Abstract
KIT is a cell surface tyrosine kinase receptor whose ligand SCF (Stem Cell Factor) triggers homodimerization and activation of downstream effector pathways involved in cell survival, proliferation, homing or differentiation. KIT activating mutations are major oncogenic drivers in subsets of acute myeloid leukemia (AML), in mast cell leukemia and in gastrointestinal stromal tumors (GIST). The overexpression of SCF and/or wild type (WT) KIT is also observed in a number of cancers including 50% of AML and small cell lung cancer. The use of tyrosine kinase inhibitors (TKI) in these pathologies is however hampered by initial or acquired resistance following treatment. Using antibody phage display, we obtained 2 antibodies (2D1 and 3G1) specific for the most membrane proximal extracellular immunoglobulin domain (D5) of KIT which is implicated in KIT homodimerization. Produced as single chain variable antibody fragments fused to the Fc fragment of a human IgG1, bivalent 2D1-Fc and 3G1-Fc inhibited KIT-dependent growth of leukemic cell lines expressing WT KIT (UT7/Epo) or constitutively active KIT mutants including the TKI imatinib resistant KIT D816V mutant (HMC1.2 cell line). In all models, either expressing WT KIT or mutated KIT, 2D1 and 3G1-Fc induced KIT internalization and sustained surface down regulation. However, interestingly, KIT degradation was only observed in leukemic cell lines with oncogenic KIT, a property likely to limit the toxicity of these antibodies in patients. These fully human antibody formats may represent therapeutic tools to target KIT signaling in leukemia or GIST, and to bypass TKI resistance of certain KIT mutants.
Keywords: KIT/CD117, SCF, monoclonal antibody, leukemia, GIST, antibody phage-display
Introduction
KIT is a 145 kDa type III tyrosine kinase receptor that functions as a growth factor receptor (1). Type III tyrosine kinase receptors are characterized by five N-glycosylated immunoglobulin (Ig) domains (D1 to D5) in the N-terminal extra-cellular region and by a split tyrosine kinase intra-cellular domain. D2 and D3 domains are involved in the binding of KIT ligand, SCF, while the fourth (D4) and fifth (D5) domains are implicated in receptor dimerization following SCF binding (2, 3). KIT intracellular region contains the catalytic domain composed of the ATP binding site and the phosphotransferase domain. KIT stimulation by SCF induces KIT dimerization and transphosphorylation which activates downstream effector pathways (1). SCF is a major cytokine for self-renewal, proliferation and differentiation of hematopoietic lineage, germ cells, melanocytes, gut and central nervous system in embryo (4). In adult, KIT is expressed in a limited number of tissues and its defects induce impaired hematopoiesis, decreased number of tissue mast cells, decreased fertility and pigmentation, and defective development of the interstitial cells of Cajal responsible for intestinal pacemaker activity, for review see (5).
Abnormal KIT signaling is observed in cancer due either to overexpression of SCF and/or KIT itself or to gain-of-function KIT mutations. These mutations of KIT are generally heterozygous (6) and are major oncogenic drivers in gastrointestinal stromal tumors (GIST) (7), that derive from Cajal cells, in subsets of acute myeloid leukemia (Core-binding factor acute myeloid leukemia, CBF-AML) (8), in mast cell leukemia (9), in melanoma (10) and less frequently, in other cancers. Consequently, oncogenic KIT inhibition with specific tyrosine kinase inhibitors (TKI) has proven successful in those pathologies (11).
KIT mutations can be classified in two main categories corresponding to distinct structural and functional locations (12). Catalytic mutants display point mutations located in the kinase domain, mainly in the activation loop. For instance, the mutation D816V is keeping the activation loop in extended conformation corresponding to constitutive phosphotransferase activity (13). Catalytic KIT mutants are found in CBF-AML (8), germ cell tumors (14), mast cell tumors and mastocytosis (9). Regulatory mutants, found in GIST (12), display point mutations or short deletions in the juxtamembrane domain that induce constitutive dimerization therefore permanent activation of the receptor (15).
Wild type KIT is also involved in a number of malignant diseases mainly derived from cell-types that express KIT transiently during embryogenesis (16). The SCF/KIT axis works as an autocrine or paracrine loop sustaining proliferation and/or migration. Indeed, KIT is present on 50% of AML (17) and AML blasts frequently respond to SCF stimulation by increased proliferation (18). Consistently, AML patients refractory to chemotherapy can be cured by KIT inhibition by TKI (19). A number of solid tumors also frequently express KIT and/or SCF including small cell lung cancer (SCLC), melanoma, semimoma. Seventy % of SCLC co-express SCF and KIT (20) and high KIT levels are associated with poor prognosis (21). Other solid tumors have been reported to express KIT including breast cancers, neuroblastomas, colon cancers, gynecological tumors, gliomas and non SCLC (11).
Pharmacologic inhibition of KIT is an efficient targeted approach to treat malignancies that are partly or completely dependent on KIT signaling. In GIST patients, ATP-competitive type II TKI (as defined by TKI binding to the inactive conformation of the kinase) imatinib (22) has proven successful. However, some KIT catalytic mutants exhibit initial resistance to imatinib (23) and long term TKI treatment with imatinib induces secondary mutations and resistance to the drug (24). ATP-competitive type I TKI (as defined by TKI binding to the active conformation of the kinase) have been developed like dasatinib (25) but their use in the clinic is impaired by toxic side effects due to broad specificity (26).
Because of their high specificity combined to their natural ability to recruit immune effectors or their conjugation to toxic drugs or radio-isotopes, monoclonal antibodies (mAbs) offer an attractive approach to target KIT (27). Independently of effector recruitment to KIT positive cells, inhibitory effect on KIT signaling could be obtained with mAbs by direct blocking SCF binding to KIT, inhibition of KIT dimerization and/or induction of KIT degradation. Recent studies reported pre-clinical data in GIST using naked anti-KIT mAb either competing with SCF (28) or targeting KIT D4 dimerization domain (29). A radiolabelled anti-KIT antibody was also shown to decrease SCLC progression in a mouse model (30).
We report here the generation and characterization of a panel of fully human anti-KIT single-chain variable antibody fragment (scFv) selected by phage-display on a recombinant form of KIT. Among the 8 scFv obtained, 5 bound to endogenous KIT and were produced in fusion to the Fc fragment of a human IgG1 as dimeric scFv-Fc (31). Two of the scFv-Fc, namely 2D1 and 3G1, were found to target KIT D5 membrane most proximal domain and to efficiently inhibit KIT-dependent signaling and cell growth in various cells models of leukemia dependent of KIT signaling for their growth and survival. In all the models, either expressing WT KIT or various KIT mutants, mAbs induce KIT internalization. However, interestingly, KIT degradation is only observed in leukemic cell lines with mutated KIT, a property likely to limit the toxicity of these mAbs in patients.
Materials and Methods
Cells and cell culture conditions
None of the cell lines were authenticated. Ba/F3, Ba/F3-KIT and TF-1 cell lines (a gift from Patrice Dubreuil, Institut Paoli-Calmettes, Marseille, France, in 2009) were grown in complete (10%FBS + antibiotics) RPMI. Ba/F3 and Ba/F3-KIT cells were supplemented with 0.1 % conditioned medium from X63-IL-3 (gift from Patrice Dubreuil) and TF-1 with GM-CSF (10 ng/mL) (Invitrogen). UT-7/Epo cell line, a subline of UT-7 cells (gift from Isabelle Dusanter-Fourt, Institut Cochin, Paris, France, in 2010) were grown in complete IMDM supplemented either with EPO (2 U/mL, Abnova) or SCF. Recombinant human SCF (Cell Signaling) or CHO-KL (a recombinant CHO cell line expressing mouse SCF) culture supernatant (gift from Michel Arock, ENS Cachan, France) was used as a source of SCF. HMC1.1 and HMC1.2 cell lines, that harbor a V560G KIT or a V560G D816V KIT allele, respectively (gift from Michel Arock, in 2010) were grown in complete IMDM. HEK-T cell line was grown in complete DMEM. All these cells were cultured at 37°C in a humidified atmosphere enriched in CO2 (5 %), were checked for absence of mycoplasma infection, dependency to cytokines sensitivity to TKI dasatinib and imatinib. Insect Sf9 and High Five cells (Invitrogen) were grown at 27°C in respectively Grace medium - 10 % FBS - or Express Five medium (both from Fisher Scientific) supplemented with 20 mM of L-Glutamine.
