Abstract
Diurnal preference (e.g. being an “owl” or “lark”) has been associated with several psychiatric disorders including bipolar disorder (BP), major depressive disorder, and substance use disorders. Previous large-scale genome-wide association studies (GWAS) aimed at identifying genetic influences on diurnal preference have exclusively included subjects of European ancestry. This study examined the genetic architecture of diurnal preference in two minority samples: a young adult sample of Mexican Americans (MAs), and a family-based sample of American Indians (AIs). Typed or imputed variants from exome chip data from the MA sample and low pass whole-genome sequencing from the AI cohort were analyzed using a mixed linear model approach for association with being an owl, as defined by a usual bedtime after 23:00 hrs. Genetic risk score (GRS) profiling detected shared genetic risk between evening preference and related disorders. Four variants in KIAA1549 like (KIAA1549L), a gene previously associated with attempted suicide in bipolar patients, were suggestively associated with being an owl at p<1.82E-05; post hoc analyses showed the top variant trending in both the MA and AI cohorts at p=2.50E-05 and p=0.030, respectively. Variants associated with BP at p<0.03 from the Psychiatric Genomics Consortium nominally predicted being an owl in the MA/AI cohort at p=0.012. This study provides some additional evidence that genetic risk factors for BP also confer risk for being an owl in MAs/AIs and that evening preference may be a useful endophenotype for future studies of BP.
Keywords: chronotype, diurnal preference, Hispanic, Native American, genome-wide association study, polygenic risk score
The timing of the sleep-wake cycle is regulated by the body’s endogenous circadian rhythm system as well as sleep homeostatic processes (Panda et al., 2002). Although environmental cues entrain phase and length of sleep, there is substantial variability in the timing of the biological clock in individuals. This “temporal phenotype of an organism” is now commonly referred to as a chronotype (Ehret, 1974). An element of chronotype, diurnal preference, is believed to reflect endogenous circadian rhythms. People colloquially referred to as a morning-person (or “lark”) are more alert in the morning and fall asleep easily at night, and an evening-person (or “owl”) is more alert in the evening and can go to bed and wake later.
Natural variation in circadian rhythmicity can contribute to disparities in fitness within species; variations in circadian period length may influence a variety of physiological and behavioral characteristics. Lifespan decreases as the magnitude of circadian period length deviation from the 24-hour cycle increases in rodents, and primate species (Kerkhof, 1985; Partonen, 2013; Wyse et al., 2010). Evening preference is associated with several psychosocial and psychopathological issues (Fabbian et al., 2016; Fares et al., 2015; Haregu et al., 2015; Merikanto et al., 2016; Rose et al., 2015; Voinescu et al., 2012), including depression (Abe et al., 2011; Antypa et al., 2016; Chan et al., 2014; Chelminski et al., 1999; Drennan et al., 1991; Gaspar-Barba et al., 2009; Hidalgo et al., 2009; Kitamura et al., 2010; Levandovski et al., 2011; Merikanto et al., 2015; Merikanto et al., 2013; Muller et al., 2015; Reid et al., 2012), bipolar disorder (Ahn et al., 2008; Giglio et al., 2010; Jeong Jeong et al., 2015; Mansour et al., 2005; Melo et al., 2016; Seleem et al., 2015; Wood et al., 2009), and substance use (Adan, 1994; Barclay et al., 2013; Broms et al., 2011; Hasler et al., 2013; Kervran et al., 2015; Lemoine et al., 2013; Reid et al., 2012; Sivertsen et al., 2015).
Genetic epidemiologic estimates of the additive heritability of diurnal preferences range from 21% to 54% (Barclay et al., 2010; Evans et al., 2011; Heath et al., 1990; Hur et al., 1998; Klei et al., 2005; Koskenvuo et al., 2007; Toomey et al., 2015; Vink et al., 2001; von Schantz et al., 2015; Watson et al., 2013). The results of several Genome-Wide Association Studies (GWAS) on sleep duration, sleep latency, sleep depth, sleep quality, insomnia, daytime sleepiness, sleep apnea, and actigraphic measurements are available (Allebrandt et al., 2013; Amin et al., 2016; Byrne et al., 2013; Cade, Chen, et al., 2016; Cade, Gottlieb, et al., 2016; Gottlieb et al., 2015; Gottlieb et al., 2007; Lane et al., 2017; Marinelli et al., 2016; Ollila et al., 2014; Scheinfeldt et al., 2015; Spada et al., 2016), yet many of these phenotypes are independent of chronotype (Jones et al., 2016; Lane et al., 2017; Lehnkering & Siegmund, 2007). Two early GWAS identified several single nucleotide polymorphisms (SNPs) associated with the chronotype measure bedtime. Only an intronic variant in erythrocyte membrane protein band 4.1, EPB41, was nominally associated in both studies. The EPB41 linked variant is not significantly associated after correction for multiple testing (Byrne et al., 2013; Gottlieb et al., 2007). Many loci associated with chronotype have been detected and replicated by three large-scale GWAS. Hu and colleagues identified 15 genome-wide significant loci using 89,283 subjects from the 23andMe cohort (Hu et al., 2016). Eight of the 15 loci were subsequently replicated in a sample of 100,420 subjects from the UK Biobank (Lane et al., 2016). Among these loci, the linked genes for regulator of G-protein signaling 16 (RGS16), vasoactive intestinal peptide (VIP), aph-1 homolog A, gamma-secretase subunit (APH1A), F-box and leucine rich repeat protein 13 (FBXL13), and the period genes PER2 and PER3 have plausible roles sleep biology. Jones et al. (2016) used a larger cohort from the UK Biobank (N = 128,266) in their GWAS. This work replicated these findings and confirmed associations detected with the 23andMe cohort (linked to adenylate kinase 5 (AK5) and hypocretin receptor 2 (HCRTR2)) (Hu et al., 2016), and identified 7 additional loci that were genome-wide significant in both the UK Biobank cohort and meta-analysis of the UK Biobank and 23andME cohorts (linked to exonuclease 3′-5′ domain containing 3 (EXD3), 5-hydroxytryptamine receptor 6 (HTR6), FK506 binding protein 1B (FKBP1B), calbindin 1 (CALB1), PATJ crumbs cell polarity complex component (INADL), and acylphosphatase 2 (ACYP2)) (Jones et al., 2016). While these studies have shed light onto the molecular basis of diurnal preference in human populations, they have solely looked at populations of European ancestry. Several GWAS on sleep phenotypes have included subjects of diverse ethnic background (Cade, Gottlieb, et al., 2016; Scheinfeldt et al., 2015), including a solely Hispanic sample (Cade, Chen, et al., 2016), yet these studies did not assess chronotype.
Over the last decade there has been a growing body of literature that has explored the potential overlap between health disparities research and sleep medicine (Jean-Louis & Grandner, 2016). A number of epidemiological studies have reported poorer quality sleep and a higher prevalence of short and/or long sleep in non-White adults, particularly from lower socioeconomic status groups, when compared to White adults (Carnethon et al., 2016; Hale & Do, 2007; Krueger & Friedman, 2009; Stamatakis et al., 2007). Little is known concerning genetic factors that might influence sleep in different ethnic groups.
