Abstract
A method for a highly selective and sensitive identification and quantitation of lysophosphatidic acid (LPA) and phosphatidic acid (PA) molecular species was developed using hydrophilic interaction liquid chromatography (HILIC) followed by negative-ion electrospray ionization high resolution mass spectrometry.
Different extraction methods for the polar LPA and PA species were compared and a modified Bligh & Dyer extraction by addition of 0.1 M hydrochloric acid resulted in a ≈ 1.2-fold increase of recovery for the 7 PA and a more than 15-fold increase for the 6 LPA molecular species of a commercially available natural mix compared to conventional Bligh & Dyer extraction. This modified Bligh & Dyer extraction did not show any artefacts resulting from hydrolysis of natural abundant phospholipids.
The developed HILIC method is able to separate all PA and LPA species from major polar membrane lipid classes which might have suppressive effects on the minor abundant lipid classes of interest. The elemental compositions of intact lipid species are provided by the high mass resolution of 100,000 and high mass accuracy below 3 ppm of the Orbitrap instrument. Additionally, tandem mass spectra were generated in a parallel data dependent acquisition mode in the linear ion trap to provide structural information at molecular level. Limits of quantitation were identified at 45 fmol on column and the dynamic range reaches 20 pmol on column, covering the range of natural abundance well.
By applying the developed method to mouse brain it can be shown that phosphatidic acid contains less unsaturated fatty acids with PA 34:1 and PA 36:1 as the major species. In contrast, for LPA species a high content of polyunsaturated fatty acids (LPA 20:4 and LPA 22:6) was quantified.
Keywords: phosphatidic acid, lysophosphatidic acid, high-resolution mass spectrometry, hydrophilic interaction liquid chromatography, modified Bligh & Dyer
1. Introduction
Phosphatidic acid (PA, Supplemental Fig. 1A) is the simplest membrane phospholipid class and has a key function as an intermediate in phospholipid and glycerolipid metabolism. Due to its intermediate function, the steady state concentration of PA is very low in cellular systems [1]. Beside these functions, phosphatidic acid is involved in multiple regulatory cascades such as signal transduction, membrane trafficking, secretion and cytoskeletal rearrangement [2]. Lysophosphatidic acid (LPA, Supplemental Fig. 1B), a degradation product of phosphatidic acid, is a bioactive lipid which is important in a multitude of cellular processes, like bone remodeling, inflammation and migration [3–6]. The action of LPA is transduced by G protein-coupled receptors at the cells surface [7]. These receptors are expressed in cells of the nervous, immune and cardiovascular system [8, 9] where they cause atherosclerosis, inflammation and cancer. Hence, LPA can be regarded as an important biomarker for several diseases such as acute coronary syndrome or ovarian cancer [10, 11].
Due to its high selectivity and sensitivity, mass spectrometry is frequently used for analysis of lipids [12–14]. Nevertheless, PA and LPA are often not detected by standard lipidomic platforms [15, 16] for several reasons. In contrast to most other phospholipid classes, PA and LPA do not have any lipid class specific fragment ions, which could be used in class specific precursor ion scans at sufficient sensitivity, because the fragment ion at m/z 153 can also be observed for phosphatidylglycerol (PG) or phosphatidylinositol (PI). Furthermore, the phosphate group of PA and LPA needs to be protonated to avoid smearing high pressure liquid chromatography (HPLC) peaks (unpublished observation), which is incompatible with the pH of many solvents typically used for negative-ion electrospray ionization (ESI). Finally, the most demanding challenge is the very low concentration of PA and even more so of LPA compared to other phospholipids in most biological systems [17].
The majority of published methods for analysis of PA and LPA relies on triple quadrupole mass spectrometry coupled to reversed phase HPLC [18–22]. Other examples for chromatographic approaches coupled to triple quadrupole or ion trap mass spectrometry would be normal phase [23, 24], hydrophilic interaction liquid chromatography (HILIC) [25] or supercritical fluid chromatography after derivatization [26, 27]. Furthermore, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry without previous chromatographic pre-separation was successfully used for analysis of LPA [28, 29].
In contrast to these low resolution methods, our approach takes advantage of the ultrahigh mass resolution (100,000) and high mass accuracy (≤ 3 ppm) available from an Orbitrap instrument, which greatly enhances identification certainty. Additionally, tandem mass spectra acquired at a linear ion trap deliver valuable fragments corroborating structure proposals for molecular species. HILIC separation of individual phospholipid classes for as little ion suppression effects as possible completes the setup.