Antibodies and pharmacological reagents
For Western blot analysis, anti-KIT-extracellular domain (ECD) (H300) and anti-Erk 2 antibodies were obtained from Santa Cruz, anti-phospho-Akt (Ser 473) and anti-phospho-Erk (Thr 202/Tyr 204) from Cell Signaling, anti-Akt and anti-GAPDH from Millipore and anti-phospho-KIT (Y823 or Y568-570) from Invitrogen. HRP-conjugated anti-mouse or rabbit IgG antibodies were from Sigma. PE/Cy5 or PE/Cy7-conjugated anti-human KIT 104D2 mAb (that does not compete with SCF for binding to KIT (information provided by the manufacturer; Biolegend), anti-SV5 mAb (Invitrogen), conjugated to Alexa-647 (Labeling Kit Molecular Probes) and anti-c-myc peptide mAb 9E10 (Sigma) were used for FACS analysis. PE-anti-mouse Ig and anti-human Fc F(ab')2 fragment were from Beckton Dickinson and Rockland, respectively. Imatinib was a gift from Serge Roche (CRBM, Montpellier, France) and dasatinib was obtained from Santa Cruz.
Recombinant soluble KIT extra cellular domain expression and purification
Soluble recombinant KIT-ECD (a.a 26 to 509, P10721 UniProtKB/Swiss-Prot) was produced using baculovirus/insect cell system. C-terminal His6-tagged KIT-ECD cDNA was generated by PCR amplification from plasmid pSKΔSapI-Kit (a gift from Frédéric Subra, ENS Cachan, France) and cloned into transfer vector pVL1393 (Beckton Dickinson). Recombinant Kit-ECD viruses were produced in Sf9 cells and recombinant protein KIT-ECD in High Five cells following the BD Baculogold™ protocol. The culture medium, containing the secreted 6His-tagged KIT-ECD, was collected and dialyzed in PBS before purification with Ni-NTA affinity chromatography (Qiagen). The eluted fraction (elution buffer PBS-Imidazole 250 mM) was concentrated and buffered into PBS using Amicon Ultra-4 30K centrifugal Filter Units (Millipore). Total protein concentration was determined by spectrophotometry (1 uA280 nm = 0.83 g/L) and purified products analyzed by SDS-PAGE.
Selection of KIT-ECD specific phage antibodies
General steps for the selection have been described elsewhere (32, 33). Briefly, 100 μL of KIT-ECD at 50 μg/mL were used to coat a well of a maxisorp 96-well plate (Nunc) at 4°C overnight. The well was saturated with PBS-2 % milk for 2 h at RT before adding 1011 cfu of the phage antibody library (34) diluted in PBS-2 % milk. After a 2 h incubation at RT, the plate was washed 20 times with PBS-Tween 0.1 % and 10 times with PBS. Bound phages (output phages) were eluted with triethylamine 100 mM, neutralized with a solution of Tris 1M pH 7 and amplified for the next round of selection. Selection was monitored by titration of input/output phages by infection of Escherichia coli TG1 for each round of selection. Screening for KIT binders was performed by soluble scFv-ELISA. Monoclonal scFvs were expressed from single colonies grown in 96-well plates and induced with IPTG 1 mM for 12-16 h. Crude culture supernatant containing soluble scFv was tested by ELISA for binding to recombinant KIT-ECD or bovine serum albumin (BSA) (Sigma). The diversity of ELISA positive clones was determined by sequencing. For FACS staining, 5 mL inducted supernatants were concentrated 10 times and buffered in PBS using Amicon Ultra-4 10K centrifugal Filter Units.
Human scFv-Fc expression and purification
To express scFv in fusion with human Fcγ1, scFv cDNA was Nco I/Not I subcloned from the library phagemid into pFUSE-hFc2(IL2ss) vector (31), a gift from Frank Perez, Institut Curie, Paris, France. Soluble 110 kDa scFv-Fc were produced by transient transfection of HEK-T cells (supplemental material and methods). ScFv-Fc binding botulinum neurotoxin (Bot-Fc) and TfR1 (transferrine receptor-1) (H7-Fc) (35) were used as controls.
Quantification of scFv-Fc binding to recombinant KIT-ECD by ELISA
A maxisorp 96-well plate was coated with KIT-ECD (or BSA) 5 μg/mL in PBS overnight at 4°C. Wells were saturated with PBS-1 % BSA for 2 h at RT. ScFv-Fc were diluted in PBS-1% BSA and incubated 2 h at RT. Bound scFv-Fc were detected with HRP-conjugated anti-human IgG (Fc specific) secondary antibody (Sigma) followed by ABTS enzymatic substrate solution incubation (Roche). Fits and EC50 were determined with GraphPad software.
Flow cytometry
Staining was performed at 4°C. Two hundred thousand mammalian cells were resuspended in 100 μL of FACS buffer (PBS-0.5%BSA) and incubated with crude concentrated bacteria supernatant containing myc-tagged scFv or purified scFv-Fc then with secondary antibodies. For competition experiments, cells were preincubated with SCF (1 μg/mL) during 40 min before adding the scFv-Fc (10 μg/mL). For internalization experiments, cells were seeded at 5 × 105 in 700 μL complete growth medium per well and incubated with antibodies (2D1-Fc, 3G1-Fc or Bot-Fc, 10 μg/mL) at 37°C. KIT cell surface expression was detected on cells returned at 4°C with anti-KIT 104D2 mAb. Yeast cells (1 million) displaying SV5 epitope-tagged KIT truncated forms (supplemental material and methods) were stained with anti-KIT scFv-Fc followed by secondary antibodies or with PE-conjugated 104D2 mAb directly. Cells were analyzed with a FACS Quanta (Beckman Coulter).
Western blot analysis
Cells were washed with stop buffer [150 mM NaCl, 50 mM Tris-HCl (pH 6.7), 1 mM EDTA, 2 mM Na3VO4, 7.5 mM, Na4P2O7, 100 mM NaF] and lysed in stop buffer complemented with 1 mM PMSF, 0.25 % DOC, 1 % NP40 and protease inhibitor cocktail (Roche Diagnostic). Lysates were cleared by centrifugation at 13 000 × g for 10 min at 4°C, quantified with BCA assay (Interchim) and 30 μg of proteins were separated by SDS-PAGE (10% polyacrylamide) for analysis by Western Blot.
Proliferation assay
UT-7/Epo were seeded at 104 per well and treated for 4 days with scFv-Fc at 5 or 50 μg/mL in presence of EPO or CHO-KL supernatant diluted 500 times. HMC1.1 and 1.2 were seeded at 2.105 cells per well and treated the same but for 7 days and with 2% FBS and without cytokines. Cell viability was estimated with MTS assay (Promega) as recommended by the manufacturer.