The purpose of this study was to investigate genetic influences on chronotype in two admixed populations: a young adult sample of Hispanics who predominately identify as Mexican American (MA), and a family-based sample of American Indians (AIs). Recent work from our laboratory has identified specific demographic, clinical, and cultural factors associated with sleep characteristics in these samples (Ehlers et al., 2010; Ehlers et al., 2016). Briefly, in the AI sample, higher degrees of AI Heritage, but not cultural identification, being over the age of 30, and having a high school diploma were all predictive of short sleep duration (>6 hrs). The global score on the Pittsburgh Sleep Quality Index (PSQI) (Buysse et al., 1989) was significantly higher in those participants with a lifetime diagnosis of substance use disorders, anxiety disorders, and affective disorders. Alcohol use disorders and affective disorders were significant predictors of sleep latency whereas anxiety and affective disorders were correlated with waking more often in the night/early morning. Nicotine dependence was associated with having trouble breathing and alcohol use disorders and anxiety disorders with bad dreams (Ehlers et al., 2017). In the MA sample lifetime diagnoses of alcohol-use disorders, family history of alcohol dependence, acculturation stress, and lifetime diagnoses of major depressive disorder were all correlated with significantly poorer quality sleep as indexed by the global score on the PSQI. Regression analyses also revealed that gender was correlated with habitual bedtime, whereas drug dependence (cannabis, stimulants, and/or opiates) was significantly correlated with how long it took to fall asleep, major depressive disorder with the number of hours spent sleeping a night, and anxiety disorders and major depressive disorder with waking up in the early morning or middle of the night (Ehlers et al., 2010).
Previous GWAS on chronotype have exclusively included subjects of European ancestry; hence, two specific analyses in these understudied populations were undertaken in this study. First, we conducted a GWAS on diurnal preference. In existing studies of Europeans only common sequence variants with small effect have been identified through GWAS. In the second analysis, polygenic risk score profiling was used to determine whether a larger proportion of variance in diurnal preference could be attributed to variants that have been associated with other complex disorders. The following phenotypes were analyzed for overlapping genetic influences with chronotype: bipolar disorder and depression, given the robust link between these psychiatric disorders and chronotype (Abe et al., 2011; Ahn et al., 2008; Antypa et al., 2016; Chan et al., 2014; Chelminski et al., 1999; Drennan et al., 1991; Gaspar-Barba et al., 2009; Giglio et al., 2010; Hidalgo et al., 2009; Jeong Jeong et al., 2015; Kitamura et al., 2010; Levandovski et al., 2011; Mansour et al., 2005; Melo et al., 2016; Merikanto et al., 2015; Merikanto et al., 2013; Muller et al., 2015; Reid et al., 2012; Seleem et al., 2015; Wood et al., 2009); and schizophrenia and body mass index, given previously identified genetic correlations between these phenotypes and chronotype (Lane et al., 2016).
Materials and methods
Samples
Six hundred and nineteen young adult MAs were recruited using a commercial mailing list. The sample consists of primarily 2nd generation MAs, as previously described (Ehlers et al., 2010). Each subject was required to be of MA heritage, between the ages of 18 and 30, residing in the United States legally, and able to read and write in English to be included in the study; subjects who were pregnant, nursing, or currently had a major medical or neurological disorder were excluded. Hispanic heritage was assessed based on the origin (Caribbean or West Indian, Chicano, Cuban, Mexican, Mexican-American, Puerto Rican, South American, other Spanish, or Mexican Indian) of each subject’s 8 grandparents. 83.8% of the sample self-identified as having 50% or more Mexican heritage alone.
A family-based sample of AI participants recruited from eight geographically continuous reservations on which roughly 3,000 individuals reside comprised the second sample. To be included in the study, each participant had to be of AI heritage, between the ages of 18 and 82, and be mobile enough to be transported from his or her home to The Scripps Research Institute (TSRI), as previously described (Ehlers et al., 2016).
The primary phenotype, whether each subject was an “owl” or not, was indexed by the PSQI (Buysse et al., 1989), the psychometric properties of which have been described elsewhere (Carpenter & Andrykowski, 1998). The PSQI has over 12,000 citations and has been used in many cultures. The PSQI consists of 19 individual items such as usual bedtime, wakeup time, actual hours slept, number of minutes to fall asleep, and nighttime awakenings. If a subject answered after 11pm when asked the question “when (what time) have you usually gone to bed?” they were considered an owl, based on Dr. Buysse’s recommendation of what is standard to use for the definition of evening preference on the PSQI. Extraneous phenotypic information on substance use and various affective disorders for these samples was obtained through interviews using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994), which has exhibited reliability and validity for diagnosing these disorders (Bucholz et al., 1994; Hesselbrock et al., 1999).
All work was carried out in accordance with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. Written consent was obtained for all participants. The Institutional Review Board (IRB) at TSRI approved the protocol for this study and the Indian Health Council, a tribal review group overseeing health issues for the reservations in which recruitment took place, additionally approved the AI study protocol.
Genotyping
DNA was extracted from consenting subjects from blood samples. In the MA cohort, DNA was prepared and genotyped using the Affymetrix Exome1A chip according to the Affymetrix Axiom 2.0 Assay Manual Workflow documentation. As described previously (Norden-Krichmar et al., 2014), quality control on the markers was initially performed according to Affymetrix best practices. Additional quality control measures included: removing SNPs out of Hardy-Weinberg equilibrium (HWE, p<1E-10), as calculated on a set of maximally unrelated subjects using PLINK (Purcell et al., 2007); excluding SNPs with bad genotype clusters using snpChecker (a Java program with a graphical interface that allows the investigator to visually check and compare the clustering characteristics of each variants and either re-cluster bad calls or remove variants with poor overall genotype calls accordingly); removing subjects of high hidden relatedness using Genome-wide Complex Trait Analysis (GCTA, with the genetic relationship matrix, GRM, cutoff 0.125) (Yang et al., 2011); and pruning out SNPs with low minor allele frequency (MAF<1%). Imputation was performed using the Michigan Imputation Server (https://imputationserver.sph.umich.edu/index.html#!pages/home); the ShapeIT software was used to phase the data and the 1000 Genomes (1000G) Phase 3 v5 American (AMR) population as a reference panel. Post-imputation quality control steps were performed as follows: removing duplicates, retaining variants with very high imputation accuracy (Rsq≥0.9), removing variants with low MAF (<1%) and variants out of HWE (p<0.001). 301,019 variants remained for analysis.
DNA from the AI cohort was also extracted from blood samples. DNA sequencing was done using Illumina low-coverage whole-genome sequencing (WGS) and samples were additionally genotyped using an Affymetrix Exome1A chip, as previously described (Bizon et al., 2014). Paired-end sequencing was performed using the HiSeq2000 sequencers. For roughly 80% of samples the sequencing coverage was approximately evenly distributed at 3–12x. WGS reads were aligned using BWA (H. Li & Durbin, 2009) and realigned around indels using GATK (DePristo et al., 2011). Variants were called using two pipelines for comparison: the GATK Unified Genotyper, following best practices for low-coverage samples (Van der Auwera et al., 2013); and the linkage disequilibrium (LD) aware variant caller Thunder (Y. Li et al., 2011). Variant calls from Thunder were used in these analyses. The sequencing calls and exome genotypes were in good agreement (Bizon et al., 2014). Variant calls for the 301,019 variants imputed in the MA sample were extracted from the AI sequence calls; 269,453 total variants were selected based on base pair location. Quality control steps for this subset of variants included removing variants with low MAF (<1%) and variants out of HWE (p<0.001, using a set of maximally unrelated subjects).