2. Material and Methods
2.1. Chemicals
Methanol, acetonitrile, 1 butanol, methyl-tert-butyl ether (MTBE) and disodium hydrogen phosphate were purchased from Sigma-Aldrich (St. Louis, MO, USA), 2-propanol, ammonium formate and citric acid monohydrate from Fluka Analytical (Buchs, Switzerland) and chloroform, formic acid and hydrochloric acid 37 % from Merck (Darmstadt, Germany). LIPID MAPS quantitative lipid standards (1,2-dilauroyl-sn-glycero-3-phosphate, 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphate, 1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phosphate, 1-heneicosanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phosphate, 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-phosphate (ammonium salt), 1-(10Z-heptadecenoyl)-sn-glycero-3-phosphate, 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphocholine, 1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phosphocholine, 1-heneicosanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phosphocholine, 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-phosphocholine, 1-(10Z-heptadecenoyl)-sn-glycero-3-phosphocholine, 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphoethanolamine, 1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phosphoethanolamine, 1-heneicosanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phosphoethanolamine, 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-phosphoethanolamine, 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phospho-L-serine (ammonium salt), 1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phospho-L-serine (ammonium salt), 1-heneicosanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phospho-L-serine (ammonium salt), 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-phospho-L-serine (ammonium salt), 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phospho-(1'-myo-inositol) (ammonium salt), 1-heptadecanoyl-2-(5Z,8Z,11Z,14Z-eicosatetraenoyl)-sn-glycero-3-phospho-(1'-myo-inositol) (ammonium salt), 1-heneicosanoyl-2-(4Z,7Z,10Z,13Z,16Z,19Z-docosahexaenoyl)-sn-glycero-3-phospho-(1'-myo-inositol) (ammonium salt) and 1-heptadecanoyl-2-(9Z-tetradecenoyl)-sn-glycero-3-phospho-(1'-myo-inositol) (ammonium salt)) were purchased from Avanti Polar Lipids (Alabaster, AL, USA); natural phospholipids (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate, L-α-phosphatidic acid (egg PC), L-α-phosphatidylinositol (bovine heart), L-α-phosphatidylcholine (bovine liver), L-α-lysophosphatidylcholine (bovine liver), L-α-phosphatidylethanolamine (corn germ), L-α-lysophosphatidylethanolamine (egg yolk), L-α-phosphatidylglycerol (egg PC), L-α-phosphatidylserine (porcine brain) and cardiolipin (ammonium salt) (heart, bovine)) were purchased from Larodan Fine Chemicals AB (Malmö, Sweden). Deionized water was obtained from a MilliQ Gradient A10 system (Millipore, Billerica, MA, USA).
Lipid shorthand nomenclature is used according to Liebisch et al. [30].
2.2. Mouse information and tissue homogenization
A male C57BL/6JRj mouse was euthanized and the brain was dissected and flash frozen in liquid nitrogen. The brain was homogenized in a BioPulverizer (BioSpec Products, Bartlesville, OK) and triplicates of approximately 25 mg of tissue homogenate were weighed and extracted using the modified Bligh & Dyer HCl method described below.
2.3. Lipid Extraction
Evaluation of extraction efficiency was performed on standard mixtures of naturally occurring phospholipid standards by comparing the extract to the unextracted standard mixture. Prior to extraction of tissue samples, 20 pmol of PA 24:0 were added to the sample to account for analyte losses during extraction. After extraction, the organic phase was dried in a vacuum centrifuge (Thermo Fisher Scientific Inc., Waltham, MA)
2.3.1. Bligh & Dyer (B&D, modified after [31])
To 10 µL of phospholipid standard mixture were added 2.5 mL of methanol and 2.5 mL of chloroform and the mixture was then incubated for 60 min in an overhead shaker at room temperature. 2.25 mL of deionized water was added to induce phase separation and after additional 10 min of shaking the mixture was centrifuged for 3 min at 2,000 g and the lower phase was collected.
2.3.2. Bligh & Dyer HCl (B&D HCl) [32]
To phospholipid standard mixture or tissue homogenate were added 800 µL of a 1:1 (v/v) mixture of 0.1 M HCl and methanol and the mixture was vortexed for 1 min. After addition of 400 µL of chloroform and vortexing for 1 min, the mixture was centrifuged for 3 min at 2,000 g and the lower phase was collected.
2.3.3. MTBE [33]
To phospholipid standard mixture were added 1.5 mL of methanol and 5 mL of MTBE and the mixture was incubated for 60 min in an overhead shaker at room temperature. After addition of 1.25 mL of deionized water and 10 min of additional incubation the mixture was centrifuged for 3 min at 2,000 g and the upper phase was collected.
2.3.4. Butanol [34]
To phospholipid standard mixture were added 500 µL of citrate-phosphate buffer (200 mM citric acid, 280 mM disodium hydrogen phosphate) and 1 mL of 1 butanol. After vortexing for 10 s, 500 µL of water-saturated 1-butanol was added and the mixture was incubated for 10 min at room temperature using an overhead shaker. After centrifugation for 3 min at 2,000 g the upper phase was collected.
2.4. Critical evaluation of extraction method
To determine whether the extraction with 0.1 M HCl produces method relevant artefacts through hydrolysis of phospholipids, a mixture containing PC, LPC, PE, PS and PI LIPID MAPS quantitative lipid standards was extracted using the B&D HCl method described above and compared to the unextracted standard mixtures. To evaluate also the influence of increased extraction time on hydrolysis, two mixtures, one containing PC, LPC, PE, LPE, PS, PG, PI and CL natural phospholipid standards and one containing PA natural standards were extracted. Both mixtures were processed with extraction times of 60 s and 80 s respectively and compared to the unextracted standard mixtures. All experiments described above were performed in triplicate.
