Abstract
A long-standing debate in the Cu-O2 field has revolved around the relevance of the Cu(III) oxidation state in biological redox processes. The proposal of Cu(III) in biology is generally challenged as no spectroscopic or structural evidence exists currently for its presence. The reaction of synthetic Cu(I) complexes with O2 at low temperature in aprotic solvents provides the opportunity to investigate and define the chemical landscape of Cu-O2 species at a small molecule level of detail; eight different types are characterized structurally, three of which contain at least one Cu(III) center. Simple imidazole or histamine ligands are competent in these oxygenation reactions to form Cu(III) complexes. The combination of synthetic structural and reactivity data suggests (i) that Cu(I) should be considered as either a one or two electron reductant reacting with O2, (ii) that Cu(III) reduction potentials of these formed complexes are modest and well within the limits of a protein matrix and (iii) that primary amine and imidazole ligands are surprisingly good at stabilizing Cu(III) centers. These Cu(III) complexes are efficient oxidants for hydroxylating phenolate substrates with reaction hallmarks similar to that performed in biological systems. The remarkable ligation similarity of the synthetic and biological systems makes it difficult to continue to exclude Cu(III) from biological discussions.
Biology & Cu-O2
Copper (Cu) is one the most frequently used metals in enzymes to activate dioxygen (O2) and the mushroom Cu enzyme Tyrosinase (Ty) provided the first example in 1955 of direct integration of an oxygen-atom from O2 into an organic product by an enzyme [1]. Given the diverse oxidative function of these enzymes, many different assemblies have evolved. Mononuclear to trinuclear copper active sites that activate O2 are now known, which are capable of oxidase transformations (e.g. alcohols to aldehydes), monooxygenase transformations (e.g. alkanes to alcohols), and full reduction of O2 to water [2–4]. Each enzyme is tuned remarkably to perform its specific reaction by exploiting the oxidizing power of O2 [4]. It is the controlled and efficient nature of these oxidative processes with O2 that has captured the attention of the synthetic community with a vision of selective Cu-O2 catalysts, using an earth-abundant metal and the most prevalent oxidant on the planet.
Copper in biology is considered currently to be a one electron reservoir shuttling between the Cu(I) and Cu(II) oxidation states, whether mediating an electron transfer (ET) process or activating O2 in an enzyme to oxidize a substrate. In the past two decades, it has become clear from synthetic systems with ligation similar, if not identical, to the biological sites that Cu(I) complexes react with O2 to yield not only Cu(II) but also Cu(III) species – Cu(III)-O2 species are thermodynamically stable relative to their Cu(I) and O2 starting materials. Yet, even with their characteristic Cu(III) spectroscopic signatures, no experimental evidence exists currently for its role in biology.
The scope of this review focuses on spectroscopically or structurally characterized high-valent Cu-O2 complexes (i.e., Cu(III)) formed in the homogeneous reaction of Cu(I) with O2 with nitrogen and oxygen ligation, and their subsequent reactivity with exogenous substrates. This review will concentrate primarily on cases where definitive spectroscopic and structural characterization of Cu(III) exist, though systems exhibiting reactivity hallmarks of Cu(III) species and lacking spectroscopic detection will be introduced, as interconverting valence isomers may be operative. While intramolecular ligand oxidation by these species defines reactivity possibilities, this review concentrates on intermolecular substrate reactivities, which, in the opinion of the reviewers, provide greater insights into development of useful stoichiometric or catalytic oxidants derived from O2. Furthermore, catalytic Cu-O2 systems for organic oxidations have been reported [5–8], but the involvement of Cu(III)-O2 intermediates is unknown.
Structural Types of Cu-O2 Species
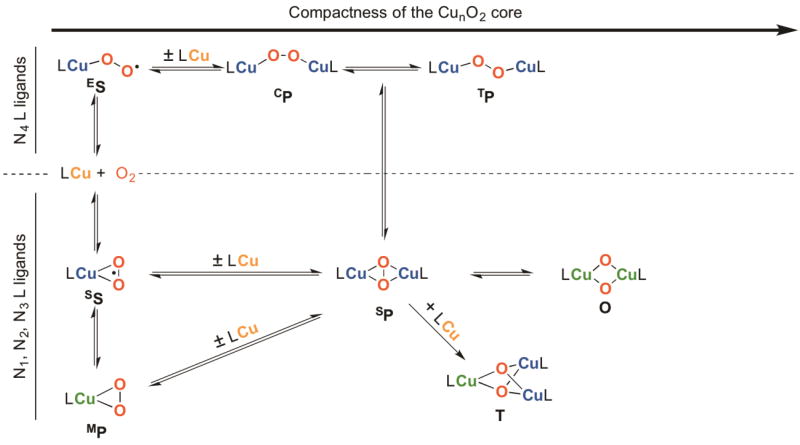
The pursuit of the types of Cu-O2 species that can be formed in the reaction of simple Cu(I) complexes and O2 has fascinated synthetic and mechanistic chemists for more than seventy years. Their ligation lability and oxidative power confound this research, which has trended to low temperatures (−80 °C to −145 °C) to stabilize the complexes. The resulting structural diversity is remarkable, with 1:1 to 3:1 Cu:O2 stoichiometries, including several isomers within a given stoichiometry. Currently, eight different Cu-O2 types have been characterized structurally from the reaction of Cu(I) and O2: η1-end-on Cu(II) superoxide ES (2006) [9], η2-side-on Cu(II) superoxide SS (1994) [10], η2-side-on Cu(III) peroxide MP (2002) [11,12], dimeric cis-μ-η1:η1 Cu(II) peroxide CP (2014) [13], dimeric trans-η-1,2 Cu(II) peroxide TP (1988) [14], dimeric μ-η2:η2-side-on Cu(II) peroxide SP (1989) [15], dimeric Cu(III) bis-μ2-oxide O (1996) [16], and trinuclear Cu(III)Cu(II)Cu(II) bis-μ3-oxide T (1996) [17] (Fig. 1). Three of the eight types contain a Cu(III) center derived from the reaction of Cu(I) and O2.
Fig. 1.
Formation of the eight structurally characterized Cu-O2 species generated in the reaction of Cu(I) and O2 with mono-, bi-, tri-, and tetradentate ligation (modified from Mirica et al.) [18].
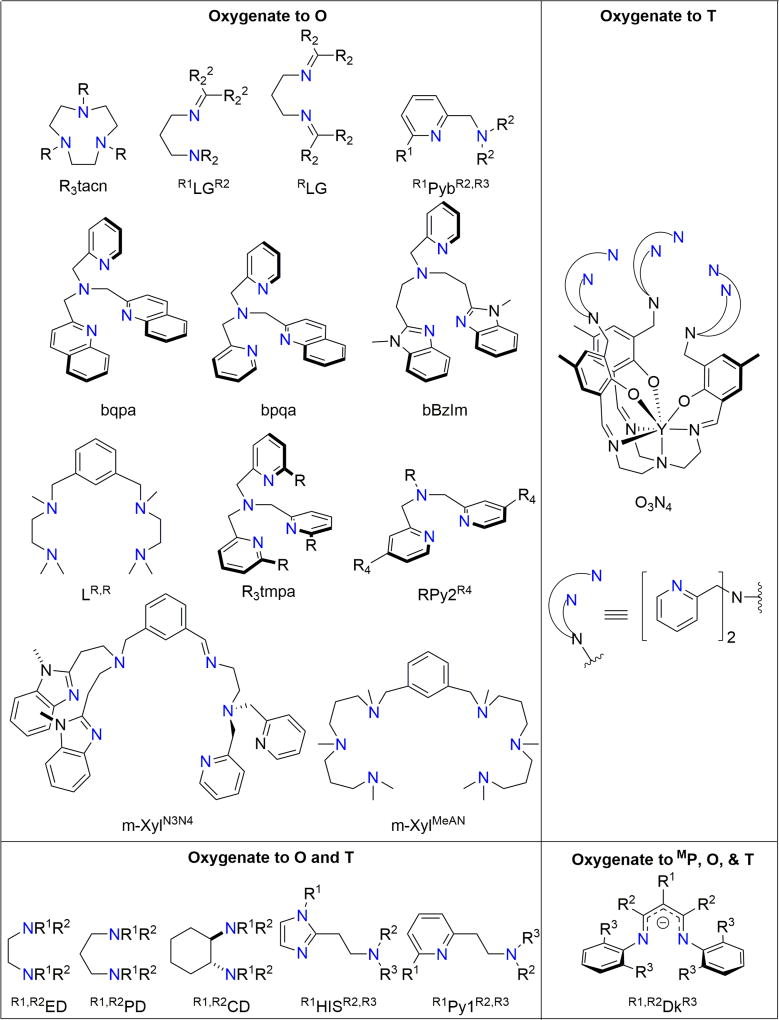
This structural diversity is associated with copper nuclearity and the reduced O2 binding mode. The stoichiometries of these complexes are determined primarily by ligand steric demands and ligand denticity, which is succinctly summarized through a ligand steric continuum [19] (Fig. 1). The most sterically demanding ligands, which effectively preclude copper dimerization, yield mononuclear superoxide or peroxide species, with the superoxide ligated either in a side-on (ES) or end-on fashion (SS) [20]. Less demanding ligands, if tetradentate, preferentially stabilize a TP species (Cu-Cu, ca. 4.4 Å) or a CP species (Cu-Cu, ca. 3.8 Å), if the ligand enforces a cis geometry. Tri- or bi-dentate ligands collapse to SP species (Cu-Cu, ca. 3.5 Å) or a Cu(III)-containing O species (Cu-Cu, ca. 2.8 Å). Bidentate amine ligands (e.g., diamines, histamines, pyridylamines) almost exclusively form O species, which constitute the largest family of synthetic Cu-O2 species to date [19,21,22]. Finally, the least sterically demanding bidentate ligands (e.g., primary amines, methylated amines, histamines) form the most compact Cu-O2 core, a trinuclear T cluster (Cu-Cu ca. 2.7 Å) [2,23]. Ligands that support the Cu(III) species of interest for this review are shown in Fig. 2.
Fig. 2.
Top left: Ligands that stabilize Cu(I) for reaction with O2 to produce Cu(III)-containing O species exclusively. Top right: Ligands that oxygenate to T species exclusively. Bottom left: Sufficiently compact ligands allow formation of both O and T species, dependent on their substituents and reaction conditions. Bottom right: β-diketiminate ligands that produce all three Cu(III)-containing species (MP, O, and T).
While ligand steric demands are a dominant force in determining the Cu-O2 binding mode, the nature of the ligating groups and the Cu(I) geometry are also important. Strongly donating, basic amines or anionic nitrogen donors not only stabilize the contracted Cu(III) center but also create more reducing Cu(I) precursor complexes, facilitating their reaction with O2 [2,24]. If a productive reaction is to occur, the Cu(I) precursor must provide inner-sphere binding access for O2 to allow association and ET; Cu(I) complexes in a tetrahedral geometry are typically not reactive with O2. Three-coordinate trigonal or T-shape complexes or four-coordinate trigonal monopyramidal geometries are generally the most reactive Cu(I) precursor geometries, as O2, an impotent ligand, is not required to compete with a weakly associating solvent or counteranion.