The MA and AI data were merged using PLINK (Purcell et al., 2007), resulting in 258,423 markers, and cleaned as previously: removing variants with low MAF (<1%), and removing variants out of HWE (p<0.001, using a set of maximally unrelated subjects). The final number of variants for analysis was 258,411. Lastly, GCTA (Yang et al., 2011) was used to generate a GRM and 20 principal components (PCs) after the genotype data were pruned for LD.
Genome-wide association analysis
A mixed linear model association (MLMA) (Yang et al., 2014) was run using GCTA (Yang et al., 2011). This model tests the additive effect of each SNP for association with the phenotype, given the relationship matrix between subjects (i.e. the GRM calculated in GCTA). Gender, study, age, and 20 PCs were included as covariates. Allele frequencies were calculated within GCTA. Manhattan and Quantile-Quantile (QQ) plots was generated in R (R Development Core Team 2012) using the Manhattan package (Turner, 2014). The genomic inflation factor (lambda, λ) was also calculated in R using the commands outlined in http://genometoolbox.blogspot.com/2014/08/how-to-calculate-genomic-inflation.html. Locus zoom plots were generated with the LocusZoom software (http://locuszoom.sph.umich.edu/locuszoom/) (Pruim et al., 2010) using the 1000G AMR population as a reference population and the hg19 build. Variant annotation was performed with ANNOVAR (Wang et al., 2010). Power calculations were executed using the case-control variance components module for discrete traits based upon a formula derived in (Sham et al., 2000), as implemented in the Genetic Power Calculator (Purcell et al., 2003).
Because much fewer variants were tested than in a standard GWAS (which typically includes roughly a million SNPs), the standard Bonferroni correction threshold of p<5E-08 would be too stringent. Thus suggestive and genome-wide significance thresholds were calculated based on the effective number of independent tests using the Genetic Type 1 Error Calculator (GEC) software (M. X. Li et al., 2012). This software uses LD information from the sample to calculate the number of independent SNPs being tested and calculates suggestive and genome-wide significance thresholds given the number of independent tests. Suggestive and genome-wide significance thresholds were estimated at p<1.82E-05 and p<9.11E-07, respectively.
Genetic risk score profiling
Genetic Risk Scores (GRSs) were generated using PRSice (Euesden et al., 2015). Briefly, to detect shared genetic etiology between traits, GRSs are calculated on a base phenotype using large-scale GWAS results and used as predictors of a target phenotype in individuals from an independent data set. As LD between variants can inflate GRSs, PRSice prunes the base data for LD based on the genotypic structure of the target data (using PLINK 1.9 (Chang et al., 2015)). PRSice then performs “high-resolution” scoring in which a number of p-value thresholds are used to construct a GRS at each cutoff. Only variants exceeding the set p-value threshold (PT) are included in the GRS, and each is weighted by its effect size in the base data. Each GRS, which represents a sum of trait-associated alleles in the base data for each p-value cutoff, is calculated in the independent target data for each subject. Although by default these scores are then regressed on the phenotype in the target data, for our purposes all scores were written out in a file (i.e. using the commands “no.regression T report.best.score F report.individual.scores T” in PRSice) and regressed on the phenotype using a generalized linear mixed model fit by maximum likelihood. This model was fit using the pedigreemm package (https://cran.r-project.org/web/packages/pedigreemm/pedigreemm.pdf). All default options were used, with the exception of specifying “family=binomial” (to indicate a binary outcome), and a pedigree to be included in the model. Gender, study, age, and 20 PCs were included as fixed effects, with subject fitted as random effects.
Cross-phenotype GRSs were computed for four phenotypes and used as predictors in the model for the owl phenotype: bipolar disorder (BP), depressive symptoms (DS), schizophrenia (SCZ), and body mass index (BMI). All of these data, described briefly below, were meta-analyses of large-scale GWAS. Summary statistics on BP (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011) were made available through the Psychiatric Genomics Consortium (PGC) download page (https://www.med.unc.edu/pgc/results-and-downloads) and were based on a GWAS of 7,481 cases and 9,250 controls. GWAS results of DS in 161,460 subjects (Okbay et al., 2016) were downloaded from the Social Science Genetic Association Consortium (http://www.thessgac.org/data). The SCZ data (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) were also downloaded from the PGC and consisted of up to 36,989 cases and 113,075 controls. Finally, multi-ethnic summary statistics on BMI from 339,224 individuals were made available through the Genetic Investigation of ANthropometric Traits (GIANT) consortium (Locke et al., 2015). For both the BP and SCZ results, risk score training sets were available in which the summary data had already been pruned for LD. For these analyses PRSice’s internal clumping procedures were turned off (i.e. “clump.snps F prune.snps F”). Default clumping procedures in PRSice were used for the DS and BMI results, using the LD structure of the MA/AI target data. GRSs were calculated between p-value thresholds of 0.01 and 0.5 at increments of 0.01. The major histocompatibility complex (MHC) region, owing to its complex LD structure, was removed from calculation of the GRS. The suggested significance threshold for a single-phenotype high-resolution GRS analyses after correction for multiple testing is p=0.001 (Euesden et al., 2015).
Results
The analyses included 299 AIs (159 cases and 140 controls) and 535 MAs (371 cases and 164 controls) for a total of 834 subjects (Table 1). Generally, the AIs were older, had higher rates of AUDs, higher BMI, fewer years of education, and had a higher percentage of the sample earning less than $20,000 per year compared to the MA cohort. The decision was made to combine the cohorts due to the small sample size of each.
Table 1.
Phenotypic characteristics of the samples.
