Abstract
The Huntington's disease (HD) mutation is a polyglutamine expansion in the N-terminal region of huntingtin (N-htt). How neurons die in HD is unclear. Mutant N-htt aggregates in neurons in the HD brain; expression of mutant N-htt in vitro causes cell death. Other in vitro studies show that proteolysis by caspase 3 could be important in regulating mutant N-htt function, but there has been no direct evidence for caspase 3-cleaved N-htt fragments in brain. Here, we show that N-htt fragments consistent with the size produced by caspase 3 cleavage in vitro are resident in the cortex, striatum, and cerebellum of normal and adult onset HD brain and are similar in size to the fragments seen after exogenous expression of human huntingtin in mouse clonal striatal neurons. HD brain extracts treated with active caspase 3 had increased levels of N-htt fragments. Compared with the full-length huntingtin, the caspase 3-cleaved N-htt fragments, especially the mutant fragment, preferentially segregated with the membrane fraction. Partial proteolysis of the human caspase 3-cleaved N-htt fragment by calpain occurred in vitro and resulted in smaller N-terminal products; products of similar size appeared when mouse brain protein extracts were treated with calpain. Results support the idea that sequential proteolysis by caspase 3 and calpain may regulate huntingtin function at membranes and produce N-terminal mutant fragments that aggregate and cause cellular dysfunction in HD.
A polyglutamine expansion located in the N terminus of huntingtin (N-htt) causes Huntington's disease (HD). How the mutation causes cell death is unknown. Several recent observations implicate altered huntingtin (htt) processing in the pathogenesis of HD. In the HD brain, mutant N-htt fragments aggregate in the nucleus and cytoplasm (1); the expression of mutant N-htt fragments in vitro causes cell death (2, 3). These findings suggest that proteolysis in the N-terminal region of htt may be important in HD pathogenesis. Furthermore, htt cleavage by caspase 3 could contribute to neurodegeneration in HD. Htt can serve as a substrate for caspase activity. Two proximate caspase 3-sensitive sites and one caspase 6-sensitive site are distal to the polyglutamine tract at aspartate residues 513, 552, and 589, respectively, in the wild-type (wt) protein (4–6). Caspase 3-cleaved N-htt products have been observed in vitro after exogenous expression of human htt in HEK 293 cells (4) and clonal striatal neurons (7). Treatment with the broad acting caspase inhibitor Z-VAD-FMK attenuates caspase 3 cleavage of htt and increases cell survival (6, 7). Enhanced immunoreactivity for caspases has been reported in HD striatal neurons compared with control brain,§ supporting the involvement of caspase activation in HD pathogenesis. However, there is no evidence for htt proteolysis by caspases in the brain. No non-caspase proteases have been identified that produce a limited proteolysis of htt. Thus, despite the presence of N-htt fragments in adult and juvenile HD brain (1), the proteolytic pathway involved in the production of mutant N-htt fragments in the brain is unknown. That caspase 3 or other proteases cleave the N terminus of mutant htt in the HD brain would provide strong support that N-terminal htt proteolysis is a critical factor in HD pathogenesis.
Here, we demonstrate that caspase 3-cleaved N-htt fragments occur in the control and HD brain and are similar in size to fragments produced in vitro after exogenous expression of human wt and mutant htt in mouse clonal striatal cells (7, 8). The wt and mutant caspase 3-cleaved N-htt fragments in brain varied in size with polyglutamine length and were preferentially enriched in membrane fractions. Partial proteolysis of the caspase 3-cleaved N-htt fragments by calpain produced smaller N-terminal fragments. We speculate that caspase 3 cleavage regulates the proteolysis of wt and mutant htt in the brain and increases the association of N-terminal htt with membranes. Calpain-induced proteolysis of the caspase 3-cleaved mutant N-htt fragment may lead to the formation of mutant N-htt fragments that can aggregate and form inclusions.
Materials and Methods
Cell Culture and Transfections.
The culturing and transfection of mouse clonal striatal cells (X57 cells) have been described in our recent publications (7, 8, 9). MCF-7 cells were grown according to the suppliers' recommendations [American Tissue Culture Collection (ATCC)]. DNA was introduced by using Superfect Transfection Reagent (Qiagen, Valencia, CA). Cells were harvested for analysis 24 h after transfection.
Expression Plasmids.
Plasmids encoding FLAG-htt have been previously described (7, 8). A series of pcDNA3 plasmids encode FLAG followed in frame by the first 3,221 bp of human htt with 18, 46, or 100 glutamines. These constructs are denoted as FH3221-18, FH3221-46, and FH3221-100, respectively. The FH3221-18 encodes the first 969 aa in htt. A pcDNA3 plasmid encoding FLAG only or no plasmid was used for mock transfections.
Preparation of Protein Extracts from Human Postmortem Brain, Mouse Brain, and Cell Culture.
Frozen human autopsy brain from the cortex, striatum, and cerebellum of controls (C2, C8, C18), adult onset HD patients (A3, A4, A12), and juvenile onset HD patients (J6, J11) were used in this study. Postmortem interval for the control and HD brains ranged from 4 to 40 h. For analysis of total protein extracts, blocks (about 0.1 g) of frozen human autopsy brain were dissected into 5 vol of buffer [20 mM Tris (pH 7.4) plus protease inhibitors; Boehringer Mannheim], homogenized on ice, and centrifuged at 100,000 × g for 30 min. The supernatant was analyzed by SDS/PAGE and Western blot. For subcellular fractionation, brain was placed in 5 vol of buffer (20 mM Tris/0.9% sucrose/1 mM EDTA, pH 7.4, plus protease inhibitors), homogenized on ice (CH), and centrifuged at 2,000 × g for 10 min at 4°C. The pellet was resuspended in buffer (P1), and the supernatant (S1) was further centrifuged at 100,000 × g for 1 h at 4°C to obtain a pellet, which was resuspended in buffer (P2), and a supernatant (S2).
