyzed respective regulatory sequences, were amplified from the genomic DNA (Col-0) by PCR and cloned into the pENTR/D-TOPO vector (Invitrogen) (Lebel et al. 1998, Utsugi et al. 1998). Primers used for the cloning are listed in Supplementary Table S1. The promoter regions were recombined with the aid of Gateway technology into the binary pBGYN vector (Kubo et al. 2005). The resulting pBGYN-pPR1-YFP-NLS and pBGYN-pVSP1-YFP-NLS vectors were introduced into Agrabocterium tumefaciens GV3101::pMP90 and then into A. thaliana Col-0 wild-type plants using the floral dip method (Clough and Bent 1998). The characteristic spatial patterns of pPR1 and pVSP1 activities upon Pst_a2 infiltration, shown in this study, were confirmed in multiple T1 plants. Three (pPR1-YFP-NLS) and two (pVSP1-YFP-NLS) homozygous lines were selected by segregation analysis in the following generations. A representative homozygous line was selected for each for further detailed analyses.
Pseudomonas inoculation
Pst_a2 was previously described (Aarts et al. 1998). The bacterial cells were harvested and resuspended in 10 mM MgCl2 to appropriate optical densities measured by a DU640 spectrophotometer (Beckman). The bacterial suspensions were infiltrated by hand into leaves using a 1 ml needleless syringe (Terumo).
Time-lapse imaging
Time-lapse imaging was performed using an M205FA automated stereomicroscope controlled by AF6000 software (Leica Microsystems). A DFC365FX camera (Leica Microsystems) was used in 12-bit mode. Chl autofluorescence and YFP were detected through TXR and YFP filters, respectively (both Leica Microsystems). The TXR filter enabled us to reduce almost fully non-specific autofluorescence from dead cells. Bright field, YFP and TXR pictures were taken every 3 min and the intervals were programmed to expose the plant specimen to the light from the light-emitting diode (LED). Data analyses were performed with AF6000 (intensity plots) and Fiji (intensity plots, kymographs and Temporal-Color Code, ver. 2.0.0-rc-12/1.49g, build. 2352160d02) software.
Confocal microscopy
Confocal images were taken using a confocal microscope FV1200 equipped with UPLSAPO10 �2, NA: 0.40 (Olympus). Z-projected pictures were generated by the FV10-ASW (Olympus). Chl autofluorescence was captured through propidium iodide (PI) and red fluorescent protein (RFP) pre-setting. Enhanced green fluorescent proteim (eGFP) pre-setting was used for YFP imaging.
Gene expression analysis
Total RNA was isolated using an RNeasy Plant Mini Kit (Qiagen). For the first-strand cDNA synthesis using Superscript III, 100 ng (leaf disk samples) or 150 ng (whole-leaf samples) of total RNA were used. Quantitative reverse transcription–PCR (qRT–PCR) analysis was performed with a LightCycler TaqMan Master (Roche Applied Science) on a LightCycler 480 instrument II (Roche Applied Science). Relative mRNA levels were determined using ACT2 as a reference gene. PR1, VSP1 and ACT2 expression was measured using UPLs #135, #91 and #30, respectively, with the primers listed in Supplementary Table S1.
Quantification of hormone levels
The quantification of SA and SA glucoside (SAG) was performed as described previously with a minor modification (Seo et al. 1995). Briefly, three leaf disks were frozen and ground using liquid nitrogen. SA and SAG were extracted with 90% methanol, and SAG was converted to SA by β-glucosidase treatment. After separation by HPLC (Shimadzu) with an ODS column (μ-Bondasphere C18, 150 mm�ID3.9 mm, 5 μm, 100A; Waters), SA levels were determined using a fluorescence detector (RF-20A; Shimadzu) with an excitation wavelength of 313 nm and an emission wavelength of 405 nm.
JA quantification was performed as described previously (Kojima et al. 2009, Shinozaki et al. 2015). Briefly, leaf samples (one leaf per sample) were frozen and ground using liquid nitrogen, and freeze dried. JA was extracted and determined using an ultra-HPLC-Q-Exactive™ system (Thermo Scientific) using an ODS column (AQUITY UPLC BEH C18, 1.7 μm, 2.1�100 mm; Waters) as described (Shinozaki et al. 2015).
Supplementary Data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Science and Technology Agency [PRESTO117665 to S.B., ERATOJPMJER1502 to N.N.] and by the Japan Society for the Promotion of Science [Grants-in-Aid for Research Activity Start-up (22880008 to S.B.), for Young Scientists (B) (23780040 to S.B.) and for Young Scientists (A) (23688005 to S.K.)].