| American Indians | Mexican Americans | |||
|---|---|---|---|---|
|
| ||||
| Cases | Controls | Cases | Controls | |
|
| ||||
| N | 159 | 140 | 371 | 164 |
|
| ||||
| % Male | 44.03% | 46.43% | 45.01% | 33.54% |
|
| ||||
| Age (SDa) | 29.74 (13.46) | 34.89 (15.74) | 23.39 (3.78) | 24.41 (3.93) |
|
| ||||
| % with AUDb | 63.52% | 65.71% | 52.02% | 40.24% |
|
| ||||
| % with affective disorder | 40.25% | 35.00% | 38.54% | 34.76% |
|
| ||||
| BMIc (SD) | 32.57 (8.16) | 31.78 (7.27) | 26.73 (6.49) | 28.01 (6.68) |
|
| ||||
| % with 50% or more respective ancestry | 30.82% | 31.43% | 82.75% | 87.20% |
|
| ||||
| % making <$20,000/year | 39.57% | 43.90% | 20.34% | 24.03% |
|
| ||||
| Education (SD) | 11.79 (1.62) | 11.41 (1.54) | 13.50 (1.74) | 13.36 (1.95) |
Standard deviation
Alcohol use disorder (mild, moderate, or severe by DSM-5)
Body mass index
Genome-wide association test
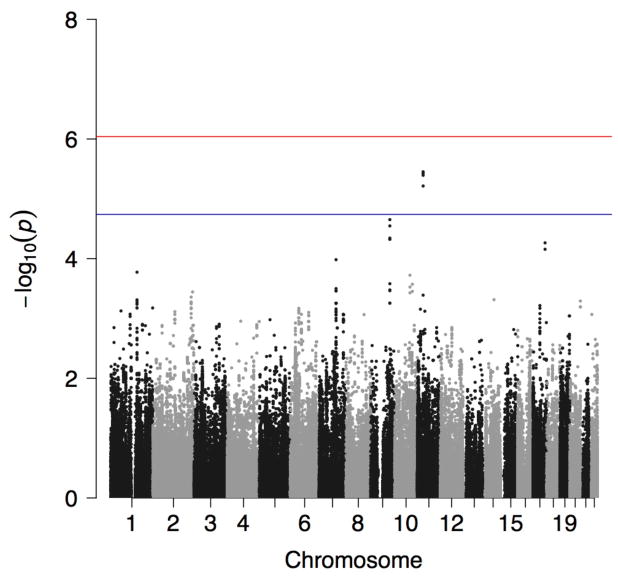
Four variants reached the threshold for suggestive evidence for association with being an owl at p<1.82E-05 (Figure 1). A genomic inflation factor of λ=0.989 indicated no inflation of test scores (Supplementary Figure S1). All four suggestively significant SNPs were in KIAA1549 like (KIAA1549L), the minor alleles of which showed a consistently negative effect on being an owl (Table 2), and were in high LD (Supplementary Figure S2). A list of the top 50 associations is provided in Supplementary Table S1. Post hoc analyses were performed separately in each cohort for the most highly significant SNP. Results indicated that, adjusting for gender, age, 20 PCs, and the GRM, rs10768009 was associated at p<0.05 in the same direction in both individual cohorts in a MLMA (for the minor A allele: SNP effect = −0.120, standard error [se]=0.029, p=2.50E-05 in the MAs; and SNP effect = −0.092, se=0.043, p=0.030 in the AIs). Alcohol use has a well-characterized link to sleep (reviewed in (Chakravorty et al., 2016)). The GWAS was repeated including AUD as a covariate (specifically having any AUD, mild, moderate, or severe, by DSM-5); the results were unchanged.
Figure 1.
Manhattan plot showing the p-value for the owl phenotype in the combined American Indian and Mexican American cohorts. Suggestive (blue line) and genome-wide (red line) significance thresholds are p<1.82E-05 and p<9.11E-07, respectively, calculated using the Genetic Type 1 Error Calculator (GEC) software.
Table 2.
Suggestively significant hits for the owl phenotype in the combined American Indian and Mexican American sample (additional information is available in Supplementary Table S1 and Supplementary Figure S2).
| Chra | dbSNP 138 | BP (hg19) | Effect allele | Freqb | SNP Effect | sec | p | Refd | Alte | X1000g2014oct_allf |
|---|---|---|---|---|---|---|---|---|---|---|
| 11 | rs10768009 | 33632390 | A | 0.4227 | −0.1104 | 0.0238 | 3.53E-06 | C | A | 0.3466 |
| 11 | rs7115532 | 33633523 | T | 0.4239 | −0.1103 | 0.0238 | 3.77E-06 | A | T | 0.3908 |
| 11 | rs2073174 | 33630891 | T | 0.4233 | −0.1095 | 0.0238 | 4.04E-06 | A | T | 0.3397 |
| 11 | rs2076622 | 33631423 | A | 0.4305 | −0.1078 | 0.0238 | 6.11E-06 | G | A | 0.4517 |
Chromosome
Frequency of the effect allele
Standard error
Reference allele
Alternate allele
Allele frequency of the alternate allele from the 1000 Genomes Project (all populations)
Power calculations for the high risk C allele were performed with the following criteria: high-risk allele frequency 0.5773; prevalence 0.25; 530 cases; and a 0.574 ratio of controls to cases. For genotype relative risk ratios of 1.5 (heterozygotes) and 1.75 (homozygotes for the risk allele), our study is 12.15% powered to detect an association at p<1.82E-05. To be 80% powered to detect an effect, 1,432 cases would be needed at p<1.82E-05 and 1,803 cases would be needed at p<9.11E-07.
Replication of previous associations
Association summary statistics are displayed for SNPs annotated to some of the genes significantly associated with chronotype from previous studies in Supplementary Table S2: RGS16, AK5, HCRTR2, and FBXL13 (Hu et al., 2016; Jones et al., 2016; Lane et al., 2016). No SNPs were mapped to APH1A. Of the SNPs previously associated with chronotype, only one was directly assessed in the current study: rs1144566 in RGS16 was associated at p<1.3E-08 in the study by Lane et al. (2016) but failed to reach nominal significance in our study with p>0.1. In fact, none of the SNPs mapped to these genes were associated with being an owl at p<0.05; however, evaluation of LD using SNP Annotation and Proxy Search (SNAP) (Johnson et al., 2008) showed that none of the SNPs that mapped to AK5 and HCRTR2 in the current study were in LD at r2>0.5 with those SNPs associated in previous GWAS (notably, this was limited to SNPs that were in the 1000 Genomes Pilot 1 data and only evaluated LD for subjects of Northern and Western European (CEU) ancestry; thus evaluation of LD proxies for some SNPs was impossible, particularly the previously associated SNPs in FBXL13 as those were not in the 1000 Genomes Pilot 1 data, and LD estimations for our admixed population may be different regardless).
Genetic risk score profiling
Cross-phenotype GRS analysis provided evidence that variants associated with BP and DS nominally predicted some of the risk for being an owl (BP: PT=0.03, z = −2.505, p=0.012; and DS: PT=0.49, z = −2.148, p=0.032). These results do not survive correction for multiple testing at p<0.001. Variants associated with SCZ or BMI did not predict being an owl at p<0.05. Plots showing the number of SNPs used in generating each GRS given the p-value cutoff are shown in Supplementary Figure S3.
Discussion
We present the first investigation of genetic risk factors for evening preference in a minority sample of AIs and MAs. Over the last decade several epidemiological studies have reported poorer quality sleep and a higher prevalence of short and/or long sleep in non-White adults, particularly from lower socioeconomic status groups, when compared to White adults (Carnethon et al., 2016; Hale & Do, 2007; Krueger & Friedman, 2009; Stamatakis et al., 2007). While not a debilitating disease in itself, evening preference has been associated with several psychiatric disorders. Previous GWAS on chronotype have included predominantly European American samples; two minority samples were used in this study, for whom substance use, anxiety, and depressive disorders have been shown to differentially affect various aspects of sleep (Ehlers et al., 2010; Ehlers et al., 2016). Specifically, in the AI sample the global score on the PSQI was significantly higher in those participants with a lifetime diagnosis of substance use disorders, anxiety disorders, and affective disorders (Ehlers et al., 2017), and in the MA sample lifetime diagnoses of alcohol-use disorders, family history of alcohol dependence, acculturation stress, and lifetime diagnoses of major depressive disorder were all correlated with significantly poorer quality sleep as indexed by the global score on the PSQI (Ehlers et al. 2010). The results presented herein suggest that variants previously associated with BP confer risk for being an owl in this minority sample of AIs and MAs.