Mouse brain was removed rapidly from the deeply anesthetized mouse, hemisectioned, and rapidly frozen on dry ice. There was essentially no postmortem delay. For preparation of protein extracts from mouse brain, one hemisphere of a mouse brain was homogenized on ice in 20 mM Tris buffer (pH 7.4). The homogenate was centrifuged at 100,000 × g for 30 min, and the supernatant was collected for analysis. Protein extraction from cultured cells was performed as previously described (7, 8). Briefly, cells were washed twice in ice-cold PBS and then harvested in 500 μl of homogenization buffer with protease inhibitors. For the collection of debris, media from the cultures was centrifuged at 2000 × g at 4°C for 10 min. Supernatants were removed, and the pellets were washed once and resuspended in ice-cold PBS. After centrifugation at 10,000 × g for another 10 min at 4°C, supernatant was removed, and pellets were lysed with 20 μl of the homogenization buffer with protease inhibitors. Samples were stored at −80°C.
Western Blots.
Protein concentrations were determined by the Bradford method (Bio-Rad). Samples (20–30 μg) were loaded for SDS/PAGE by using gels made with 7.5% acrylamide, 10% acrylamide, and 0.05% bis-acrylamide, or as 4–20% gradients (Bio-Rad). Proteins were separated by using 160 V for 1 h, then transferred to 0.45 μm nitrocellulose at 100 V for 1 h. Blots were blocked in 5% nonfat dry milk (Bio-Rad) in TBS + 0.1% Tween 20 (TBST) for 1 h at room temperature and then incubated overnight in anti-htt antibody ab1 (ref. 10; 0.25–0.5 μg/ml) or monoclonal antibody 2166 (Chemicon, 1:2000; diluted in 5% milk/TBST) at 4°C. Ab1 recognizes amino acids 1–17, and antibody 2166 detects an epitope between 181–812 in htt. Peroxidase-labeled secondary antibody (Vector Laboratories) was diluted 1:10,000 in 5% milk/TBST and incubated for 1 h at room temperature. Protein bands were visualized by using enhanced chemiluminescence (ECL kit, Amersham Pharmacia) and exposed to Hyperfilm (Amersham Pharmacia). Some blots were reprobed with antisera to synaptophysin or alpha-tubulin.
Immunoprecipitation Assays.
Human cortex (0.1–0.12 g) was homogenized in 10 vol of immunoprecipitation buffer (PBS with 1% Triton X-100 and protease inhibitors) on ice. The mixture was centrifuged at 14,000 × g for 15 min, and some of the supernatant was mixed 1:1 with sample buffer, boiled for 5 min, and reserved as the “total” extract. Protein G agarose beads were preblocked by addition of supernatant in a 1:1 ratio by volume. For the immunoprecipitation, anti-htt antisera ab1 (3 μg; ref. 10) or 2166 (Chemicon, 4 μl) was added to supernatant (800 μl) for 2 h at 4°C. Preblocked protein G (20 μl) or protein A agarose beads (30 μl) were added for 1 h at 4°C, and then spun at 14,000 × g for 5 min. The supernatant was discarded, and the beads were washed with PBS + 1% Triton X-100, PBS + 0.1% Triton X-100, and PBS, twice each. Thirty microliters of 2× sample buffer (100 μM Tris⋅HCL (pH 6.8)/4% SDS/0.2% bromophenol blue/20% glycerol/100 mM DTT) was added to the agarose beads and boiled for 5 min. The immunoprecipitates were stored at −80°C.
Caspase Cleavage Assay.
X57 cells from a 60-mm culture dish were washed in PBS and harvested in 1 ml of PBS, spun, and separated from the supernatant, which was discarded. To get crude homogenates, cells were resuspended in lysis buffer (20 mM Tris, pH 7.4/0.9% sucrose/1 mM EDTA), to which protease inhibitors (aprotinin 1 μl/ml and pefabloc 50 μl/ml) were added, and homogenized by using a Dounce homogenizer for at least 30 strokes or until more than 90% of cells were lysed based on microscopic observation. The protein concentration was determined by the Bradford assay. Samples were aliquotted into several tubes and saved at −70°C. For the reaction, 15 μg of protein was diluted 1:10 in reaction buffer (50 mM Hepes, pH 7.4/100 mM NaCl/1 mM EDTA/10% glycerol/0.1% CHAPS/10 mM DTT) and incubated with 500 ng of active caspase 3 enzyme (PharMingen) at 37°C for 1 h. Sample buffer (4×) and DTT (0.1 M final concentration) were added, and the mixture was boiled for 15 min.
Calpain Inhibition and Calpain Cleavage Assays.
