Abstract
Atypical chemokine receptors are expressed by discrete populations of stromal cells at specific anatomical locations where they control leukocyte migration by scavenging or transporting chemokines. ACKR4 is an atypical receptor for CCL19, CCL21 and CCL25. In skin, ACKR4 plays indispensable roles in regulating CCR7-dependent APC migration, and there is a paucity of migratory APCs in the skin-draining lymph nodes of Ackr4-deficient mice under steady state and inflammatory conditions. This is caused by loss of ACKR4-mediated CCL19/21 scavenging by keratinocytes and lymphatic endothelial cells. In contrast, we show here that Ackr4 deficiency does not affect dendritic cell abundance in the small intestine and mesenteric lymph nodes, at steady state or after R848-induced mobilisation. Moreover, Ackr4 expression is largely restricted to mesenchymal cells in the intestine, where it identifies a previously uncharacterised population of fibroblasts residing exclusively in the submucosa. Compared to related Ackr4- mesenchymal cells, these Ackr4+ fibroblasts have elevated expression of genes encoding endothelial cell regulators and lie in close proximity to submucosal blood and lymphatic vessels. We also provide evidence that Ackr4+ fibroblasts form physical interactions with lymphatic endothelial cells, and engage in molecular interactions with these cells via the VEGFD/VEGFR3 and CCL21/ACKR4 pathways. Thus, intestinal submucosal fibroblasts in mice are a distinct population of intestinal mesenchymal cells that can be identified by their expression of Ackr4, and which have transcriptional and anatomical properties that strongly suggest roles in endothelial cell regulation.
Keywords: chemokine, cell trafficking, atypical chemokine receptor, intestine, fibroblast, vasculature, lymphatics
Introduction
The intestine is a complex and dynamic organ requiring continuous interactions between many different haemopoietic and non-haemopoietic (stromal) cell types. These include resident and migratory leukocytes, epithelial cells, blood vessel endothelial cells (BECs)2, lymphatic endothelial cells (LECs), neurons, and various populations of mesenchymal cells, such as pericytes, smooth muscle cells, fibroblasts and myofibroblasts. Mesenchymal cells play many important roles in the physiological and immunological functions of the intestine (1–3). In addition to providing the 3-dimensional matrix of the tissue, they supply growth factors critical for epithelial cell renewal, regulate endothelial cell function, contribute to innate and adaptive immune responses, and provide surfaces upon which interstitial leukocytes can move and migrate (1–3). These processes are essential for tissue development and repair, maintenance of homeostasis, adaptation to environmental challenges, and response to disease.
Leukocyte migration in the intestine and elsewhere is dependent on chemokines, which engage conventional G-protein coupled chemokine receptors on leukocytes (4). There are also four ‘atypical’ chemokines receptors (ACKRs) that regulate chemokine-driven migration by transcytosing, sequestering or scavenging chemokines (5). ACKRs are typically expressed by stromal cells in highly defined microanatomical niches, where they have been shown to be capable of regulating leukocyte trafficking into, within and out of tissues (5). ACKR4, previously known as CCRL1 or CCX-CKR, binds with high affinity to CCL19 and CCL21, which direct leukocyte migration through CCR7, and to CCL25, the sole ligand for CCR9 (6–9). By internalising and degrading its ligands, ACKR4 can regulate their abundance and shape chemokine gradients (10–12). In vivo, this has been most studied in the skin where, under steady state and inflammatory conditions, the CCR7-dependent trafficking of Langerhans cells (LCs) and dendritic cells (DCs) to draining lymph nodes (LNs) is controlled by ACKR4 expression on keratinocytes, a subset of dermal LECs, and by LECs lining the ceiling of the LN subcapsular sinus (SCS) (11–15). CCR7 is also essential for the migration of DCs from the small intestine (SI) to the mesenteric LNs (MLNs) (9), but the function of ACKR4 in this context has not been explored. Ackr4 transcripts are detectable in the SI and colon of mice (7), and GFP+ cells are present in the SI of Ackr4gfp/+ reporter mice (13), although the identity of these cells is not clear.
Here we report that Ackr4 deficiency does not alter migratory DC abundance in the SI or mesenteric LNs (MLNs), either at steady state or after R848-induced DC mobilisation. Thus ACKR4 in the SI, in contrast to the skin, serves no detectable indispensable role in regulating DC trafficking to draining LNs. We also find that Ackr4 expression in the SI and colon, unlike the skin, is largely restricted to a subset of mesenchymal cells. These Ackr4+ cells represent a novel and discrete population of fibroblasts that reside exclusively in the intestinal submucosa, and which, to our knowledge, have not been characterised before. We show that these Ackr4+ fibroblasts preferentially express an array of genes encoding endothelial cell regulators, and that they lie in close proximity to blood and lymphatic vessels in submucosa. We also provide evidence that LECs and Ackr4+ fibroblasts physically interact, and that these two cell types are equipped to engage in unique reciprocal molecular interactions through the VEGFD/VEGFR3 and CCL21/ACKR4 axes.
Materials and Methods
Mice
Wild type, Ackr4-/- (Ackr4tm1.1Rjbn: www.informatics.jax.org/allele/key/625599 (16)), Ackr4gfp/+ and Ackr4gfp/gfp (Ackr4tm1Ccbl: www.informatics.jax.org/allele/key/817740) (13) mice, all on a C57BL/6 background, were bred and maintained in specific pathogen-free conditions in the Central Research Facility at the University of Glasgow. Female mice 6-11 weeks of age were used in all experiments, which were performed under the auspices of UK Home Office Licences.
Preparation of single cell suspensions
Single cell suspensions were generated from intestinal tissue by enzymatic digestion as described previously (17–19). Briefly, excised SI and colons were washed in 2mM EDTA in HBSS for 3 x 20min at 37°C. Tissue was then digested in R10 medium (10% FCS, 2mM EDTA in RPMI 1640 (Thermo Fisher, Waltham, MA, USA)) containing 1mg/ml Collagenase VIII (Sigma Aldrich, St Louis, MO, USA) for SI digests, or 0.65mg/ml Collagenase V (Sigma Aldrich), 0.45mg/ml Collagenase D (Roche Diagnostics, Mannheim, Germany), 1mg/ml Dispase (Thermo Fisher) and 30μg/ml DNase I (Roche Diagnostics) for colon digests. Digests were then passed through a 40μm cell strainer, and the cells pelleted at 400g for 5min.
In some experiments cells from the intestinal muscularis externa (ME) containing the longitudinal muscle (LM) and circular muscle (CM) were isolated as described previously (20). Briefly, ME from small intestine and colon was dissected from the mucosa and enzymatically digested in MEMα medium (Lonza, Verviers, Belgium) containing 100µg/ml of Penicillin, 100µg/ml of Streptomycin, 50µM β-mercaptoethanol, 5% fetal calf serum, 0.5mg/ml protease type I (Sigma-Aldrich), 0.25mg/ml collagenase type IV (Sigma-Aldrich) and 5U/ml DNase I (Sigma-Aldrich) for 15min at 37°C. Cell suspensions were filtered through a 40µm cell strainer, and the cells were pelleted at 1500 rpm for 5min.
Flow cytometry and cell sorting
1-6 x 106 cells were blocked using anti-CD16/CD32 Ab (Biolegend, San Diego, CA) and labelled with fluorescently conjugated Abs in the dark as described previously (17–19). The following anti-mouse Abs were purchased from Biolegend: CD103-PE (2E7), CD11c-PE/Cy7 (N418), CD146-PerCP/Cy5.5 and CD146-APC (ME-9F1), B220-PerCP/Cy5.5 (RA3-6B2), CD3ε-PerCP/Cy5.5 (145-2C11), CD31-BV605 and CD31-PE (390), CD45-PerCP/Cy5.5, CD55-PE (RIKO-3), CD66a-APC (Mab-CC1), CD105-APC (MJ7/18), CD106-PE (429), CD9-Alexa Fluor® (AF) 647 (MZ3), CD90.2-PE (30-H12), EPCAM-PerCP/Cy5.5 and EPCAM-APC/Cy7 (G8.8), F4/80-BV605 and F4/80-APC (BM8), GP38-PE/Cy7 (8.1.1), ICAM1-PE (YN1/1.7.4), Ly6G-PE (1A8), MHCII-BV510 (M5/114.15.2), NK1.1-PerCP/Cy5.5 (PK136), PDGFRα-PE (APA5), SCA1-APC (D7). The following anti-mouse Abs were purchased from eBioscience (San Diego, CA): CD11b-AF700 and CD11b-eFluor 450 (M1/70), CD45-AF700 and CD45-eFluor 450 (30-F11), CD166-PE (ALC48), Ly6C-eFluor 450 (HK1.4), Ter119-eFluor 450 (TER-119). BD Biosciences (Franklin Lakes, NJ) supplied CD29-PE (HM β1-1), CD34-AF647 (RAM34) and Siglec F-PE (E50-2440). Dead cells were excluded using 7AAD (Biolegend) or Fixable Viability Dye eFluor 780 (eBioscience, San Diego, CA). Stained cells were analysed or sorted using an LSR II (BD Biosciences), or a FACS Aria I, IIu or III (BD Biosciences). The purity of all sorted cell populations was >95%. Analysis was performed using FlowJo v10 (Tree star, Ashland, OR).
CCL19AF647 uptake assay
Prior to staining of surface markers, 1-2 x 106 cells were incubated in 250μg/ml of AF647-labelled CCL19 (CCL19AF647; Almac, Craigavon, UK) in R10 medium containing 20mM HEPES (Thermo Fisher) for 1h at 37°C, as described previously (21–23).
DSS-induced colitis
Mice received 2% dextran sodium sulfate (DSS) (MP Biomedicals, Santa Ana, CA) in sterile distilled water ad libitum for 5d. Control animals received sterile water. Colitis scoring was performed as described previously (24).
R848-induced DC mobilisation
Wild type and Ackr4-/- mice received an i.p. injection of 100μg of R848 (Invivogen, San Diego, CA) in 100μl of sterile PBS. Control animals were injected with 100μl of sterile PBS alone. Mice were culled 12h or 24h following injection.
Immunofluorescent microscopy
Tissues were fixed for 3h in 1% methanol-free paraformaldehyde (Thermo Fisher) in PBS, washed for 1h in PBS and then frozen in Optimal Cutting Temperature compound (Cell Path, Newton, UK). Embedded tissue was then cut into 7μm sections, which were blocked using PBS containing 1% BSA (Sigma Aldrich) and 10% donkey serum (Sigma Aldrich) before being stained in the dark for >8h with a primary Ab cocktail. The following anti-mouse monoclonal Abs were used: CD45-eFluor 450 (30-F11), unlabelled Endomucin (rat-derived; V.7C7), and LYVE1-eFluor 660 (ALY7) from eBioscience; PDGFRα-biotinylated (APA5) (Biolegend); and CD31-purified (MEC 13.3) (BD Bioscience). Polyclonal anti-mouse Abs were also used: anti-CCL21 and anti-VEGFR3/Flt4 (both generated in goat; R&D systems (Minneapolis, MN)), and anti-alpha smooth muscle actin (αSMA) (rabbit-derived; Abcam (Cambridge, UK)). Unlabelled primary Abs were visualised using polyclonal, directly conjugated Abs including donkey anti-goat AF555, donkey anti-goat AF647, donkey anti-rat AF594, goat anti-rat AF555, and goat anti-rabbit AF647, all from Thermo Fisher. Biotinylated primary Abs were visualised using Streptavidin DyLight 549 (Vector Labs, Burlingame, CA). When biotinylated Abs were used, tissue sections were also blocked using an Avidin/Biotin Blocking kit (VectorLabs) in accordance with the manufacturer’s instructions. GFP expression in samples from Ackr4gfp/+ mice was directly detected by fluorescence microscopy without using anti-GFP antibodies. Images were acquired on a Leica SP5 confocal laser-scanning microscope or an EVOS FL Cell Imaging System (Thermo Fisher).