Results
Selection and initial characterization of KIT-ECD-specific single chain Fv antibodies
The goal of this work was to isolate fully human KIT-specific antibodies that could inhibit KIT signaling to interfere with the growth of KIT dependent tumors. We proceeded by antibody phage-display selection on recombinant human KIT-ECD produced in insect cells. The purified C-terminal 6His-tagged recombinant KIT-ECD analyzed on a reducing SDS-PAGE gel showed a major protein of 60 kDa also detected by Western blot with an antibody directed against KIT-ECD which validated the production (Fig.S1). For selections, phages were prepared from a human scFv phage naïve antibody library (34). After 3 rounds of selection for enrichment of KIT ECD binders (TableS1), 8 different scFv anti-KIT ECD were obtained, 2D1 being the most frequent (Fig.S1). Because these scFv had been selected based on their binding to recombinant human KIT produced in insect cells, that have distinct N-glycosylation enzymatic machineries compared with mammalian cells (36), we tested whether they could also bind to endogenous KIT. Only the antibodies scFv that detected endogenous KIT on HMC1.2 cells (2D1, 2A3, 3G1, 2B12, 2A6) (Fig.S1) were kept for further analysis.
Conversion into a bivalent scFv-Fc formats
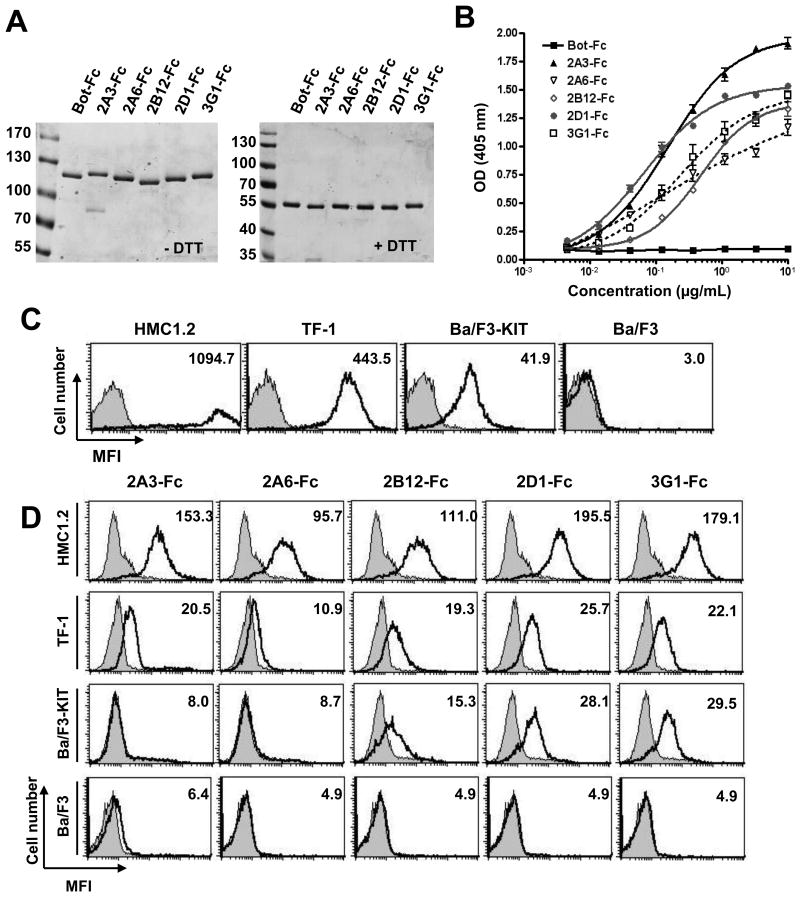
Five anti-KIT monovalent scFv were converted into a bivalent scFv-Fc format (31) consisting of a fusion at the C-terminal end of the scFv of the hinge domain and the 2 constant domains of the human γ1 Ig heavy chain, and produced in HEK-T cells. SDS-PAGE analysis of protein A affinity purified products showed proteins with the expected 110 kDa molecular weight corresponding to covalently di-sulfide linked scFv-Fc homodimers (55 kDa each) (Fig.1A). The reformatting conserved antibody specificity since all scFv-Fc bound to the receptor, as shown by ELISA on recombinant KIT-ECD, while an irrelevant antibody fragment Bot-Fc in the same format produced in the same conditions, did not (Fig.1B, Table1). Concentrations to reach half of the saturation ELISA signal (EC50) were in the nmolar range except for 2A6-Fc for which signal saturation was not reached suggesting a lower affinity. Within the antibodies tested bivalent 2D1-Fc displayed the lowest EC50 (0.66 nM or 72 ng/mL) therefore had the better apparent affinity for recombinant KIT. We then tested the binding of the 5 anti-KIT scFv-Fc by FACS to cell lines expressing different levels of surface KIT, as assessed by using the commercial 104D2 mouse mAb (37) (Fig.1C), including human HMC1.2 (high levels of KIT) and erythroleukemic TF-1 (medium levels of KIT) cell lines and murine pro-B Ba/F3 cell line modified to express human KIT (low levels of KIT). The parental Ba/F3 cell line, that does not express endogenous mouse KIT, was used as a negative control. KIT was detected on HMC1.2 and TF-1 cells by all 5 anti-KIT scFv-Fc (used at 10 μg/mL), 2A6-Fc showing lower staining, consistent with lower signal observed (Fig.1B) on ELISA on recombinant KIT-ECD. 2B12, 2D1 and 3G1-Fc, but not 2A6-Fc nor 2A3-Fc, detected low level of KIT on Ba/F3-KIT cell line, (Fig.1D).
Figure 1. Characterization of KIT binding of dimeric anti-KIT scFv-Fc antibody formats.
A, SDS-PAGE analysis of the 5 anti-KIT-ECD scFv-Fc and the irrelevant scFv-Fc Bot-Fc in non-reducing condition (left panel) versus reducing conditions (right panel). ScFv-Fc are produced in HEK-T cells and purified on protein A-agarose. B, ELISA analysis of purified scFv-Fc binding to recombinant KIT-ECD. C, Cells were resuspended in FACS buffer and were incubated for 15 min at 4°C with 104D2 anti-KIT antibody conjugated to APC to determine KIT cell surface expression (black lane), isotype control (shaded). D, Cells were resuspended in FACS buffer and were incubated scFv-Fc (10 μg/mL) for 2 hrs at 4°C. Antibody binding to cells (black line) was detected with anti-human Fc fragment coupled to PE. Negative control experiment with Bot-Fc (shaded).
Table 1. EC50 of the scFv-Fc binding to KIT-ECD by ELISA.
Concentrations (nM) of scFv-Fc antibody format giving 50% of maximal OD at 405 nm are given. Fits and EC50 were determined with GraphPad software. NA, non appropriate, SD, standard deviation.