Spectroscopic Characterization
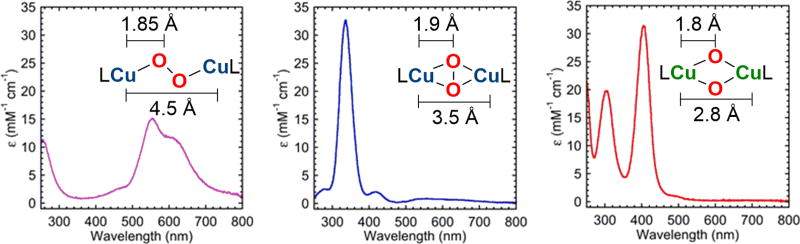
The spectroscopic and metrical parameters for the eight structurally characterized Cu-O2 species are surprisingly distinct, which allows for solution characterization by a combination of optical, vibrational, and x-ray absorption spectroscopies (XAS). The reaction of d10 Cu(I) complexes with O2 generally lead to significant changes in the optical spectra with the appearance of intense ligand to metal charge transfer (LMCT) bands, generally characterized as oxygen to copper charge transfers. The optical comparison in Fig 3. of the three most common 2:1 species (TP, SP, O) effectively allows for their unambiguous characterization, assuming full formation of a single Cu-O2 species. Redox titrations with weak H-atom donors such as ascorbic acid or carboxyferrocene provide critical insights into formation yields [21,23,25]. Of course, the energies of the LMCT bands are sensitive to the nature of the ligand, but the optical features are generally consistent. Recent studies on O and T species with varied amine and imidazole ligation correlate more donating ligation to a systematic blue-shifting of the LMCT optical bands for complexes of the same structural type [21,22]. As many of these complexes are thermally sensitive and can exist as an equilibrium of isomers in solution (vide infra), resonance Raman (rR) is generally preferred over infrared spectroscopic characterization, as the former enables optical and vibrational correlations [26–28]. A superoxide stretching frequency (960–1130 cm−1) [4] is distinguished readily from that of a peroxide (720–960 cm−1) [2,24,13] or that of the bis-oxide core (590–620 cm−1) [29,30]. The combination of optical and rR spectroscopy is extremely powerful for distinguishing among these species. Solution Cu K-edge XAS is also a valuable tool to differentiate between the SP (Cu-Cu, ca. 3.6 Å), O (ca. 2.8 Å), and T (ca. 2.7 Å) species through their Cu-Cu separations; acceptable fits to their Extended X-ray Absorption Fine Structure (EXAFS) require a Cu-Cu scattering distance that is effectively invariant among all model fits for a single complex. The copper pre-edge feature of these absorptions, albeit weak, also has characteristic oxidation information encoded in their energies, with Cu(I) absorbing at 8984 ± 1 eV, Cu(II) at 8979 ± 0.5 eV, and Cu(III) at 8981 ± 0.5 eV [31,32]. The intensity of Cu(II) and Cu(III) features generally increase with distortion from a planar coordination geometry [33]. However, the Cu(II) pre-edge intensity is usually more intense than that of Cu(III) for a given ligation environment. Consequently, the pre-edge of a T complex, composed of two-thirds Cu(II) and one-third Cu(III), only reveals Cu(II) features; the higher energy feature associated with Cu(III) is obscured by the Cu(II) pre-edge absorption along with the rising edge [33].
Fig. 3.
Optical spectra of a bis-Cu(II) TP (left), bis-Cu(II) SP (middle), and a bis-Cu(III) O species (right).
General Reactivity
Beyond the wealth of spectroscopic and structural data provided by these synthetic complexes, detailed mechanistic studies provide insights into their reactivity tendencies with external substrates. The oxidative reactivity of these reagents can vary from electrophilic to nucleophilic. Generally, the superoxide species are limited in their oxidizing abilities; if reactivity is observed, it is usually only with weak O-H and C-H bonds [34]. Reactivity characterization of the CP species is incomplete currently as reactivity with weak C-H bonds is only observed upon warming, leaving the possibility of forming a different active species upon decomposition [13]. The end-on peroxide of TP has nucleophilic character, as it is protonated readily, does not oxidize PPh3, and oxidizes benzaldehyde to benzoic acid [35,36]. By contrast, the side-on peroxide of SP, while susceptible to protonation by acid [35,37,38], is capable of PPh3 oxidation and can oxidize phenolates to catechols and catechols to quinones through an electrophilic mechanism [34,37]. The O species are the most potent oxidants, capable of phenolate hydroxylation, C-F activation, and C-H activation [2,18,25,39–41]. The extremely compact nature of the T species precludes most reactivity, with the exceptions of proton-coupled electron transfer (PCET) from weak O-H substrates and oxidation of PPh3 [23,42–44].
Cu(III) Geometric Preferences and Redox Potentials
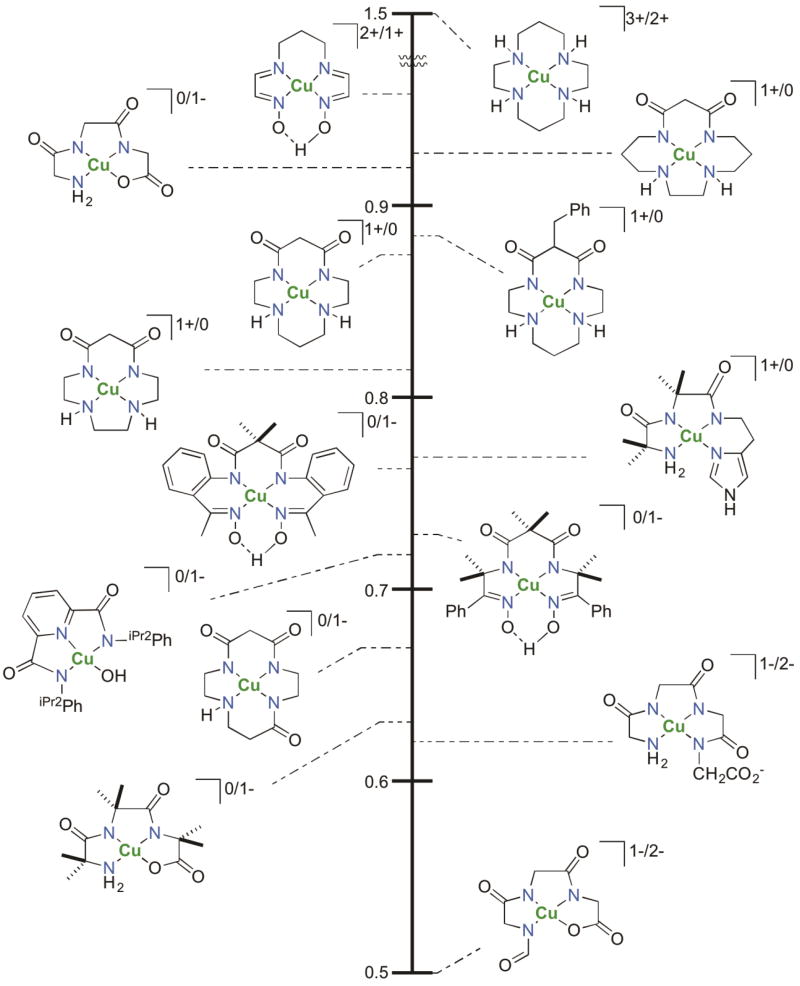
Cu(III) has been excluded generally from biological discussions, as the positive reduction potentials of most mononuclear Cu(III) complexes are significantly above 1 V vs NHE, which would lead to oxidation of proximal protein residues. The highest reported reversible copper potential is that of the blue copper site of fungal laccases, a Cu(II)/(I) process, at ca. 0.8 V [45]. By comparison, the reduction of [Cu(III)(cyclam)]3+ occurs above 1.5 V, with a macrocyclic tetraamine ligand almost ideally suited to match the planar coordination preference of a d8 Cu(III) center [46]. However, the addition of anionic ligation significantly attenuates the positive reduction potentials. The Margerum group demonstrated clearly that monomeric Cu(III)/Cu(II) potentials can be moved within the biological range by including strong anionic ligation in a planar coordination geometry: a triply deprotonated glycine tripeptide Cu(III) complex with two amidate ligands has a potential in water near 0.9 V, and even more donating ligands lower the reduction potentials to near 0.4 V [47,48]. This principle is further demonstrated by a dianionic pyridinedicarboxamide ligand in conjunction with a ligated hydroxide that creates a neutrally charged Cu(III) species with a reduction potential of ca. 0.7 V [49,50], which is reactive with moderately strong C-H bonds. Though deprotonated amide ligands are now reported to ligate Fe, Co, and Ni centers in enzymes [51–54], no such ligation is known currently for any copper enzymes. The redox potentials of a variety of mononuclear Cu complexes with neutral to trianionic ligands, presented by the Krüger group [55], clearly show a correlation of anionic ligation to lower Cu(III) reduction potentials (Fig. 4). Copper centers that react with O2 have an advantage in stabilizing Cu(III), as the reduced O2 species generate anionic ligation. In the extreme of four electron reduction of O2, two dianionic oxide ligands are formed possessing the ability to bind the copper centers in a planar geometry, thereby stabilizing the Cu(III) oxidation state. This analysis suggests that if Cu(III) plays a mechanistic role in a copper, O2-activating enzyme, the bi- and trinuclear sites would be foremost candidates.
Fig. 4.
Comparison of Cu(III/II) potentials for Cu(III) complexes with varied donor strength and overall complex charge. Adapted from Hanss et al. [55].
Mononuclear Cu(III) Examples
Examples of mononuclear Cu-O2 species are less common than their multinuclear counterparts presumably due to their tendency to dimerize rapidly in homogenous solutions. This transient existence often confounds their characterization. While several examples of mononuclear Cu(II)-superoxide complexes exist [9,56–58,20], only a single example of a mononuclear, Cu(III)-peroxide, MP, is known: the Cu(I) complex of a sterically demanding β-diketiminate (Dk) ligand directly oxygenates to a mononuclear Cu-O2 species [11]. X-ray crystallography reveals short Cu-O/N distances of ca. 1.8 Å with an O-O separation of ca. 1.4 Å, consistent with a peroxide level O2 species [12]. Spectroscopic studies of three MP derivatives show rR O-O vibrations in the 750–930 cm−1 range and a pre-edge Cu K-edge XAS feature at 8981 eV, both of which support a Cu(III) peroxide description [24]. This contrasts with the reaction of O2 with sterically bulky tris-pyrazolylborate Cu(I) complexes, an overall neutral complex similar to Cu(I)-Dk, that yields a side-on Cu(II)-superoxide species, SS [10].
Despite the tendency of Cu(III) complexes to be avid oxidants, the MP complexes exhibit limited reactivity with exogenous substrates; no reaction is reported with weak O-H or C-H substrates, and PPh3 displaces the bonded O2 to yield a Cu(I)-PPh3 complex, suggesting weak nucleophilic activity. Density functional theory (DFT) calculations indicate that these Cu(III)-peroxides possess modest reduction potentials and are weakly basic upon reduction, supporting the limited PCET abilities. This stabilized Cu(III) center presumably results from the strong anionic donation from the Dk ligand. While this sterically demanding anionic ligation promotes the reactivity of the ligated Cu(I) with O2 and prevents dimerization, its stabilizing effect attenuates reactivity [24,59].