RNA isolation
Purified cells were pelleted and lysed in 500μl RLT lysis buffer (Qiagen, Hilden, Germany). Lysed cells were homogenised using QIAshredders (Qiagen). Under RNase-free conditions, RNA was isolated using an RNeasy Micro kit (Qiagen). Genomic DNA contaminants were digested on column using the RNase-free DNase kit (Qiagen).
Transcriptomics
Microarray assays were performed on GeneChip® Mouse Transcriptome Arrays 1.0 by Almac Diagnostics Ltd. Briefly, purified total RNA was converted into sense strand cDNA and amplified using an Ovation Pico WTA System V2 kit (NuGEN, San Carlos, CA). Amplified cDNA was then fragmented and labelled using an Encore Biotin module (NuGEN). Fragmented cDNA was then hybridised to GeneChip Mouse Transcriptome Arrays 1.0 (Affymetrix, Santa Clara, CA, USA). Procedures were carried out as described by the manufacturers. Datasets were analysed using GeneSpring GX software. Data were normalised using a variant of the robust multichip average method (RMA16). Normalised data were analysed using unpaired t tests to determine the significance of gene expression differences. The resulting P values were adjusted for multiple comparisons using the Benjamini-Hochberg multiple testing correction at a false discovery rate (FDR) of 0.1. Differentially expressed genes were assigned gene ontology terms and grouped into biological processes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources v6.8 (https://david.ncifcrf.gov). Analysis was performed using protocols developed by Huang and colleagues (25, 26). Significance of enrichment was determined using a modified Fisher’s exact test and a Benjamini-Hochberg multiple testing correction was used to correct for the rate of type I errors. Enrichment of biological processes was considered significant if P ≤ 0.05. The microarray data have been deposited in NCBI’s Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/), and are accessible through GEO Series accession number GSE113665.
QPCR
Total RNA was reverse transcribed with random primers using Quantitect Reverse Transcription kit (Qiagen). Quantitative real-time polymerase chain reaction (QPCR) amplifications were performed in triplicate using PerfeCTa® SYBR® Green FastMix® (Quanta Biosystems, Gaithersburg, MD, USA) as described previously (27). A 500μM mix of forward and reverse primers was used per reaction. Primers were designed using Primer3 Input software (version 0.4.0) and generated by IDT technologies. Primer sequences were as follows (5’ to 3’): Adm: TCT TGG ACT TTG GGG TTT TG and ATT CTG TGG CGA TGC TCT G; Ccl21: CCA ACT CAC AGG CAA AGA GG and GCC AGG TAA GAA AGG GAT GG; Ctsh: TCG CCA GTG AAA AAT CAG G and CCC GTG GTA GAG AAA GTC CA; Fap: CGC ACA GAC CAA GAA ATA CAA G and CAA ACT GAG GAG GCA GAA TCA; Figf: AGC CAG GAG AAC CCT TGA TT and GGG CAA CAG TGA CAG CAA C; Grem1: CCT TTC AGT CTT GCT CCT TCT G and CGT GTG ACC CTT TTC TTC TTG; Il6: TTC CAT CCA GTT GCC TTC TT and ATT TCC ACG ATT TCC CAG AG; Serpine1: GTA AAC GAG AGC GGC ACA G and CCG AAC CAC AAA GAG AAA GG; and Tek: TGA GAT GCC TGC GTT TAC TG and TTG TAG TTC TGT CTG CCG TTG T. QPCR reactions were performed using a Prism® 7900HT Sequence Detection System (Life Technologies, Invitrogen, CA, USA) for 40 cycles. Target gene expression was normalised to expression of a reference gene, Tbp, which encodes the TATA-box binding protein: preliminary data indicated that this gene had very similar levels of expression between different iMC populations. The sequences of the Tbp-specific primers were TGC TGT TGG TGA TTG TTG GT and AAC TGG CTT GTG TGG GAA AG. Fold change values were calculated using the equation: Fold Change = 2-ΔΔCT. The mean ΔCT of the control samples was used as a calibrator.
Statistical analysis
Data were analysed using Prism 6 software (GraphPad, San Diego, CA) and are represented as mean ±1 SD. Statistical tests used are indicated in the figure legends.
Results
Ackr4 is expressed by MLN LECs and a subset of intestinal mesenchymal cells in steady state and during inflammation
To characterise Ackr4 expression in MLN and intestine, we used flow cytometry to examine GFP expression by CD45+ leukocytes and CD45- stromal cells in Ackr4gfp/+ reporter mice (Ackr4tm1Ccbl) (13), using wild-type (WT) mice as controls. As in peripheral lymphoid tissues, blood and skin (11–13), CD45+ cells in the MLN, SI or colon were entirely GFP- (Fig. 1A). However, GFP+ cells were present in the CD45- compartment of all tissues (Fig. 1A).
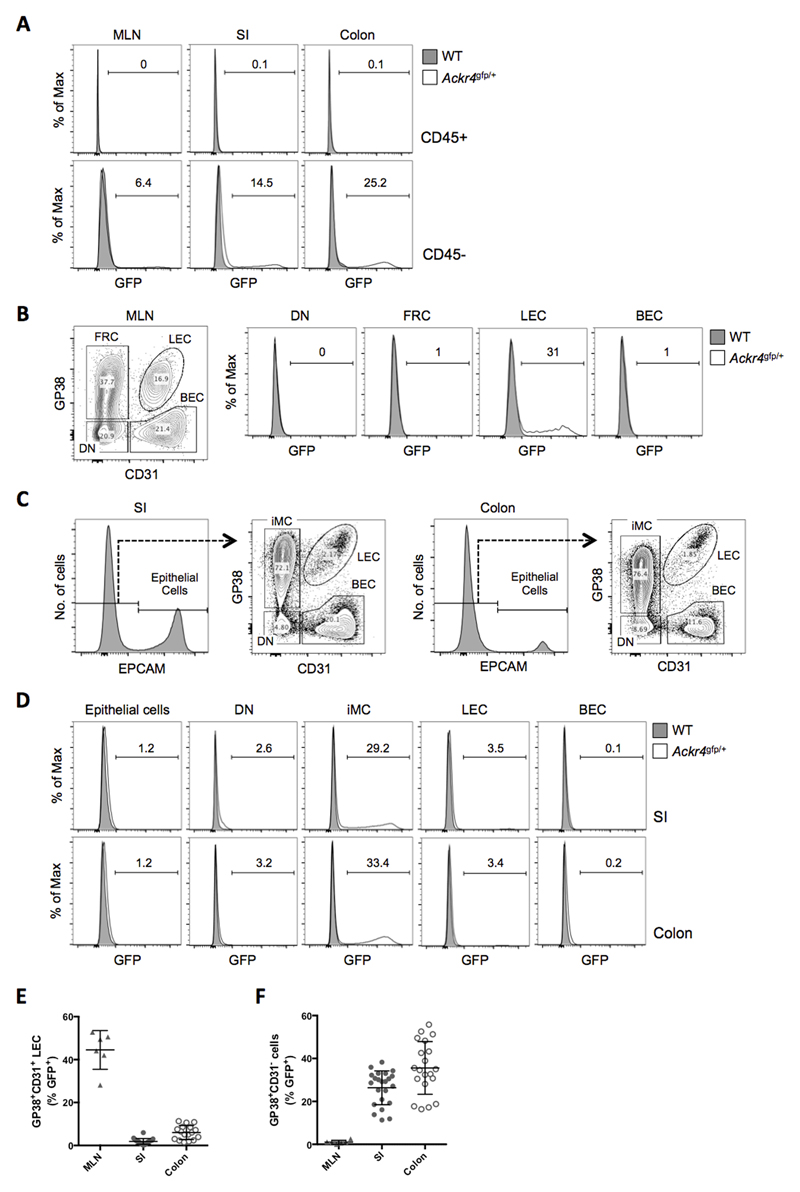
Figure 1. Ackr4 is expressed by LECs in the MLN and mesenchymal cells in the intestine.
(A) Overlaid histogram flow cytometry plots showing GFP expression by CD45+ (top panels) and CD45- cells (bottom panels) among single, live, Ter119- cells in cell suspensions of the MLN, small intestine (SI) and colon of WT and Ackr4gfp/+ (Ackr4tm1Ccbl) mice. (B) Left panel: flow cytometry contour plot of stromal cell populations among single, live, CD45-Ter119- cells in the MLN, identifying GP38+CD31- fibroblastic reticular cells (FRC), GP38+CD31+ lymphatic endothelial cells (LEC), GP38-CD31+ blood endothelial cells (BEC) and GP38-CD31- ‘double negative’ (DN) cells. Right panels: representative overlaid histogram plots showing GFP expression by stromal cell populations in the MLN of WT and Ackr4gfp/+ mice. (C) Flow cytometric analysis of stromal cell populations among single, live, CD45-Ter119- cells in the SI and colon, identifying EpCAM+ epithelial cells (histograms), and GP38+CD31- intestinal mesenchymal cells (iMC), LECs, BECs and DN cells in the EpCAM- population (contour plots). (D) Representative overlaid histograms showing GFP expression by stromal cell populations in the SI and colon of WT and Ackr4gfp/+ mice. In A, B and D, the numbers on the histogram plots indicate the percentage of GFP+ cells in Ackr4gfp/+ samples. Flow cytometry plots in A-D are representative of those from at least three individual experiments involving at least three mice of each genotype. (E, F) Percentage of cells expressing GFP in LEC (E) and GP38+CD31- cell (F) populations in the MLN (n=6), SI (n=23) and colon (n=20) of Ackr4gfp/+ mice. Mean±1SD is indicated. Data are pooled from two or more individual experiments.
LN stromal cells can be subdivided according to expression of CD31 and GP38 into GP38-CD31+ BECs, GP38+CD31+ LECs, GP38+CD31- fibroblastic reticular cells (FRCs), and a ‘double negative’ (DN) population containing pericytes and other rare stromal cell types (28). A substantial proportion (28-52%) of LECs were GFP+ in the MLN of Ackr4gfp/+ mice, while all other stromal cells lacked GFP (Fig. 1B and E).
Amongst CD45- cells in digests of SI and colon, epithelial cells were identified as EPCAM+, while EPCAM- cells were subdivided into GP38-CD31+ BECs, GP38+CD31+ LECs, GP38+CD31- cells (referred to hereafter as intestinal mesenchymal cells (iMCs)), and a GP38-CD31- DN population (Fig. 1C). In contrast to the MLN, virtually all LECs in the SI and colon of Ackr4gfp/+ mice were GFP- (Fig. 1D-E). GFP was also barely detectable in epithelial cells, BECs and DN cells in the SI and colon of Ackr4gfp/+ mice (Fig. 1D). The vast majority of GFP+ cells in these tissues were in the iMC population (Fig. 1D), with on average ~25% and ~35% of iMCs expressing GFP in the SI and colon, respectively (Fig. 1F).