| 2A3-Fc | 2A6-Fc | 2B12-Fc | 2D1-Fc | 3G1-Fc | |
|---|---|---|---|---|---|
| nM | 1,40 | NA | 4.56 | 0.66 | 2.04 |
| SD +/- | 0.11 | NA | 0.35 | 0.06 | 0.28 |
Antibody interference with WT KIT
Signaling interference
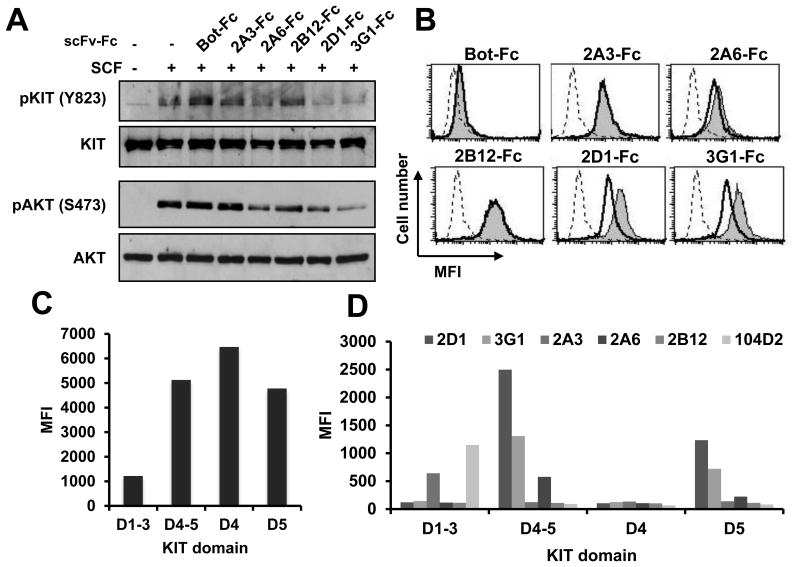
Anti-KIT antibodies interference with WT KIT signaling was first tested using the TF-1 cell line. Starved TF-1 cells were incubated with scFv-Fc before stimulation with recombinant SCF. Western blot showed that 2A6, 2D1 and 3G1-Fc inhibited SCF induced KIT phosphorylation on tyrosine 823 (Y823), 2D1 and 3G1-Fc being the most effective while irrelevant antibody Bot-Fc did not (Fig.2A). Downstream AKT phosphorylation was also reduced by the same antibodies, with stronger effect with 2D1 and 3G1-Fc. Preincubation with SCF at 4°C reduced the binding of 2A6, 2D1 and 3G1-Fc to HMC1.2 cells while 2B12 and 2A3-Fc binding was not affected (Fig.2B). To narrow the binding site on KIT, truncated forms of KIT were displayed on yeast (Fig.2C, TableS2) and tested for their detection by the anti-KIT antibodies (Fig.2D). 2D1, 3G1 and 2A6-Fc bound to KIT D4-D5 and to KIT D5 but not to KIT D4 and KIT-D1-D3 constructs, while 2A3 only detected KIT D1-D3, similarly to the 104D2 commercial antibody. 2B12 binding site on KIT was not found using this method. Moreover, 3G1 but not 2A6 competed with 2D1 for binding to KIT (Fig.S2), suggesting an overlapping epitope for 2D1 and 3G1 and a distinct epitope for 2A6 on KIT D5. In the rest of the study, we focused on the strongest KIT signaling inhibitory anti-KIT-D5 2D1 and 3G1 antibodies.
Figure 2. Anti-KIT scFv-Fc Interference with WT KIT signaling and epitope mapping.
A, TF-1 cells were starved overnight in serum-free medium and incubated with scFv-Fc (10 μg/mL) before 5 min stimulation with SCF (100 ng/mL). KIT and AKT phosphorylation were analyzed by Western blot with phospho-specific antibodies. Total KIT and AKT levels were visualized after stripping the membranes and reprobing with anti-KIT and anti-AKT antibodies. B, Cells were incubated with recombinant SCF (1 μg/mL) at 4°C before incubation with scFv-Fc (10 μg/mL) and revelation as in Fig. 1D. Dashed lane, secondary antibody only, shaded, scFv-Fc binding and black lane, scFv-binding with SCF pre-incubation. C, represents the MFI (mean fluorescent intensity) (FL4-H) of yeast displaying various KIT truncated forms as indicated with anti-SV5 mAb conjugated to Alexa 647. D, Yeast expressing KIT truncated forms are co-stained with SV5 mAb conjugated to Alexa 647 and 100 nM scFv-Fc followed by anti-human Fc-PE Ig or 104D2-PE-Cy7 mAb and MFI (FL2-H) of SV5 positive yeast is represented.
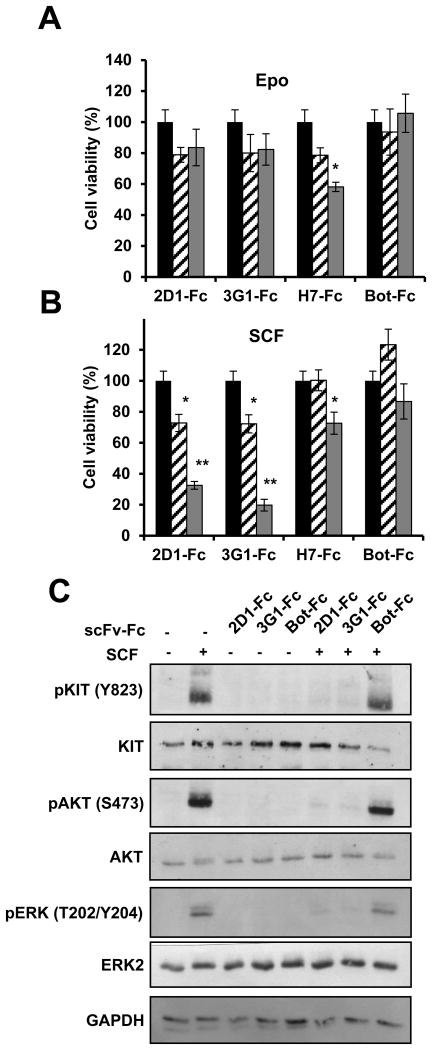
To investigate if the antagonistic effect of 2D1 and 3G1 scFv-Fc antibody formats on SCF-dependent KIT signaling translated into functional effects, we used the megakaryoblastic leukemic cell line UT-7/Epo which is dependent on either Epo or SCF cytokines for its proliferation in vitro ((38) and Fig.S3)). UT-7/Epo cells were incubated 4 days with antibodies and the effect on cell viability was measured. 2D1-Fc and 3G1-Fc alone had no effect on cells grown with Epo (Fig.3A). 2D1-Fc and 3G1-Fc decreased cell viability in a dose dependent manner when cells were cultivated with SCF (Fig.3B), while H7-Fc, an anti-TfR1 antibody (35) in the same format, decreased cell viability both in Epo and SCF cultivated cells and irrelevant antibody Bot-Fc had no effect. Like in TF-1 cells, the anti-KIT antibodies inhibited downstream KIT signaling pathways, with inhibition of phosphorylation of KIT on Y823, of phosphorylation of AKT and ERK2, and when used alone did not stimulate KIT signaling (Fig.3C).
Figure 3. Effect of anti-KIT D5 domain scFv-Fc 2D1 and 3G1 on UT-7/Epo cell line.
A, B, UT-7/Epo cells were incubated 4 days in Epo (A) or CHO-KL supernatant (B) in the presence of anti-KIT antibodies or control antibody as indicated at 5 μg/mL (dashed) or 50 μg/mL (grey). Cell viability was measured by MTS assay and is expressed in % compared to non-treated cells (black). Data were analyzed with a Kruskal Wallis test. Differences were considered significant when p < 0.05: *, p < 0.05; **, p < 0.01, versus non treated cells. C, UT-7/Epo cells were starved overnight in serum-free medium and incubated with scFv-Fc (10 μg/mL) before 5 min stimulation with SCF (100 ng/mL). KIT, AKT and ERK1/2 phosphorylation were analyzed by Western blot with phospho-specific antibodies. Total KIT and ERK2 levels were visualized after stripping the membranes and reprobing with anti-KIT, anti-AKT and anti-ERK2 antibodies.
Antibody modulation of KIT levels
In a therapeutic orientated strategy, one way to reduce KIT signaling in cancer cells would be to decrease KIT levels at the cell surface in order to reduce its access to SCF. SCF binding to KIT at the cell surface has been shown to rapidly induce internalization and degradation of KIT (39) while re-expression of KIT at the cell surface after internalization requires neosynthesis (40). We analyzed the effect of 2D1 and 3G1 antibody treatment on KIT surface level by FACS using the 104D2 anti-KIT mAb, that does not interfere with 2D1 and 3G1-Fc for KIT binding on cells (Fig.S4).