2.5. Chromatography
Chromatographic separation was performed on a Phenomenex Kinetex HILIC column (2.1 x 100 mm, 2.6 µm), thermostatted to 50 °C in a Thermo Accela HPLC system. Mobile phase A was deionized water containing 10 mM ammonium formate and 0.5 % formic acid. Mobile phase B was 2 propanol/acetonitrile 5:2 (v/v) containing 10 mM ammonium formate and 0.5 % formic acid. Gradient elution began at 5 % A with a linear increase to 50 % A over 12 min; the 50 % A were held for 3 min and lastly the column was reequilibrated for 15 min. Sample solvent was chloroform/methanol 1:1 (v/v), of which 5 µL were injected. The autosampler tray temperature was kept at 5 °C.
2.6. Mass Spectrometry
Detection was performed on a Thermo Orbitrap Velos Pro (Thermo Fisher Scientific Inc., Waltham, MA) hybrid mass spectrometer, using a HESI II Probe in negative ionization mode with the following parameters: source voltage 3.5 kV, capillary temperature 300 °C, source heater temperature 350 °C, sheath gas 20 arbitrary units, auxiliary gas 5 arbitrary units, sweep gas 1 arbitrary unit. The Orbitrap hybrid mass spectrometer operated in data dependent analysis mode; a full scan spectrum was obtained in the Orbitrap mass analyzer at R=100,000 while simultaneously tandem mass spectra of the ten most abundant full scan masses (selected from an inclusion list containing phosphatidic acid and lysophosphatidic acid molecular species) were obtained in the linear ion trap at a normalized collision energy of 35 % and an exclusion duration of 20 s. LPA, PA, PG, PI, phosphatidylethanolamine (PE), lysophosphatidylethanolamine (LPE), phosphatidylserine (PS) and lysophosphatidylserine (LPS) were detected as deprotonated ([M–H]–) ions, phosphatidylcholine (PC) and lysophosphatidylcholine (LPC) and sphingomyelin (SM) as formate adducts ([M+HCOO]–).
2.7. Data Processing
Identification and quantitation of lipid molecular species was performed by Lipid Data Analyzer, as previously reported [35]. Briefly, the algorithm determines peaks in three dimensions (time, intensity and m/z). Furthermore, the use of HILIC chromatography necessitates the use of an isotope correction algorithm also implemented in the program. Quantitation of naturally occurring lipid molecular species was performed utilizing LPA and PA LIPID MAPS quantitative lipid standards in known concentrations. Statistical calculations were performed using Microsoft Excel and the statistical software R (R Development Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria). All data were subjected to Kolmogorov–Smirnov test prior to ANOVA or t-test.
3. Results and Discussion
3.1. Lipid extraction
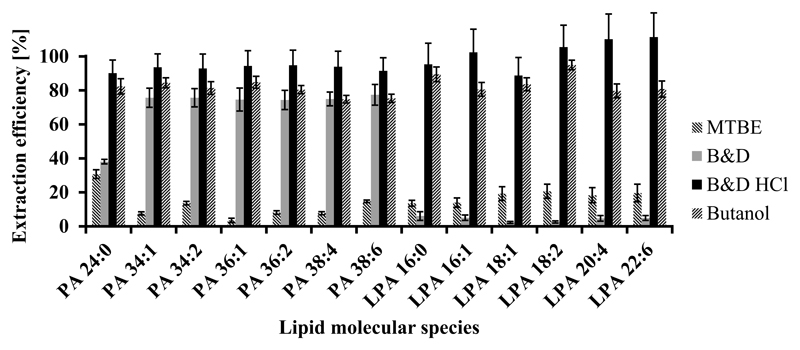
The first and often most important step to achieve superior sensitivity in lipid analysis by mass spectrometric methods is sample preparation. Therefore, lipid extraction from the biological matrix is one of the major sources for quantitative errors and has to be evaluated carefully for every lipid class. We used three literature-known extraction methods: MTBE [33], Bligh & Dyer [31] and extraction with 1-butanol [34]. For evaluation of lipid extraction efficiencies we used a commercially avaliable natural PA and LPA mixture (Fig. 1). Lipid extraction with MTBE did not work well for any of the PA or LPA species available in the mixture and produced losses of 69 % or more. While the Bligh & Dyer protocol did result in acceptable recoveries of about 75 % for most PA species the recoveries for LPA species were very poor. This behavior might be accounted for by the higher polarity of LPA compared to PA. In contrast, the extraction method with 1-butanol containing citric acid resulted in extraction efficiencies between 75 % and 95 % for PA species as well as for LPA species. Since acidic conditions (pH 4) are of importance in order to suppress dissociation and enhance extraction efficiency of PA and LPA, we also modified the Bligh & Dyer protocol by addition of HCl. Some literature known sample preparation methods for PA and particularly LPA use acidified Bligh & Dyer extraction with 6 M HCl [21, 36]. Our results indicate that a concentration of 0.1 M is already sufficient to achieve recoveries of at least 90 % for all PA and LPA species. ANOVA tests for PA and LPA classes showed a very significant difference (p<0.01) in extraction efficiency between the groups (ANOVA was performed for each individual lipid class, after Kolmogorov–Smirnov testing for normal distribution of each extraction method). T-tests between each individual extraction method indicated that their efficiencies differ very significantly (p<0.01) from the each other. Since B&D HCl yielded the highest extraction efficiency for PA and LPA (Fig. 1) and because of the shorter time of sample preparation (evaporation of butanol is time-consuming), B&D HCl extraction method was chosen for this study.
Fig. 1.