X57 cells transfected with htt cDNA FH3221-100 were treated for 12 h at 37°C with one of the following calpain inhibitors (purchased from CalBiochem): calpain inhibitor I (ALLN, 10–20 μM), calpain inhibitor II (ALLM, 10 μM), or calpeptin (20 μM). Control cultures received no treatment. Debris from the culture media was collected, and protein extracts were prepared as described above. For calpain cleavage, mouse brain lysates (20 μg of protein) were incubated with 0.5 units of purified calpain II in a buffer (50 mM Tris, pH 7.5/100 mM NaCl/2 mM DTT/1 mM EDTA/5 mM CaCl2) at 30°C for 15 min. Sample buffer was added, and the mixture was boiled for 15 min.
Results
Characteristics of N-Terminal htt Fragments in X57 Cells and in Mouse Brain.
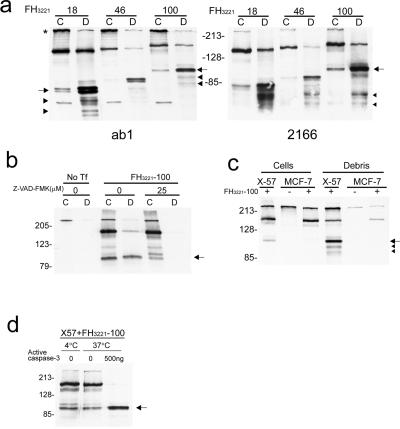
cDNAs encoding the first one third of human htt (FH3221) were transiently transfected into X57 cells. Protein extracts were made from the transfected cells and examined by SDS/PAGE and Western blot. N-htt fragments migrated between 80–100 kDa depending on the polyglutamine length in htt (18, 46, or 100 glutamines) and were seen in blots probed with anti-htt antisera ab1 or 2166 (Fig. 1a). The N-htt fragments migrate to the position expected by caspase 3 cleavage based on previous studies using site directed mutagenesis (6). Hereafter, these N-htt fragments are referred to as N-httcasp3 fragments. In Western blots, the N-httcasp3 fragment resolved as a doublet with antibody ab1; antibody 2166 detected the upper band of the doublet more than the lower band. It is unclear whether the doublet arises from cleavage at the two nearby caspase 3 sites in htt (6) or represents a modification of a single N-htt fragment produced by cleavage at one of the caspase 3 sites. X57 cells were transiently transfected with mutant htt cDNA FH3221-100 and treated with Z-VAD-FMK. Z-VAD-FMK treatment markedly reduced the level of mutant N-httcasp3 fragment in the cells and increased the level of a precursor fragment (Fig. 1b). No mutant N-httcasp3 fragment was detected when htt cDNA FH3221-100 was transfected into MCF-7 cells (Fig. 1c), which lack caspase 3 activity (11). Protein extracts from X57 cells expressing exogenous mutant htt were treated with active caspase 3 enzyme for 1 h at 37°C. Treatment with active caspase 3 significantly increased the level of the mutant N-httcasp3 fragment; the uncleaved htt was undetectable after treatment (Fig. 1d).
Figure 1.
Western blots show caspase 3 cleavage of htt in transfected cells and in mouse brain. (a) X57 cells were transfected with FLAG-htt cDNAs (FH3221) with 18, 46, or 100 glutamines. Blots are from different gels run in parallel and probed with anti-htt antisera ab1 (Left) and 2166 (Right).The N-httcasp3 fragment varies in size from about 80–100 kDa (arrows) depending on polyglutamine length in htt and is present in cells (C) and in debris (D). The N-httcasp3 fragment appears as a doublet (left arrow). Proteolytic fragments of the N-httcasp3 fragment are identified at the level of the arrowheads. The uncleaved protein expressed from FH3221 migrates between 140 and 160 kDa depending on polyglutamine length in htt. Native mouse full-length htt (*) and a small mouse N-htt fragment is seen in cells in blot on left. (b) X57 cells were transfected with FH3221-100 or not transfected (No Tf) and cells (C) and debris (D) were assayed for htt expression with Ab1. In the transfected cells, treatment with Z-VAD-FMK (25 μM) attenuates the level of the N-httcasp3 fragment (arrow), and there is an increase in the level of a precursor fragment. The N-httcasp3 fragment is not present in debris (D) from cells treated with Z-VAD-FMK. (c) X57 cells and MCF-7 cells were transfected with FLAG-htt cDNA FH3221-100. The N-httcasp3 fragment (arrow) appears in cells and debris of X57 cells but is not present in the caspase 3 deficient MCF-7 cells. Proteolytic products of the N-httcasp3 fragment are identified at arrowheads in the debris of X57 cells. The blot was probed with Ab1. Native mouse htt is at the top. (d) Cleavage of human htt by active caspase 3. Protein extracts from X57 cells transfected with mutant htt cDNA FH3221-100 were treated with active caspase 3 (500 ng) for 1 h at 37°C. There is an increase in the level of the N-httcasp3 fragment (arrow) after exposure to active caspase 3. The blot was probed with Ab1. In a—d, 10 μg protein were loaded per lane for cell extracts, and equal volumes were loaded per lane for debris.
Wt and mutant N-httcasp3 fragments were also detected in protein extracts collected from debris 24 h after transfection of htt cDNAs (Fig. 1 a–c). The debris contains dying cells that have detached from the culture dish after transfection. Compared with cells, which had lower levels of N-httcasp3 fragments than of uncleaved htt, the debris had greater levels of N-httcasp3 fragments than of uncleaved htt. Treatment with Z-VAD FMK (25 μM) eliminated the mutant N-httcasp3 fragment in debris (Fig. 1b). Thus, the detached dying cells in debris had relatively high levels of mutant N-httcasp3 fragment compared with the cells still attached to the culture dish; treatment with Z-VAD-FMK reduced the accumulation of the mutant N-httcasp3 fragment in dying cells. We tested a variety of non-caspase protease inhibitors including lysosomal enzyme inhibitors and the proteasome inhibitor lactacystin. These inhibitors had no effect on the levels of the N-httcasp3 fragment.