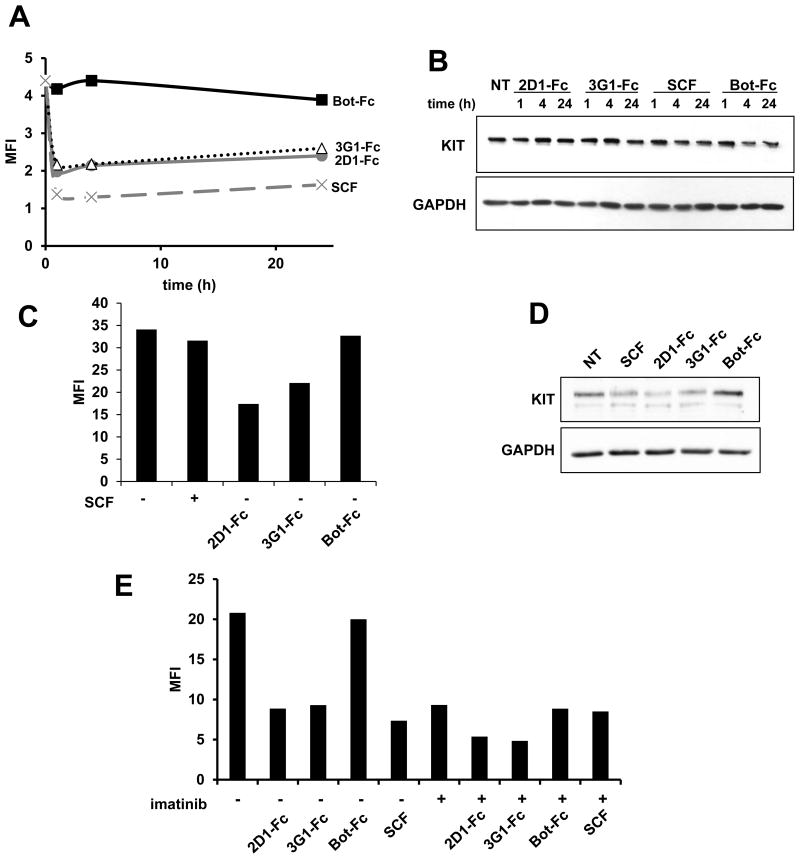
Treatment of UT-7/Epo cells for 1 to 24 h with 2D1 or 3G1-Fc induced KIT cell surface down-regulation with a 50% decrease but did not affect total KIT levels (Fig.4A, B). Kinetics of KIT surface level decrease were similar when cells were treated with SCF from 1 to 24 h, but after 72 h KIT surface levels stayed low with antibodies while going back to initial levels with SCF (Fig.4C), both treatments decreasing similarly slightly total KIT levels (Fig.4D). Since in the same cell model, TKI imatinib was also recently shown to decrease KIT surface levels (41), we tested the effect of a combination treatment of anti-KIT antibody and imatinib on UT-7/Epo cells cultivated in the presence of EPO as a source of cytokine. Interestingly, the combination with TKI had an additive effect with 2D1 or 3G1 antibody treatment on KIT surface level reduction (Fig.4E).
Figure 4. WT KIT modulation by anti-KIT D5 scFv 2D1 and 3G1 in UT-7/Epo cell line.
UT-7/Epo cells were incubated in IMDM 10% FCS and Epo and treated for (A, B) 1, 4, 24 hrs (C, D), 72 hrs or (E) 4 hrs with scFv-Fc antibodies (10 μg/mL), CHO-KL (diluted 500 times) and imatinib (1 μM) as indicated. (A, C, E) KIT surface levels were estimated by FACS upon treatment. Results expressed in MFI. (B, D) Total KIT levels were determined by Western blot using KIT and GAPDH specific antibodies as indicated. Data shown are representative of 3 experiments.
To sum up, these data show that bivalent 2D1 and 3G1 scFv-Fc antibodies, that target KIT D5 membrane proximal region do not induce KIT phosphorylation and downsteam signaling but inhibit SCF induced KIT signaling resulting in reduced growth of KIT-dependent cell lines. The antibodies induce the internalization but limited down regulation of KIT and combination treatment with imatinib increases this effect.
Antibody interference with oncogenic KIT
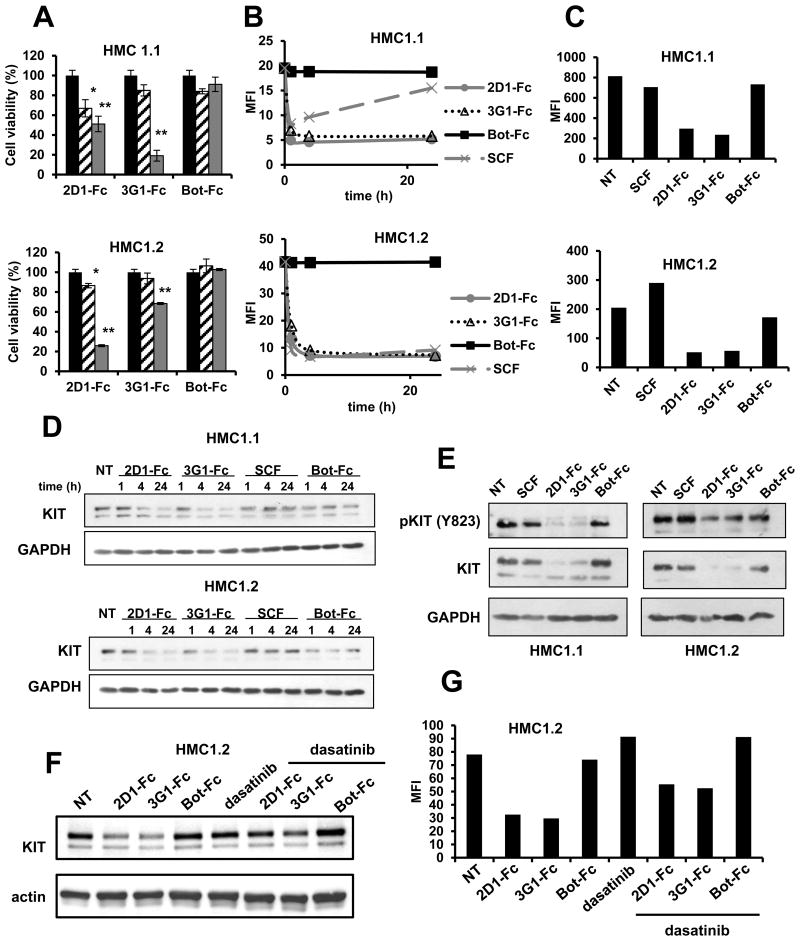
To test the effect of anti-KIT D5 antibodies on oncogenic KIT activity, we used the mast cell leukemic cell lines HMC1.1 and HMC1.2 that express constitutively active KIT V560G regulatory or KIT D816V catalytic oncogenic KIT mutants, respectively, and depend on KIT signaling (but not on SCF) for their growth and survival (42). As expected, HMC1.1 cells were sensitive to TKI imatinib and dasatinib but HMC1.2 cells were only sensitive to dasatinib due to the resistance of the KIT D816V mutant to imatinib (23) (Fig.S3). When HMC1.1 and HMC1.2 cell lines were treated with 2D1 and 3G1-Fc at doses of 5 and 50 μg/mL for 7 days we found that both anti-KIT antibodies decreased the viability of both cell lines (Fig.5A). Antibody 3G1-Fc was less active than 2D1 on HMC1.2 cells and irrelevant antibody Bot-Fc had no effect.