To determine if Ackr4 expression in the intestine might be modified by inflammation, we induced colitis in Ackr4gfp/+ mice by administering 2% DSS in the drinking water. All DSS-treated mice had developed clinical colitis by day 5 of treatment (Supplementary Fig. 1A-C) and this led to a decrease in the proportion of colonic iMCs that were GFP+ from an average of ~30% to ~20%. However, the overall pattern of GFP expression in Ackr4gfp/+ mice was unchanged, with the dominant GFP+ populations being iMCs in SI and colon, and LECs in the MLN (Supplementary Fig. 1D-G, and data not shown). Thus, colonic inflammation is not associated with a change in the distribution of Ackr4 expression.
iMCs and MLN LECs express functional ACKR4 protein
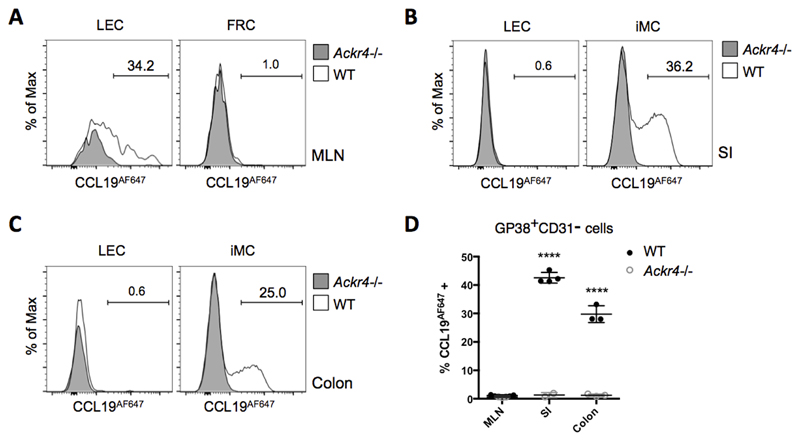
Next, we sought to identify cells expressing ACKR4 protein. Using samples from Ackr4-deficient (Ackr4tm1.1Rjbn (16)) mice as controls, commercially available anti-ACKR4 antibodies repeatedly failed to provide convincing detection of ACKR4 in the intestine of WT mice by flow cytometry or immunofluorescence microscopy (data not shown). We therefore used fluorescent chemokine uptake assays, a technique that we have used to successfully and sensitively detect ACKR expression in other contexts (12, 21–23). Single cell suspensions of MLN, SI and colon from WT and Ackr4-deficient mice were incubated ex vivo with CCL19AF647 and analysed by flow cytometry (Fig. 2). Consistent with the analysis of GFP expression in Ackr4gfp/+ mice (Fig. 1), ACKR4-dependent CCL19AF647 uptake was restricted to LECs in MLN (Fig. 2A) and iMCs in SI and colon (Figs. 2B-D). Thus, iMCs and MLN LECs express ACKR4 protein capable of internalising CCL19.
Figure 2. ACKR4 is expressed as a functional protein on a subset of iMCs and MLN LECs.
Single cell suspensions prepared from WT and Ackr4-deficient (Ackr4-/-) mice (Ackr4tm1.1Rjbn) were incubated in medium containing CCL19AF647 for 1h at 37°C, then labelled with fluorescent Abs, and analysed by flow cytometry. Stromal cell subsets were identified from among live, single, CD45-Ter119- cells. (A-C) Representative overlaid histogram plots showing uptake of CCL19AF647 by lymphatic endothelial cells (LEC) and GP38+CD31- cells (i.e. fibroblast reticular cells (FRC) or intestinal mesenchymal cells (iMC)) from (A) MLN (B) small intestine (SI), and (C) colon of WT and Ackr4-/- mice. The numbers on the plots indicate the percentage of CCL19AF647-positive cells in the WT samples. (D) Mean percentage of CCL19AF647-positive cells (±1SD) in the GP38+CD31- population in the MLN, SI and colon of WT and Ackr4-/- mice (n=3/4 per group). Data are representative of two individual experiments. ****p<0.0001, unpaired Student’s t test, comparing data from the same tissue from WT vs Ackr4-/- mice.
Ackr4 deficiency does not affect DC migration from the intestine to the MLN
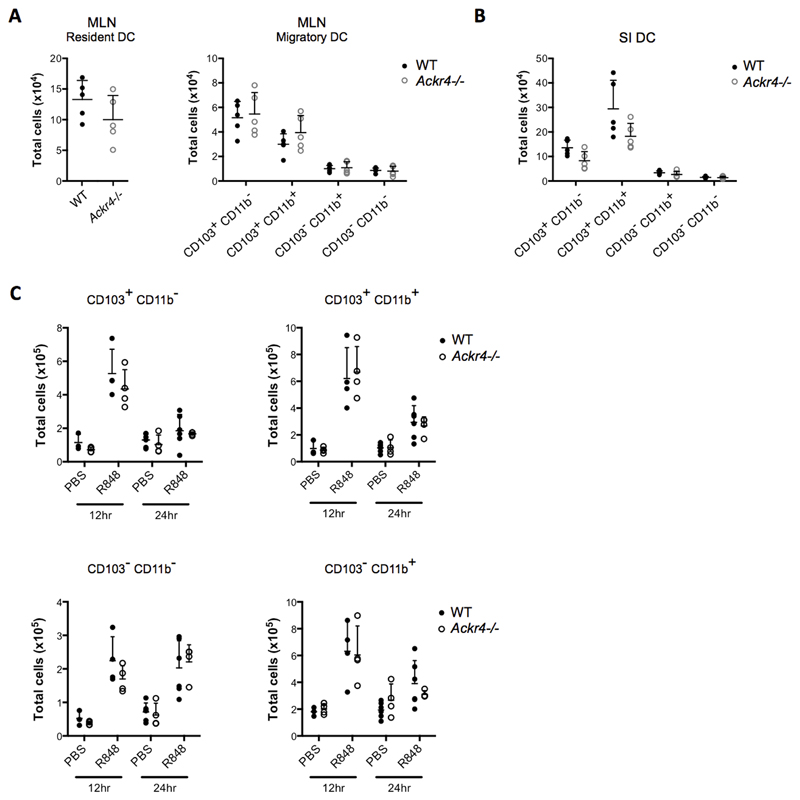
ACKR4 regulates CCR7-dependent trafficking of DCs and LCs from the skin under steady state and inflammatory conditions, and Ackr4-deficient skin-draining LNs contain fewer DCs/LCs than WT controls, while the skin contains more (11–13). DC trafficking from the SI to MLN is also CCR7-dependent (29–31). However, we found no significant differences in the numbers or relative proportions of total DC, or their subsets, in the SI LP or MLN of WT and Ackr4-deficient mice (Fig. 3A-B and data not shown; DC gating strategy in Supplementary Fig 2A-B). This was also the case after injection of R848, a synthetic TLR7/8 agonist that stimulates rapid CCR7-dependent mobilisation of DC from the SI LP to the MLN (30, 32) (Fig. 3C and Supplementary Fig 2C). ACKR4 is therefore dispensable for steady state and R848-induced DC migration from SI to MLN.
Figure 3. Ackr4 deficiency has no detectable effect on steady state or R848-induced migration of intestinal DC to the MLN.
At steady state, or after R848 treatment, DC subsets in the MLN and small intestine (SI) of WT and Ackr4-deficient (Ackr4-/-) mice (Ackr4tm1.1Rjbn) were identified by flow cytometry and quantified (for gating, see Supplementary Fig. 2A-B). (A) Numbers of CD11c+MHCII+F4/80- resident DC and CD11c+MHCIIhiF4/80- migratory DC subsets in the MLN of steady state WT and Ackr4-/- mice. (B) Numbers of CD11c+MHCII+F4/80- DC subtypes in the SI of steady state WT and Ackr4-/- mice. (C) Numbers of CD103+CD11b- and CD103+CD11b+ DC in the CD11c+MHCIIhiF4/80- migratory DC population in the MLN of WT and Ackr4-/- mice, 12 and 24 hours after i.p. administration of 100μg R848 and in PBS-treated controls. (D) Numbers of CD103+CD11b- and CD103+CD11b+ cells within the CD11c+MHCII+F4/80- DC population of the siLP of WT and Ackr4-/- mice 24 hours after administration of R848 and in PBS treated controls. In all graphs, data from individual mice are shown, along with the mean (+1SD) for each group. No statistically significant differences are present between WT and Ackr4-/- data, and this was also seen in two or more repeat experiments.
Ackr4 expression defines a discrete subset of intestinal fibroblasts
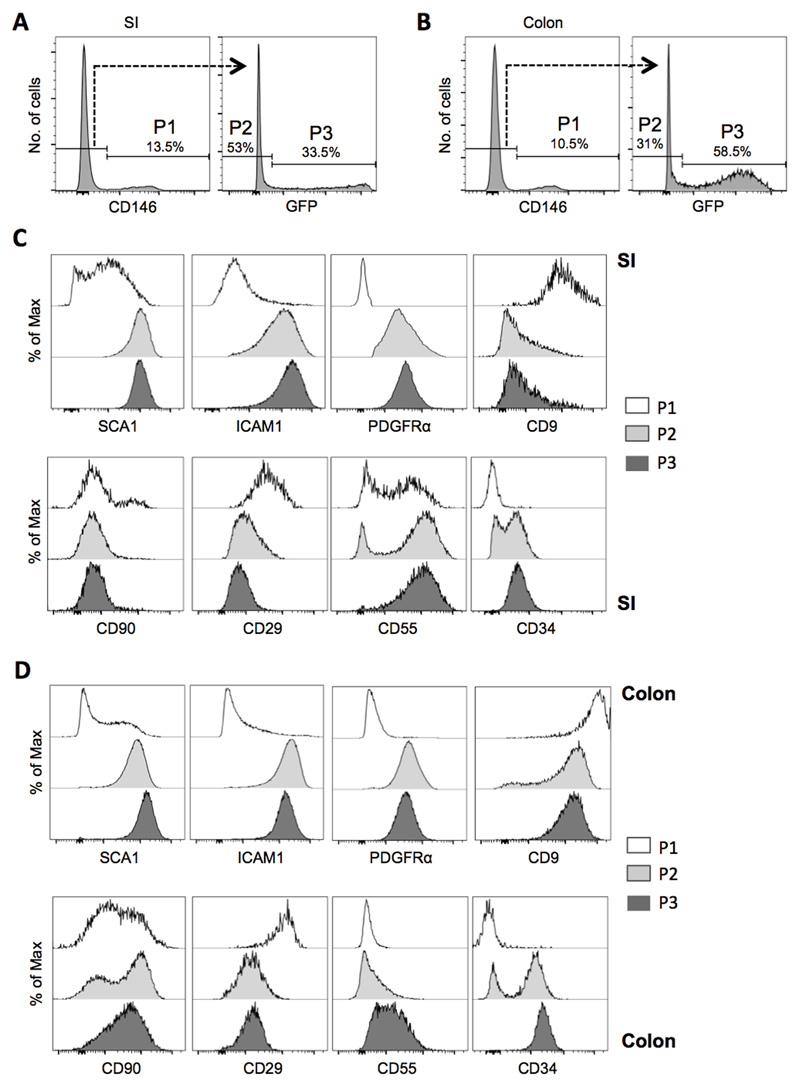
We next explored iMC heterogeneity and considered whether Ackr4 expression identifies a specific subset of iMCs. This was done by analysing iMCs from Ackr4gfp/+ mice for expression of twelve mesenchymal cell markers: CD9, CD29 (integrin β1), CD54 (ICAM1), CD55 (DAF), CD66a (CEACAM-1), CD90 (Thy1), CD105 (endoglin), CD106 (VCAM1), CD140a (PDGFRα), CD146 (MCAM), CD166 (ALCAM) and SCA1.