Fragments smaller than the N-httcasp3 fragment were detected in the transfected x57 cells, especially in the debris (Fig. 1 a and c). Two proteolytic N-htt fragments, each running as a doublet, were recognized by antibody ab1. These N-htt products varied in size with polyglutamine length (Fig. 1a) and migrated at about 55 kDa and 65 kDa when wt htt was expressed from cDNA FH3221-18 and at about 80 kDa and 90 kDa when mutant htt was expressed from cDNA FH3221-100. The larger of the two proteolytic fragments had a stronger signal on Western blot, suggesting that the smaller fragment was cleaved from the larger fragment. Antibody 2166 also recognized two fragments on Western blot that were smaller than the N-httcasp3 fragment. These fragments migrated at about 65 kDa and 75 kDa, regardless of polyglutamine-length, indicating that they might be C-terminal products arising from the proteolysis of N-httcasp3.
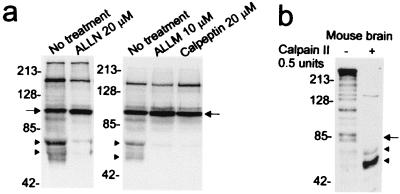
Treatment of transfected X57 cells with different calpain inhibitors including ALLN (10–20 μM), ALLM (10 μM), and calpeptin (20 μM) blocked the formation of the smaller fragments in debris and increased the level of the N-httcasp3 fragment (Fig. 2a). The increase in N-httcasp3 further suggested that the smaller fragments were products of the N-httcasp3 fragments. Treatment of mouse brain protein extracts with calpain reduced endogenous full-length mouse htt and the N-httcasp3 fragments of mouse htt and produced two N-htt products (Fig. 2b). These results suggested that the calpain-cleaved N-htt fragments could arise from full-length htt and the N-httcasp3 fragments. The calpain-cleaved N-htt products were similar in size to the N-htt fragments seen in the debris of X57 cells exogenously expressing human wt htt (Fig. 1a). These findings suggested that the N-htt fragments seen in debris of transfected X57 cells and in mouse brain extracts treated with calpain arose from the proteolysis of the N-httcasp3 fragment.
Figure 2.
Calpain cleavage of human and mouse htt. (a) Calpain inhibitors ALLN, ALLM, and calpeptin block formation of the proteolytic products of the human N-httcasp3 fragment. X57 cells were transfected and treated with inhibitors as described in Materials and Methods. Western blots (10 μg protein per lane) were probed with 2166 antibody. Arrows identify the level of the N-httcasp3 fragment, and arrowheads mark the level of the “C-terminal” portions of the calpain-cleaved fragments that are detected by antibody 2166. (b) Mouse brain homogenates treated with calpain II show increase in the levels of two N-htt fragments at arrowheads migrating at about 65 kDa and 55 kDa. The arrow identifies the level of the caspase-cleaved fragments in mouse htt. Blot was probed with Ab1. Twenty micrograms of protein was loaded per lane. The numbers beside blots in a and b are molecular mass markers.
N-Terminal htt Fragments in Human Control and HD Brain Correspond to Cleavage Products Observed in Vitro.
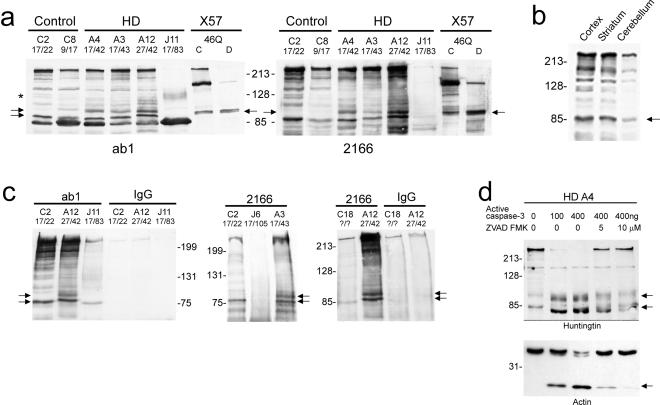
Protein homogenates from the cortex of controls and patients with adult or juvenile onset HD were analyzed by Western blot with ab1 and 2166 antibodies (Fig. 3a). A wt N-htt fragment appeared in the controls and the adult onset HD cases. A slower migrating mutant N-htt fragment was present in the adult onset HD brains. The HD genes in the adult onset HD patients had CAG repeats of 42 (A4 and A12) and 43 (A3). The mutant N-htt fragments from these patients migrated to about the same position as the N-httcasp3 fragment formed in vitro after exogenous expression of human htt with 46 glutamines (FH3221-46; Fig. 3a). The mutant N-htt was also recognized by the 1C2 antibody, which detects only the expanded polyglutamine epitope (results not shown). In the juvenile HD brain (J11), a broad band migrating at about 100–120 kDa was detected with the ab1 antibody. We interpret this band to be the juvenile form of the mutant N-httcasp3. The postmortem intervals for obtaining the control and HD patient brains included 4 h (A4), 24–26 h (C2, C18, A3 and A12, J11), and 40 h (C8, J6). There was no correlation between postmortem interval (PMI) and levels of N-htt fragments, suggesting that the fragments were not caused by postmortem proteolysis. A comparison of the cortex, striatum, and cerebellum in one control brain (C20, PMI = 24 h) showed the presence of the N-htt fragment in all three regions (Fig. 3b). The full-length htt and the N-htt fragments were present at higher levels in cortex and striatum than in cerebellum, which has relatively low levels of htt compared with forebrain regions. Protein extracts from the striatum and cerebellum of other control and HD patients showed the presence of wt and mutant N-htt fragments. In all cases, these fragments migrated at a molecular weight consistent with N-httcasp3 observed in vitro.