Figure 5. Oncogenic KIT modulation by anti-KIT D5 scFv 2D1 and 3G1 in HMC1 cell lines.
A, HMC1 cell lines were incubated for 7 days in IMDM 1% FCS in presence of anti-KIT (2D1 and 3G1-Fc) or control scFv-Fc antibody (Bot-Fc) at 5 μg/mL (dashed) or 50 μg/mL (grey). Cell viability is expressed in % compared to non-treated cells (black). Data were analyzed with a Kruskal Wallis test. Differences were considered significant when p < 0.05: *, p < 0.05; **, p < 0.01, versus non treated cells. (B-G) HMC1 cells were incubated in IMDM 10% FCS in presence of scFv-Fc antibodies (10 μg/mL), SCF and dasatinib (1 μM) as indicated for 1, 2 and 24 hrs (B, D), 72 hrs (C, E) or (F, G) 4 hrs. Treated cells were analyzed by FACS using 104D2 anti-KIT antibody (B, C, G) or by Western blot using anti-KIT and GAPDH antibodies (D, E, F). Representative data of 3 independent experiments each time.
To understand the mechanism of inhibition of anti-KIT antibodies, surface KIT level modulation kinetics were analyzed like on the UT-7/Epo cell line. In both mast cell leukemic cell lines, after 1 h, both 2D1 and 3G1-Fc induced a strong decrease of KIT levels similar to SCF treatment (Fig. 5B). Upon a 72 h treatment, the down-regulation of surface KIT was maintained (Fig.5C). Unlike in UT-7/Epo cells, surface down-regulation of KIT was associated with a strong decrease of total KIT levels after 4 h (Fig.5D) which was conserved after 72 h in both cell lines (Fig.5E). Notably this decrease was accompanied by a decrease of phosphorylated KIT levels, stronger in HMC1.1 than in HMC1.2 cell line (Fig.5E). Additionally, when tested on the HMC1.2 cell line, the inhibition of kinase activity by TKI dasatinib reversed partially the antibody-induced KIT degradation and surface down regulation, suggesting that degradation is dependent on KIT phosphorylation (Fig.5F,G). In contrast with antibody treatment, SCF treatment induced only a transient decrease of KIT surface levels on both cell line (Fig.5C) and total KIT levels were not affected (Fig.5D, E).
Altogether these data show that the targeting of KIT D5 membrane proximal region reduces the viability of HMC1 mast cell leukemic cell lines. On these cells, that are dependent on oncogenic KIT for their growth and survival, anti-KIT D5 antibodies induce a strong KIT degradation and reduction of phosphorylated KIT levels contributing to reduce KIT downstream signaling. Interestingly, HMC1.2 cells that are resistant to TKI imatinib due to the D816V mutation are sensitive to 2D1-Fc and, to a lesser extent to 3G1-Fc.
Discussion
In this work we have generated anti-KIT antibodies using antibody phage display to select for binders to recombinant KIT-ECD (Fig.S1, TableS1). Eight distinct KIT-ECD phage binders were isolated but when expressed as soluble monovalent scFv only 5 detected endogenous KIT on HMC1.2 cells, maybe due to a low affinity for KIT or to the recognition N-glycosylation motifs specific to insect cells (36) for the 3 negative scFvs. When reformated into bivalent scFv-Fc formats and produced in mammalian cells, all 5 endogenous KIT binders still bound to high KIT expressing HMC1.2 cells. Nevertheless 2 of them (2A3 and 2A6) did not detect KIT on mouse Ba/F3 cells transfected to express WT human KIT (Fig.1D). This could be due to a low affinity for KIT in the case of 2A6 (Table1, Fig.1B) but the reason is unclear for 2A3 since its affinity for KIT and staining intensity of TF-1 and HMC1.2 cells by FACS were similar to that of 2B12, 2D1 and 3G1 that indeed also stained KIT on Ba/F3-KIT cells (Table1, Fig.1D). This approach therefore allowed the rapid selection of a panel of 5 distinct human antibodies, isolated from a natural naïve antibody phage library (34) that bind endogenous KIT.
KIT-ECD is structured in 5 Ig domains. From the crystal structure, it is know that the binding of dimeric SCF to domain 2 and 3 of KIT favors KIT homodimerization at the cell surface. This results in homotypic interactions between domain 4 and domain 5 of each monomer that induce a twist in the intracellular domain, leading to the intracellular kinase domain activation (3). We used yeast display to identify the domain bound by the various antibodies. KIT truncated forms were successfully displayed on yeast (Fig2C-D). Lower molecular weight truncated forms were better displayed than larger constructs, as previously observed (43). Anti-KIT 2B12 scFv-Fc did not detect any KIT truncated form despite strong detection of KIT on all mammalian cell lines tested but, using this technique, we determined that 2A3 binds to KIT D1-D3 ligand binding region and that 2D1, 3G1 and 2A6 bind to KIT D5 membrane most proximal domain. 2D1 and 3G1, but not 2A6, binding to KIT was mutually exclusive, suggesting a shared epitope for the formers and a distinct one for the latter. Because SCF blocked partially all three anti-KIT D5 antibodies binding to KIT (Fig.2B), it is likely that these antibodies bind a region involved in D5 homotypic interactions implicated in SCF induced KIT homodimerization and that the binding of SCF renders their epitope inaccessible.
Because of their dimeric format, we expected a possible agonistic activity of the anti-KIT scFv-Fc suggested by their potential induction of KIT dimerization, but none of the 5 anti-KIT scFv-Fc induced activation of KIT downstream signaling nor were able to maintain the growth of SCF-dependent cell lines in cytokine free medium. On the contrary, interestingly, all 3 anti-KIT D5 antibodies (2A6-Fc, 2D1-Fc and 3G1-Fc) inhibited SCF-dependent KIT activation (Fig. 2A, 3C) and the most potent, 2D1 and 3G1, also reduced cell viability of leukemic cell lines dependent on SCF and KIT signaling for their survival (Fig.3B).
One major effect of targeting KIT D5 domain with 2D1 and 3G1 antibodies was the rapid induction of KIT internalization and the long term decrease of KIT cell surface levels, both in leukemic cells expressing only WT KIT (UT-7/Epo) and in cells expressing a WT and a constitutively mutated KIT allele (HMC1.1 and HMC1.2). KIT is described as a rapidly renewed receptor that is not recycled at the cell surface after ligand-induced internalization (40, 44). Intrinsic kinase activity and phosphorylation of KIT are required for efficient SCF-induced degradation of KIT and involve the recruitment to KIT of the Cbl E3-ubiquitin ligase, ubiquitinylation of KIT and degradation by both lysosomal and proteasomal pathways (39). As previously observed by others (45), we found that KIT turnover rate was in the same range whether KIT was constitutively active or not (Fig.S3). We also showed that despite early KIT internalization induction similar to anti-KIT D5 2D1 or 3G1 antibody treatment, treatment with SCF did not change KIT surface level in the long term (Fig.4C,5C). This difference is likely due to increased KIT neosynthesis upon SCF treatment, but not upon antibody treatment.