Extraction efficiency for PA and LPA species using different extraction methods. B&D HCl extraction shows a very significant difference in extraction efficiency (p<0.01) in comparison to the butanol extraction. n=3, error bars denote one standard deviation.
Critical evaluation of the B&D HCl extraction method showed no artificial formation of LPA arising from hydrolysis of other phospholipids. Due to the fact that no LPA molecular species were detected before and after extraction, Supplemental Fig. 2A exemplarily shows XICs (extracted-ion chromatograms) from one sample after extraction. Neither LPA molecular species formed from PA (Supplemental Fig. 3) nor PA and LPA molecular species formed from other phospholipids (Supplemental Fig. 4) were significantly increased during extraction. Furthermore, we evaluated the influence of prolonged extraction time on phospholipid hydrolysis to PA and LPA and observed no significant changes in levels of PA or LPA molecular species even when extraction time was increased by 30 % (Supplemental Figs. 3 & 4).
Moreover one has to keep in mind that the natural abundance of PA in mammalian cellular systems is usually only a tiny fraction of the total phospholipid pool, which in turn makes up only a few percent of the lipidome. For LPA this ratio gets even more unfavorable, since this is a highly regulated signaling molecule occurring at very low concentration levels. This in turn means that hydrolysis of only a minor part of phospholipids to PA and/or LPA could make a huge difference in the concentration of these phospholipids. Therefore we recommend keeping the acid concentration and extraction time as low as possible.
3.2. Development of LC-MS
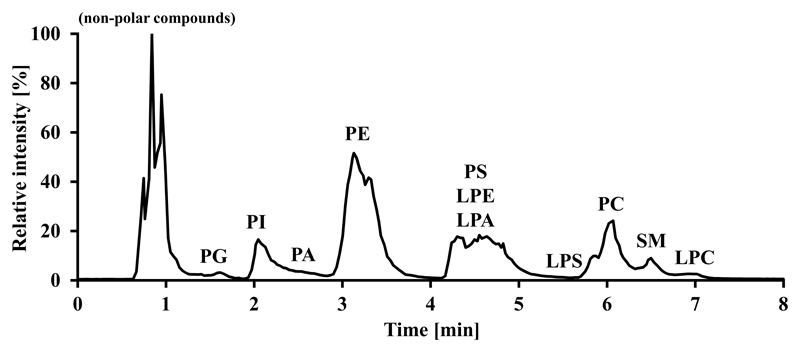
The fundamental issue in chromatography development was a neat separation of PA and LPA species from more abundant lipid classes like PC, PE or SM, because these lipid classes tend to suppress coeluting minor lipid classes due to their abundance. Although negative ionization alleviates the suppressive effects of choline headgroup containing lipids substantially, these species can still be ionized as formate or acetate adducts and therefore exert ion suppression effects due to their sheer amount compared to PA and LPA. Existing HILIC methods do not take this fact into account and show coeluting phospholipid classes [25, 37], whereas silica based normal phase HPLC methods could suffer from poor compatibility of organic solvents with the electrospray process. The C18 reversed phase system previously established by our group [15] does neither ensure complete separation of all phospholipid classes, because the separation mechanism is rather driven by the hydrophobic fatty acyl chains than by the polar head groups, nor is it able to provide baseline separation of all molecular species, particularly in complex biological samples. Hence we developed a HILIC method, which is able to separate all major polar membrane lipid classes by their head groups in a gradient run of 8 min. Another important point is that we hereby additionally circumvent false positive results due to in source fragmentation of LPS and LPC to LPA as described elsewhere [38]. While the method development was performed using a mixture of various lipid standards, the data obtained from mouse brain show the excellent applicability of this separation method to biological samples (Fig. 2). Another issue frequently encountered when performing chromatography of PA and LPA are smearing and tailing peaks because of the charged phosphate group at neutral pH. It is known that addition of phosphoric acid diminishes this problem [39], but this takes place at the cost of rapid ion source and transfer optics contamination (unpublished observation). For these reasons we decided to use 0.5 % formic acid, which has a visible positive effect on peak shape of PA and LPA species.
Fig. 2.
HILIC total ion chromatogram of mouse brain extract with lipid classes annotated.
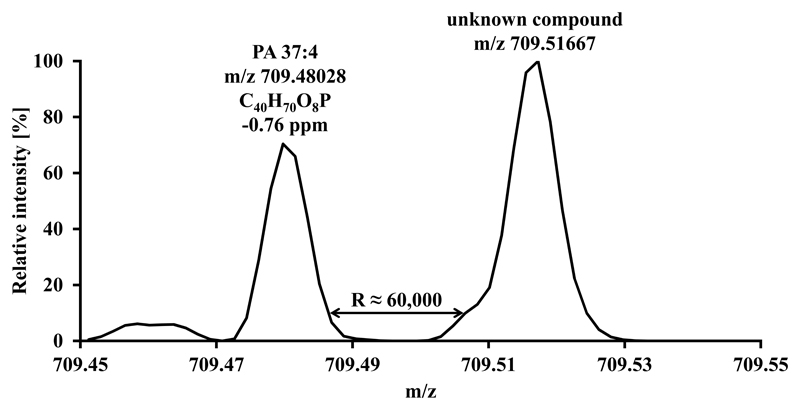
The biggest advantage offered by an Orbitrap instrument is its high mass resolution, which results in high mass accuracy and in turn enhances identification certainty. Full scan spectra acquired at a resolution of 100,000 provide elemental compositions of intact lipid species with high certainty, leaving only a couple of theoretical possibilities for elemental assignment. Furthermore, quantitation at high mass resolution can avoid misinterpretation of spectra and the resulting erroneous amounts of certain PA or LPA species as exemplified in Fig. 3. These features of high resolution equipment are of particular importance for compounds expected at low concentrations like LPA, which would otherwise eventually disappear in background noise. Therefore high mass resolution clearly has a beneficial effect on identification and quantitation of PA and LPA.