Binuclear Cu(III) Examples
The reduction of O2 to a peroxide or oxide level is more thermodynamically favorable than to a superoxide level, and four different 2:1 Cu:O2 isomers are known currently (i.e., TP, CP, SP, O) [2,60]. The distinct spectroscopy of each isomer is consistent with their strikingly different metrical parameters and substrate reactivity (Fig. 3). While the reactivity of the CP species has not been extensively explored [13], the end-on peroxide of the TP species primarily reacts as a nucleophilic oxidant [36], and the SP [61] and O [18,62] species react as electrophilic oxidants. The O species are of particular interest because of their varied substrate scope. While attributing a reactivity to a specific structural type is convenient, active oxidant identification can be complicated by the existence of facile equilibria among the 2:1 species (vide infra) [16,28,38,63,64].
O Species Discovery
The O species, initially observed in 1993 [65], was characterized in 1995 by the Tolman group using a sterically demanding triazacyclononane ligand; oxygenation of [(Bn3TACN)Cu(I)(MeCN)]+ at −80 °C produced distinct optical features, rR vibrations, and Cu-Cu distance from Cu K-edge XAS [66]. The short Cu-Cu distance (ca. 2.8 Å) was consistent with high-valent copper centers, though initially assigned as a bis-Cu(II)-bis-oxyl formulation from calculations. Subsequent crystallographic characterization confirmed Cu-O distances (ca. 1.8 Å) much shorter than previously observed for Cu(II) complexes [16]. The facial-capping trinitrogen ligation of each TACN ligand flexes to allow two short Cu-N distances (ca. 2.0 Å) in the plane defined by the Cu2O2N4 atoms and one long axial Cu-N distance (ca. 2.3 Å) creating a coordination environment suitable for Cu(III). Each copper center provides two electrons to fully reduce O2, forming two anionic oxide ligands. Intermolecular reactivity of this complex and the related iPr3TACN complex is limited given the large steric demands of the ligands. However, intramolecular dealkylation of N-alkyl ligand substituents, a two electron, two proton process, is observed upon thermal decay [16,67].
Most O species are thermally sensitive, and most investigations have been performed at −80 °C to −145 °C. The thermal decay is usually associated with ligand oxidation; in the case of N-alkyl substituents, the weakest C-H(D) bond is removed in a unimolecular rate limiting step upon warming, as evidenced by a large primary kinetic isotope effect (KIE) with deuterium substitution [68–70]. Formation of a carbinolamine intermediate is proposed generally, which subsequently decomposes to an aldehyde or ketone product and dealkylated amine. A O species can also thermally decay through oxidation of ligand aryl substituent to yield a phenolate through a direct, electrophilic oxygen-atom transfer [71,72]. These intramolecular oxidations demonstrate that the Cu2O2 O core can be reactive to either alkyl or aryl C-H bonds. Numerous examples of O species are now characterized to be stabilized by peralkylated diamines [19,21], triamines [16,73], and hybrid ligands containing alkylated amines combined with aromatic nitrogen or guanidyl groups [22,72,74]. Bidentate ligation provides a coordination environment that best matches the planar preference of a Cu(III) center with the added benefit of providing axial access of substrates to the Cu2O2 core [75].
O and SP Equilibrium
Oxygenation of [(iPr3TACN)Cu(I)(MeCN)]+ at −80 °C under specific conditions yields a remarkable finding by the Tolman group of an equilibrium of two structural types, O and SP isomers, in optically-measurable amounts. This isomeric interconversion involves the forming and breaking of the O-O bond with concurrent reduction and oxidation of the Cu(III) and Cu(II) centers, respectively [16,76]. Oxygenation of the more sterically demanding [(tBu3TACN)Cu(I)(MeCN)]+ yields exclusively a SP species [73,77]. Thus, the variation of the alkyl substituents on the TACN ligand backbone from Me [78] to tBu [73] generates optically pure O to SP species, respectively, illustrating a ligand steric continuum resulting in a Cu-O2 speciation change. The variation of Cu-O2 structural type with variation of alkyl substituents is even more extreme with bidentate, nitrogenous ligands as SP, O, and T species can all be formed as clearly demonstrated by the Stack and Itoh groups [19].
The potential energy surface (PES) of the O ↔ SP equilibrium can be relatively flat depending on the ligand choice, such that speciation is affected by many subtle variables including solvent, temperature, and counteranion. In the cases where measured, the O species is favored enthalpically (i.e., the equilibrium is biased to the O species at lower temperatures), while SP species are favored entropically [26,63,79]. More significant changes, such as axial coordination of a bridging ligand or addition of a more coordinating anion, can trigger isomerization of a O isomer to its corresponding SP species, as demonstrated with the bidentate ligand TEPD (N,N,N’,N’-tetraethylpropylenediamine) [79]. Such valence isomerization equilibria has now been extended by the Karlin group to include TP and O species, and calibrated DFT calculations support a relatively flat PES among three 2:1 Cu:O2 isomers [28].
Optically characterized O species are known to react with an array of externally added substrates including weak O-H, S-H, and C-H bonds, such as ascorbic acid derivatives, phenols, catechols, thiols, alkylated amines, and hydroxylamines at low temperatures. In most cases, these reactions result in formation of Cu(II)-hydroxide products, which are the thermodynamic products of this chemistry. This suggests that the O species is ideally suited for two proton, two electron oxidations. The O complexes can also act as an oxygen-atom transfer reagent for reactive, exogenous substrates such as PPh3 [63]. Understanding the factors that control this diverse exogenous reactivity is important for dissecting reactivity in biological systems and development of selective chemical oxidants.
Oxidase Reactivity of O Species with External Substrates
Significant insights into the exogenous substrate reactivity of the O species initially came in 1999 from the stoichiometric oxidation of alcohols to yield carbonyl products [70]. Screening greater than ten different O species ligated by alkylated diamines identified OTMPD (TMPD = N,N,N’,N’-tetramethylpropylenediamine) as the most reactive of the group [69]. The small methyl substituents of TMPD are beneficial, allowing for substrate access to the Cu2O2 core. Additionally, the relatively strong N-methyl C-H bonds generate a more stable O species, as the N-dealkylation pathway is attenuated significantly; more than 90% of the intact ligand is recovered upon thermal decomposition [69]. OTMPD is indefinitely stable at −80 °C and sufficiently stable for reactivity studies at −40 °C. OTMPD reacts with alkyl and benzylic alcohols stoichiometrically to produce aldehyde or ketone products in yields of 60–90% based on oxidant using two equivalents of alcohol per OTMPD. The isolated yields can be improved with additional equivalents of OTMPD as subsequent oxidation of product is not observed. Stoichiometric addition of Et3N greatly enhances the rate of alcohol oxidation, allowing the reaction to be performed at −80 °C in less than three hours with a dramatic increase in selectivity for primary over secondary alcohols.
The rate limiting step does not involve the Cα-H bond cleavage, as the rates of PhCH2OH and PhCD2OH oxidation at −40 °C are effectively identical, but the product determining step (PDS) does involve Cα-H(D) bond cleavage as evidenced by an intramolecular KIE of 5 for PhCHDOH [70,80]. The lithium salt of PhCHDOH reacts on mixing at −80 °C with a similar KIE of 5, suggesting that alcohol and alkoxide oxidation occurs through the same PDS. Combined, these data suggest a mechanism in which the alcohol binds to OTMPD and, upon deprotonation, rearrangement positions the alkoxide within the equatorial plane of the Cu2O2 O core enabling C-H activation and substrate oxidation. The amine ligand must be sufficiently flexible to allow for this rearrangement, an attribute of the 6-membered TMPD chelate. The four electron oxidation of primary amines to nitriles also occurs using two equivalents of OTMPD at −80 °C through the intermediacy of an imine. As with alcohol substrates, a significant primary over secondary amine selectivity exists, supporting an initial binding to the Cu2O2 core. However, OTMPD does not react in any appreciable way with the weak C-H bonds of 1,4-cyclohexanediene (CHD) or dihydroanthracene (DHA) at −80 °C over a three-hour period. Beyond performing near-quantitative oxidation of weak O-H and S-H substrates (e.g., phenols to phenoxyl radicals or bis-phenols; catechols to quinones; thiols to disulfides) through either a sequential electron, proton transfer (ETPT) or concerted PCET mechanism, an oxygen atom of OTMPD can be transferred to an exogenous substrate, initially demonstrated with PPh3, under purged O2 conditions [63]. This detailed reactivity scope clearly demonstrates the oxidizing proclivity of the Cu2O2 O core and the importance of substrate access to the copper centers.
C-H Oxidase Reactivity of O Species with External Substrates
In 2001, the Fukuzumi and Itoh groups reported the first example of a O species that reacts with exogenous substrates containing weak C-H bonds, such as N-methyl-9,10-dihydroacridine (AcrH2) and CHD to yield N-methylacridinium ion and benzene, respectively [81]. Curiously, this oxidase reactivity is second order with respect to the O species with C-H bond cleavage as rate limiting. Prior work from this group reported that a closely related bidentate ligand with appended benzylic C-H groups exhibited intramolecular monooxygenase reactivity [72,82]. Intermolecular monooxygenase activity on aliphatic C-H substrates is still not documented for an O species.
Despite the wealth of structural, spectroscopic, and reactivity data from O species, their biological significance is generally excluded given the absence of any known characteristic spectroscopic signature of Cu(III) from a biological system in conjunction with limited use of biologically relevant ligands in the synthetic investigations [4,83]. Oxygenation of Cu(I)-histamine, Cu(I)-alkylhistamine or Cu(I)-imidazole complexes do not yield spectra consistent with a pure O species even at −145 °C [23,39]. A pure O species with imidazole ligation, only disclosed in 2014, requires an indirect, synthetic pathway through a ligand exchange reaction with a preformed O species at −125 °C [22,84]. Two equivalents of Nτ-butylhistamine (Nτ ≡ Nε), Nτ,N,N-trimethylhistamine (HIS), or even propylenediamine (PD) displace the TMPD ligands from OTMPD at −125 °C to generate new O species that contain histamine ligation (OHIS) or exclusive primary amine ligation (OPD) in yields greater than 80% by redox titrations. The facile oxidation of primary amines by OTMPD is not competitive with the fast ligand exchange (ca. 10 min) at −125 °C, allowing respectable yields of OPD and OHIS.