While anti-KIT D5 2D1 and 3G1 scFv-Fc induced internalization of KIT was observed independently of the presence of KIT activating mutations, KIT degradation was only observed in cells harboring a constitutively active mutated KIT allele (Fig.4B,D;5D-E). Indeed, antibody induced KIT degradation was dependent on effective mutant KIT kinase activity since it was reversed by treatment with TKI dasatinib (Fig.5F). Dasatinib also reversed partially the decrease of KIT surface levels induced by 2D1 and 3G1 in HMC1.2 cells (Fig.5G). Notably, an opposite effect was obtained on UT-7/Epo leukemic cells that express exclusively WT KIT. In those cells, imatinib and anti-KIT D5 antibody combination enhanced KIT cell surface reduction compared with treatment with either reagent alone (Fig.4E). This observation is in line with a recent study in the same cell model that reports that the occupancy of the KIT intracellular kinase ATP binding site by the TKI imatinib induces KIT internalization and degradation, without the requirement of KIT phosphorylation (41). Therefore, in a context where KIT is not mutated, the association of TKI and anti-D5 antibody could durably decrease KIT from the cell surface of cancer cells, avoiding its activation by SCF. To summarize, anti-KIT D5 scFv-Fc inhibit KIT signaling in leukemic cells by distinct mechanism depending on KIT status. Inhibition of cell growth is obtained by induction of the degradation of constitutively activated KIT or by intracellular sequestration of WT KIT.
Recently, two studies presenting naked anti-KIT antibodies for cancer therapy have been reported. Edris and colleagues (28) studied the effect of SCF competing anti-KIT antibody SR1 (46, 47) on GIST cell lines presenting various constitutively active KIT mutant alleles. In a mouse model, SR1 was reported to limit GIST progression essentially by a macrophage-dependent mechanism while KIT degradation upon SR1 treatment was not observed suggesting a different mechanism of inhibition than 2D1 and 3G1 antibodies. Reshetnyak and collegues presented an in vitro study with an anti-KIT antibody binding to an epitope implicated in homotypic D4-D4 KIT interactions required for KIT activation after SCF binding. Similarly to the anti-D5 antibodies of this study, the anti-KIT D4 antibody inhibited SCF induced WT KIT activation (29). It also inhibited the growth of BaF/3 cells modified to express a constitutively active KIT mutant found in 10% of GIST patients that harbors a 2 aminoacid AY502-503 insertion in KIT Domain 5. No details were provided on mutant KIT degradation or effector functions with this particular antibody.
Compared to small inhibitors, antibody based immunotherapy of tumors have the advantage to recruit immune effectors through the Fc part of the antibody and to potentially mediate ADCC (antibody-dependent cell cytotoxicity) or cell lysis through the complement cascade (CDC) (27). To our knowledge, within anti-KIT antibodies published or commercially available, of mouse or human KIT specificity, none was described to mediate ADCC or CDC (30, 48) which probably explains the mild toxicity observed in animal models where anti-KIT antibodies were injected (48, 49). Together with the fact that in GIST patients, long term treatment with imatinib does not induce profound hematological toxicity, these observations let anticipate that treatment with KIT specific inhibitory antibodies will not elicit strong toxic secondary effects. Since 2D1 and 3G1 are cross-reactive with mouse KIT, they will be helpful to explore further toxicity issues of KIT D5 targeting on mouse models.
To conclude, we have obtained 2 anti-KIT D5 antibodies that inhibit mast cell leukemic cell lines growth in vitro including cells harboring a D816V mutation on KIT that renders them resistant to imatinib. These fully human antibodies, that do not elicit immune effector function targeted to KIT expressing cells, therefore anticipating limited toxicity in vivo, represent promising molecules for killing cancer cells depending on KIT signalling for their survival.
Supplementary Material
Acknowledgments
We thank Veronique Fabre-Merssemann for help for the production of recombinant KIT-ECD
M Le Gall and R Crépin were supported by PhD fellowships from the French Government, M.A. Poul was supported by the mobility program of Cancéropôle Grand Sud Ouest, France. This work was supported by the program “Investissement d'avenir” grant agreement: Labex MabImprove, ANR-10-LABX-53-01.
Footnotes
No conflicts of interest
Bibliography
- 1.Roskoski R., Jr Signaling by Kit protein-tyrosine kinase--the stem cell factor receptor. Biochemical and biophysical research communications. 2005;337(1):1–13. doi: 10.1016/j.bbrc.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 2.Lev S, Blechman J, Nishikawa S, Givol D, Yarden Y. Interspecies molecular chimeras of kit help define the binding site of the stem cell factor. Molecular and cellular biology. 1993;13(4):2224–34. doi: 10.1128/mcb.13.4.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130(2):323–34. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 4.Orr-Urtreger A, Avivi A, Zimmer Y, Givol D, Yarden Y, Lonai P. Developmental expression of c-kit, a proto-oncogene encoded by the W locus. Development. 1990;109(4):911–23. doi: 10.1242/dev.109.4.911. [DOI] [PubMed] [Google Scholar]
- 5.Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90(4):1345–64. [PubMed] [Google Scholar]
- 6.Wallander ML, Willmore-Payne C, Layfield LJ. C-KIT and PDGFRA zygosity in gastrointestinal stromal tumors: Correlation with tumor site, tumor size, exon, and CD117 immunohistochemistry. Appl Immunohistochem Mol Morphol. 2011;19(1):21–7. doi: 10.1097/PAI.0b013e3181ec4f95. [DOI] [PubMed] [Google Scholar]
- 7.Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. The New England journal of medicine. 2002;347(7):472–80. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 8.Wang YY, Zhao LJ, Wu CF, Liu P, Shi L, Liang Y, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2450–5. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orfao A, Garcia-Montero AC, Sanchez L, Escribano L. Recent advances in the understanding of mastocytosis: the role of KIT mutations. British journal of haematology. 2007;138(1):12–30. doi: 10.1111/j.1365-2141.2007.06619.x. [DOI] [PubMed] [Google Scholar]
- 10.Hodi FS, Corless CL, Giobbie-Hurder A, Fletcher JA, Zhu M, Marino-Enriquez A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(26):3182–90. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiological reviews. 2012;92(4):1619–49. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 12.Roskoski R., Jr Structure and regulation of Kit protein-tyrosine kinase--the stem cell factor receptor. Biochemical and biophysical research communications. 2005;338(3):1307–15. doi: 10.1016/j.bbrc.2005.09.150. [DOI] [PubMed] [Google Scholar]
- 13.Mol CD, Lim KB, Sridhar V, Zou H, Chien EY, Sang BC, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. The Journal of biological chemistry. 2003;278(34):31461–4. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 14.Kemmer K, Corless CL, Fletcher JA, McGreevey L, Haley A, Griffith D, et al. KIT mutations are common in testicular seminomas. The American journal of pathology. 2004;164(1):305–13. doi: 10.1016/S0002-9440(10)63120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitayama H, Kanakura Y, Furitsu T, Tsujimura T, Oritani K, Ikeda H, et al. Constitutively activating mutations of c-kit receptor tyrosine kinase confer factor-independent growth and tumorigenicity of factor-dependent hematopoietic cell lines. Blood. 1995;85(3):790–8. [PubMed] [Google Scholar]
- 16.Bernex F, De Sepulveda P, Kress C, Elbaz C, Delouis C, Panthier JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122(10):3023–33. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda H, Kanakura Y, Tamaki T, Kuriu A, Kitayama H, Ishikawa J, et al. Expression and functional role of the proto-oncogene c-kit in acute myeloblastic leukemia cells. Blood. 1991;78(11):2962–8. [PubMed] [Google Scholar]