Fig. 3.
Unknown compound separated from PA 37:4 (internal standard) only at high mass resolution. Mass resolution was calculated according to IUPAC definition (10 % peak valley).
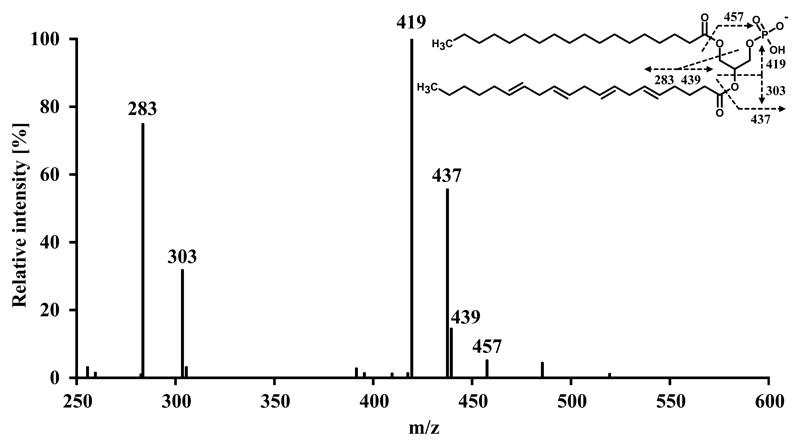
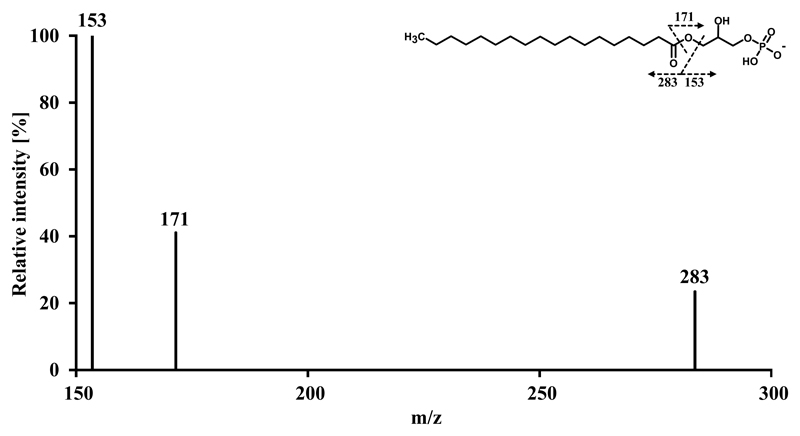
In addition to the information obtained on the identity of intact lipid molecular ions, tandem mass spectra allow unambiguous identification in cases of ambiguity. For the sake of a faster duty cycle we sacrificed the high mass resolution delivered by sequential higher-energy collisional dissociation (HCD) Orbitrap tandem mass spectra and generated collision-induced dissociation (CID) tandem mass spectra in the linear ion trap in a parallel data dependent acquisition mode. Despite the low mass cut off range inherent to any ion trap, most diagnostic ions remain in the range of stability. The only major exceptions are m/z 79 and m/z 97, which are indicative of the phosphate group and in some cases m/z 153, indicative of the glycerol phosphate. Besides m/z 153, all PA tandem mass spectra are unambiguously interpreted by the following fatty acyl specific fragment ions for both fatty acid moieties: Carboxylate anion, fatty acid neutral loss and fatty acid neutral loss as ketene. These specific anions are paradigmatically shown for PA 18:0/20:4 in Fig. 4. In contrast to PA, LPA shows a smaller number of specific fragment ions due to the fact that only one hydroxyl group on the glycerol is esterified. Exemplified by LPA 18:0, the carboxylate anion (m/z 283) identifies the fatty acid whereas m/z 153 and m/z 171 are indicative of the glycerol phosphate inherent to all LPA species (Fig. 5). In the case of LPA, the low mass cut off range of the linear ion trap does not adversely affect the stability of m/z 153.
Fig. 4.
Tandem mass spectrum of PA 18:0/20:4.
Fig. 5.
Tandem mass spectrum of LPA 18:0.