The displacement of TMPD by these ligands clearly indicates a greater thermodynamic stability of a O species with primary rather than tertiary amines, a trend that correlates to DFT calculations [21]. These calculated structures clearly display shorter Cu(III)-N distances to primary amines than to tertiary amines by ca. 0.05 Å. The one primary amine Cu(III)-N distance, crystallographically characterized at 1.9 Å, is even shorter than those predicted by DFT calculations [85]. The primary amines, though less basic than tertiary amines in the gas phase, are stronger donors to the compact Cu(III) centers, as their lesser steric demands allow closer proximity and greater orbital overlap. Indeed, the O species with primary amine ligation have LMCT bands that are blue-shifted relative to the tertiary amines. DFT calculations show the acceptor orbital is antibonding with respect to the Cu-N bonds; stronger Cu-N bonding should increase the optical transition energy, as observed experimentally [21]. More remarkable is that ligation from an imidazole is more thermodynamically stabilizing than from a primary amine for O species. From ligand exchange reactions, HIS is capable of ligand exchange with OPD to produce OHIS with a corresponding blue-shift of the LMCT band (378 to 363 nm) [22]. DFT calculations support an even shorter imidazole Cu(III)-N distance, and time-dependent DFT optical predictions correlate with the observed optical blue-shifting. These simple ligand exchange reactions provide the unanticipated result that imidazole ligation best stabilizes a O species of all neutral nitrogenous ligands examined to date.
Despite the greater thermodynamic stability of OPD compared to OTMPD, the former activates the weak C-H bond of AcrH2 and the latter does not. Isodesmic DFT reactions suggest that OPD is a stronger hydrogen-atom acceptor than OTMPD, assuming that the hydrogen-atom reduced product is a Cu(III)Cu(II)-oxide-hydroxide species. On a thermodynamic basis, OPD should be a faster hydrogen-atom abstracting agent compared to OTMPD for externally added substrates, which is observed experimentally. Additionally, this reactivity trend is consistent with greater substrate accessibility to the OPD Cu2O2 core. Using CHD as a weak C-H atom substrate, the lowest energy pathway calculated by DFT is along the O-O vector (Fig. 5). N-alkylation impedes this pathway, consistent with the 10-fold slower reactivity observed from the O species generated with the more sterically demanding N,N’-dimethylpropylenediamine with AcrH2 [21].
Fig. 5.
The lowest energy DFT calculated pathway for C-H bond activation from cyclohexanediene (CHD) along the O-O vector of OPD [21].
Monooxygenase Reactivity of O Species
While exogenous oxidase reactivity involving either an ETPT or PCET pathway is amply demonstrated in the literature for O species, exogenous monooxygenase reactivity has been demonstrated only for phenolate hydroxylation in a limited number of cases in a homogeneous solution. By contrast, a heterogenized, discrete O species on nanoparticles are also known to catalytically oxidize toluene to benzyl alcohol and benzaldehyde at elevated temperatures using O2 as the oxidant [86]. In all cases, facile substrate accessibility to the Cu2O2 core is possible [25,28,40,87].
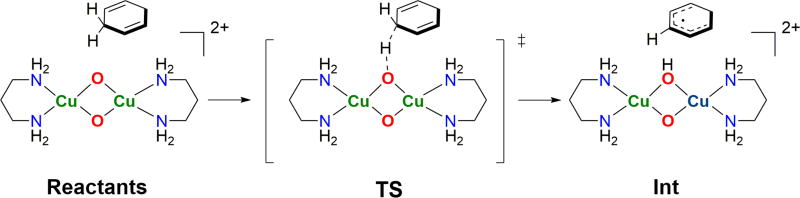
In the case of phenolate hydroxylation, initial axial approach of a phenolate to a copper center followed by rapid rearrangement into an equatorial position of the O core allows for an inner-sphere hydroxylation to occur [18]. This mechanistic postulate is different from that generally accepted for Tyrosinase enzymes (Ty), which are proposed to operate through a side-on Cu(II) peroxide active oxidant (vide infra) [88,89].
Active oxidant identification is a difficult task given the possibility of multiple Cu-O2 structural types existing in facile equilibria. An informative example that showcases these intricacies is found in the work of the Suzuki group that observes epoxidation of styrene and hydroxylation of tetrahydrofuran (THF) at −80 °C by a spectroscopically characterized SP species [64]. 18O2 experiments confirm the source of the oxygen-atom for the epoxidation, and the large H(D) KIE of 50 for THF hydroxylation support a rate limiting C-H activation. Subsequent reactivity studies report not only exogenous phenolate hydroxylation but also C-H activation with a wide range of C-H substrates including CHD, fluorene, and toluene. As this latter reactivity is generally associated with a O rather than a SP species [90], the authors did not exclude the possible role of a O isomer in these oxidations.
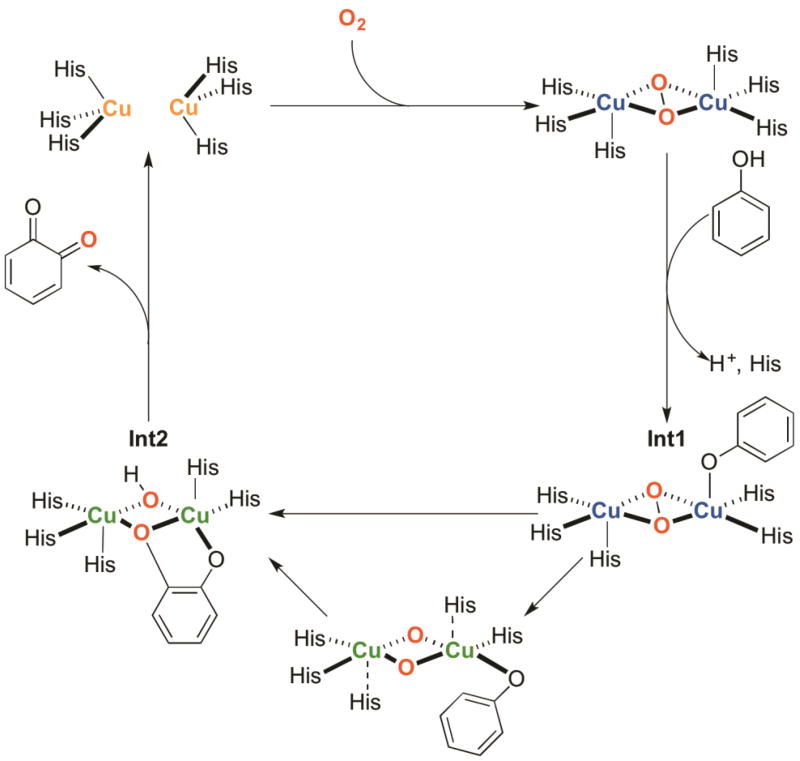
A clearer example of step-by-step hydroxylation of an exogenous phenolate by a O species is the reactivity reported by the Costas group in 2008 [40]. Oxygenation of their binucleating, Cu(I) polydentate amine complex at −80 °C yields the characteristic optical and rR spectroscopic features of a O species. Addition of 1.5 equivalents of exogenous phenolate forms a new intermediate species, A (Fig. 6a), with spectroscopic features similar to the parent O species, yet with new visible absorption features suggestive of direct phenolate ligation. This complex thermally decays by a first order process to form catechol products through an electrophilic oxidation mechanism, evidenced by slower reactivity of electron deficient phenolates. The simplest interpretation of this data is that a Cu(III) O species is capable of stoichiometric electrophilic hydroxylation through an inner-sphere mechanism.
Fig. 6.
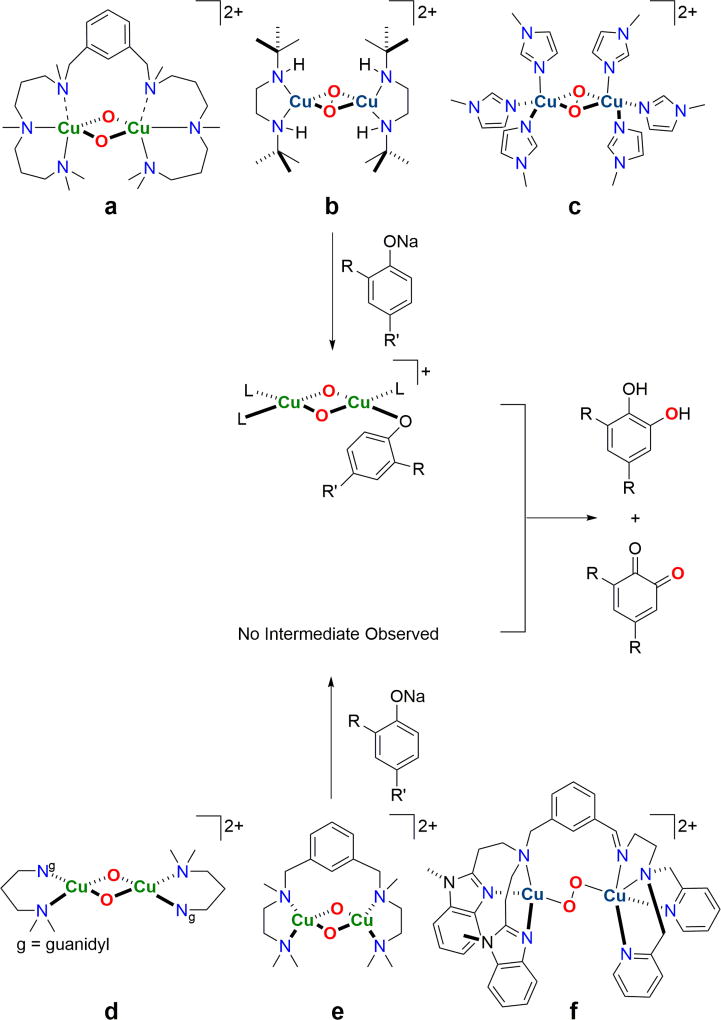
Cu-O2 complexes known to perform ortho-hydroxylation of exogenous phenolates through a O active oxidant. Reactions with complexes a, b, and c proceed through an observed, phenolate-bonded Cu(III) intermediate [18,39,40]. Complexes d, e, and f are proposed to react in a similar fashion, although no Cu(III) intermediates are detected spectroscopically [25,28,38,91].
Support for a O species acting as the active oxidant for exogenous phenolate hydroxylation was proposed initially in 2005 starting from a side-on peroxide species, SPDBED, using the sterically demanding, bidentate N,N’-di-tert-butylethylenediamine (DBED) ligand [18]. While SPDBED hydroxylates added phenolates in high yields relative to oxidant at −80 °C, no oxidation intermediate is observed. Cooling the reacting mixture to −125 °C stabilizes a transient intermediate, ADBED, with a 18O2-sensitive stretch at 590 cm−1, a 2.8 Å Cu-Cu distance, and an intense LMCT band at 418 nm, all fully consistent with a Cu(III)2O2 bis-oxide core (Fig. 6b). Additional visible absorptions are assigned as equatorially ligated phenolate to copper LMCTs through comparative rR studies with 16O and 18O substituted phenolates, features similar to those reported by Costas et al. (vide supra). ADBED undergoes first-order decay at −125 °C to produce hydroxylated products with the oxygen-atom derived from O2. A Hammett ρ-value of −2.2 and an inverse deuterium KIE for the oxidized ortho-carbon support an electrophilic oxidation mechanism. The simplest interpretation of these results is that SPDBED oxidizes phenolates through the intermediacy of a O species that forms as a result of phenolate binding to the initial SP species.