All these markers, except CD105, CD166, VCAM1 and CEACAM-1, were detected on some or all iMCs in both SI and colon (Fig. 4 and data not shown). iMCs were divided into three populations based on expression of CD146 and Ackr4 (Fig. 4A-B). A small population (P1 in Fig. 4A-B) was uniformly CD146+CD9+CD29+ and lacked Ackr4, ICAM1, PDGFRα and CD34 (Fig. 4C-D). In contrast, the larger CD146-Ackr4- and CD146-Ackr4+ populations (P2 and P3 in Fig. 4A-B) were uniformly SCA1+ICAM1+PDGFRα+, had lower expression of CD9 and CD29, and were mostly (P2), or all (P3), CD34+ (Fig. 4C-D). Colonic iMCs typically expressed less CD55, but more CD9 and CD90, than SI iMCs (Fig. 4C-D). CD146-Ackr4- and CD146-Ackr4+ iMCs from the same tissue differed in their expression of CD34 and CD55 (SI and colon) and CD90 (colon only), each of which was expressed more uniformly by Ackr4-expressing cells (Fig. 4C-D). The surface phenotype of Ackr4+ iMCs, particularly the presence of CD34 and PDGFRα, indicates that these cells are fibroblasts.
Figure 4. Ackr4 expression and surface immunophenotyping identifies discrete subsets of iMCs.
(A-B) Flow cytometric histogram plots showing CD146 and GFP expression by live, single CD45-GP38+CD31- iMCs from the (A) small intestine (SI) and (B) colon of Ackr4gfp/+ (Ackr4tm1Ccbl) mice. P1 cells are defined as CD146+GFP-; P2 as CD146-GFP-; and P3 as CD146-GFP+. The percentage of all iMCs found in the P1, P2 and P3 populations is indicated. (C-D) Histogram plots showing expression of SCA1, ICAM1, PDGFRα, CD9, CD90, CD29, CD55 and CD34 by P1, P2 and P3 in the (C) SI and (D) colon. Results are representative of at least three individual experiments.
Thus although iMCs comprise a complex mixture of different cell types, Ackr4+ iMCs in the SI and colon appear to be a discrete and relatively homogeneous subset of fibroblasts.
Ackr4+ fibroblasts are restricted to the intestinal submucosa
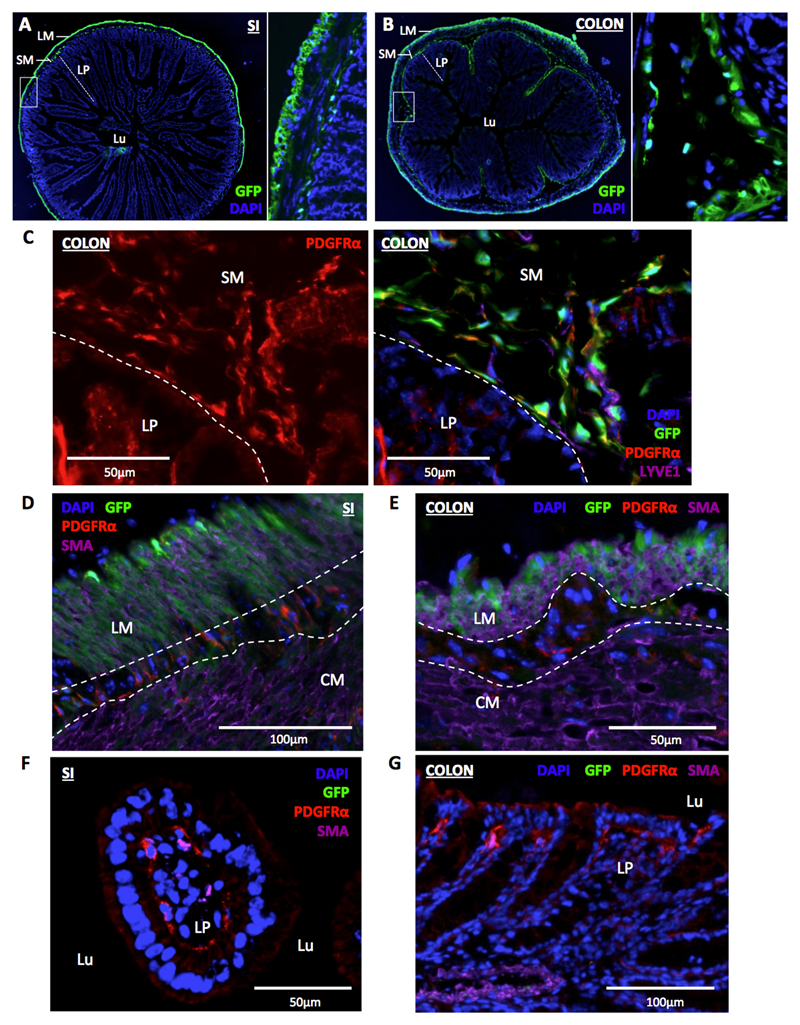
Next we sought to determine if Ackr4+ fibroblasts occupy a specific anatomical niche. Immunohistological analysis of SI and colon from Ackr4gfp/+ mice showed that GFP+ cells were absent from the lamina propria (LP), but abundant in the submucosa particularly in regions directly underlying the LP (Fig. 5A-B). Unexpectedly, cells of the longitudinal muscle (LM) layer of the SI and colon were also GFP+ (Fig. 5A-B). No green fluorescent cells were present in the submucosa or muscle layers of control WT SI and colon sections (Supplementary Fig. 3A). In both tissues, all the GFP+ cells in the submucosa were PDGFRα+, and virtually all PDGFRα+ cells in this region were GFP+ (Fig. 5C and data not shown). In contrast, GFP+ cells in the αSMA+ LM layer of the SI and colon lacked PDGFRα, although PDGFRα+ cells lacking GFP and αSMA were readily detectable between, and less frequently within, the LM and circular muscle (CM) (Fig. 5D-E). PDGFRα+ cells were also abundant in the LP, particularly in the region underneath the epithelium, but these cells were uniformly GFP- (Fig. 5C, F-G). All GFP+ iMCs detected in the flow cytometry experiments were PDGFRα+ (P3 in Fig. 4) so must all be submucosal fibroblasts: the GFP+ cells in the LM layer are presumably not liberated alive when preparing single cell suspensions. On the other hand, the GFP-PDGFRα+ iMC population identified by flow cytometry (P2 in Fig. 4) presumably contains the PDGFRα+ cells that we observed in the LP and between and within the LM and CM layers.
Figure 5. Ackr4+ fibroblasts populate the intestinal submucosa.
(A-B) Representative fluorescent microscopic images showing the location of GFP expression (green) in transverse sections of (A) small intestine (SI) and (B) colon of Ackr4gfp/+ (Ackr4tm1Ccbl) mice co-stained with DAPI (blue). The image on the right of each panel is a magnified version of the region shown in the box on the image to its left. LM, longitudinal muscle; SM, submucosa; LP, lamina propria; Lu, lumen. (C) Representative immunofluorescent microscopic image of section of colon from Ackr4gfp/+ mouse immunostained with Abs against PDGFRα (red) and LYVE1 (purple), and counterstained with DAPI (blue). Left, PDGFRα (red) only; right, composite image of all colours. The submucosa (SM) and lamina propria (LP) are indicated and separated by the dotted line. (D-G) Representative immunofluorescent microscopic images of sections of (D, F) SI or (E, G) colon of Ackr4gfp/+ mice immunostained with Abs against PDGFRα (red) and αSMA (purple), and counterstained with DAPI (blue). In D and E, longitudinal muscle (LM) and circular muscle (LM) layers are labeled and their boundaries marked with dotted lines; in F and G, lamina propria (LP) and lumen (Lu) are marked. Scale bars are shown on the images. Images are representative of at least two individual experiments each involving two or more mice.
Given the location of Ackr4-expressing cells, we were interested in whether Ackr4-deficiency was associated with any changes in the DC and lymphocyte content of the muscularis externa (ME). This structure was therefore dissected from the SI and colon of WT and Ackr4-deficient mice, digested, and analysed by flow cytometry (20) (Supplementary Fig. 2D). Lymphocytes were barely detectable in the MM of either the colon or the SI (data not shown), but DCs (CD45+CD64-CD11c+MHCIIhi) were present. However, neither the overall number of MM DCs, nor the size of individual MM DC subsets, was different between WT and Ackr4-deficient mice.
Ackr4 expression therefore shows exquisite anatomical restriction in the intestine, and defines a discrete population of fibroblasts resident in the submucosa, but Ackr4 deficiency does not lead to any detectable change in DC abundance in the ME.
Ackr4+ fibroblasts show elevated expression of genes encoding regulators of endothelial cells
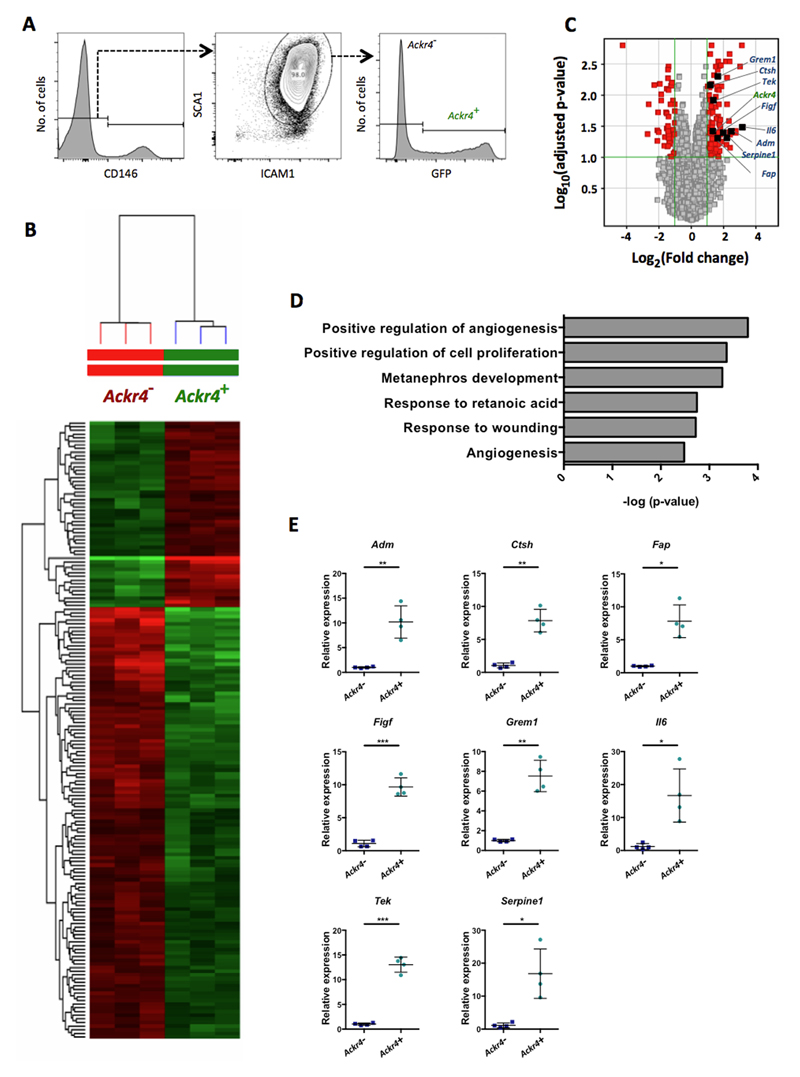
To identify genes specifically expressed by submucosal fibroblasts, we FACS-purified Ackr4+ fibroblasts and Ackr4- iMCs from the SI of Ackr4gfp/+ mice (Fig. 6A) and compared their transcriptomes using Mouse Transcriptome Arrays (Fig. 6B-D). In addition to Ackr4, 169 entities encompassing ~125 characterised genes were differentially expressed between the two groups by at least 2-fold (p<0.05; FDR 0.1) (Fig. 6B-C, Supplementary Table 1). Interestingly, more than half of the genes elevated in Ackr4+ fibroblasts encode secreted or cell surface proteins (Supplementary Table 1). Many of these are components, or regulators, of the extracellular matrix. Cytokines and their regulators were also present. It appears likely therefore that Ackr4+ fibroblasts shape the submucosal microenvironment to regulate the behaviour of cells resident in, or migrating through, this region of the intestine.