3.3. Method validation
We validated our method in mouse brain by addition of non-endogenous PA and LPA internal standards from 45 to 20,000 fmol on column. Mouse brain was chosen because it is a very lipid rich biological matrix where dire suppression effects are to be expected. Since the high mass resolution of an Orbitrap instrument results in extraordinarily narrow mass peaks, the background noise in a spectrum is usually separated from the compound under investigation, making unambiguous identification possible even in the presence of much higher neighboring mass peaks. The dynamic range for the chosen reference standards shows a linear relationship between amount of analyte injected and signal intensity over 3 orders of magnitude, covering the range of naturally occurring species well (Table 1). Signal-to-noise ratios at the lowest point of the calibration curve exceed 10:1, a value generally referred to in literature, thus defining the limit of quantitation (LOQ) at 45 fmol on column. Accuracy and precision of the method were determined in triplicate at 1.2 pmol on column without any internal standard correction and vary between different reference standards. Generally, lipid species with a higher degree of unsaturation show a higher value of imprecision, but still precision variances for 3 spiked and individually processed samples do not exceed 15 % of the signal intensities. Accuracy is above 84 % for all lipid species investigated. Since it can always happen that some samples need to be reacquired after overnight acquisition of a sample batch, we evaluated the stability of samples after 24 h in the autosampler. The average signal intensity observed after 24 h was 96 % of the signal intensity observed the day before.
Table 1. Validation data.
All calculated values are means of n=3
| analyte | slope | intercept | R2 | accuracy [%] | precision [%] |
|---|---|---|---|---|---|
| LPA 17:0 | 1714.1 | 2000000 | 0.9960 | 88 | 2 |
| LPA 17:1 | 1644.7 | -381423 | 0.9977 | 95 | 4 |
| PA 24:0 | 2886.8 | 217614 | 0.9967 | 97 | 4 |
| PA 25:0 | 3608.8 | 164423 | 0.9966 | 97 | 3 |
| PA 31:1 | 1713.5 | 103885 | 0.9971 | 85 | 6 |
| PA 37:4 | 1770.4 | 63431 | 0.9959 | 84 | 5 |
| PA 43:6 | 637.57 | -120706 | 0.9972 | 90 | 12 |
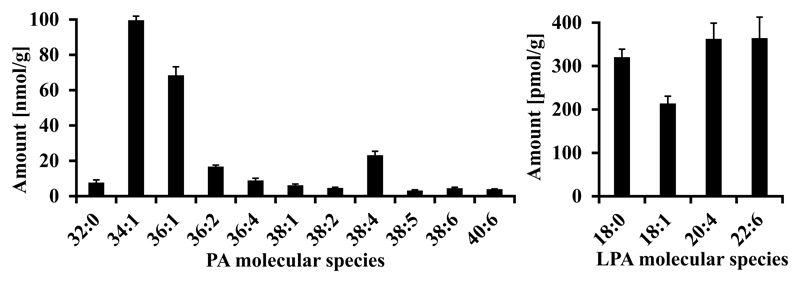
In contrast to other major lipid classes in mouse brain like phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine or plasmalogens (unpublished data & [40]), phosphatidic acid does contain less unsaturated fatty acids whereas LPA in brain shows, as expected, a high relative content of polyunsaturated fatty acids (PUFAS) (Table 2, Fig. 6). The major species (65 %) of phosphatidic acid are PA 34:1 and PA 36:1, both containing oleic acid.
Table 2. Molecular species of PA and LPA detected in mouse brain extract.
Retention time (RT) and all calculated values are means of n=3. Concentrations are given as nmol/g brain tissue. If fatty acid composition is not given, no sufficient fragment spectra were available.
| molecular species | fatty acid composition | concentration [nmol/g] | standard deviation | ‰ of class amount | RT [min] | theoretical m/z | mass accuracy [ppm] | |
|---|---|---|---|---|---|---|---|---|
| PA | 30:0 | - | 0.1 | 0.0 | 0.3 | 2.88 | 619.43439 | 1.6 |
| PA | 32:0 | 16:0_16:0 | 7.7 | 1.5 | 29.7 | 2.88 | 647.46569 | 2.2 |
| PA | 32:1 | 16:0_16:1 | 1.2 | 0.1 | 4.6 | 2.86 | 645.45004 | 1.4 |
| PA | 34:1 | 16:0_18:1 | 99.6 | 2.3 | 383.6 | 2.76 | 673.48134 | 3.2 |
| 16:1_18:0 | ||||||||
| PA | 34:2 | 16:1_18:1 | 0.8 | 0.1 | 3.1 | 2.69 | 671.46569 | 0.9 |
| PA | 36:1 | 18:0_18:1 | 68.4 | 4.8 | 263.3 | 2.73 | 701.51264 | 3.0 |
| PA | 36:2 | 18:1_18:1 | 16.7 | 0.9 | 64.4 | 2.69 | 699.49699 | 2.7 |
| 18:0_18:2 | ||||||||
| PA | 36:3 | - | 0.3 | 0.1 | 1.0 | 2.65 | 697.48134 | 0.3 |
| PA | 36:4 | 16:0_20:4 | 8.9 | 1.2 | 34.3 | 2.56 | 695.46569 | 2.3 |
| PA | 38:1 | 18:0_20:1 | 6.2 | 0.7 | 23.7 | 2.72 | 729.54394 | 1.7 |
| PA | 38:2 | 18:1_20:1 | 4.6 | 0.4 | 17.7 | 2.69 | 727.52829 | 1.4 |
| PA | 38:4 | 18:0_20:4 | 23.2 | 2.2 | 89.3 | 2.52 | 723.49699 | 1.9 |
| 18:1_20:3 | ||||||||