- 18.Pietsch T, Kyas U, Steffens U, Yakisan E, Hadam MR, Ludwig WD, et al. Effects of human stem cell factor (c-kit ligand) on proliferation of myeloid leukemia cells: heterogeneity in response and synergy with other hematopoietic growth factors. Blood. 1992;80:1199–206. [PubMed] [Google Scholar]
- 19.Schittenhelm M, Aichele O, Krober SM, Brummendorf T, Kanz L, Denzlinger C. Complete remission of third recurrence of acute myeloid leukemia after treatment with imatinib (STI-571) Leukemia & lymphoma. 2003;44(7):1251–3. doi: 10.1080/1042819031000077034. [DOI] [PubMed] [Google Scholar]
- 20.Krystal GW, Hines SJ, Organ CP. Autocrine growth of small cell lung cancer mediated by coexpression of c-kit and stem cell factor. Cancer research. 1996;56(2):370–6. [PubMed] [Google Scholar]
- 21.Micke P, Basrai M, Faldum A, Bittinger F, Ronnstrand L, Blaukat A, et al. Characterization of c-kit expression in small cell lung cancer: prognostic and therapeutic implications. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9(1):188–94. [PubMed] [Google Scholar]
- 22.Heinrich MC, Blanke CD, Druker BJ, Corless CL. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2002;20(6):1692–703. doi: 10.1200/JCO.2002.20.6.1692. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Zeng S, Metcalfe DD, Akin C, Dimitrijevic S, Butterfield JH, et al. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood. 2002;99(5):1741–4. doi: 10.1182/blood.v99.5.1741. [DOI] [PubMed] [Google Scholar]
- 24.Antonescu CR, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(11):4182–90. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- 25.Schittenhelm MM, Shiraga S, Schroeder A, Corbin AS, Griffith D, Lee FY, et al. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer research. 2006;66(1):473–81. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 26.Breccia M, Molica M, Alimena G. How tyrosine kinase inhibitors impair metabolism and endocrine system function: A systematic updated review. Leukemia research. 2014;38(12):1392–8. doi: 10.1016/j.leukres.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nature biotechnology. 2005;23(9):1147–57. doi: 10.1038/nbt1137. [DOI] [PubMed] [Google Scholar]
- 28.Edris B, Willingham SB, Weiskopf K, Volkmer AK, Volkmer JP, Muhlenberg T, et al. Anti-KIT monoclonal antibody inhibits imatinib-resistant gastrointestinal stromal tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(9):3501–6. doi: 10.1073/pnas.1222893110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reshetnyak AV, Nelson B, Shi X, Boggon TJ, Pavlenco A, Mandel-Bausch EM, et al. Structural basis for KIT receptor tyrosine kinase inhibition by antibodies targeting the D4 membrane-proximal region. Proceedings of the National Academy of Sciences of the United States of America. 2013;10(44):17832–7. doi: 10.1073/pnas.1317118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida C, Tsuji AB, Sudo H, Sugyo A, Kikuchi T, Koizumi M, et al. Therapeutic efficacy of c-kit-targeted radioimmunotherapy using 90Y-labeled anti-c-kit antibodies in a mouse model of small cell lung cancer. PloS one. 2013;8(3):e59248. doi: 10.1371/journal.pone.0059248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moutel S, El Marjou A, Vielemeyer O, Nizak C, Benaroch P, Dubel S, et al. A multi-Fc-species system for recombinant antibody production. BMC biotechnology. 2009;9:14. doi: 10.1186/1472-6750-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. Bypassing immunization. Human antibodies from V-gene libraries displayed on phage. Journal of molecular biology. 1991;222(3):581–97. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 33.Schier R, Bye J, Apell G, McCall A, Adams GP, Malmqvist M, et al. Isolation of high-affinity monomeric human anti-c-erbB-2 single chain Fv using affinity-driven selection. Journal of molecular biology. 1996;255(1):28–43. doi: 10.1006/jmbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 34.Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindquist E, Schier R, et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):6157–62. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crepin R, Goenaga AL, Jullienne B, Bougherara H, Legay C, Benihoud K, et al. Development of human single-chain antibodies to the transferrin receptor that effectively antagonize the growth of leukemias and lymphomas. Cancer research. 2010;70(13):5497–506. doi: 10.1158/0008-5472.CAN-10-0938. [DOI] [PubMed] [Google Scholar]
- 36.Shi X, Jarvis DL. Protein N-glycosylation in the baculovirus-insect cell system. Current drug targets. 2007;8(10):1116–25. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Broudy VC, Lin NL, Buhring HJ, Komatsu N, Kavanagh TJ. Analysis of c-kit receptor dimerization by fluorescence resonance energy transfer. Blood. 1998;91(3):898–906. [PubMed] [Google Scholar]
- 38.Erickson-Miller CL, Pelus LM, Lord KA. Signaling induced by erythropoietin and stem cell factor in UT-7/Epo cells: transient versus sustained proliferation. Stem Cells. 2000;18(5):366–73. doi: 10.1634/stemcells.18-5-366. [DOI] [PubMed] [Google Scholar]
- 39.Masson K, Heiss E, Band H, Ronnstrand L. Direct binding of Cbl to Tyr568 and Tyr936 of the stem cell factor receptor/c-Kit is required for ligand-induced ubiquitination, internalization and degradation. Biochem J. 2006;399(1):59–67. doi: 10.1042/BJ20060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu Y, Ashman LK, Du Z, Schwartz LB. Internalization of Kit together with stem cell factor on human fetal liver-derived mast cells: new protein and RNA synthesis are required for reappearance of Kit. J Immunol. 1996;156(9):3443–9. [PubMed] [Google Scholar]
- 41.D'allard D, Gay J, Descarpentries C, Frisan E, Adam K, Verdier F, et al. Tyrosine kinase inhibitors induce down-regulation of c-Kit by targeting the ATP pocket. PloS one. 2013;8(4):e60961. doi: 10.1371/journal.pone.0060961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah NP, Lee FY, Luo R, Jiang Y, Donker M, Akin C. Dasatinib (BMS-354825) inhibits KITD816V, an imatinib-resistant activating mutation that triggers neoplastic growth in most patients with systemic mastocytosis. Blood. 2006;108(1):286–91. doi: 10.1182/blood-2005-10-3969. [DOI] [PubMed] [Google Scholar]
- 43.Zhao L, Qu L, Zhou J, Sun Z, Zou H, Chen YY, et al. High throughput identification of monoclonal antibodies to membrane bound and secreted proteins using yeast and phage display. PloS one. 2014;9(10):e111339. doi: 10.1371/journal.pone.0111339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahn T, Seipel P, Coutinho S, Urschel S, Schwarz K, Miething C, et al. Analysing c-kit internalization using a functional c-kit-EGFP chimera containing the fluorochrome within the extracellular domain. Oncogene. 2002;21(29):4508–20. doi: 10.1038/sj.onc.1205559. [DOI] [PubMed] [Google Scholar]
- 45.Babina M, Rex C, Guhl S, Thienemann F, Artuc M, Henz BM, et al. Baseline and stimulated turnover of cell surface c-Kit expression in different types of human mast cells. Exp Dermatol. 2006;15(7):530–7. doi: 10.1111/j.1600-0625.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- 46.Papayannopoulou T, Brice M, Broudy VC, Zsebo KM. Isolation of c-kit receptor-expressing cells from bone marrow, peripheral blood, and fetal liver: functional properties and composite antigenic profile. Blood. 1991;78(6):1403–12. [PubMed] [Google Scholar]
- 47.Ashman LK, Buhring HJ, Aylett GW, Broudy VC, Muller C. Epitope mapping and functional studies with three monoclonal antibodies to the c-kit receptor tyrosine kinase, YB5.B8, 17F11, and SR-1. Journal of cellular physiology. 1994;158(3):545–54. doi: 10.1002/jcp.1041580321. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa M, Matsuzaki Y, Nishikawa S, Hayashi S, Kunisada T, Sudo T, et al. Expression and function of c-kit in hemopoietic progenitor cells. The Journal of experimental medicine. 1991;174(1):63–71. doi: 10.1084/jem.174.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue X, Pech NK, Shelley WC, Srour EF, Yoder MC, Dinauer MC. Antibody targeting KIT as pretransplantation conditioning in immunocompetent mice. Blood. 2010;116(24):5419–22. doi: 10.1182/blood-2010-07-295949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.