Figure 3.
Western blot analysis of N-terminal htt fragments in control and HD brain. (a) Protein extracts are from controls (C2 and C8), HD patients (A4, A3, A12, J11), and from X57 cells transfected with FLAG-htt cDNA FH3221-46. Wt N-htt (lower arrow on left) is present in control and adult onset HD cases (A4, A3, A12). The larger N-htt fragment (upper arrows) in HD brain migrates at the same size as the N-httcasp3 fragment produced in X57 cells transfected with a human htt cDNA encoding 46 glutamines. The asterisk identifies the level of the mutant N-httcasp3 fragment in juvenile patient J11. Blots are from separate gels. Thirty micrograms of protein was loaded per lane for brain and 10 μg protein for the X57 cells. A prominent htt-immunoreactive band is detected with ab1 and runs below the N-httcasp3 fragments. This fragment is unidentified. (b) Western blot probed with Ab1 shows htt expression in the cortex, striatum, and cerebellum of the same control brain. The N-httcasp3 fragment (arrow) is present in all regions. Full-length htt and the N-httcasp3 fragment are higher in the cortex than in the cerebellum. (c) Immunoprecipitation assays by using antisera ab1 (Left) or 2166 (Right) for immunoisolation of N-httcasp3 fragments from brain extracts. Wt N-httcasp3 fragment (lower arrow) is present in controls (C2, C18) and in HD patients (A12, J11, A3), and the mutant N-httcasp3 fragment (upper arrow) is seen in adult onset HD cases (A3 and A12) but not in the juvenile HD cases (J6, J1). The three Western blots were from different experiments. The left blot was probed with 2166 and the middle and right ones with ab1. Full-length htt is at the top of the blots. Fragments are absent when primary antibody (IgG) is omitted from the assay. Twenty-five microliters was loaded per lane. (d) Cleavage of human htt by caspase 3 in homogenates prepared from the brain of adult onset HD patient A4. Protein extracts were treated with active caspase 3 (100–400 ng) for 1 h at 37°C. Wt and mutant N-httcasp3 fragments of the expected size are produced after treatment (arrows). Proteolysis was markedly attenuated with addition of 5–10 μM Z-VAD-FMK. Actin, another substrate for caspase 3, is also cleaved in the brain extracts in the presence of active caspase 3. Ten micrograms of protein was loaded per lane. Numbers on left are molecular mass markers.
Immunoprecipitation assays were performed in protein extracts of control and adult and juvenile onset HD brain by using antisera ab1 or 2166. Protein that was immunoprecipitated with antibody ab1 was examined by Western blot with antibody 2166; in companion experiments, protein that was immunoprecipitated with antibody 2166 was examined by Western blot with antibody ab1. The results were the same with either assay procedure. The wt N-httcasp3 fragment was immunoprecipitated from control and adult onset HD brains (Fig. 3c). Immunoisolation of the wt N-httcasp3 fragment was successful in one of the two juvenile HD cases examined. The mutant N-httcasp3 fragment was immunoisolated in the adult onset HD cases but not from the juvenile HD cases possibly because, in the latter cases, there was insufficient soluble htt available for the immunoisolation. The immunoprecipitation data from the control and HD brains confirmed that the fragments identified as N-httcasp3 fragments in Western blots of total brain homogenates belonged to wt and mutant htt.
To confirm that the wt and mutant N-httcasp3 fragments in the HD brain had been cleaved by caspase 3, we treated protein extracts from HD case A4 with active caspase 3 enzyme for 1 h at 37°C. Active caspase 3 reduced the level of full-length htt and increased the level of the wt and mutant N-httcasp3 fragments. Proteolysis was inhibited by treatment with Z-VAD-FMK (Fig. 3d).
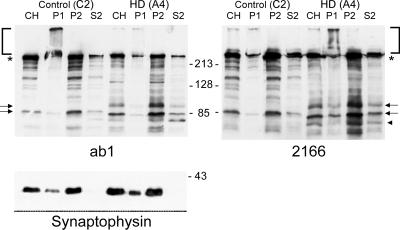
To determine the subcellular localization of N-httcasp3 fragments, subcellular fractionation of brain homogenates from two control and two adult onset HD brains was performed and examined by Western blot. P2 is the membrane fraction and is enriched with membrane-bound organelles including endoplasmic reticulum (ER), Golgi, endosomes, synaptic vesicles, lysosomes, and mitochondria. S2 is the soluble fraction and contains soluble components of the cytosol. Similar to full-length htt, the wt and mutant N-httcasp3 fragments were present in the membrane (P2) and soluble (S2) fractions of all cases (Fig. 4). Densitometry of the signal on Western blot for the N-httcasp3 fragment in the P2 and S2 fractions was performed for the two HD cases. The results were the same with both antisera and for both HD cases and were averaged. The signal intensity for full-length htt in the P2 fraction was about 2.6 times greater than in the S2 fraction. For wt and mutant N-httcasp3, the signal intensity in the P2 fraction was about 4.8 and 6.1 times greater, respectively, than in the S2 fraction. Although the overall level of the mutant N-httcasp3 fragment was less than the wt N-httcasp3 fragment in both the P2 and the S2 fractions, the mutant N-httcasp3 fragment was relatively more enriched in the P2 fraction compared with the wt N-httcasp3 fragment. These results suggested that, compared with the full-length htt, the N-httcasp3 fragments preferentially associated with membranes and the mutant N-httcasp3 fragment was more associated with membranes than the wt N-httcasp3 fragment.