Figure 6. Ackr4+ fibroblasts show elevated expression of multiple endothelial cell regulators.
(A) FACS gating strategy used to isolate Ackr4+ and Ackr4- cells from among live single CD45-GP38+CD31- cells from the small intestine of Ackr4gfp/+ (Ackr4tm1Ccbl) mice. (B) Heatmap showing the relative signal intensities of the 165 transcripts that were differentially expressed between the Ackr4+ and Ackr4- cells (Fold Change ≥2; adjusted p value <0.05; FDR 0.1). (C) Volcano plot showing differences between Ackr4+ and Ackr4- cells in their expression of all genes analyzed, against the statistical significance of those expression differences. Genes shown in red have a Fold Change ≥2 and an adjusted p value of <0.1: those on the right hand side are more highly expressed in Ackr4+ cells; those on the left are more highly expressed in Ackr4- cells. Ackr4 and genes analyzed in panel E are highlighted. Significance was calculated using a modified t-test, and adjusted using a Benjamini Hochberg multiple testing correction. (D) The biological processes most significantly upregulated in Ackr4+ cells compared to Ackr4- cells, as determined using DAVID Bioinformatics Resources. (E) Relative expression of eight genes in Ackr4+ and Ackr4- cells, as determined by QPCR analysis of cDNA from cells FACS-sorted from among live single CD45-GP38+CD31- cells from the small intestine. Gene names are indicated at the top of each graph. Data points from individual mice are shown, along with means (±1SD). Mean expression in Ackr4- cells is set to 1. *p< 0.05, **p < 0.01, ***p< 0.001; Student’s t test.
Interestingly, gene ontology clustering of genes elevated in Ackr4+ fibroblasts identified Positive regulation of angiogenesis as the most significantly enriched biological process (Fig. 6D). The genes responsible for this association were Adm (Adrenomedullin), Col8a1 (Collagen type VIII α1), Ctsh (Cathepsin H), Fap (Fibroblast activation protein-α), Figf (VEGFD), Grem1 (Gremlin-1), Il6 (IL-6), Serpine1 (Plasminogen activator inhibitor-1) and Tek (Angiopoietin receptor Tie2), all but one of which encode secreted or surface proteins (Fig. 6C, Supplementary Table 1). Moreover, several other genes significantly elevated in Ackr4+ fibroblasts encode secreted proteins that have also been implicated in endothelial cell regulation, including Pi16 (Peptidase inhibitor 16) (33), Postn (Periostin) (34–36), Ism1 (Isthmin-1) (37), Slit2 (38, 39), Ntn4 (Netrin-4) (40–42), and Chrdl1 (Chordin-like-1) (43).
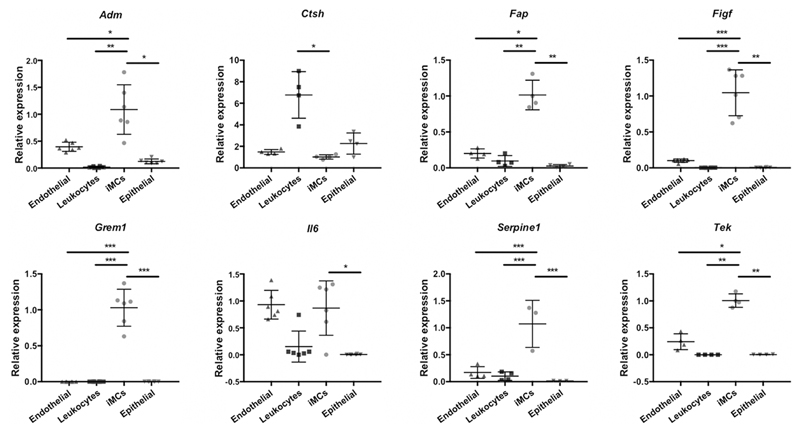
To validate the transcriptomic data we compared the expression of eight of these endothelial cell regulators (Adm, Ctsh, Fap, Figf, Grem1, Il6, Tek and Serpine1) in GFP+ and GFP- iMCs purified from the SI of Ackr4gfp/+ mice. GFP+ cells more strongly expressed all eight genes (Fig. 6E). Moreover, six of the genes we analysed (Adm, Fap, Figf, Grem1, Tek and Serpine1) were more highly expressed by SI iMCs than by endothelial cells, leukocytes or epithelial cells from the SI (Fig. 7), which, together with the data in Fig. 6E, indicate that Ackr4+ submucosal fibroblasts are the dominant source of these endothelial cell regulators in the intestine.
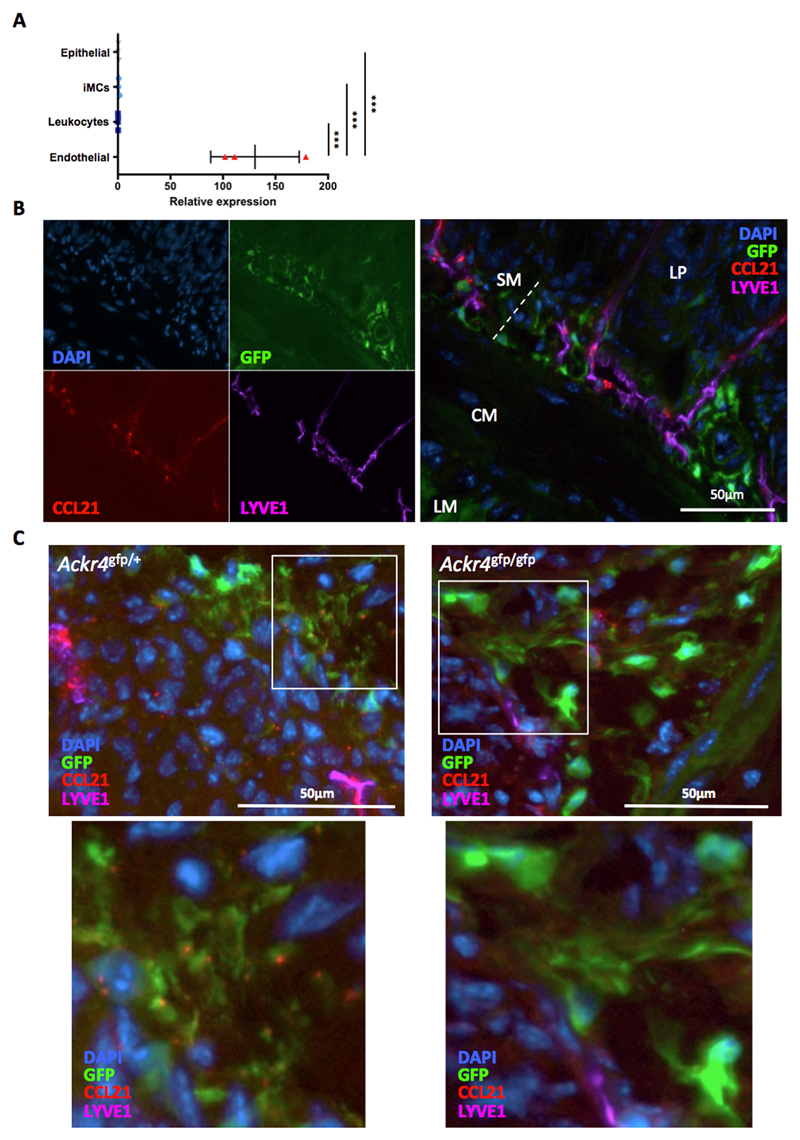
Figure 7. Expression of genes encoding regulators of endothelial cells and IESCs.
RNA was isolated from populations of cells that had been purified by FACS sorting from the small intestine of wild-type mice. cDNA was prepared and analyzed by QPCR. (A) Expression of eight genes encoding endothelial cell regulators by endothelial cells (CD45-CD31+), leukocytes (CD45+), intestinal mesenchymal cells (iMCs; CD45-GP38+CD31-) and epithelial cells (CD45-EPCAM+). Mean expression in iMCs is set to 1. *p< 0.05, **p< 0.01, ***p< 0.001; one way ANOVA. Only significant differences between iMCs and other cell types are shown. (B) Expression of genes encoding key IESC regulators by Ackr4- and Ackr4+ cells purified from the CD34+ iMC population (CD45-GP38+CD31- cells). Mean expression in CD34+Ackr4+ iMCs is set to 1. ***p< 0.001; Student’s t test. Gene names are indicated at the top of each graph. Data points from individual mice are shown, along with means (±1SD).
Collectively, these observations indicate that Ackr4+ fibroblasts have the potential to create a microenvironment that controls the biology of endothelial cells.