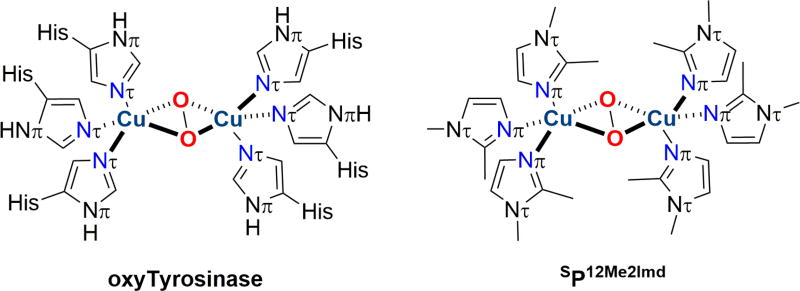
More recent results have created SP species with exclusive imidazole ligation that exhibit a similar type of SP to O isomerization triggered by phenolate ligation, followed by a first-order decay with associated phenolate hydroxylation. In 2012 the self-assembly of SP complexes from 6 monodentate imidazoles, Cu(I), and O2 in near quantitative yields was reported, albeit the imidazoles required a non-hydrogen substituent adjacent to the ligating imidazole nitrogen atom, Cu-Nπ ligation (Nπ ≡ Nδ), to be contrasted with the less sterically demanding Nτ ligation observed exclusively in oxygenated Ty (oxyTy) and oxyHc [37,88,92] (Fig. 7). These SP complexes hydroxylate electron deficient phenolates to catechol products without any intermediate detection at −125 °C, and cooling to −145 °C only slows this hydroxylation reactivity. In 2016, the biologically-relevant Nτ ligation in synthetic SP complexes became possible by working at −145 °C; 1-methylimidazole (SP1MeImd), 4-methylimidazole (SP4MeImd), and imidazole (SPNHImd) complexes are now characterized [39]. The remote positioning of each imidazole substituent in SP4MeImd and SP1MeImd results in much more exposed Cu2O2 SP cores. Indeed, SP1MeImd reacts with exogenous, electron-deficient phenolates at −145 °C to generate an intermediate, A1MeImd, with a similar optical spectrum to ADBED, which thermally decomposes to form catechols (Fig. 6c). The Cu K-edge XAS of A1MeImd reveals a pre-edge feature and a short Cu-Cu distance (ca. 2.8 Å), both consistent with a O species. The chemical precedence of a Cu(III) species in a ligand environment remarkably similar to oxyTy suggests that there is no chemical reason, thermodynamic or kinetic, to exclude Cu(III) from discussions of biological redox processes.
Fig. 7.
Left: The oxygenated form of Ty and Hc displaying exclusive Nτ imidazole ligation to the Cu centers [88,92]. Right: The SP species formed from oxygenation of Cu(I) and 1,2-dimethylimidazole showing exclusively Nπ imidazole ligation [37].
Other characterized O species with amine, imine, and hybrid guanidine/amine ligands exhibit similar stoichiometric electrophilic hydroxylation reactivity with added phenolates, albeit without detection of an A-like intermediate species (Fig. 6d & 6e). Phenolate hydroxylation by a TP species has also been proposed using an asymmetric, hexadentate ligand (Fig. 6f), a reactivity behavior not consistent with its general oxidative tendencies [38]. Motivated by a measureable O ↔ TP equilibrium, the TP phenolate reactivity is suggested now to operate through a O species [28]. Chemical precedence that Cu(III) O species are good electrophilic hydroxylation reagents is now firmly established, yet with only a single example of biological nitrogenous ligation, the relationship to the active oxidant in Ty is still tenuous.
Trinuclear
The highest nuclearity synthetic cluster known to form directly from the reaction of Cu(I) and O2 is the trinuclear T cluster with a single Cu(III) center. These synthetic clusters are much less prevalent than O species. In part, this dearth is due to the more stringent constraints of formation of the extremely compact T clusters, coupled with greater detection difficulties of their higher energy and lower intensity optical features compared to other Cu-O2 species.
The disclosure of a T cluster in 1996 by the Stack group resulted from the oxygenation of [(TMCD)Cu(MeCN)]+ (TMCD = (1R,2R)-tetramethylcyclohexanediamine) at −80 °C. The 3:1 Cu:O2 stoichiometry by manometry and the unique optical spectrum defined a new structural type, which was confirmed by X-ray diffraction [17]. The metrical parameters of the two-fold symmetric, tricationic L3Cu3O2 cluster clearly support a valence-localized Cu(III)Cu(II)Cu(II) bis-μ3-oxide cluster. Each copper center maintains a planar geometry, with two ligands from the diamine and two from the oxide ligands. The unique Cu site has shorter Cu-O bonds (ca. 1.8 Å) than the symmetric Cu sites (ca. 2.0 Å), indicative of a higher oxidation state. Ferromagnetic coupling of two Cu(II) centers, dictated by the acute Cu(II)-O-Cu(II) angle, leads to a paramagnetic, triplet ground state. While EXAFS modeling corroborate the metrical parameters of the X-ray structure, the more intense Cu(II) pre-edge feature at 8979 eV masks any potential lower intensity Cu(III) feature at 8981 eV [31,32]. A limited number of other T complexes formed with non-sterically demanding, bidentate ligation are known, primarily characterized through their optical spectra [42,43,93,94].
Reactivity from T clusters remains limited. Generally, T clusters react with weak hydrogen-atom donors such as phenols, catechols, and thiols similar to O species, except that the reactivity has a 1:1 rather than a 2:1 stoichiometry. Redox-titrations indicate reactivity with ferrocene, but not with acetylferrocene, in line with a mild one-electron oxidant [17,23,93]. Similar to the O species, T clusters exhibit thermal decomposition through ligand N-dealkylation, albeit at much higher temperatures suggesting a more stable complex [44]. The TTMCD cluster decays at −40 °C to produce formaldehyde and dealkylated ligand with a measured H(D) KIE of 8 supporting C-H activation as the rate limiting step. Oxygen-atom transfer to PPh3 is also possible, albeit performed at a higher temperature than with O species and presumably associated with a Cu(I) formation. Recently, trinuclear Cu clusters formed with a trinucleating ligand were reported by the Chan group to effect catalytic hydroxylation of hydrocarbon substrates including methane, using O2 and a sacrificial reductant. More productive hydroxylation reactivity is found using H2O2 as the oxidant in both homogeneous and heterogenized solutions [95,96]. The active oxidant is proposed as a localized valence Cu(III)Cu(II)Cu(II) mono-oxide cluster, as opposed to the μ2-oxide of the T clusters.
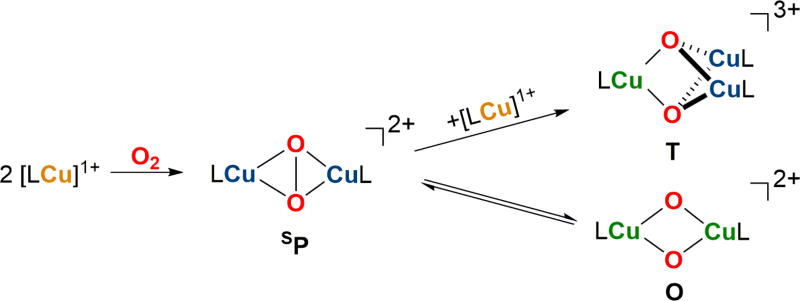
Recently, histamine ligated Cu(I) complexes were reported to react with O2 to yield trinuclear T clusters as the thermodynamic product, the first example of the direct formation of a Cu(III)-containing cluster from Cu(I) and O2 using a biologically relevant histamine chelate [23]. By modification of reaction conditions, non-equilibrating mixtures of O and T clusters could be formed from the same Cu(I)-histamine precursor. Formation kinetics suggest that the low energy pathway to the trinuclear T species does not involve the reaction of a Cu(I) species with a O species initially proposed [42], but is better understood to occur through the intermediacy of an unobserved 2:1 species, presumed to be a SP species (Fig. 8).
Fig. 8.
Proposed formation mechanism of the trinuclear cluster given observed formation kinetics [23].
The ability to form both T and O species with the same bidentate ligand also allows for a direct comparison of reactivity between the two cluster types. The reaction of each species with ascorbic acid as a hydrogen-atom donor shows a stark difference: the T clusters perform the hydrogen-atom abstraction 106 times slower than the corresponding O species at parity of ligand. As DFT calculations suggest that the T and O clusters should have comparable hydrogen-atom affinities, the observed anemic reactivity of the T cluster is attributed to its limited accessibility, given that influence of steric demands from ligand substituents on reactivity is known in other Cu-O2 species [22].
Synthetic and Biological Worlds
Synthetic systems have certain advantages over biological systems in that working with simpler ligation reduces characterization complexity, along with the potential of working at lower temperatures in aprotic solvents to trap intermediates in chemical transformations. Low temperatures not only attenuate entropic costs of assembly, but also raise effective barriers to deleterious decay pathways [84]. In effect, one can consider low temperatures and aprotic polar solvents as surrogates for the protein matrix [37,84], especially considering the dielectric inside of a protein matrix is far closer to that of an organic solvent than water [97,98]. As such, a synergy exists between synthetic Cu-O2 chemistry and chemistry of biological systems; that gleaned from one is generally applicable to the other, assuming a limited number of thermodynamically stable Cu-O2 structural types exist. While no enzymes have been observed to contain Cu(III), several enzymes have been postulated at some point to use a Cu(III) species for oxidative function [18,99–101].
Oxygenases vs Oxidases
Copper enzymes that directly activate O2 can generally be separated into two groups based on reactivity: monooxygenases and oxidases. Monooxygenases (i.e., dopamine-β-monooxygenase (DBM), peptidylglycine-α-hydroxylating monooxygenase (PHM), lytic polysaccharide monooxygenases (LPM), Ty, and particulate methane monooxygenase (pMMO)) are a class of enzyme that hydroxylate organic molecules [4]. In this reaction, dioxygen is reduced to one equivalent of H2O, and the other oxygen atom is inserted into the C-H bond of an organic substrate. Oxidases (i.e., galactose oxidase (GO), glyoxal oxidase (GLOX), amine oxidase (AO), lysyl oxidase (LO), cytochrome c oxidase, and multicopper oxidases (MCO)) perform oxidations that are coupled to the reduction of O2 without incorporation of an oxygen-atom into the product. Depending on the enzyme, O2 can be reduced to either H2O2 or H2O. These groups can be further divided by nuclearity to allow for easier categorization.
Mononuclear
Mononuclear copper sites that directly activate O2 in biology are capable of hydroxylation reactivity (i.e., DBM, PHM, LPM) and oxidase reactivity (i.e., GO, GLOX, AO, LO) [4]. The reaction of O2 with reduced Cu(I) forms is postulated generally to form an initial ES species [20,24]. For the monooxygenase enzymes, the presumed neutral Cu(I) coordination yields a monocationic species that reacts with O2 to generate a non-planar Cu(II) ES species, crystallographically confirmed for PHM [102,103]. This geometry and hydroxylating reactivity is not consistent with the synthetic Cu(III) species, MP.
The oxidases, on the other hand, are proposed to react with O2 through a trigonally ligated Cu(I) monocationic form with two imidazoles and a ligated phenol to form a 4-coordinate end-on superoxide Cu(II) species. The superoxide is proposed to abstract the phenolic hydrogen-atom to form a hydroperoxide ligated to either one of the two valence isomers: a Cu(II)-phenoxyl radical or a Cu(III)-phenolate species. Assuming the former as a strongly antiferromagnetically coupled species, both valence isomers would be EPR silent, as observed experimentally [102,104]. Though initially postulated in 1973 as a Cu(III) species [105], subsequent spectroscopic analyses are more consistent with a Cu(II)-phenoxyl formulation [106].