| PA | 38:5 | 18:1_20:4 | 3.2 | 0.4 | 12.3 | 2.47 | 721.48134 | 1.0 |
| PA | 38:6 | 16:0_22:6 | 4.5 | 0.5 | 17.2 | 2.49 | 719.46569 | 1.3 |
| PA | 38:7 | - | 0.1 | 0.0 | 0.4 | 2.97 | 717.45004 | 1.8 |
| PA | 40:1 | - | 1.4 | 0.1 | 5.4 | 2.65 | 757.57524 | 1.1 |
| PA | 40:2 | - | 1.0 | 0.5 | 4.0 | 2.64 | 755.55959 | 1.1 |
| PA | 40:4 | 18:0_22:4 | 1.8 | 0.5 | 6.9 | 2.54 | 751.52829 | 1.4 |
| PA | 40:5 | - | 0.5 | 0.3 | 1.7 | 2.41 | 749.51264 | 1.1 |
| PA | 40:6 | 18:0_22:6 | 3.9 | 0.1 | 15.2 | 2.44 | 747.49699 | 1.4 |
| 18:1_22:5 | ||||||||
| PA | 40:7 | - | 0.9 | 0.1 | 3.3 | 2.41 | 745.48134 | 0.6 |
| PA | 42:2 | - | 0.6 | 0.2 | 2.1 | 2.59 | 783.59089 | 0.4 |
| PA | 44:7 | - | 2.4 | 0.2 | 9.1 | 3.64 | 801.54394 | 2.4 |
| PA | 46:7 | - | 1.7 | 0.1 | 6.7 | 3.61 | 829.57524 | 1.8 |
| PA | 48:10 | - | 0.2 | 0.1 | 0.9 | 3.49 | 851.55959 | 1.4 |
| LPA | 18:0 | 0.3 | 18.1 | 254.2 | 4.31 | 437.26733 | 1.4 | |
| LPA | 18:1 | 0.2 | 16.5 | 169.5 | 4.20 | 435.25168 | 2.7 | |
| LPA | 20:4 | 0.4 | 36.2 | 287.5 | 4.30 | 457.23603 | 3.1 | |
| LPA | 22:6 | 0.4 | 48.4 | 288.8 | 4.61 | 481.23603 | 3.7 | |
Fig. 6.
PA and LPA molecular species detected in mouse brain extract, normalized to tissue net weight. Only species contributing to more than 1 % of total class amount are depicted.
4. Conclusions
We developed a sensitive and highly reliable method for the detection and quantitation of LPA and PA molecular species. Elemental compositions of intact [M–H]– ions are delivered by high resolution and high mass accuracy of an Orbitrap mass spectrometer, while tandem mass spectra acquired in a linear ion trap provide information about the fatty acid composition and corroborate molecular identity. Additional selectivity is achieved by chromatographic HILIC separation. Finally, the use of lipid extraction protocols strictly defined in terms of acidity and extraction time has to be taken into account in order to avoid either substantial extraction losses or production of artefacts resulting from hydrolysis of phospholipids.
Supplementary Material
Acknowledgement
This work was supported by the Austrian Science Fund (FWF) [P 26148-N19].
References
- [1].Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H-E, Rajashekar CB, Williams TD, Wang X. J Biol Chem. 2002;277:31994. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- [2].Wang X, Devaiah SP, Zhang W, Welti R. Prog Lipid Res. 2006;45:250. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- [3].Mansell JP, Blackburn J. Biochim Biophys Acta. 2013;1831:105. doi: 10.1016/j.bbalip.2012.04.005. [DOI] [PubMed] [Google Scholar]
- [4].Sims SM, Panupinthu N, Lapierre DM, Pereverzev A, Dixon SJ. Biochim Biophys Acta. 2013;1831:109. doi: 10.1016/j.bbalip.2012.08.001. [DOI] [PubMed] [Google Scholar]
- [5].Zhao Y, Natarajan V. Biochim Biophys Acta. 2013;1831:86. doi: 10.1016/j.bbalip.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Meeteren La, Moolenaar WH. Prog Lipid Res. 2007;46:145. doi: 10.1016/j.plipres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- [7].Xiang SY, Dusaban SS, Brown JH. Biochim Biophys Acta. 2013;1831:213. doi: 10.1016/j.bbalip.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schober A, Siess W. Br J Pharmacol. 2012;167:465. doi: 10.1111/j.1476-5381.2012.02021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ueda H, Matsunaga H, Olaposi OI, Nagai J. Biochim Biophys Acta. 2013;1831:61. doi: 10.1016/j.bbalip.2012.08.014. [DOI] [PubMed] [Google Scholar]
- [10].Dohi T, Miyauchi K, Ohkawa R, Nakamura K, Kishimoto T, Miyazaki T, Nishino A, Nakajima N, Yaginuma K, Tamura H, Kojima T, et al. Clin Chim Acta. 2012;413:207. doi: 10.1016/j.cca.2011.09.027. [DOI] [PubMed] [Google Scholar]
- [11].Bese T, Barbaros M, Baykara E, Guralp O, Cengiz S, Demirkiran F, Sanioglu C, Arvas M. J Gynecol Oncol. 2010;21:248. doi: 10.3802/jgo.2010.21.4.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Köfeler HC, Fauland A, Rechberger GN, Trötzmüller M. Metabolites. 2012;2:19. doi: 10.3390/metabo2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cífková E, Holčapek M, Lísa M, Ovčačíková M, Lyčka A, Lynen F, Sandra P. Anal Chem. 2012;84:10064. doi: 10.1021/ac3024476. [DOI] [PubMed] [Google Scholar]