Supplementary Material
Acknowledgments
We thank A. Senzaki, Y. Suzuki, Y. Sugisawa, T. Hosaka and E. Betsuyaku for excellent technical assistance, J. Parker for providing the Pst_a2 strain, and T. Demura for the pBGYN vector. S.B. is deeply grateful to K. Shimamoto for his continuous encouragement at the very early stage of this study. We thank J. Parker, M. Sato and K. Shirasu for critically reading the manuscript.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- Act2
Actin 2
- Chl
chlorophyll
- ETI
effector-triggered immunity
- h.p.i
hours post-inoculation
- HR
hypersensitive response
- JA
jasmonic acid
- MPK
mitogen-activated protein kinase
- pPR1
PR1 promoter
- PR1
Pathogenesis-Related Gene 1
- Pst_a2
Pseudomonas syringae pv. tomato DC3000 carrying the AvrRpt2 effector
- pVSP1
VSP1 promoter
- qRT–PCR
quantitative reverse transcription–PCR
- ROI
region of interest
- RPS2
Resistance to Pseudomonas syringae 2
- SA
salicylic acid
- SID2
Salicylic acid Induction-Deficient 2
- VSP1
Vegetative Storage Protein 1
- YFP
yellow fluorescent protein
- YFP–NLS
YFP fused to the nuclear localization signal
References
- Aarts N., Metz M., Holub E., Staskawicz B.J., Daniels M.J., Parker J.E. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M., Gallagher M., Fagg J., Bestwick C., Paul T., Beale M., et al. (1996) The hypersensitive reaction, membrane damage and accumulation of autofluorescent phenolics in lettuce cells challenged by Bremia lactucae. Plant J. 9: 851–865. [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J.E. (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27: 933–942. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010) Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Dorey S., Baillieul F., Pierrel M.A. (1997) Spatial and temporal induction of cell death, defense genes, and accumulation of salicylic acid in tobacco leaves reacting hypersensitively to a fungal glycoprotein elicitor. Mol. Plant 10: 646–655. [Google Scholar]
- Enyedi A.J., Yalpani N., Silverman P., Raskin I. (1992) Localization, conjugation, and function of salicylic acid in tobacco during the hypersensitive reaction to tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 89: 2480–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Guadagno C.R., Ewers B.E., Speckman H.N., Aston T.L., Huhn B.J., DeVore S.B., et al. (2017) Dead or alive? Using membrane failure and chlorophyll fluorescence to predict mortality from drought. Plant Physiol. 175: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau F., Benjdia M., Zhou D.X. (2003) A jasmonate-responsive element within the A. thaliana vsp1 promoter. J. Exp. Bot. 54: 1153–1162. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013) MYC2: the master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Kenton P., Mur L.A.J., Atzorn R., Wasternack C., Draper J. (1999) (–)-Jasmonic acid accumulation in tobacco hypersensitive response lesions. Mol. Plant Microbe Interact. 12: 74–78. [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., et al. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E., Heifetz P., Thorne L., Uknes S., Ryals J., Ward E. (1998) Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16: 223–233. [DOI] [PubMed] [Google Scholar]
- Liu L., Sonbol F.-M., Huot B., Gu Y., Withers J., Mwimba M., et al. (2016) Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7: 13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A., Sato M., Tsuda K. (2014) Toward a systems understanding of plant–microbe interactions. Front. Plant Sci. 5: 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S.L., Thomson C., Chini A., Read N.D., Loake G.J. (2002) Characterization of a novel, defense-related Arabidopsis mutant, cir1, isolated by luciferase imaging. Mol. Plant Microbe Interact. 15: 557–566. [DOI] [PubMed] [Google Scholar]
- Ohshima M., Itoh H., Matsuoka M., Murakami T., Ohashi Y. (1990) Analysis of stress-induced or salicylic acid-induced expression of the pathogenesis-related 1a protein gene in transgenic tobacco. Plant Cell 2: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E., Kruger-Lebus S., Hahlbrock K. (1989) Temporal and spatial patterns of gene expression around sites of attempted fungal infection in parsley leaves. Plant Cell 1: 993–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Okamoto M., Seto H., Ishizuka K., Sano H., Ohashi Y. (1995) Tobacco MAP kinase: a possible mediator in wound signal transduction pathways. Science 270: 1988–1992. [DOI] [PubMed] [Google Scholar]
- Shinozaki Y., Hao S., Kojima M., Sakakibara H., Ozeki-Iida Y., Zheng Y., et al. (2015) Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83: 237–251. [DOI] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Koornneef A., Claessens S.M., Korzelius J.P., Van Pelt J.A., Mueller M.J., et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Somssich I.E. (2015) Transcriptional networks in plant immunity. New Phytol. 206: 932–947. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Mine A., Bethke G., Igarashi D., Botanga C.J., Tsuda Y., et al. (2013) Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genet. 9: e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009) Network properties of robust immunity in plants. PLoS Genet. 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi S., Sakamoto W., Murata M., Motoyoshi F. (1998) Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol. Biol. 38: 565–576. [DOI] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A., Klessig D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206. [DOI] [PubMed] [Google Scholar]
- Wolpert L. (1969) Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol.25: 1–47. [DOI] [PubMed] [Google Scholar]
- Yan S., Dong X. (2014) Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20: 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.L., Katagiri F., Ausubel F.M. (1993) Arabidopsis mutations at the RPS2 locus result in loss of resistance to Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Mol. Plant Microbe Interact. 6: 434–443. [DOI] [PubMed] [Google Scholar]
- Zeier J., Pink B., Mueller M.J., Berger S. (2004) Light conditions influence specific defence responses in incompatible plant–pathogen interactions: uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 219: 673–683. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.