Ackr4+ fibroblasts are intimately associated with blood and lymphatic vessels in the submucosa
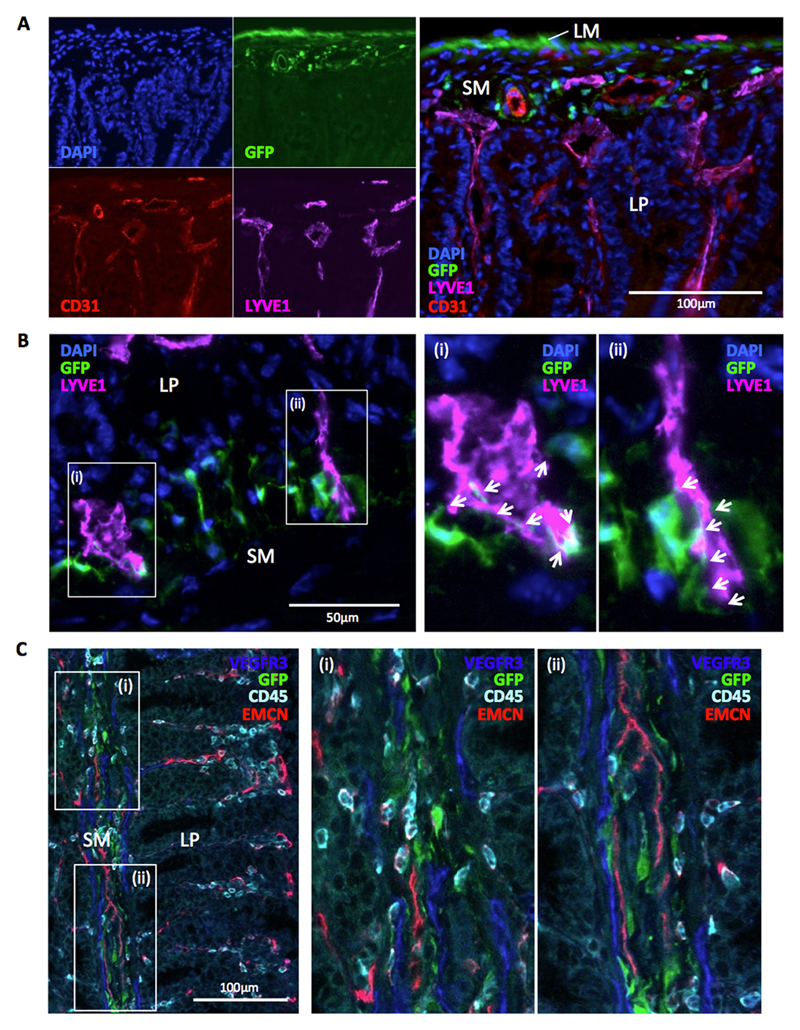
In light of the transcriptomic and QPCR data, we performed fluorescence microscopy on sections of intestine from Ackr4gfp/+ mice to examine the physical relationship between Ackr4+ fibroblasts and endothelial cells lining blood and lymphatic vessels. This revealed that Ackr4+ fibroblasts reside in close proximity to both CD31+LYVE1- blood vessels and CD31intLYVE1+ lymphatic vessels in the submucosa of the SI (Fig. 8A) and colon (Supplementary Fig. 3B). Many Ackr4+ fibroblasts showed particularly close physical interactions with lymphatic vessels, and were often seen projecting extensions onto the LYVE1+ LECs lining these structures (Fig. 8B). Moreover, LECs in the submucosa expressed VEGFR3 (Fig. 8C and Supplementary Fig. 3C), a receptor for VEGFD. VEGFD is encoded by the Figf gene, which shows strong preferential expression by Ackr4+ fibroblasts in the steady state intestine (Fig. 6E and 7). Endothelial cells (Fig. 9A), and specifically LYVE1+ LECs (Fig. 9B), were also the principal source of the ACKR4 ligand CCL21 in the intestine. Although Ccl21 transcripts were undetectable in iMCs (Fig. 9A), under high magnification small deposits of CCL21 protein could be seen associated with GFP+ fibroblasts in the intestine of Ackr4gfp/+ mice (Fig. 9C). These were virtually absent from GFP+ submucosal fibroblasts in Ackr4-deficient Ackr4gfp/gfp mice (Fig. 9C), indicating that they are likely generated by ACKR4-mediated uptake of LEC-derived CCL21.
Figure 8. Intestinal Ackr4+ fibroblasts interact with the submucosal vasculature.
(A-B) Representative images captured by fluorescence microscopy from sections of small intestine from Ackr4gfp/+ (Ackr4tm1Ccbl) mice immunostained with fluorescent Abs against (A) CD31 (red) and LYVE-1 (purple) or (B) LYVE1 only (purple), and co-stained with DAPI (blue). In A, the images in the left panel show individual colours; a composite image is shown in the right panel. In B, images in (i) and (ii) show high power views of the areas highlighted in the left panel, with arrows indicating sites of physical interaction between LYVE1+ cells and GFP+ cells. LM, longitudinal muscle; SM, submucosa; LP, lamina propria. (C) Image of section of colon from Ackr4gfp/+ mouse immunostained with Abs against VEGFR3 (blue), CD45 (cyan) and blood vessel protein, endomucin (EMCN, red). Right panels show high power views of the areas (i) and (ii) highlighted in left panel. Scale bars are shown. Images are representative of those taken from at least three independent experiments.
Figure 9. ACKR4-dependent association of LEC-derived CCL21 with submucosal Ackr4+ fibroblast.
(A) cDNA was prepared from RNA extracted from endothelial cells (CD45-CD31+), leukocytes (CD45+), intestinal mesenchymal cells (iMCs; CD45-GP38+CD31-) and epithelial cells (CD45-EPCAM+) that had been FACS-sorted from the small intestine. Expression of Ccl21 was analyzed by QPCR. Mean expression by iMCs is set to 1. Data points from individual mice are shown, along with means (±1SD). ***p< 0.001; one way ANOVA. (B-C) Representative images of sections of small intestine from (B, C) Ackr4gfp/+ or (C) Ackr4-deficient Ackr4gfp/gfp (Ackr4tm1Ccbl) mouse immunostained with Abs against CCL21 (red) and LYVE1 (purple), and co-stained with DAPI (blue). In B, the images in the left panel show individual colors; a composite image is shown in the right panel. LM, longitudinal muscle; CM, circular muscle; SM, submucosa; LP, lamina propria. In C, lower panels show high power views of the areas highlighted in the panel above. Scale bars are shown. Images are representative of two or more individual experiments.
Thus, Ackr4+ fibroblasts are a rich source of endothelial cell regulators and reside in close proximity to blood and lymphatic vessels in the submucosa. They form intimate physical associations with LECs, and are likely to regulate the function of these cells by producing the VEGFR3 ligand VEGFD and by scavenging LEC-derived CCL21 using ACKR4.
Discussion
Our study provides the first detailed analysis of ACKR4 in the intestine and reveals fundamental differences in the expression and function of this ACKR in the skin and intestine. Furthermore we have identified a previously uncharacterised population of intestinal fibroblasts that expresses ACKR4 together with genes encoding regulators of endothelial cell function. These Ackr4+ fibroblasts associate closely with the submucosal vasculature, and we have provided evidence that they engage in physical and molecular interactions with LECs. Together these data provide novel insights into the complexity and heterogeneity of the intestinal mesenchymal cell compartment, and suggest a specific role for Ackr4+ fibroblasts in regulating blood and lymphatic vessels.
Like keratinocytes, dermal LECs, and LECs on the SCS of the skin-draining LNs (SLNs) (12), subsets of iMCs and MLN LECs were capable of mediating ACKR4-dependent uptake of ACKR4 ligands. This was evident in CCL19AF647 uptake experiments performed ex vivo, and by the comparison of anti-CCL21 immunostaining of GFP+ cells in the intestines of Ackr4gfp/+ mice and Ackr4-deficient Ackr4gfp/gfp mice. However, the functional significance of ACKR4-mediated chemokine scavenging in the intestine remains unclear. Ackr4 deficiency disrupts steady state and inflammation-driven APC trafficking from skin to SLN (11–13), but we could find no evidence for this in the intestinal immune system, despite APC migration to draining LNs being profoundly dependent on CCR7 in both tissues (8, 9, 14, 15, 29–31). This likely reflects differences in the anatomical location of the ACKR4-expressing cells in the two tissues. In skin, ACKR4 is strongly expressed by keratinocytes, which may explain why the migration of epidermal LCs is particularly sensitive to Ackr4 deficiency (11, 12). In contrast, the vast majority of migratory intestinal DCs are in the mucosal LP and will use CCR7 to exit via LP-resident CCL21-expressing lymphatic vessels that are some distance from the Ackr4+ cells. We considered the possibility that ACKR4 might regulate the migratory behaviour of CCR7+ cells in the deeper layers of the intestine in response to CCL21 derived from submucosal LECs. However, the rare DC populations present in the ME under steady state conditions were not detectably affected by the absence of ACKR4.
A further activity of ACKR4 specifically in the mucosa of the SI could be to regulate the function of CCL25. CCL25 contributes to the recruitment of activated CCR9+ T and B cells into the LP and epithelium of the SI (9). It is expressed by intestinal epithelial cells, most prominently those at the base of the crypts (44), and it is conceivable that ACKR4-mediated scavenging by neighbouring submucosal fibroblasts could help maintain CCL25 gradients in this region. However, plasmacytoid DCs (pDCs), whose presence in the SI LP is dependent on CCR9 (45), are present in normal numbers in the SI of Ackr4-deficient mice, and we have been unable to detect any effect of the absence of Ackr4. Furthermore, we have found normal numbers of T and B cells in the SI LP of steady state Ackr4-deficient mice, while activated intestinal T cells home normally to this site in these animals and they have unaltered susceptibility to DSS colitis (our unpublished observations). In parallel, Ackr4-deficient mice have normal architecture and composition of intestinal secondary lymphoid organs such as the mesenteric lymph nodes and Peyer’s patches. While we cannot formally exclude a role for ACKR4 in the development of smaller organised lymphoid structures, such as cryptopatches and isolated lymphoid follicles (46), this seems unlikely in view of the normal numbers of T and B cells present in the lamina propria of Ackr4-deficient mice. Further studies are therefore required to determine the immunological functions of ACKR4 in the intestine.
GFP expression in Ackr4gfp/+ mice has been used to identify discrete populations of stromal cells, including subpopulations of thymic epithelial cells (47, 48) and LECs on the SCS ceiling of LNs (11). A key finding of our study is that GFP expression in Ackr4gfp/+ mice also identifies a discrete population of fibroblasts in the mouse intestine. To our knowledge, these cells have not been described previously, and indeed, prior to our study, the submucosal stromal compartment was very poorly characterised.
Ackr4+ fibroblasts, which our surface immunophenotyping suggests are a relatively homogeneous population of cells, are clearly anatomically distinct from mesenchymal cells located in the LP, such as myofibroblasts, LP fibroblasts, crypt stromal cells (49), PDGFRα+ subepithelial cells (50), pericytes and smooth muscle cells (1). They are also distinct from the PDGFRα+ fibroblast-like cells that we observed in, and between, the LM and CM layers, and which have been reported previously by others (51). In our FACS analysis, these various Ackr4- mesenchymal cells were either in the small GP38-CD31- DN population or in the GP38+CD31- cell subsets that we designated P1 and P2. P2 contained all PDGFRα+ iMCs that lacked Ackr4 expression. These cells appear to share many features with Ackr4+ iMCs, as evident from their surface immunophenotype and the relatively small number of differentially expressed genes identified in the transcriptomic analysis. The surface immunophenotype of the P1 population suggests that these cells are less closely related to other GP38+ iMCs. Indeed, transcriptomic analysis indicated that many hundreds of genes are differentially expressed between P1 cells and other GP38+ iMCs (data not shown). Particularly prominent amongst genes preferentially expressed by P1 cells were those encoding markers of enteric glial cells, including Plp1, S100b, Gfap and Sox10 (52) (data not shown).