- [14].Schuhmann K, Herzog R, Schwudke D, Metelmann-Strupat W, Bornstein SR, Shevchenko A. Anal Chem. 2011;83:5480. doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- [15].Fauland A, Köfeler H, Trötzmüller M, Knopf A, Hartler J, Eberl A, Chitraju C, Lankmayr E, Spener F. J Lipid Res. 2011;52:2314. doi: 10.1194/jlr.D016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, Shevchenko A. J Mass Spectrom. 2012;47:96. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- [17].van Meer G, Voelker DR, Feigenson GW. Nat Rev Mol Cell Biol. 2008;9:112. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aaltonen N, Laitinen JT, Lehtonen M. J Chromatogr B. 2010;878:1145. doi: 10.1016/j.jchromb.2010.03.030. [DOI] [PubMed] [Google Scholar]
- [19].Bathena SP, Huang J, Nunn ME, Miyamoto T, Parrish LC, Lang MS, McVaney TP, Toews ML, Cerutis DR, Alnouti Y. J Pharm Biomed Anal. 2011;56:402. doi: 10.1016/j.jpba.2011.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee JY, Min HK, Moon MH. Anal Bioanal Chem. 2011;400:2953. doi: 10.1007/s00216-011-4958-7. [DOI] [PubMed] [Google Scholar]
- [21].Shan L, Jaffe K, Li S, Davis L. J Chromatogr B. 2008;864:22. doi: 10.1016/j.jchromb.2008.01.031. [DOI] [PubMed] [Google Scholar]
- [22].Wijesinghe DS, Mayton EK, Mietla JA, Mukherjee A, Wu J, Fang X, Chalfant CE. Anal Methods. 2011;3:2822. doi: 10.1039/C1AY05459G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Baker DL, Desiderio DM, Miller DD, Tolley B, Tigyi GJ. Anal Biochem. 2001;292:287. doi: 10.1006/abio.2001.5063. [DOI] [PubMed] [Google Scholar]
- [24].Bollinger JG, Ii H, Sadilek M, Gelb MH. J Lipid Res. 2010;51:440. doi: 10.1194/jlr.D000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Scherer M, Schmitz G, Liebisch G. Clin Chem. 2009;55:1218. doi: 10.1373/clinchem.2008.113779. [DOI] [PubMed] [Google Scholar]
- [26].Lee JW, Yamamoto T, Uchikata T, Matsubara A, Fukusaki E, Bamba T. J Sep Sci. 2011;34:3553. doi: 10.1002/jssc.201100539. [DOI] [PubMed] [Google Scholar]
- [27].Lee JW, Nishiumi S, Yoshida M, Fukusaki E, Bamba T. J Chromatogr A. 2013;1279:98. doi: 10.1016/j.chroma.2013.01.020. [DOI] [PubMed] [Google Scholar]
- [28].Tanaka T, Tsutsui H, Hirano K, Koike T, Tokumura A, Satouchi K. J Lipid Res. 2004;45:2145. doi: 10.1194/jlr.D400010-JLR200. [DOI] [PubMed] [Google Scholar]
- [29].Morishige J, Urikura M, Takagi H, Hirano K, Koike T, Tanaka T, Satouchi K. Rapid Commun Mass Spectrom. 2010;24:1075. doi: 10.1002/rcm.4484. [DOI] [PubMed] [Google Scholar]
- [30].Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, Spener F, Wakelam MJO. J Lipid Res. 2013;54:1523. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bligh E, Dyer W. Can J Biochem Physiol. 1959;37:911. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- [32].Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, Wongsiriroj N, Nagy HM, Ivanova PT, Scott Sa, Knittelfelder O, et al. Cell Metab. 2012;15:691. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. J Lipid Res. 2008;49:1137. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Scherer M, Schmitz G, Liebisch G. Anal Chem. 2010;82:8794. doi: 10.1021/ac1021826. [DOI] [PubMed] [Google Scholar]
- [35].Hartler J, Trötzmüller M, Chitraju C, Spener F, Köfeler HC, Thallinger GG. Bioinformatics. 2011;27:572. doi: 10.1093/bioinformatics/btq699. [DOI] [PubMed] [Google Scholar]
- [36].Yoon H-R, Kim H, Cho S-H. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;788:85. doi: 10.1016/s1570-0232(02)01031-0. [DOI] [PubMed] [Google Scholar]
- [37].Liebisch G, Scherer M. J Chromatogr B. 2012;883–884:141. doi: 10.1016/j.jchromb.2011.10.037. [DOI] [PubMed] [Google Scholar]
- [38].Zhao Z, Xu Y. J Chromatogr B. 2009;877:3739. doi: 10.1016/j.jchromb.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Asakawa Y, Tokida N, Ozawa C, Ishiba M, Tagaya O, Asakawa N. J Chromatogr A. 2008;1198–1199:80. doi: 10.1016/j.chroma.2008.05.015. [DOI] [PubMed] [Google Scholar]
- [40].Köfeler HC, Tiboldi A, Hoeger H, Lubec G. Neurochem Int. 2010;57:935. doi: 10.1016/j.neuint.2010.09.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.