Figure 4.
Western blots of htt expression are shown in subcellular fractions prepared from the cortex of a control brain (C2) and an adult onset HD brain (A4). Wt N-httcasp3 fragment appears in control brain (lower arrow) and wt N-httcasp3 and mutant N-httcasp3 fragments (upper arrow) are present in HD brain. N-httcasp3 fragments are most abundant in the membrane fraction (P2). A smaller product (arrowhead) possibly arising from proteolysis of mutant N-httcasp3 is detected by 2166 in HD brain. Blots are from separate gels and were probed with ab1 (left) and 2166 (right). Full-length htt (*) appears in all lanes. The stacking gels (brackets) are clear except for the P1 fraction, which was hard to load. Twenty micrograms was loaded per lane. Synaptophysin, a vesicle membrane marker, appears in the P2 membrane fraction and is absent from the S2 fraction. Alpha tubulin was equally present in all fractions (not shown). Numbers beside blots are molecular mass markers.
Immunoreactive bands smaller than the caspase 3-cleaved N-htt fragments were seen in the control and HD brain (Fig. 4). Some of these htt products may include calpain-cleaved proteolytic products of the N-httcasp3 fragment. Antibody 2166 detected a product of about 75 kDa in HD brain that was not seen in the control brain (Fig. 4 Right, arrowhead). Further studies of human control and HD brain will be needed to determine whether smaller products seen on Western blot in human brain are N-terminal and C-terminal products arising from calpain cleavage of the N-httcasp3 fragment.
Discussion
In this study, we provide biochemical evidence that N-terminal fragments migrating at a size consistent with cleavage by caspase 3 are resident products in normal and HD patient brains in the cortex, striatum, and cerebellum. We provide parallel data from in vitro and in vivo studies of human htt protein by using immunoprecipitation assays and cleavage by active caspase 3 to support our interpretation. Whether one or both caspase 3 sensitive sites are activated in brain is unclear from our results. Considering the previous detailed mutational analysis of caspase 3 cleavage of human htt performed by Wellington et al. (5, 6) and the data we present here, it is unlikely that a non-caspase protease is responsible for the fragments we observed in human brain. However, it is possible that another caspase with shared specificity for one or both caspase 3 sensitive-sites in htt contributes to htt proteolysis in the brain. Caspases 2 and 7 share similarities with caspase 3 in their preferred substrates (12). Caspase 7 does not cleave htt (6), but caspase 2, which is activated in parallel with caspase 3 in models of neuronal injury and in Alzheimer's disease brain (13, 14), could be active at caspase 3 cleavage sites in htt in brain. Thus, there is the possibility that the fragments referred to in this manuscript as N-httcasp3 might be products of caspase 2 cleavage. It is unlikely that the N-htt fragments seen in human brain arise after death because of postmortem delay. The N-httcasp3 fragments were seen in mouse brain that was rapidly removed without postmortem delay and in HD brain with a very short postmortem interval (4 h); longer postmortem delays up to 40 h did not change the levels of N-htt fragments detected in human brain.
Caspase 3 activation is known to regulate many cellular changes during apoptosis by targeting critical substrates for degradation (10). The identification of N-httcasp3 fragment in the human control brain suggests that caspase 3-mediated proteolysis participates in the normal enzymatic processing of full-length htt in neurons. Although specific accounts of nonapoptotic roles of caspase 3 are few, a recent study in hippocampal neurons showed that caspase 3 activity participates in long-term spatial memory storage (15). One possibility is that the level of locally active caspase 3 at membranes may be enough to cleave substrates like htt as part of normal processing but not be sufficient to cause apoptotic cell death. The detrimental effect of mutant N-httcasp3 in HD might not be a caspase cascade leading to apoptosis but rather a production of N-htt mutant fragments that are toxic in other ways. The mutant N-htt fragment may oligomerize into insoluble complexes that are not detected by our assay or be degraded by calpain into a smaller N-htt mutant fragment that is more prone to aggregation. Further study of these possibilities is underway. One way caspases might be activated in the HD brain is through damaged autophagosomes. Mutant htt accumulates on autophagosomes (8), a lysosome-like organelle that is used by cells to remove excess protein from the cytosol. Increased accumulation of mutant htt on autophagosomes could actuate the rupture of autophagosome membranes, the leakage of lysosomal proteases, and an activation of caspases (16). Future experiments to identify the initiators of caspase and calpain processing of htt in brain, especially to examine a possible influence of expanded polyglutamines on their recruitment, will be an important direction of our observations.