The primary finding from the GWAS was a suggestively significant signal in KIAA1549L associated with being an owl. One cohort did not drive this association as post hoc analyses indicated the top SNP was associated at p<0.05 in the same direction in both cohorts. The KIAA1549L protein, spanning 1,849 amino acids, contains highly conserved regions and had been shown to act as a fusion partner of paired box 5 (PAX5), a transcription factor that is critical for B-cell activation and maintenance (Anderl et al., 2015; Medvedovic et al., 2011). PAX5 was one of the most associated genes with chronotype in a gene-based test from Lane and colleagues’ (2016) study of the UK Biobank sample. KIAA1549L has previously been associated with attempted suicide in bipolar patients; in a meta-analysis of a discovery sample (1,201 suicide attempters and 1,497 non-attempters) and replication sample (1,295 attempters and 1,822 non-attempters), a signal in C11orf41 (aka KIAA1549L) was associated at p=3.77E-06 (Willour et al., 2012). Although this SNP, rs10437629, was not significantly associated with being an owl in the current study (p=0.663; results not shown), the discovery sample used by Willour et al. (2012) was of European ancestry. It is likely that neither the signal identified in our study nor the signal identified by Willour and colleagues is the causal signal and, furthermore, that LD patterns between each signal differ between populations. These factors could explain the lack of agreement across studies. Suicidal ideation was positively correlated with evening tendencies in a sample of patients with depression (Bahk et al., 2014) and given the robust link between evening preference and bipolar disorder (Ahn et al., 2008; Giglio et al., 2010; Jeong Jeong et al., 2015; Mansour et al., 2005; Melo et al., 2016; Seleem et al., 2015; Wood et al., 2009), this is a prime gene for follow-up studies. Finally, although not suggestively significant, the 2nd two most associated genes in our GWAS, sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (SVEP1) and sidekick cell adhesion molecule 2 (SDK2), were nominally associated in previous GWAS with bipolar disorder and panic disorder, respectively (Ferreira et al., 2008; Otowa et al., 2009).
Tangentially, the F-box protein gene FBXO3 is downstream of KIAA1549L (see Supplementary Figure S2). The F-box proteins constitute one of the four subunits of the SKP1-cullin-F-box ubiquitin protein ligase complex which functions in phosphorylation-dependent ubiquitination. This is notable in that previous GWAS have identified SNPs in FBXL13 and FBXL3 (Hu et al., 2016; Jones et al., 2016; Lane et al., 2016) associated with chronotype, thus highlighting the potential role of F-box proteins in diurnal preference.
Although a full investigation of the variants previously associated with chronotype was not possible given the nature of these data, overall there was limited convergence in results between this study and previous GWAS. There are a number of potential explanations for this phenomenon. First, our owl phenotype was loosely defined; many previous studies have used the Horne-Östberg Morningness-Eveningness Questionnaire (Horne & Ostberg, 1976), or adaptations of it, to assess diurnal preference. Our case/control phenotype also differed from recent large-scale GWAS which assessed chronotype a number of ways, including using extreme owls as compared to extreme larks and creating a continuous score of diurnal preference (Hu et al., 2016; Jones et al., 2016; Lane et al., 2016). These GWAS were limited to subjects of European ancestry; differences in LD and MAF between ethnicities could further account for discrepancies. Finally, this study was not fully powered to investigate genetic influences, particularly those of small effect, on being an owl, which may have led to false negatives.
Cross-phenotype GRS profiling suggested that being a night owl shares some genetic risk with BP and depression, while no significant genetic risk scores from SCZ or BMI predicted being an owl in this population. Previous work has found no evidence for a genetic correlation between chronotype and BP (Jones et al., 2016; Lane et al., 2016). Although it did not survive correction for multiple testing, a GRS from PT=0.03 indicated only those variants more highly associated with BP predict being an owl in our data, rather than an overall genetic correlation. Conversely, a GRS from the much higher PT=0.49 using summary statistics on DS nominally predicted being an owl in the MA/AI sample. This is roughly in agreement with previous work: a twin study previously demonstrated a significant genetic correlation of −0.21 between diurnal preference and depression (Toomey et al., 2015); and GWAS in the UK Biobank found nominally significant genetic correlations between major depressive disorder and chronotype (Jones et al., 2016; Lane et al., 2016), although neither of those p-values, nor our own, survived correction for multiple testing.
The UK Biobank GWAS also found significant positive genetic correlations between eveningness and SCZ, and nominal genetic correlations between chronotype and BMI that were lower in magnitude; these findings were not replicated in this study. It is not clear what these negative findings mean, as these results may reflect underlying genetic architecture between populations and environmental factors. With the exception of the summary data on BMI, which was from a multi-ethnic sample, all GWAS summary statistics were derived from European populations. While these negative results may in part be due to the factors listed above (e.g. phenotype definition, limited power, and limited number of imputed variants), the predictive ability of GRSs are sensitive to differences in LD and MAF across base and target samples and previous work on our AI sample indicated that GRSs for smoking behaviors from a European population were only predictive in our sample for those subjects with a high proportion of European ancestry (Otto et al., 2016). The findings presented above highlight the need for both replication and large-scale studies of these minority populations to probe for ethnic-specific genetic risk factors and endophenotypes for complex diseases.
This study provides evidence that genetic risk factors for BP also confer risk to being an owl in a sample of MAs and AIs, suggesting that evening preference may be a useful endophenotype for future studies of BP. This study was underpowered to fully evaluate the phenotype given the small sample size but a signal in KIAA1549L, a gene previously associated with attempted suicide in bipolar patients, was associated with being an owl in both cohorts and, furthermore, a GRS derived from a large-scale meta-analysis of BP nominally predicted being an owl at p=0.012. These findings are in line with a body of literature supporting a link between BP and evening preference (Ahn et al., 2008; Giglio et al., 2010; Jeong Jeong et al., 2015; Mansour et al., 2005; Melo et al., 2016; Seleem et al., 2015; Wood et al., 2009). Although these results warrant replication, the growing body of literature exploring the potential overlap between health disparities research and sleep medicine (Jean-Louis & Grandner, 2016) and lack of a potential replication sample comprised of MAs and/or AIs simultaneously highlights the importance of conducting studies on these understudied populations.
Supplementary Material
Acknowledgments
The authors would like to thank: Drs. Chris Bizon, Scott Chase, and Jeffrey Tilson for their genotyping efforts; Derek Willis, Dr. Qian Peng and Dr. Trina Norden-Krichmar for assistance with data organization and analysis; and Dr. David Gilder, Evie Philips, Gina Stouffer, and Corinne Kim for phenotypic data collection.
Financial support
This research was supported by grants from the National Institute on Alcoholism and Alcohol Abuse (NIAAA; grants AA010201 and AA006420 to C.L.E. and AA013525 to E.P.R.) and the National Institute on Drug Abuse (NIDA; grant DA030976 to K.C.W. and C.L.E.) of the National Institutes of Health. Neither NIAAA nor NIDA had any role in the study design, collection, analysis, or interpretation of the data, nor in the writing and submission of this work.
Footnotes
Conflict of interest
None.
Supplementary material is available on the Cambridge Journals Online website.