Our combined transcriptomic, QPCR, and microscopic analyses strongly suggest that an important function of Ackr4+ fibroblasts is to regulate endothelial cells. The submucosa is rich in blood and lymphatic vessels, and capillaries in the LP originate from these vessels. Thus Ackr4+ fibroblasts are well positioned to regulate capillary growth and endothelial cell function in both arms of the vasculature. Many of the genes preferentially expressed by Ackr4+ fibroblasts encode proteins that regulate BECs. Interestingly, in thymus, ACKR4 also appears to be expressed by stromal cells adjacent to blood vessels, specifically those at the corticomedullary junction where thymocyte progenitors enter and mature thymocytes leave the thymus (48). However, the close physical association between Ackr4+ fibroblasts and LECs in the submucosa was particularly striking, and we found strong evidence of potential crosstalk between these two cell types through the CCL21/ACKR4 and VEGFD/VEGFR3 pathways. Moreover, several other genes preferentially expressed by Ackr4+ fibroblasts encode proteins that regulate LECs, including Tie2 (53, 54), adrenomedullin (55), netrin-4 (41) and periostin (35).
The lymphatic vessel network of the intestine is unique. Not only does it act as a crucial conduit for tissue fluid and migratory DCs, it also plays a crucial role in metabolism by mediating the transport of dietary fat out of the intestine in the form of chylomicrons. These lipoprotein particles enter the lymphatic capillaries in SI villi and travel in the lymph to the bloodstream. Moreover, under steady state conditions lymphatic vessels in the intestine, unlike those in other tissues, are in a permanent state of regeneration and proliferation (56, 57). This is dependent on VEGFR2- and VEGFR3-induced expression of the Notch ligand delta-like 4 (DLL4) (57). Smooth muscle cell-derived VEGFC is the key VEGFR3 ligand in this context, but VEGFD also makes a contribution (56), and Gremlin1, which is a VEGFR2 agonist (58), could play a role too. The strong and restricted expression of Adm by Ackr4+ fibroblasts is also particularly interesting given that adrenomedullin is critical for the development and maintenance of lymphatic vessels (55). Indeed, recent elegant work has shown that inducing the conditional deletion of Calcrl (which encodes the adrenomedullin receptor) exclusively in the LECs of adult mice leads to intestinal lymphangiectasia, impaired lipid uptake, defective LEC expression of DLL4 and junctional proteins, and failure to resolve intestinal inflammation (59). Together these findings indicate that a number of the genes we have shown as being highly and specifically expressed by submucosal Ackr4+ fibroblasts encode mediators that control homeostasis of the local lymphatic vasculature.
Their high expression of Grem1 raises the possibility that Ackr4+ fibroblasts may also be capable of regulating epithelial cell turnover in the intestine. In addition to acting as a VEGFR2 agonist (58), Gremlin1 is an antagonist of bone morphogenetic proteins (BMPs) 2, 4 and 7 (60) and this activity is thought to contribute to the maintenance of intestinal epithelial stem cells (IESCs) (49, 61). Indeed, previous work has shown that CD34+GP38+ iMCs are a major source of Grem1 transcripts in the mouse intestine and can induce Gremlin1-dependent expansion of IESCs in intestinal organoids in vitro (49). These CD34+GP38+ iMCs were reported to be present in the LP (49), but since we have found that all Ackr4+ fibroblasts express CD34 and GP38, a large fraction of CD34+GP38+ iMCs reside in the submucosa. Moreover, our data suggest that these submucosal cells express more Grem1. Indeed, when we purified Ackr4+ and Ackr4- cells direct from the CD34+GP38+ iMC population of the SI, transcripts encoding Gremlin1 were >4-fold more abundant in the Ackr4+ cells (data not shown). The Ackr4+ cells also showed higher expression of genes encoding Wnt2b and R-spondin1 (data not shown), two other important IESC regulators reported to be specifically expressed by CD34+GP38+ iMCs (49, 62–64). In addition, our transcriptomic data show that Ackr4+ fibroblasts have elevated expression of Chrdl1, which encodes the BMP4 antagonist Chordin-like 1 protein (61), while Bmp3, Bmp5 and Bmp7 were amongst the small number of genes whose expression was significantly lower in Ackr4+ cells (Supplementary Table 1). Collectively, these observations suggest that Ackr4+ fibroblasts are well equipped to regulate the IESC niche, and although, like GREM1+ mesenchymal cells in the human colon (61), they do not appear to lie immediately adjacent to the crypt bases where the most primitive IESCs are found, it remains possible that by producing Wnt2b, R-spondin1 and BMP antagonists these cells can contribute to IESC regulation and epithelial cell turnover.
To our knowledge, our studies are the first to characterise submucosal fibroblasts in mice. It will now be of interest to dissect their contribution to the maintenance of homeostasis in the SI and colon, and to characterise their function in other contexts, such as during development, injury, inflammation or tumorigenesis when angiogenesis and lymphangiogenesis are prominent and vitally important processes. It will also be important to characterise, in the context of health and disease, the phenotype and function of submucosal mesenchymal cells in the human intestine. Ultimately, a deeper understanding of the molecular and cellular properties of submucosal fibroblasts and other mesenchymal cell populations could reveal novel therapeutic approaches to treat diseases affecting the intestine.
Supplementary Material
Acknowledgements
The authors thank staff at Glasgow University’s Central Research Facility for animal husbandry.
Footnotes
This work was supported by a project grant (MR/L000598) from the United Kingdom Medical Research Council (CAT, AMM, RJBN). SAvdP was supported by an A*MIDEX Chaire d’Excellence grant (ANR-11-IDEX-0001-002), FRM Jeunes Equipes (AJE20150633331), and France Bio Imaging (ANR-10-INBS-04-01); and GM, EL and MS were supported by a Research Foundation Flanders grant (G0D8317N) and PhD fellowship (to MS (ZKD1563)).
Non-standard abbreviations used in this paper: ACKR, atypical chemokine receptor; AF, Alexa Fluor®; αSMA, alpha smooth muscle actin; BEC, blood vessel endothelial cell; BMP, bone morphogenetic protein; CM, circular muscle; DAVID, Database for Annotation, Visualization and Integrated Discovery; DC, dendritic cell; DN, double negative; DSS, dextran sodium sulfate; FDR, false discovery rate; FRC, fibroblastic reticular cell; IESC, intestinal epithelial stem cell; iMC, intestinal mesenchymal cell; LC, Langerhans cells; LEC, lymphatic endothelial cell; LM, longitudinal muscle; LN, lymph node; LP, lamina propria; Lu, lumen; ME, muscularis externa; MLN, mesenteric lymph node; QPCR, quantitative real-time polymerase chain reaction; SCS, subcapsular sinus; SI, small intestine; SM, submucosa; WT, wild-type.
The authors declare that they have no conflict of interest.
References
- 1.Powell DW, Pinchuk IV, Saada JI, Chen X, Mifflin RC. Mesenchymal cells of the intestinal lamina propria. Annu Rev Physiol. 2011;73:213–237. doi: 10.1146/annurev.physiol.70.113006.100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens BMJ, Simmons A. Intestinal stromal cells in mucosal immunity and homeostasis. Mucosal Immunol. 2013;6:224–234. doi: 10.1038/mi.2012.125. [DOI] [PubMed] [Google Scholar]
- 3.Owens BMJ. Inflammation, Innate Immunity, and the Intestinal Stromal Cell Niche: Opportunities and Challenges. Front Immunol. 2015;6:319. doi: 10.3389/fimmu.2015.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadière C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, Mantovani A, et al. International Union of Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nibbs RJB, Graham JG. Immune regulation by atypical chemokine receptors. Nat Rev Immunol. 13:815–829. doi: 10.1038/nri3544. [DOI] [PubMed] [Google Scholar]
- 6.Gosling J, Dairaghi DJ, Wang Y, Hanley M, Talbot D, Miao Z, Schall JT. Cutting edge: identification of a novel chemokine receptor that binds dendritic cell- and T cell-active chemokines including ELC, SLC, and TECK. J Immunol. 2000;164:2851–2856. doi: 10.4049/jimmunol.164.6.2851. [DOI] [PubMed] [Google Scholar]
- 7.Townson JR, Nibbs RJB. Characterization of mouse CCX-CKR, a receptor for the lymphocyte-attracting chemokines TECK/mCCL25, SLC/mCCL21 and MIP-3beta/mCCL19: comparison to human CCX-CKR. Eur J Immunol. 2002;32:1230–1241. doi: 10.1002/1521-4141(200205)32:5<1230::AID-IMMU1230>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 8.Girard J-P, Moussion C, Förster R. HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol. 2012;12:762–773. doi: 10.1038/nri3298. [DOI] [PubMed] [Google Scholar]
- 9.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 10.Comerford I, Milasta S, Morrow V, Milligan G, Nibbis R. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur J Immunol. 2006;36:1904–1916. doi: 10.1002/eji.200535716. [DOI] [PubMed] [Google Scholar]
- 11.Ulvmar MH, Werth K, Braun A, Kelay P, Hub E, Eller K, Chan L, Lucas B, Novitzky-Basso I, Nakamura K, Rulicke T, et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat Immunol. 2014;15:623–630. doi: 10.1038/ni.2889. [DOI] [PubMed] [Google Scholar]
- 12.Bryce SA, Wilson RAM, Tiplady EM, Asquith DL, Bromley SK, Luster AD, Graham GJ, Nibbs RJB. ACKR4 on Stromal Cells Scavenges CCL19 To Enable CCR7-Dependent Trafficking of APCs from Inflamed Skin to Lymph Nodes. J Immunol. 2016;196:3341–3353. doi: 10.4049/jimmunol.1501542. [DOI] [PubMed] [Google Scholar]
- 13.Heinzel K, Benz C, Bleul CC. A silent chemokine receptor regulates steady-state leukocyte homing in vivo. Proc Natl Acad Sci USA. 2007;104:8421–8426. doi: 10.1073/pnas.0608274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohl L, Mohaupt M, Czeloth N, Hintzen G, Kiafard Z, Zwirner J, Blankenstein T, Henning G, Förster R. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity. 2004;21:279–288. doi: 10.1016/j.immuni.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Braun A, Worbs T, Moschovakis GL, Halle S, Hoffmann K, Bölter J, Münk A, Förster R. Afferent lymph-derived T cells and DCs use different chemokine receptor CCR7-dependent routes for entry into the lymph node and intranodal migration. Nat Immunol. 2011;12:879–887. doi: 10.1038/ni.2085. [DOI] [PubMed] [Google Scholar]
- 16.Comerford I, Nibbs RJ, Litchfield W, Bunting M, Harata-Lee Y, Haylock-Jacobs S, Forrow S, Körner H, McColl SR. The atypical chemokine receptor CCX-CKR scavenges homeostatic chemokines in circulation and tissues and suppresses Th17 responses. Blood. 2010;116:4130–4140. doi: 10.1182/blood-2010-01-264390. [DOI] [PubMed] [Google Scholar]
- 17.Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J Immunol. 2010;184:6843–6854. doi: 10.4049/jimmunol.0903987. [DOI] [PubMed] [Google Scholar]