Binuclear
Binuclear Cu-O2 sites are also known to act as both monooxygenases (i.e., Ty, Hc, pMMO) and oxidases (i.e., Ty, CO). Unlike the mononuclear enzymes above, Hc, Ty, and CO are known to share a similar oxygenated form, a SP species, despite exhibiting different reactivity. Protein x-ray structures of the oxygenated forms indicate each Cu center is ligated in what is best described as a square pyramidal coordination with three imidazole ligands of histidine residues and two oxygen atoms of the peroxide moiety in a side-on fashion [88,92]. Each imidazole ligates through an Nτ atom with the connecting methylene substituent positioned remotely, creating an accessible Cu2O2 core. However, as an O2 transport protein, Hc does not react with exogenous substrates under native conditions, while CO is capable of catecholase activity (i.e., catechols to quinones), and Ty is capable of both hydroxylase (i.e., phenols to catechols) and catecholase reactivity [89]. The difference in reactivity between all these proteins is attributed largely to variation in substrate access to the Cu2O2 core, evidenced by Hc showing slow monooxygenase reactivity under mildly denaturing conditions [107,108]. Extensive reactivity and spectroscopic studies on SP-containing enzymes and model systems suggest that if Cu(III) is operative in any of these binuclear copper enzymes, it would be involved in the monooxygenase reactivity, primarily exhibited by Ty.
Ty are found in all aerobic life forms and are responsible for pigment biosynthesis through regioselective tyrosine hydroxylation to a catechol. Subsequent catechol oxidation to a quinone yields the pigment polymer precursor. While the products of Ty reactivity are well defined, the intimate details of the hydroxylation are far from complete [109–111]. The Itoh group have shown that the Ty hydroxylation activity can be decoupled from the catecholase reactivity under borate buffer conditions, where the catechol is trapped as a borate ester, consistent with catechol release from the binuclear site [61]. Under these conditions, the rate-limiting step of the hydroxylation reaction is slowed by electron deficient substrates (Hammett ρ = −2.4), in line with an electrophilic oxidation mechanism. The KIE of 1.1 for mushroom Ty with deuterium substitution on the oxidized carbon of the phenol suggests that the C-H(D) bond cleavage is not rate limiting [61,112].
The generally accepted hydroxylation mechanism of Ty from these and other investigations are deprotonation of the exogenous phenol by an active-site base, inner-sphere copper coordination by the phenolate, electrophilic peroxide attack on the aromatic π-system, followed by proton rearrangement to yield a μ-hydroxo-μ-catecholato intermediate [113]. Protonation of the catecholate releases catechol, generating a dicopper(II)-μ-hydroxo complex that can further react with the released catechol to produce quinone [89]. While no Cu(III) intermediate has been detected spectroscopically in biology, substantial synthetic experimental evidence support Cu(III)-containing O species as chemically competent to perform phenolate hydroxylation with reaction hallmarks similar to those of Ty. An alternative mechanism to that generally accepted for Ty is that peroxide bond cleavage occurs prior to formation of the C-O bond, which would make the O species the active oxidant [18,39,40] (Fig. 9).
Fig. 9.
Two potential phenol hydroxylation mechanisms by Ty. Oxygenation to a SP species, followed by phenolate ligation with imidazole dissociation creates Int1. The generally accepted biological mechanism involves a rate limiting concerted C-O bond formation with C-H bond cleavage to Int2. Alternatively, phenolate rearrangement could trigger O-O bond cleavage, followed by a rate limiting C-H insertion to Int2.
The remaining binuclear enzyme capable of monooxygenase reactivity is pMMO, which has a unique proposed active site. Its structure was unknown until structural and reactivity studies supported a binuclear Cu active site as the site oxidation. pMMO mediates the remarkable transformation of methane to methanol in methanotrophic bacteria under ambient conditions. X-ray crystallographic and XAS investigations by the Rosenzweig group identify an asymmetric binuclear copper site, in which one copper is coordinated by two imidazole groups of histidine residues and the other is chelated by an imidazole and primary amine of an N-terminal histidine, termed a histidine brace [114]. The resting state of the enzyme is a mixed-valence Cu(I)-Cu(II) state from XAS and EPR. However, the active oxidant formed with O2 is unknown currently [115,116].
In 2005, the first crystal structure of bacterial pMMO at 2.8 Å resolution revealed three metal sites: a Zn site, a mononuclear Cu site, and an asymmetric binuclear Cu site as described above [114]. Cu K-edge EXAFS modeling show a short Cu-Cu vector (ca. 2.6 Å), consistent with the crystal structure and supporting the postulate of a binuclear site. The geometry at each Cu atom in this binuclear site is nearly planar. The bidentate, planar, nitrogenous ligation and short Cu-Cu distance better matches the geometry and coordination preference of a Cu(III) O species than that of a Cu(II) SP species.
Trinuclear
The trinuclear Cu-O2 enzymes (i.e., laccase, ascorbate oxidase, Fet3p), generally referred to as MCO, function as oxidases removing weakly reducing electrons from substrates and reducing O2 by four electrons to water. The fungal laccases are remarkable for their small overpotential (ca. 100 mV) in the reduction of O2 [117,118]. The structure of the trinuclear sites are conserved largely across all MCO with the reduction site of O2 possessing three Cu centers within 4.5 Å [119,120], and a blue copper ET site 12 Å distant. The copper sites of the fully reduced trinuclear cluster are ligated differentially: two have three imidazole ligands while the other has two imidazoles and a water or hydroxide; seven of the eight imidazole ligands are Nτ ligated. The crystallographic data from the Sakurai group on a small MCO mutant show that oxygenation of a reduced crystal yields a tri-Cu μ3-oxide-μ2-hydroxide species as the first and only intermediate, which upon further x-ray exposure degrades to the fully reduced enzyme [121]. The connectivity and metrical parameters are consistent with a paramagnetic Cu(II)Cu(II)Cu(II) μ3-oxide-μ2-hydroxide intermediate, or native intermediate, determined from extensive spectroscopic investigations by the Solomon group [122]. Spectroscopic investigations of several different MCO lacking their blue copper sites, including several from the Sakurai group, detect two intermediates, the first as a diamagnetic species [123–125] and the second as the native intermediate. The first intermediate is postulated by the Solomon group to be a Cu(I)Cu(II)Cu(II) peroxide species, yet no direct spectroscopic evidence of a Cu(I) center exists [121] and no 18O sensitive rR vibrational features in the 600–1200 cm−1 range are observed by excitation into its visible absorptions [121,123,126]. An alternative postulate that may merge the spectroscopic and x-ray data is the inclusion of Cu(III) into the discussion.
Conclusions
Our knowledge of Cu-O2 chemistry over the past decades has increased rapidly since the discovery of Cu activating O2 in hemocyanin in 1878 [127,128]. The continued development of technology for the formation and characterization of Cu-O2 species (e.g., cooling baths, stopped-flow systems, X-ray techniques) has allowed for the understanding of general reactivity and characteristics of each distinct species. With these advancements comes clearer insight into the processes that control Cu-O2 complex formation and subsequent oxidative reactivity. For example, imidazole-ligated Cu(I) species should be considered as one or two electron reductants in reacting with O2 if a binuclear or trinuclear cluster is formed and the redox potential of the resulting species should lie within the biological window.
The characterization of Cu(III) species that form from Cu(I) and O2 provide important chemical precedence to define the conditions and ligation environments for stabilization of this uncommon oxidation state. Defining spectroscopic and reactivity hallmarks whereby the presence of Cu(III) might be observed or inferred in a biological system is an exciting prospect. With the synthetic ligation environments moving ever closer to that observed in biology and knowing the tendency of these system to stabilize Cu(III), ignoring its role in biology becomes increasingly difficult.
Multinuclear systems have advantages over mononuclear systems for stabilizing Cu(III) as multiple Cu sites enable the four electron reduction of O2 to two strongly anionic oxide ligands. Notably, multiple Cu-O2 complexes perform Ty-like hydroxylation chemistry through an observable O intermediate [18,25,39,40]. The well-defined, facile SP ↔ O equilibrium, observed Cu(III)-containing intermediates, and nearly identical kinetic and thermodynamic reaction parameters (e.g., KIEs, Hammett ρ-values) support the notion of Cu(III) as an active player in phenol hydroxylation reactivity akin to Ty, a concept first proposed in 1990 [129]. For these reasons we suggest that the greatest probability of detection of Cu(III) in a biological system exists in a multinuclear Cu system that activates O2.
Acknowledgments
This work was supposed by the National Institutes of Health (GM120187). J.B.G. acknowledges the National Institutes of Health Ruth L. Kirchstein National Research Service Fellowship (5F32GM103071) for financial support.