References
- Abe T, Inoue Y, Komada Y, Nakamura M, Asaoka S, Kanno M, … Takahashi K. Relation between morningness-eveningness score and depressive symptoms among patients with delayed sleep phase syndrome. Sleep Medicine. 2011;12(7):680–684. doi: 10.1016/j.sleep.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Adan A. Chronotype and personality factors in the daily consumption of alcohol and psychostimulants. Addiction. 1994;89(4):455–462. doi: 10.1111/j.1360-0443.1994.tb00926.x. [DOI] [PubMed] [Google Scholar]
- Ahn YM, Chang J, Joo YH, Kim SC, Lee KY, Kim YS. Chronotype distribution in bipolar I disorder and schizophrenia in a Korean sample. Bipolar Disorders. 2008;10(2):271–275. doi: 10.1111/j.1399-5618.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- Allebrandt KV, Amin N, Muller-Myhsok B, Esko T, Teder-Laving M, Azevedo RV, … Roenneberg T. A K(ATP) channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Molecular Psychiatry. 2013;18(1):122–132. doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- Amin N, Allebrandt KV, van der Spek A, Muller-Myhsok B, Hek K, Teder-Laving M, … van Duijn CM. Genetic variants in RBFOX3 are associated with sleep latency. European Journal of Human Genetics. 2016;24(10):1488–1495. doi: 10.1038/ejhg.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl S, Konig M, Attarbaschi A, Strehl S. PAX5-KIAA1549L: a novel fusion gene in a case of pediatric B-cell precursor acute lymphoblastic leukemia. Molecular Cytogenetics. 2015;8:48. doi: 10.1186/s13039-015-0138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antypa N, Vogelzangs N, Meesters Y, Schoevers R, Penninx BW. Chronotype Associations with Depression and Anxiety Disorders in a Large Cohort Study. Depression and Anxiety. 2016;33(1):75–83. doi: 10.1002/da.22422. [DOI] [PubMed] [Google Scholar]
- Bahk YC, Han E, Lee SH. Biological rhythm differences and suicidal ideation in patients with major depressive disorder. Journal of Affective Disorders. 2014;168:294–297. doi: 10.1016/j.jad.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Barclay NL, Eley TC, Buysse DJ, Archer SN, Gregory AM. Diurnal preference and sleep quality: same genes? A study of young adult twins. Chronobiology International. 2010;27(2):278–296. doi: 10.3109/07420521003663801. [DOI] [PubMed] [Google Scholar]
- Barclay NL, Eley TC, Parsons MJ, Willis TA, Gregory AM. Monozygotic twin differences in non-shared environmental factors associated with chronotype. Journal of Biological Rhythms. 2013;28(1):51–61. doi: 10.1177/0748730412468698. [DOI] [PubMed] [Google Scholar]
- Bizon C, Spiegel M, Chasse SA, Gizer IR, Li Y, Malc EP, … Wilhelmsen KC. Variant calling in low-coverage whole genome sequencing of a Native American population sample. BMC Genomics. 2014;15:85. doi: 10.1186/1471-2164-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms U, Kaprio J, Hublin C, Partinen M, Madden PA, Koskenvuo M. Evening types are more often current smokers and nicotine-dependent-a study of Finnish adult twins. Addiction. 2011;106(1):170–177. doi: 10.1111/j.1360-0443.2010.03112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, … Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol and Drugs. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Buysse DJ, Reynolds CF, 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Research. 1989;28(2):193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- Byrne EM, Gehrman PR, Medland SE, Nyholt DR, Heath AC, Madden PA … Chronogen Consortium. A genome-wide association study of sleep habits and insomnia. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2013;162B(5):439–451. doi: 10.1002/ajmg.b.32168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade BE, Chen H, Stilp AM, Gleason KJ, Sofer T, Ancoli-Israel S, … Redline S. Genetic Associations with Obstructive Sleep Apnea Traits in Hispanic/Latino Americans. American Journal of Respiratory and Critical Care Medicine. 2016a;194(7):886–897. doi: 10.1164/rccm.201512-2431OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade BE, Gottlieb DJ, Lauderdale DS, Bennett DA, Buchman AS, Buxbaum SG, … Saxena R. Common variants in DRD2 are associated with sleep duration: the CARe consortium. Human Molecular Genetics. 2016b;25(1):167–179. doi: 10.1093/hmg/ddv434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnethon MR, De Chavez PJ, Zee PC, Kim KY, Liu K, Goldberger JJ, … Knutson KL. Disparities in sleep characteristics by race/ethnicity in a population-based sample: Chicago Area Sleep Study. Sleep Medicine. 2016;18:50–55. doi: 10.1016/j.sleep.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter JS, Andrykowski MA. Psychometric evaluation of the Pittsburgh Sleep Quality Index. Journal of Psychosomatic Research. 1998;45(1):5–13. doi: 10.1016/s0022-3999(97)00298-5. [DOI] [PubMed] [Google Scholar]
- Chakravorty S, Chaudhary NS, Brower KJ. Alcohol Dependence and Its Relationship With Insomnia and Other Sleep Disorders. Alcoholism: Clinical and Experimental Research. 2016;40(11):2271–2282. doi: 10.1111/acer.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JW, Lam SP, Li SX, Yu MW, Chan NY, Zhang J, Wing YK. Eveningness and insomnia: independent risk factors of nonremission in major depressive disorder. Sleep. 2014;37(5):911–917. doi: 10.5665/sleep.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelminski I, Ferraro FR, Petros TV, Plaud JJ. An analysis of the “eveningness-morningness” dimension in “depressive” college students. Journal of Affective Disorders. 1999;52(1–3):19–29. doi: 10.1016/s0165-0327(98)00051-2. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, … Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan MD, Klauber MR, Kripke DF, Goyette LM. The effects of depression and age on the Horne-Ostberg morningness-eveningness score. Journal of Affective Disorders. 1991;23(2):93–98. doi: 10.1016/0165-0327(91)90096-b. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Criado JR, Caetano R. Sleep quality and alcohol-use disorders in a select population of young-adult Mexican Americans. Journal of Studies on Alcohol and Drugs. 2010;71(6):879–884. doi: 10.15288/jsad.2010.71.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Wills DN, Lau P, Gilder DA. Sleep Quality in an Adult American Indian Community Sample. Journal of Clinical Sleep Medicine. 2017;13:385–391. doi: 10.5664/jcsm.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret CF. The sense of time: evidence for its molecular basis in the eukaryotic gene-action system. Advances in Biological and Medical Physics. 1974;15(0):47–77. doi: 10.1016/b978-0-12-005215-8.50009-7. [DOI] [PubMed] [Google Scholar]
- Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31(9):1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DS, Snitker S, Wu SH, Mody A, Njajou OT, Perlis ML, … Hsueh WC. Habitual sleep/wake patterns in the Old Order Amish: heritability and association with non-genetic factors. Sleep. 2011;34(5):661–669. doi: 10.1093/sleep/34.5.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbian F, Zucchi B, De Giorgi A, Tiseo R, Boari B, Salmi R, … Manfredini R. Chronotype, gender and general health. Chronobiology International. 2016;33(7):863–882. doi: 10.1080/07420528.2016.1176927. [DOI] [PubMed] [Google Scholar]