After cleavage by caspase 3, the N-httcasp3 fragment can be processed further by calpain, a calcium activated non-caspase cysteine protease that is enriched in brain and associated mainly with membranes. The caspase 3-calpain sequence produces partial proteolysis, to generate two N-terminal htt fragments smaller than the N-httcasp3 fragment. Further study is needed to determine whether calpain degrades full-length htt independent of caspase cleavage. Because proteins with increased polyglutamines show an atypical mobility on SDS/PAGE (17), it is difficult to estimate the actual size of the products arising from the cleavage of the N-httcasp3 fragment and to speculate about the sites of calpain cleavage. Calpain activation contributes to multiple neuronal activities: differentiation, synaptic plasticity, and necrotic cell death (18, 19). Together, caspases and calpains can influence how neurons respond to normal and noxious stimuli (18–20). A number of cytoskeletal and membrane-associated regulatory proteins including α-spectrin and focal adhesion kinase are substrates for cleavage by caspases and calpains (20, 21). Mutant N-htt fragments produced by calpain cleavage might undergo further proteolysis in the cytoplasm. N-htt fragments smaller in size than the calpain-cleaved N-htt fragments were found elevated in HD striatum compared with controls (22).
There is currently no compelling explanation for selective vulnerability in HD—why striatal and cortical neurons are especially affected. If the mutant N-httcasp3 fragment were harmful to neurons because of its accumulation in the brain, we might expect levels of the fragment to be greater than wt or to find mutant N-httcasp3 fragment only in affected brain regions. In the HD brain, mutant N-httcasp3 was detected at the same or lower levels than was the wt N-httcasp3 fragment. It is possible that some of the mutant N-httcasp3 fragment in the HD cortex eludes immunodetection because of its insolubility and exclusion from SDS/PAGE or sequestration by autophagosomes or inclusions (1, 8). Furthermore, as with transfected X57 cells with excess N-httcasp3 fragments that detach from the cell culture dish, neurons in the brain with the most mutant N-httcasp3 fragment might escape measurement because they rapidly die and are removed by phagocytosis. Instead of accumulating, the mutant N-httcasp3 fragment may alter its properties subsequent to caspase 3 cleavage. Changes might include an abnormal protein interaction at membranes and aggregation of N-terminal fragments produced by calpain dependent proteolysis.
Levels of the N-httcasp3 fragments, especially the mutant fragment, were greater in the adult onset HD brains than in the juvenile onset HD brains. Some of this difference may be accounted for by the more extensive neurodegeneration in the juvenile HD brain compared with the adult onset HD brain. Another reason may be that the brain homogenates we analyzed included the cytoplasm and not the nuclei of cells. In the adult onset HD brain, the mutant protein accumulates more in the cytoplasm and forms cytoplasmic inclusions whereas, in the juvenile HD brain, mutant htt accumulates more in the nucleus and forms nuclear inclusions (1). Thus, it is likely that, in the adult onset HD brain, the mutant N-httcasp3 fragment, or the calpain-dependent proteolytic products arising from it, contribute to the formation of cytoplasmic aggregates.
Full-length wt and mutant htt localize to a variety of intracellular membranes, including endosomes and synaptic vesicles (23–25), and the N-terminal region of htt interacts with numerous proteins involved in membrane trafficking (26–29). The implication of these observations is that htt or its proteolytic products contribute to vesicle transport. The wt and mutant N-httcasp3 fragments in brain were enriched in the membrane fraction compared with full-length htt, consistent with the possibility that cleaved htt binds to membrane bound organelles. Furthermore, the mutant N-httcasp3 fragment was more abundant in the membrane fraction than the wt N-httcasp3 fragment. Calpain is activated at membranes. The mutant N-httcasp3 fragment is therefore at favorable sites to influence membrane trafficking and to serve as a substrate for calpain.
In summary, we show that caspase 3-cleavable wt and mutant N-htt fragments occur in the HD brain and undergo partial proteolysis by calpain. We speculate that a proteolytic pathway involving serial cleavage by caspase 3 and calpain produces mutant N-terminal htt fragments that cause cellular dysfunction and aggregate in the cytoplasm (1, 2, 30). The enrichment of mutant N-httcasp3 fragments with the membrane fraction supports our hypothesis that there might be altered vesicle trafficking in HD. Therapeutic strategies that attenuate htt cleavage by caspase 3 and calpain may be useful in the treatment of HD.
Acknowledgments
We thank Dr. Alfred Heller for providing us with the X57 cells. This work was supported by National Institutes of Health Grants NS16367 and NS35711 (to M.D.) and NS38194, and Department of Defense U.S. Army Medical Research Acquisition Activity 98292059 (to N.A.) and by a grant from the Huntington's Disease Society of America (to M.D.).
Abbreviations
- HD
Huntington's disease
- htt
huntingtin
- N-htt
N-terminal region of htt
- N-httcasp3
caspase 3-cleaved N-terminal htt
- wt
wild type
Footnotes
Ellerby, L. M., Propp, S. S., Leavitt, C. L., Wellington, A. S., Hackam, A. V., Logvinova, S., Chen, F., Krajewski, S., Goldsmith, P., Hayden, R. R. & Bredesen, D. E. (2000) Soc. Neurosci. Abstr. 26, 1556.