References
- 1.Mason HS, Fowlks WL, Peterson E. J Am Chem Soc. 1955;77:2914–2915. [Google Scholar]
- 2.Mirica LM, Ottenwaelder X, Stack TDP. Chem Rev. 2004;104:1013–1045. doi: 10.1021/cr020632z. [DOI] [PubMed] [Google Scholar]
- 3.Lewis EA, Tolman WB. Chem Rev. 2004;104:1047–1076. doi: 10.1021/cr020633r. [DOI] [PubMed] [Google Scholar]
- 4.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. Chem Rev. 2014;114:3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann A, Citek C, Binder S, Goos A, Rübhausen M, Troeppner O, Ivanović-Burmazović I, Wasinger EC, Stack TDP, Herres-Pawlis S. Angew Chem Int Ed. 2013;52:5398–5401. doi: 10.1002/anie.201301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esguerra KVN, Fall Y, Lumb JP. Angew Chem Int Ed. 2014;53:5877–5881. doi: 10.1002/anie.201311103. [DOI] [PubMed] [Google Scholar]
- 7.Xu B, Lumb J-P, Arndtsen BA. Angew Chem Int Ed. 2015;54:4208–4211. doi: 10.1002/anie.201411483. [DOI] [PubMed] [Google Scholar]
- 8.Hoover JM, Steves JE, Stahl SS. Nat Protocols. 2012;7:1161–1166. doi: 10.1038/nprot.2012.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurtele C, Gaoutchenova E, Harms K, Holthausen MC, Sundermeyer J, Schindler S. Angew Chem Int Ed. 2006;45:3867–3869. doi: 10.1002/anie.200600351. [DOI] [PubMed] [Google Scholar]
- 10.Fujisawa K, Tanaka M, Moro-Oka Y, Kitajima N. J Am Chem Soc. 1994;116:12079–12080. [Google Scholar]
- 11.Aboelella NW, Lewis EA, Reynolds AM, Brennessel WW, Cramer CJ, Tolman WB. J Am Chem Soc. 2002;124:10660–10661. doi: 10.1021/ja027164v. [DOI] [PubMed] [Google Scholar]
- 12.Aboelella NW, Kryatov SV, Gherman BF, Brennessel WW, Young VG, Sarangi R, Rybak-Akimova EV, Hodgson KO, Hedman B, Solomon EI, Cramer CJ, Tolman WB. J Am Chem Soc. 2004;126:16896–16911. doi: 10.1021/ja045678j. [DOI] [PubMed] [Google Scholar]
- 13.Dalle KE, Gruene T, Dechert S, Demeshko S, Meyer F. J Am Chem Soc. 2014;136:7428–7434. doi: 10.1021/ja5025047. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson RR, Tyeklár Z, Farooq A, Karlin KD, Liu S, Zubieta J. J Am Chem Soc. 1988;110:3690–3692. [Google Scholar]
- 15.Kitajima N, Fujisawa K, Moro-Oka Y, Toriumi K. J Am Chem Soc. 1989;111:8975–8976. [Google Scholar]
- 16.Halfen JA, Mahapatra S, Wilkinson EC, Kaderli S, Young VG, Que L, Zuberbuhler AD, Tolman WB. Science. 1996;271:1397–1400. doi: 10.1126/science.271.5254.1397. [DOI] [PubMed] [Google Scholar]
- 17.Cole AP, Root DE, Mukherjee P, Solomon EI, Stack TDP. Science. 1996;273:1848–1850. doi: 10.1126/science.273.5283.1848. [DOI] [PubMed] [Google Scholar]
- 18.Mirica LM, Vance M, Rudd DJ, Hedman B, Hodgson KO, Solomon EI, Stack TDP. Science. 2005;308:1890–1892. doi: 10.1126/science.1112081. [DOI] [PubMed] [Google Scholar]
- 19.Stack TDP. Dalton Trans. 2003;10:1881–1889. [Google Scholar]
- 20.Itoh S. Acc Chem Res. 2015;48:2066–2074. doi: 10.1021/acs.accounts.5b00140. [DOI] [PubMed] [Google Scholar]
- 21.Citek C, Lin B-L, Phelps TE, Wasinger EC, Stack TDP. J Am Chem Soc. 2014;136:14405–14408. doi: 10.1021/ja508630d. [DOI] [PubMed] [Google Scholar]
- 22.Citek C, Gary JB, Wasinger EC, Stack TDP. J Am Chem Soc. 2015;137:6991–6994. doi: 10.1021/jacs.5b02157. [DOI] [PubMed] [Google Scholar]
- 23.Gary JB, Citek C, Brown TA, Zare RN, Wasinger EC, Stack TDP. J Am Chem Soc. 2016;138:9986–9995. doi: 10.1021/jacs.6b05538. [DOI] [PubMed] [Google Scholar]
- 24.Cramer CJ, Tolman WB. Acc Chem Res. 2007;40:601–608. doi: 10.1021/ar700008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herres-Pawlis S, Verma P, Haase R, Kang P, Lyons CT, Wasinger EC, Florke U, Henkel G, Stack TDP. J Am Chem Soc. 2009;131:1154–1169. doi: 10.1021/ja807809x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahoy J, Holland PL, Tolman WB. Inorg Chem. 1999;38:2161–2168. doi: 10.1021/ic990095+. [DOI] [PubMed] [Google Scholar]
- 27.Op't Holt BT, Vance MA, Mirica LM, Heppner DE, Stack TDP, Solomon EI. J Am Chem Soc. 2009;131:6421–6438. doi: 10.1021/ja807898h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieber-Emmons MT, Ginsbach JW, Wick PK, Lucas HR, Helton ME, Lucchese B, Suzuki M, Zuberbühler AD, Karlin KD, Solomon EI. Angew Chem Int Ed. 2014;53:4935–4939. doi: 10.1002/anie.201402166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henson MJ, Mukherjee P, Root DE, Stack TDP, Solomon EI. J Am Chem Soc. 1999;121:10332–10345. [Google Scholar]
- 30.Woertink JS, Smeets PJ, Groothaert MH, Vance MA, Sels BF, Schoonheydt RA, Solomon EI. Proc Natl Acad Sci. 2009;106:18908–18913. doi: 10.1073/pnas.0910461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 32.DuBois JL, Mukherjee P, Collier AM, Mayer JM, Solomon EI, Hedman B, Stack TDP, Hodgson KO. J Am Chem Soc. 1997;119:8578–8579. [Google Scholar]
- 33.DuBois JL, Mukherjee P, Stack TDP, Hedman B, Solomon EI, Hodgson KO. J Am Chem Soc. 2000;122:5775–5787. [Google Scholar]
- 34.Itoh S, Fukuzumi S. Acc Chem Res. 2007;40:592–600. doi: 10.1021/ar6000395. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Bosch I, Cowley RE, Díaz DE, Siegler MA, Nam W, Solomon EI, Karlin KD. Chemistry – A European Journal. 2016;22:5133–5137. doi: 10.1002/chem.201600551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul PP, Tyeklar Z, Jacobson RR, Karlin KD. J Am Chem Soc. 1991;113:5322–5332. [Google Scholar]
- 37.Citek C, Lyons CT, Wasinger EC, Stack TDP. Nature Chem. 2012;4:317–322. doi: 10.1038/nchem.1284. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Bosch I, Company A, Frisch JR, Torrent-Sucarrat M, Cardellach M, Gamba I, Guell M, Casella L, Que L, Ribas X, Luis JM, Costas M. Angew Chem Int Ed. 2010;49:2406–2409. doi: 10.1002/anie.200906749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang L, Keown W, Citek C, Wasinger EC, Stack TDP. Angew Chem Int Ed. 2016;55:10453–10457. doi: 10.1002/anie.201605159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Company A, Palavicini S, Garcia-Bosch I, Mas-Balleste R, Que L, Rybak-Akimova EV, Casella L, Ribas X, Costas M. Chem Eur J. 2008;14:3535–3538. doi: 10.1002/chem.200800229. [DOI] [PubMed] [Google Scholar]
- 41.Serrano-Plana J, Garcia-Bosch I, Miyake R, Costas M, Company A. Angew Chem Int Ed. 2014;53:9608–9612. doi: 10.1002/anie.201405060. [DOI] [PubMed] [Google Scholar]
- 42.Taki M, Teramae S, Nagatomo S, Tachi Y, Kitagawa T, Itoh S, Fukuzumi S. J Am Chem Soc. 2002;124:6367–6377. doi: 10.1021/ja026047x. [DOI] [PubMed] [Google Scholar]
- 43.Gupta AK, Tolman WB. Inorg Chem. 2012;51:1881–1888. doi: 10.1021/ic202214c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukherjee P. PhD Thesis. Stanford University; 2000. [Google Scholar]
- 45.Jones SM, Solomon EI. Cell Mol Life Sci. 2015;72:869–883. doi: 10.1007/s00018-014-1826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deming RL, Allred AL, Dahl AR, Herlinger AW, Kestner MO. J Am Chem Soc. 1976;98:4132–4137. [Google Scholar]
- 47.Bossu FP, Chellappa KL, Margerum DW. J Am Chem Soc. 1977;99:2195–2203. doi: 10.1021/ja00449a028. [DOI] [PubMed] [Google Scholar]
- 48.McDonald MR, Scheper WM, Lee HD, Margerum DW. Inorg Chem. 1995;34:229–237. [Google Scholar]
- 49.Donoghue PJ, Tehranchi J, Cramer CJ, Sarangi R, Solomon EI, Tolman WB. J Am Chem Soc. 2011;133:17602–17605. doi: 10.1021/ja207882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhar D, Tolman WB. J Am Chem Soc. 2015;137:1322–1329. doi: 10.1021/ja512014z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyanaga A, Fushinobu S, Ito K, Wakagi T. Biochem Biophys Res Commun. 2001;288:1169–1174. doi: 10.1006/bbrc.2001.5897. [DOI] [PubMed] [Google Scholar]
- 52.Huang WJ, Jia J, Cummings J, Nelson M, Schneider G, Lindqvist Y. Structure. 1997;5:691–699. doi: 10.1016/s0969-2126(97)00223-2. [DOI] [PubMed] [Google Scholar]
- 53.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 54.Neupane KP, Shearer J. Inorg Chem. 2006;45:10552–10566. doi: 10.1021/ic061156o. [DOI] [PubMed] [Google Scholar]
- 55.Hanss J, Beckmann A, Krüger H-J. Eur J Inorg Chem. 1999:163–172. [Google Scholar]
- 56.Schatz M, Raab V, Foxon SP, Brehm G, Schneider S, Reiher M, Holthausen MC, Sundermeyer J, Schindler S. Angew Chem Int Ed. 2004;43:4360–4363. doi: 10.1002/anie.200454125. [DOI] [PubMed] [Google Scholar]
- 57.Maiti D, Fry HC, Woertink JS, Vance MA, Solomon EI, Karlin KD. J Am Chem Soc. 2007;129:264–265. doi: 10.1021/ja067411l. [DOI] [PubMed] [Google Scholar]
- 58.Spencer DJE, Aboelella NW, Reynolds AM, Holland PL, Tolman WB. J Am Chem Soc. 2002;124:2108–2109. doi: 10.1021/ja017820b. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds AM, Lewis EA, Aboelella NW, Tolman WB. Chem Commun. 2005:2014–2016. doi: 10.1039/b418939f. [DOI] [PubMed] [Google Scholar]
- 60.Solomon EI, Chen P, Metz M, Lee SK, Palmer AE. Angew Chem Int Ed. 2001;40:4570–4590. doi: 10.1002/1521-3773(20011217)40:24<4570::aid-anie4570>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 61.Yamazaki S, Itoh S. J Am Chem Soc. 2003;125:13034–13035. doi: 10.1021/ja036425d. [DOI] [PubMed] [Google Scholar]
- 62.Spuhler P, Holthausen MC. Angew Chem Int Ed. 2003;42:5961–5965. doi: 10.1002/anie.200352231. [DOI] [PubMed] [Google Scholar]
- 63.Mahadevan V, Henson MJ, Solomon EI, Stack TDP. J Am Chem Soc. 2000;122:10249–10250. [Google Scholar]