- Fares S, Hermens DF, Naismith SL, White D, Hickie IB, Robillard R. Clinical correlates of chronotypes in young persons with mental disorders. Chronobiology International. 2015;32(9):1183–1191. doi: 10.3109/07420528.2015.1078346. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L … Wellcome Trust Case Control Consortium. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nature Genetics. 2008;40(9):1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Barba E, Calati R, Cruz-Fuentes CS, Ontiveros-Uribe MP, Natale V, De Ronchi D, Serretti A. Depressive symptomatology is influenced by chronotypes. Journal of Affective Disorders. 2009;119(1–3):100–106. doi: 10.1016/j.jad.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Giglio LM, Magalhaes PV, Andersen ML, Walz JC, Jakobson L, Kapczinski F. Circadian preference in bipolar disorder. Sleep and Breathing. 2010;14(2):153–155. doi: 10.1007/s11325-009-0301-3. [DOI] [PubMed] [Google Scholar]
- Gottlieb DJ, Hek K, Chen TH, Watson NF, Eiriksdottir G, Byrne EM, … Tiemeier H. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Molecular Psychiatry. 2015;20(10):1232–1239. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb DJ, O’Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Medical Genetics. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale L, Do DP. Racial differences in self-reports of sleep duration in a population-based study. Sleep. 2007;30(9):1096–1103. doi: 10.1093/sleep/30.9.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haregu A, Gelaye B, Pensuksan WC, Lohsoonthorn V, Lertmaharit S, Rattananupong T, … Williams MA. Circadian rhythm characteristics, poor sleep quality, daytime sleepiness and common psychiatric disorders among Thai college students. Asia-Pacific Psychiatry. 2015;7(2):182–189. doi: 10.1111/appy.12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasler BP, Sitnick SL, Shaw DS, Forbes EE. An altered neural response to reward may contribute to alcohol problems among late adolescents with an evening chronotype. Psychiatry Research. 2013;214(3):357–364. doi: 10.1016/j.pscychresns.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Kendler KS, Eaves LJ, Martin NG. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13(4):318–335. doi: 10.1093/sleep/13.4.318. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA--a comparison with the SCAN. Addiction. 1999;94(9):1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo MP, Caumo W, Posser M, Coccaro SB, Camozzato AL, Chaves ML. Relationship between depressive mood and chronotype in healthy subjects. Psychiatry and Clinical Neurosciences. 2009;63(3):283–290. doi: 10.1111/j.1440-1819.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. International Journal of Chronobiology. 1976;4(2):97–110. [PubMed] [Google Scholar]
- Hu Y, Shmygelska A, Tran D, Eriksson N, Tung JY, Hinds DA. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nature Communications. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y, Bouchard TJ, Lykken DT. Genetic and environmental influence on morningness–eveningness. Personality and Individual Differences. 1998;25(5):917–925. [Google Scholar]
- Jean-Louis G, Grandner M. Importance of recognizing sleep health disparities and implementing innovative interventions to reduce these disparities. Sleep Medicine. 2016;18:1–2. doi: 10.1016/j.sleep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Jeong H, Moon E, Min Park J, Dae Lee B, Min Lee Y, Choi Y, In Chung Y. The relationship between chronotype and mood fluctuation in the general population. Psychiatry Research. 2015;229(3):867–871. doi: 10.1016/j.psychres.2015.07.067. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, … Weedon MN. Genome-Wide Association Analyses in 128,266 Individuals Identifies New Morningness and Sleep Duration Loci. PLoS Genetics. 2016;12(8):e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhof GA. Inter-individual differences in the human circadian system: a review. Biological Psychol. 1985;20(2):83–112. doi: 10.1016/0301-0511(85)90019-5. [DOI] [PubMed] [Google Scholar]
- Kervran C, Fatseas M, Serre F, Taillard J, Beltran V, Leboucher J, … Auriacombe M. Association between morningness/eveningness, addiction severity and psychiatric disorders among individuals with addictions. Psychiatry Research. 2015;229(3):1024–1030. doi: 10.1016/j.psychres.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Hida A, Watanabe M, Enomoto M, Aritake-Okada S, Moriguchi Y, … Mishima K. Evening preference is related to the incidence of depressive states independent of sleep-wake conditions. Chronobiology International. 2010;27(9–10):1797–1812. doi: 10.3109/07420528.2010.516705. [DOI] [PubMed] [Google Scholar]
- Klei L, Reitz P, Miller M, Wood J, Maendel S, Gross D, … Nimgaonkar VL. Heritability of morningness-eveningness and self-report sleep measures in a family-based sample of 521 hutterites. Chronobiology International. 2005;22(6):1041–1054. doi: 10.1080/07420520500397959. [DOI] [PubMed] [Google Scholar]
- Koskenvuo M, Hublin C, Partinen M, Heikkila K, Kaprio J. Heritability of diurnal type: a nationwide study of 8753 adult twin pairs. Journal of Sleep Research. 2007;16(2):156–162. doi: 10.1111/j.1365-2869.2007.00580.x. [DOI] [PubMed] [Google Scholar]
- Krueger PM, Friedman EM. Sleep duration in the United States: a cross-sectional population-based study. American Journal of Epidemiology. 2009;169(9):1052–1063. doi: 10.1093/aje/kwp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, … Saxena R. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nature Genetics. 2017;49(2):274–281. doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JM, Vlasac I, Anderson SG, Kyle SD, Dixon WG, Bechtold DA, … Saxena R. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nature Communications. 2016;7:10889. doi: 10.1038/ncomms10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnkering H, Siegmund R. Influence of chronotype, season, and sex of subject on sleep behavior of young adults. Chronobiology International. 2007;24(5):875–888. doi: 10.1080/07420520701648259. [DOI] [PubMed] [Google Scholar]
- Lemoine P, Zawieja P, Ohayon MM. Associations between morningness/eveningness and psychopathology: an epidemiological survey in three in-patient psychiatric clinics. Journal of Psychiatric Research. 2013;47(8):1095–1098. doi: 10.1016/j.jpsychires.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Levandovski R, Dantas G, Fernandes LC, Caumo W, Torres I, Roenneberg T, … Allebrandt KV. Depression scores associate with chronotype and social jetlag in a rural population. Chronobiology International. 2011;28(9):771–778. doi: 10.3109/07420528.2011.602445. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MX, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics. 2012;131(5):747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Research. 2011;21(6):940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, … Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour HA, Wood J, Chowdari KV, Dayal M, Thase ME, Kupfer DJ, … Nimgaonkar VL. Circadian phase variation in bipolar I disorder. Chronobiology International. 2005;22(3):571–584. doi: 10.1081/CBI-200062413. [DOI] [PubMed] [Google Scholar]
- Marinelli M, Pappa I, Bustamante M, Bonilla C, Suarez A, Tiesler CM, … Sunyer J. Heritability and Genome-Wide Association Analyses of Sleep Duration in Children: The EAGLE Consortium. Sleep. 2016;39(10):1859–1869. doi: 10.5665/sleep.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Advances in Immunology. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- Melo MC, Abreu RL, Linhares Neto VB, de Bruin PF, de Bruin VM. Chronotype and circadian rhythm in bipolar disorder: A systematic review. Sleep Medicine Reviews. 2016 doi: 10.1016/j.smrv.2016.06.007. [DOI] [PubMed] [Google Scholar]