References
- 1.DiFiglia M, Sapp E, Chase K, Davies S W, Bates G P, VonSattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 2.Hackam A S, Hodgson J G, Singaraja R, Zhang T, Gan L, Gutekunst C A, Hersch S M, Hayden M R. Philos Trans R Soc London B Biol Sci. 1999;354:1047–1055. doi: 10.1098/rstb.1999.0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg Y P, Nicholson D W, Rasper D M, Kalchman M A, Doide H B, Graham R K, Bromm M, Kazemi-Esfarjani P, Thornberry N A, Vaillancourt J P, Hayden M R. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 5.Wellington C L, Ellerby L M, Hackam A S, Margolis R L, Trifiro M A, Singaraja R, McCutcheon K, Salvesen G S, Propp S S, Bromm M, et al. J Biol Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 6.Wellington C L, Singaraja R, Ellerby L, Savill J, Roy S, Leavitt B, Cattaneo E, Hackam A, Sharp A, Thornberry N, Nicholson D W, Bredesen D E, Hayden M R. J Biol Chem. 2000;275:19831–19838. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Lee H S, LaForet G, McIntyre C, Martin E J, Chang P, Kim T W, Williams M, Reddy P H, Tagle D, Boyce F M, Won L, Heller A, Aronin N, DiFiglia M. J Neurosci. 1999;19:964–973. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kegel K, Kim M, Sapp S, McIntyre C, Castaño J G, Aronin N, DiFiglia M. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wainwright M S, Perr B D, Won L A, O'Malley K L, Wang W Y, Ehrlich M E, Heller A. J Neurosci. 1995;15:676–688. doi: 10.1523/JNEUROSCI.15-01-00676.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson D W. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 11.Janicke R U, Ng P, Sprengart M L, Porter A G. J Biol Chem. 1998;273:15540–15545. doi: 10.1074/jbc.273.25.15540. [DOI] [PubMed] [Google Scholar]
- 12.Talanian R V, Quinlan C, Trautz S, Hackett M C, Mankovich J A, Banach D, Ghayur T, Brady K D, Wong W W. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 13.Singh M, Savitz S I, Hoque R, Gupta G, Roth S, Rosenbaum P S, Rosenbaum D M. J Neurochem. 2001;77:466–475. doi: 10.1046/j.1471-4159.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- 14.Shimonhama S, Tanino H, Fujimoto S. Biochem Biophys Res Commun. 1999;256:381–384. doi: 10.1006/bbrc.1999.0344. [DOI] [PubMed] [Google Scholar]
- 15.Dash P K, Blum S, Moore A N. Neuroreport. 2000;11:2811–2816. doi: 10.1097/00001756-200008210-00040. [DOI] [PubMed] [Google Scholar]
- 16.Ohsawa Y, Isahara K, Kanamori S, Shibata M, Kametaka S, Gotow T, Watanabe T, Kominami E, Uchiyama Y. Arch Histol Cytol. 1998;61:395–403. doi: 10.1679/aohc.61.395. [DOI] [PubMed] [Google Scholar]
- 17.Preisinger E, Jordan B M, Kazantsev A, Housman D. In: Glutamine Repeats and Neurodegenerative Diseases: Molecular Aspects. Harper P, Perutz M, editors. New York: Oxford Univ. Press; 2001. pp. 127–139. [Google Scholar]
- 18.Wang K K W. Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- 19.Yamashima T. Prog Neurobiol. 2000;62:273–295. doi: 10.1016/s0301-0082(00)00006-x. [DOI] [PubMed] [Google Scholar]
- 20.Carragher N O, Fincham F J, Riley D, Frame M C. J Biol Chem. 2001;276:4270–4275. doi: 10.1074/jbc.M008972200. [DOI] [PubMed] [Google Scholar]
- 21.Pike B R, Zhao O, Newcomb J K, Posmantur R M, Wang K K, Hayes R L. Neuroreport. 1998;9:2437–2442. doi: 10.1097/00001756-199808030-00002. [DOI] [PubMed] [Google Scholar]
- 22.Mende-Mueller L M, Toneff T, Hwang S-R, Chesselet M-F, Hook V Y H. J Neurosci. 2001;21:1830–1837. doi: 10.1523/JNEUROSCI.21-06-01830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel J P, Carraway R, Boyce F M, Aronin N. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 24.Velier J, Kim M, Schwarz C, Kim T W, Sapp E, Chase K, Aronin N, DiFiglia M. Exp Neurol. 1998;152:34–40. doi: 10.1006/exnr.1998.6832. [DOI] [PubMed] [Google Scholar]
- 25.Kim M, Velier J, Chase K, Laforet G, Kalchman M, Hayden M R, Won L, Heller A, Aronin N, DiFiglia M. Neuroscience. 1999;89:1159–1167. doi: 10.1016/s0306-4522(98)00400-x. [DOI] [PubMed] [Google Scholar]
- 26.Kalchman M A, Koide H B, McCutcheon K, Graham R K, Nichol K, Nishiyama K, Kazemi-Esfarjani P, Lynn R C, Wellington C, Metzler M, Goldberg Y P, Kanazawa I, Gietz R C, Hayden M R. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- 27.Li X J, Li S H, Sharp A H, Nucifora F C, Jr, Schilling G, Lanahan A, Worley P, Snyder S H, Ross C A. Nature (London) 1995;368:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- 28.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates G P, Lehrach H, Wanker E E. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 29.Faber P W, Barnes G T, Srinidhi F, Chen J, Gusella J F, MacDonald M. Hum Mol Genet. 1998;7:1463–1474. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 30.Aronin N, Chase K, Young C, Sapp E, Schwarz C, Matta N, Kornreich R, Landwehrmeyer B, Bird E, Beal M F, et al. Neuron. 1995;15:1193–1201. doi: 10.1016/0896-6273(95)90106-x. [DOI] [PubMed] [Google Scholar]