- 64.Matsumoto T, Furutachi H, Kobino M, Tomii M, Nagatomo S, Tosha T, Osako T, Fujinami S, Itoh S, Kitagawa T, Suzuki M. J Am Chem Soc. 2006;128:3874–3875. doi: 10.1021/ja058117g. [DOI] [PubMed] [Google Scholar]
- 65.Karlin KD, Wei N, Jung B, Kaderli S, Niklaus P, Zuberbühler AD. J Am Chem Soc. 1993;115:9506–9514. [Google Scholar]
- 66.Mahapatra S, Halfen J, Wilkinson E, Pan G, Cramer CJ, Que L, Jr, Tolman WB. J Am Chem Soc. 1995;117:8865–8866. [Google Scholar]
- 67.Mahapatra S, Young VG, Kaderli S, Zuberbühler AD, Tolman WB. Angew Chem Int Ed. 1997;36:130–133. [Google Scholar]
- 68.Mahapatra S, Halfen JA, Tolman WB. J Am Chem Soc. 1996;118:11575–11586. [Google Scholar]
- 69.Cole AP, Mahadevan V, Mirica LM, Ottenwaelder X, Stack TDP. Inorg Chem. 2005;44:7345–7364. doi: 10.1021/ic050331i. [DOI] [PubMed] [Google Scholar]
- 70.Mahadevan V, DuBois JL, Hedman B, Hodgson KO, Stack TDP. J Am Chem Soc. 1999;121:5583–5584. [Google Scholar]
- 71.Holland PL, Rodgers KR, Tolman WB. Angew Chem Int Ed. 1999;38:1139–1142. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1139::AID-ANIE1139>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 72.Itoh S, Taki M, Nakao H, Holland PL, Tolman WB, Que L, Jr, Fukuzumi S. Angew Chem Int Ed. 2000;39:398–400. doi: 10.1002/(sici)1521-3773(20000117)39:2<398::aid-anie398>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 73.Thangavel A, Wieliczko M, Bacsa J, Scarborough CC. Inorg Chem. 2013;52:13282–13287. doi: 10.1021/ic402626p. [DOI] [PubMed] [Google Scholar]
- 74.Herres-Pawlis S, Haase R, Verma P, Hoffmann A, Kang P, Stack TDP. Eur J Inorg Chem. 2015;2015:5426–5436. doi: 10.1002/ejic.201500884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walli A, Dechert S, Bauer M, Demeshko S, Meyer F. Eur J Inorg Chem. 2014;2014:4660–4676. [Google Scholar]
- 76.Itoh S, Tachi Y. Dalton Trans. 2006:4531–4538. doi: 10.1039/b607964d. [DOI] [PubMed] [Google Scholar]
- 77.Karahalis GJ, Thangavel A, Chica B, Bacsa J, Dyer RB, Scarborough CC. Inorg Chem. 2016;55:1102–1107. doi: 10.1021/acs.inorgchem.5b02205. [DOI] [PubMed] [Google Scholar]
- 78.Mahadevan V, Hou ZG, Cole AP, Root DE, Lal TK, Solomon EI, Stack TDP. J Am Chem Soc. 1997;119:11996–11997. [Google Scholar]
- 79.Ottenwaelder X, Rudd DJ, Corbett MC, Hodgson KO, Hedman B, Stack TDP. J Am Chem Soc. 2006;128:9268–9269. doi: 10.1021/ja061132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahadevan V. PhD. Thesis. Stanford University; 2001. [Google Scholar]
- 81.Taki M, Itoh S, Fukuzumi S. J Am Chem Soc. 2001;123:6203–6204. doi: 10.1021/ja015721s. [DOI] [PubMed] [Google Scholar]
- 82.Itoh S, Nakao H, Berreau LM, Kondo T, Komatsu M, Fukuzumi S. J Am Chem Soc. 1998;120:2890–2899. [Google Scholar]
- 83.Chen P, Solomon EI. J Am Chem Soc. 2004;126:4991–5000. doi: 10.1021/ja031564g. [DOI] [PubMed] [Google Scholar]
- 84.Citek C, Herres-Pawlis S, Stack TDP. Acc Chem Res. 2015;48:2424–2433. doi: 10.1021/acs.accounts.5b00220. [DOI] [PubMed] [Google Scholar]
- 85.Diaddario LL, Robinson WR, Margerum DW. Inorg Chem. 1983;22:1021–1025. [Google Scholar]
- 86.Liu C-C, Lin T-S, Chan SI, Mou C-Y. J Catal. 2015;322:139–151. [Google Scholar]
- 87.Mandal S, Mukherjee J, Lloret F, Mukherjee R. Inorg Chem. 2012;51:13148–13161. doi: 10.1021/ic3013848. [DOI] [PubMed] [Google Scholar]
- 88.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 89.Rolff M, Schottenheim J, Decker H, Tuczek F. Chem Soc Rev. 2011;40:4077–4098. doi: 10.1039/c0cs00202j. [DOI] [PubMed] [Google Scholar]
- 90.Matsumoto T, Ohkubo K, Honda K, Yazawa A, Furutachi H, Fujinami S, Fukuzumi S, Suzuki M. J Am Chem Soc. 2009;131:9258–9267. doi: 10.1021/ja809822c. [DOI] [PubMed] [Google Scholar]
- 91.Mukherjee S, Stull JA, Yano J, Stamatatos TC, Pringouri K, Stich TA, Abboud KA, Britt RD, Yachandra VK, Christou G. Proc Natl Acad Sci. 2012;109:2257–2262. doi: 10.1073/pnas.1115290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cuff ME, Miller KI, Vanholde KE, Hendrickson WA. J Mol Biol. 1998;278:855–870. doi: 10.1006/jmbi.1998.1647. [DOI] [PubMed] [Google Scholar]
- 93.Lionetti D, Day MW, Agapie T. Chem Sci. 2013;4:785–790. doi: 10.1039/C2SC21758A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Machonkin T, Mukherjee P, Stack TDP, Solomon EI. Inorg Chim Acta. 2002;341:39–44. [Google Scholar]
- 95.Liu C-C, Mou C-Y, Yu SSF, Chan SI. Energy & Environmental Science. 2016;9:1361–1374. [Google Scholar]
- 96.Chan SI, Lu Y-J, Nagababu P, Maji S, Hung M-C, Lee MM, Hsu IJ, Minh PD, Lai JCH, Ng KY, Ramalingam S, Yu SSF, Chan MK. Angew Chem Int Ed. 2013;52:3731–3735. doi: 10.1002/anie.201209846. [DOI] [PubMed] [Google Scholar]
- 97.Karp DA, Gittis AG, Stahley MR, Fitch CA, Stites WE, Garcia-Moreno EB. Biophys J. 2007;92:2041–2053. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li L, Li C, Zhang Z, Alexov E. Journal of Chemical Theory and Computation. 2013;9:2126–2136. doi: 10.1021/ct400065j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hamilton GA, Adolf PK, de Jersey J, DuBois GC, Dyrkacz GR, Libby RD. J Am Chem Soc. 1978;100:1899–1912. [Google Scholar]
- 100.Yoshizawa K, Kihara N, Kamachi T, Shiota Y. Inorg Chem. 2006;45:3034–3041. doi: 10.1021/ic0521168. [DOI] [PubMed] [Google Scholar]
- 101.Aasa R, Brändén R, Deinum J, Malmström B, Reinhammar B, Vänngård T. FEBS Lett. 1976;61:115–119. doi: 10.1016/0014-5793(76)81016-2. [DOI] [PubMed] [Google Scholar]
- 102.Prigge ST, Eipper BA, Mains RE, Amzel LM. Science. 2004;304:864–867. doi: 10.1126/science.1094583. [DOI] [PubMed] [Google Scholar]
- 103.Klinman JP. Chem Rev. 1996;96:2541–2562. doi: 10.1021/cr950047g. [DOI] [PubMed] [Google Scholar]
- 104.Gherman BF, Heppner DE, Tolman WB, Cramer CJ. J Biol Inorg Chem. 2006;11:197–205. doi: 10.1007/s00775-005-0066-5. [DOI] [PubMed] [Google Scholar]
- 105.Hamilton GA, Libby RD, Hartzell CR. Biochem Biophys Res Commun. 1973;55:333–340. doi: 10.1016/0006-291x(73)91092-9. [DOI] [PubMed] [Google Scholar]
- 106.Whittaker MM, Whittaker JW. J Biol Chem. 1988;263:6074–6080. [PubMed] [Google Scholar]
- 107.Fujieda N, Yakiyama A, Itoh S. Dalton Trans. 2010;39:3083–3092. doi: 10.1039/c000760a. [DOI] [PubMed] [Google Scholar]
- 108.Solem E, Tuczek F, Decker H. Angew Chem Int Ed. 2016;55:2884–2888. doi: 10.1002/anie.201508534. [DOI] [PubMed] [Google Scholar]
- 109.Kahn V, Ben-Shalom N. Pigm Cell Res. 1998;11:24–33. doi: 10.1111/j.1600-0749.1998.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 110.Westerfeld WW. Biochem J. 1940;34:51–58. doi: 10.1042/bj0340051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mahadevan V, Gebbink RJMK, Stack TDP. Curr Opin Chem Biol. 2000;4:228–234. doi: 10.1016/s1367-5931(99)00080-0. [DOI] [PubMed] [Google Scholar]
- 112.Muñoz-Muñoz JL, Berna J, García-Molina MdM, Garcia-Molina F, Garcia-Ruiz PA, Varon R, Rodriguez-Lopez JN, Garcia-Canovas F. Biochem Biophys Res Commun. 2012;424:228–233. doi: 10.1016/j.bbrc.2012.06.074. [DOI] [PubMed] [Google Scholar]
- 113.Solomon EI, Sundaram UM, Machonkin TE. Chem Rev. 1996;96:2563–2606. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 114.Lieberman RL, Rosenzweig AC. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 115.Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC, Stemmler TL. Inorg Chem. 2006;45:8372–8381. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Balasubramanian R, Rosenzweig AC. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 117.Heller A. Phys Chem Chem Phys. 2004;6:209–216. [Google Scholar]
- 118.Lee CW, Gray HB, Anson FC, Malmstrom BG. J Electroanal Chem. 1984;172:289–300. [Google Scholar]
- 119.Messerschmidt A, Luecke H, Huber R. J Mol Biol. 1993;230:997–1014. doi: 10.1006/jmbi.1993.1215. [DOI] [PubMed] [Google Scholar]
- 120.Shin W, Sundaram UM, Cole JL, Zhang HH, Hedman B, Hodgson KO, Solomon EI. J Am Chem Soc. 1996;118:3202–3215. [Google Scholar]
- 121.Komori H, Sugiyama R, Kataoka K, Higuchi Y, Sakurai T. Angew Chem Int Ed. 2012;51:1861–1864. doi: 10.1002/anie.201107739. [DOI] [PubMed] [Google Scholar]
- 122.Lee SK, DeBeer George S, Antholine WE, Hedman B, Hodgson KO, Solomon EI. J Am Chem Soc. 2002;124:6180–6193. doi: 10.1021/ja0114052. [DOI] [PubMed] [Google Scholar]
- 123.Kataoka K, Kitagawa R, Inoue M, Naruse D, Sakurai T, Huang H-w. Biochemistry. 2005;44:7004–7012. doi: 10.1021/bi0476836. [DOI] [PubMed] [Google Scholar]
- 124.Palmer AE, Quintanar L, Severance S, Wang T-P, Kosman DJ, Solomon EI. Biochemistry. 2002;41:6438–6448. doi: 10.1021/bi011979j. [DOI] [PubMed] [Google Scholar]
- 125.Ueki Y, Inoue M, Kurose S, Kataoka K, Sakurai T. FEBS Lett. 2006;580:4069–4072. doi: 10.1016/j.febslet.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 126.Kataoka K, Sugiyama R, Hirota S, Inoue M, Urata K, Minagawa Y, Seo D, Sakurai T. J Biol Chem. 2009;284:14405–14413. doi: 10.1074/jbc.M808468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fredericq L. Comptes Rendus de l'Académie des Sciences. 1878;87:996–998. [Google Scholar]
- 128.Fredericq L. Archives de zoologie expérimentale. 1878;7:535–583. [Google Scholar]
- 129.Reglier M, Jorand C, Waegell B. Chem Comm. 1990:1752–1755. [Google Scholar]