- Merikanto I, Kronholm E, Peltonen M, Laatikainen T, Vartiainen E, Partonen T. Circadian preference links to depression in general adult population. Journal of Affective Disorders. 2015;188:143–148. doi: 10.1016/j.jad.2015.08.061. [DOI] [PubMed] [Google Scholar]
- Merikanto I, Lahti T, Kronholm E, Peltonen M, Laatikainen T, Vartiainen E, … Partonen T. Evening types are prone to depression. Chronobiology International. 2013;30(5):719–725. doi: 10.3109/07420528.2013.784770. [DOI] [PubMed] [Google Scholar]
- Merikanto I, Suvisaari J, Lahti T, Partonen T. Eveningness relates to burnout and seasonal sleep and mood problems among young adults. Nordic Journal of Psychiatry. 2016;70(1):72–80. doi: 10.3109/08039488.2015.1053519. [DOI] [PubMed] [Google Scholar]
- Muller MJ, Cabanel N, Olschinski C, Jochim D, Kundermann B. Chronotypes in patients with nonseasonal depressive disorder: Distribution, stability and association with clinical variables. Chronobiology International. 2015;32(10):1343–1351. doi: 10.3109/07420528.2015.1091353. [DOI] [PubMed] [Google Scholar]
- Norden-Krichmar TM, Gizer IR, Wilhelmsen KC, Schork NJ, Ehlers CL. Protective variant associated with alcohol dependence in a Mexican American cohort. BMC Medical Genetics. 2014;15:136. doi: 10.1186/s12881-014-0136-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, … Cesarini D. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nature Genetics. 2016;48(6):624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila HM, Kettunen J, Pietilainen O, Aho V, Silander K, Kronholm E, … Paunio T. Genome-wide association study of sleep duration in the Finnish population. Journal of Sleep Research. 2014;23(6):609–618. doi: 10.1111/jsr.12175. [DOI] [PubMed] [Google Scholar]
- Otowa T, Yoshida E, Sugaya N, Yasuda S, Nishimura Y, Inoue K, … Okazaki Y. Genome-wide association study of panic disorder in the Japanese population. Journal of Human Genetics. 2009;54(2):122–126. doi: 10.1038/jhg.2008.17. [DOI] [PubMed] [Google Scholar]
- Otto JM, Gizer IR, Bizon C, Wilhelmsen KC, Ehlers CL. Polygenic risk scores for cigarettes smoked per day do not generalize to a Native American population. Drug and Alcohol Dependence. 2016;167:95–102. doi: 10.1016/j.drugalcdep.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002;417(6886):329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Partonen T. Pay attention to evening owls. Annals of Medicine. 2013;45(5–6):395–396. doi: 10.3109/07853890.2013.814378. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, … Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature Genetics. 2011;43(10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2012. Retrieved from http://www.R-project.org/ [Google Scholar]
- Reid KJ, Jaksa AA, Eisengart JB, Baron KG, Lu B, Kane P, … Zee PC. Systematic evaluation of Axis-I DSM diagnoses in delayed sleep phase disorder and evening-type circadian preference. Sleep Medicine. 2012;13(9):1171–1177. doi: 10.1016/j.sleep.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D, Gelaye B, Sanchez S, Castaneda B, Sanchez E, Yanez ND, Williams MA. Morningness/eveningness chronotype, poor sleep quality, and daytime sleepiness in relation to common mental disorders among Peruvian college students. Psychology, Health & Medicine. 2015;20(3):345–352. doi: 10.1080/13548506.2014.951367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinfeldt LB, Gharani N, Kasper RS, Schmidlen TJ, Gordon ES, Jarvis JP, … Christman MF. Using the Coriell Personalized Medicine Collaborative Data to conduct a genome-wide association study of sleep duration. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2015;168(8):697–705. doi: 10.1002/ajmg.b.32362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleem MA, Merranko JA, Goldstein TR, Goldstein BI, Axelson DA, Brent DA, … Birmaher B. The longitudinal course of sleep timing and circadian preferences in adults with bipolar disorder. Bipolar Disorders. 2015;17(4):392–402. doi: 10.1111/bdi.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham PC, Cherny SS, Purcell S, Hewitt JK. Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data. American Journal of Human Genetics. 2000;66(5):1616–1630. doi: 10.1086/302891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsen B, Skogen JC, Jakobsen R, Hysing M. Sleep and use of alcohol and drug in adolescence. A large population-based study of Norwegian adolescents aged 16 to 19 years. Drug and Alcohol Dependence. 2015;149:180–186. doi: 10.1016/j.drugalcdep.2015.01.045. [DOI] [PubMed] [Google Scholar]
- Spada J, Scholz M, Kirsten H, Hensch T, Horn K, Jawinski P, … Sander C. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE Adult Study. Journal of Sleep Research. 2016;25(6):690–701. doi: 10.1111/jsr.12421. [DOI] [PubMed] [Google Scholar]
- Stamatakis KA, Kaplan GA, Roberts RE. Short sleep duration across income, education, and race/ethnic groups: population prevalence and growing disparities during 34 years of follow-up. Annals of Epidemiology. 2007;17(12):948–955. doi: 10.1016/j.annepidem.2007.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey R, Panizzon MS, Kremen WS, Franz CE, Lyons MJ. A twin-study of genetic contributions to morningness-eveningness and depression. Chronobiology International. 2015;32(3):303–309. doi: 10.3109/07420528.2014.971366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014 doi: 10.1101/005165. [DOI] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, … DePristo MA. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Current Protocols in Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Groot AS, Kerkhof GA, Boomsma DI. Genetic analysis of morningness and eveningness. Chronobiology International. 2001;18(5):809–822. doi: 10.1081/cbi-100107516. [DOI] [PubMed] [Google Scholar]
- Voinescu BI, Szentagotai A, David D. Sleep disturbance, circadian preference and symptoms of adult attention deficit hyperactivity disorder (ADHD) Journal of Neural Transmission (Vienna) 2012;119(10):1195–1204. doi: 10.1007/s00702-012-0862-3. [DOI] [PubMed] [Google Scholar]
- von Schantz M, Taporoski TP, Horimoto AR, Duarte NE, Vallada H, Krieger JE, … Pereira AC. Distribution and heritability of diurnal preference (chronotype) in a rural Brazilian family-based cohort, the Baependi study. Scientific Reports. 2015;5:9214. doi: 10.1038/srep09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson NF, Buchwald D, Harden KP. A twin study of genetic influences on diurnal preference and risk for alcohol use outcomes. Journal of Clinical Sleep Medicine. 2013;9(12):1333–1339. doi: 10.5664/jcsm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Seifuddin F, Mahon PB, Jancic D, Pirooznia M, Steele J, … Potash JB. A genome-wide association study of attempted suicide. Molecular Psychiatry. 2012;17(4):433–444. doi: 10.1038/mp.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, Birmaher B, Axelson D, Ehmann M, Kalas C, Monk K, … Nimgainkar VL. Replicable differences in preferred circadian phase between bipolar disorder patients and control individuals. Psychiatry Research. 2009;166(2–3):201–209. doi: 10.1016/j.psychres.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyse CA, Coogan AN, Selman C, Hazlerigg DG, Speakman JR. Association between mammalian lifespan and circadian free-running period: the circadian resonance hypothesis revisited. Biology Letters. 2010;6(5):696–698. doi: 10.1098/rsbl.2010.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American Jouranl of Human Genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics. 2014;46(2):100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.