- 18.Scott CL, Bain CC, Mowat AM. Isolation and Identification of Intestinal Myeloid Cells. Methods Mol Biol. 2017;1559:223–239. doi: 10.1007/978-1-4939-6786-5_15. [DOI] [PubMed] [Google Scholar]
- 19.Bain CC, Mowat AM. CD200 receptor and macrophage function in the intestine. Immunobiology. 2012;217:643–651. doi: 10.1016/j.imbio.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Farro G, Stakenborg M, Gomez-Pinilla PJ, Labeeuw E, Goverse G, Giovangiulio Di, Stakenborg N, Meroni E, D'Errico F, Elkrim Y, Laoui D, et al. CCR2-dependent monocyte-derived macrophages resolve inflammation and restore gut motility in postoperative ileus. Gut. 2017;66:2098–2109. doi: 10.1136/gutjnl-2016-313144. [DOI] [PubMed] [Google Scholar]
- 21.Ford LB, Cerovic V, Milling SWF, Graham GJ, Hansell CAH, Nibbs RJB. Characterization of conventional and atypical receptors for the chemokine CCL2 on mouse leukocytes. J Immunol. 2014;193:400–411. doi: 10.4049/jimmunol.1303236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansell CAH, Schiering C, Kinstrie R, Ford L, Bordon Y, McInnes IB, Goodyear CS, Nibbs RJB. Universal expression and dual function of the atypical chemokine receptor D6 on innate-like B cells in mice. Blood. 2011;117:5413–5424. doi: 10.1182/blood-2010-11-317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford LB, Hansell CAH, Nibbs RJB. Using fluorescent chemokine uptake to detect chemokine receptors by fluorescent activated cell sorting. Methods Mol Biol. 2013;1013:203–214. doi: 10.1007/978-1-62703-426-5_13. [DOI] [PubMed] [Google Scholar]
- 24.Bordon Y, Hansell CAH, Sester DP, Clarke M, Mowat AM, Nibbs RJB. The atypical chemokine receptor D6 contributes to the development of experimental colitis. J Immunol. 2009;182:5032–5040. doi: 10.4049/jimmunol.0802802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson CA, McColl A, Cavanagh J, Graham GJ. Peripheral inflammation is associated with remote global gene expression changes in the brain. J Neuroinflammation. 2014;11:73. doi: 10.1186/1742-2094-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang JE, Turley SJ. Stromal infrastructure of the lymph node and coordination of immunity. Trends Immunol. 2015;36:30–39. doi: 10.1016/j.it.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Worbs T, Bode U, Yan S, Hoffmann MW, Hintzen G, Bernhardt G, Förster R, Pabst O. Oral tolerance originates in the intestinal immune system and relies on antigen carriage by dendritic cells. J Exp Med. 2006;203:519–527. doi: 10.1084/jem.20052016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Márquez G, Förster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yrlid U, Cerovic V, Milling S, Jenkins CD, Klavinskis LS, MacPherson GG. A distinct subset of intestinal dendritic cells responds selectively to oral TLR7/8 stimulation. Eur J Immunol. 2006;36:2639–2648. doi: 10.1002/eji.200636426. [DOI] [PubMed] [Google Scholar]
- 33.Hazell GGJ, Peachey AMG, Teasdale JE, Sala-Newby GB, Angelini GD, Newby AC, White SJ. PI16 is a shear stress and inflammation-regulated inhibitor of MMP2. Sci Rep. 2016;6:39553. doi: 10.1038/srep39553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang X-F. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24:3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kudo Y, Iizuka S, Yoshida M, Nguyen PT, Siriwardena SBSM, Tsunematsu T, Ohbayashi M, Ando T, Hatakeyama D, Shibata T, Koizumi K, et al. Periostin directly and indirectly promotes tumor lymphangiogenesis of head and neck cancer. PLoS ONE. 2012;7:e44488. doi: 10.1371/journal.pone.0044488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G, Nakamura I, Dhanasekaran R, Iguchi E, Tolosa EJ, Romecin PA, Vera RE, Almada LL, Miamen AG, Chaiteerakij R, Zhou M, et al. Transcriptional Induction of Periostin by a Sulfatase 2-TGFβ1-SMAD Signaling Axis Mediates Tumor Angiogenesis in Hepatocellular Carcinoma. Cancer Res. 2017;77:632–645. doi: 10.1158/0008-5472.CAN-15-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venugopal S, Chen M, Liao W, Er SY, Wong W-SF, Ge R. Isthmin is a novel vascular permeability inducer that functions through cell-surface GRP78-mediated Src activation. Cardiovasc Res. 2015;107:131–142. doi: 10.1093/cvr/cvv142. [DOI] [PubMed] [Google Scholar]
- 38.Jones CA, Nishiya N, London NR, Zhu W, Sorensen LK, Chan AC, Lim CJ, Chen H, Zhang Q, Schultz PG, Hayallah AM, et al. Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat Cell Biol. 2009;11:1325–1331. doi: 10.1038/ncb1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rama N, Dubrac A, Mathivet T, Ní Chárthaigh R-A, Genet G, Cristofaro B, Pibouin-Fragner L, Ma L, Eichmann A, Chédotal A. Slit2 signaling through Robo1 and Robo2 is required for retinal neovascularization. Nat Med. 2015;21:483–491. doi: 10.1038/nm.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lejmi E, Leconte L, Pédron-Mazoyer S, Ropert S, Raoul W, Lavalette S, Bouras I, Feron J-G, Maitre-Boube M, Assayag F, Feumi C, et al. Netrin-4 inhibits angiogenesis via binding to neogenin and recruitment of Unc5B. Proc Natl Acad Sci USA. 2008;105:12491–12496. doi: 10.1073/pnas.0804008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larrieu-Lahargue F, Welm AL, Thomas KR, Li DY. Netrin-4 induces lymphangiogenesis in vivo. Blood. 2010;115:5418–5426. doi: 10.1182/blood-2009-11-252338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lejmi E, Bouras I, Camelo S, Roumieux M, Minet N, Leré-Déan C, Merkulova-Rainon T, Autret G, Vayssettes C, Clement O, Plouët J. Netrin-4 promotes mural cell adhesion and recruitment to endothelial cells. Vasc Cell. 2014;6:1. doi: 10.1186/2045-824X-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kane R, Godson C, O'Brien C. Chordin-like 1, a bone morphogenetic protein-4 antagonist, is upregulated by hypoxia in human retinal pericytes and plays a role in regulating angiogenesis. Mol Vis. 2008;14:1138–1148. [PMC free article] [PubMed] [Google Scholar]
- 44.Kunkel EJ, Campbell JJ, Haraldsen G, Pan J, Boisvert J, Roberts AI, Ebert EC, Vierra MA, Goodman SB, Genovese MC, Wardlaw AJ, et al. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: Epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wendland M, Czeloth N, Mach N, Malissen B, Kremmer E, Pabst O, Förster R. CCR9 is a homing receptor for plasmacytoid dendritic cells to the small intestine. Proc Natl Acad Sci USA. 2007;104:6347–6352. doi: 10.1073/pnas.0609180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pabst O, Herbrand H, Friedrichsen M, Velaga S, Dorsch M, Berhardt G, Worbs T, Macpherson AJ, Förster R. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J Immunol. 2006;177:6824–6832. doi: 10.4049/jimmunol.177.10.6824. [DOI] [PubMed] [Google Scholar]
- 47.Ribeiro AR, Meireles C, Rodrigues PM, Alves NL. Intermediate expression of CCRL1 reveals novel subpopulations of medullary thymic epithelial cells that emerge in the postnatal thymus. Eur J Immunol. 2014;44:2918–2924. doi: 10.1002/eji.201444585. [DOI] [PubMed] [Google Scholar]
- 48.Lucas B, White AJ, Ulvmar MH, Nibbs RJB, Sitnik KM, Agace WW, Jenkinson WE, Anderson G, Rot A. CCRL1/ACKR4 is expressed in key thymic microenvironments but is dispensable for T lymphopoiesis at steady state in adult mice. Eur J Immunol. 2015;45:574–583. doi: 10.1002/eji.201445015. [DOI] [PubMed] [Google Scholar]
- 49.Stzepourginski I, Nigro G, Jacob J-M, Dulauroy S, Sansonetti PJ, Eberl G, Peduto L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci USA. 2017;114:E506–E513. doi: 10.1073/pnas.1620059114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurahashi M, Nakano Y, Peri LE, Townsend JB, Ward SM, Sanders KM. A novel population of subepithelial platelet-derived growth factor receptor α-positive cells in the mouse and human colon. Am J Physiol Gastrointest Liver Physiol. 2013;304:G823–34. doi: 10.1152/ajpgi.00001.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurahashi M, Zheng H, Dwyer L, Ward SM, Koh SD, Sanders KM. A functional role for the “fibroblast-like cells” in gastrointestinal smooth muscles. J Physiol. 2011;589:697–710. doi: 10.1113/jphysiol.2010.201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao M, Nelms BD, Dong L, Salinas-Rios V, Rutlin M, Gershon MD, Corfas G. Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. Glia. 2015;63:2040–2057. doi: 10.1002/glia.22876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tammela T, Saaristo A, Lohela M, Morisada T, Tornberg J, Norrmén C, Oike Y, Pajusola K, Thurston G, Suda T, Ylä-Herttuala S, et al. Angiopoietin-1 promotes lymphatic sprouting and hyperplasia. Blood. 2005;105:4642–4648. doi: 10.1182/blood-2004-08-3327. [DOI] [PubMed] [Google Scholar]
- 54.Morisada T, Oike Y, Yamada Y, Urano T, Akao M, Kubota Y, Maekawa H, Kimura Y, Ohmura M, Miyamoto T, Nozawa S, et al. Angiopoietin-1 promotes LYVE-1-positive lymphatic vessel formation. Blood. 2005;105:4649–4656. doi: 10.1182/blood-2004-08-3382. [DOI] [PubMed] [Google Scholar]
- 55.Klein KR, Caron KM. Adrenomedullin in lymphangiogenesis: from development to disease. Cell Mol Life Sci. 2015;72:3115–3126. doi: 10.1007/s00018-015-1921-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurmi H, Saharinen P, Zarkada G, Zheng W, Robciuc MR, Alitalo K. VEGF-C is required for intestinal lymphatic vessel maintenance and lipid absorption. EMBO Mol Med. 2015;7:1418–1425. doi: 10.15252/emmm.201505731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernier-Latmani J, Cisarovsky C, Demir CS, Bruand M, Jaquet M, Davanture S, Ragusa S, Siegert S, Dormond O, Benedito R, Radtke F, et al. DLL4 promotes continuous adult intestinal lacteal regeneration and dietary fat transport. J Clin Invest. 2015;125:4572–4586. doi: 10.1172/JCI82045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitola S, Ravelli C, Moroni E, Salvi V, Leali D, Ballmer-Hofer K, Zammataro L, Presta M. Gremlin is a novel agonist of the major proangiogenic receptor VEGFR2. Blood. 2010;116:3677–3680. doi: 10.1182/blood-2010-06-291930. [DOI] [PubMed] [Google Scholar]
- 59.Davis RB, Kechele DO, Blakeney ES, Pawlak JB, Caron KM. Lymphatic deletion of calcitonin receptor-like receptor exacerbates intestinal inflammation. JCI Insight. 2017;2:e92465. doi: 10.1172/jci.insight.92465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brazil DP, Church RH, Surae S, Godson C, Martin F. BMP signalling: agony and antagony in the family. Trends Cell Biol. 2015;25:249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 61.Kosinski C, Li VSW, Chan ASY, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci USA. 2007;104:15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 63.Kim K-A, Kakitani M, Zhao J, Oshima T, Tang T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, Funk WD, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 64.Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantù C, Aguet M, Basler K. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep. 2016;15:911–918. doi: 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.